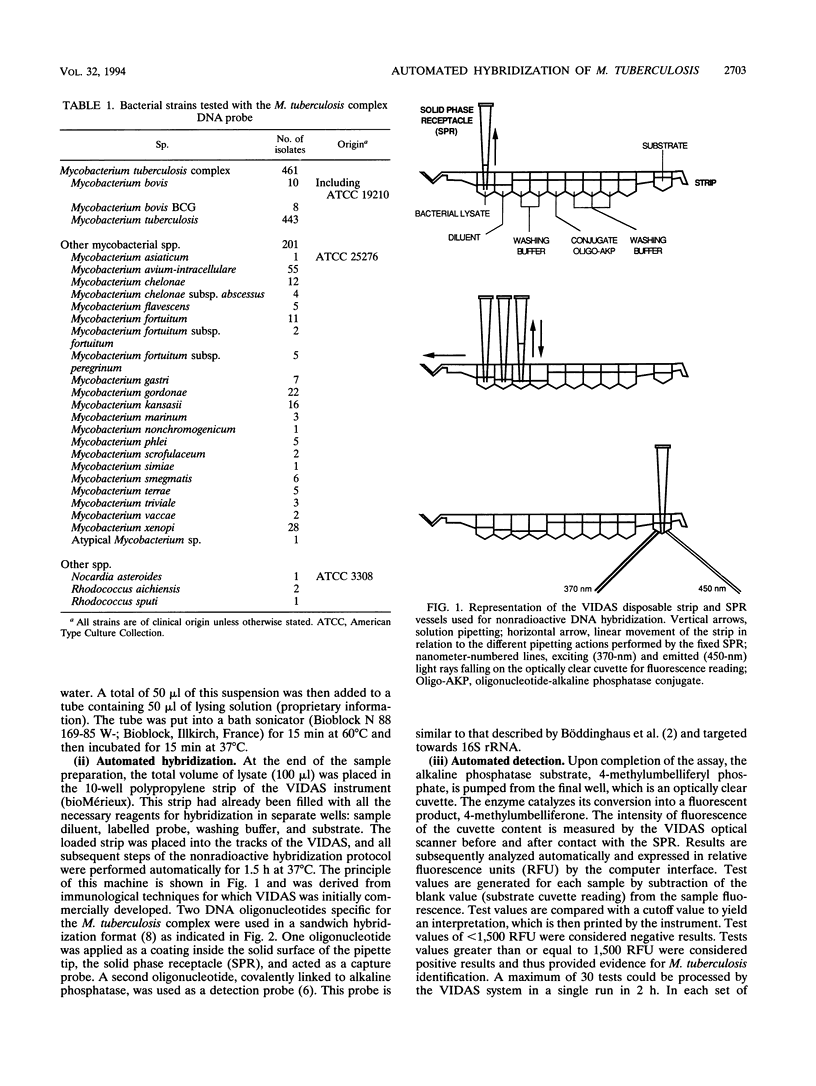
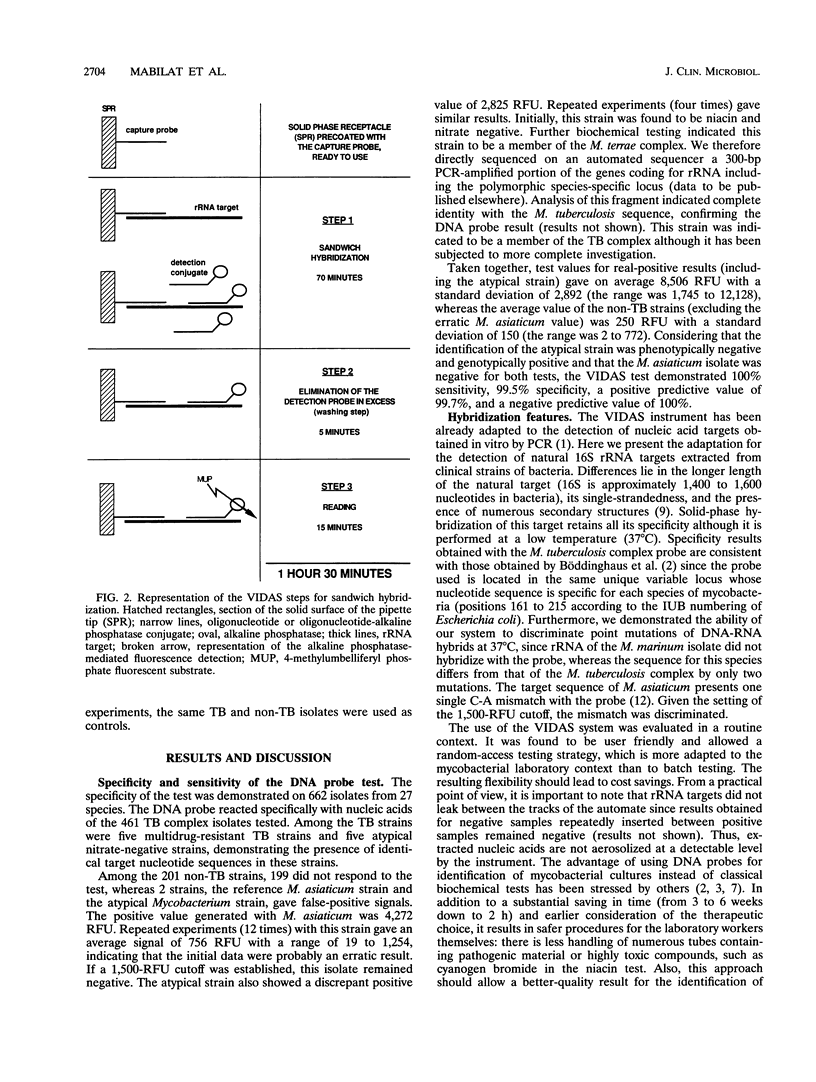
Abstract
Methodologies for biochemical identification of mycobacteria isolated from clinical samples are still cumbersome, taking skilled technicians 3 to 6 weeks. We describe here a 2-h identification system for mycobacterial isolates belonging to the Mycobacterium tuberculosis complex using a DNA probe. After 30 min of hands-off sample preparation, the 1.5-h hybridization test is totally automated in the newly developed VIDAS system (bioMérieux, Marcyl'Etoile, France), which performs solid-phase specific hybridization of 16S rRNA at 37 degrees C. The strain collection of actinomycetes tested was composed of 662 isolates from 27 species: 461 members of the M. tuberculosis complex (443 M. tuberculosis, 10 M. bovis, and 8 M. bovis BCG isolates) and 201 isolates of other species, including 55 M. avium-intracellulare isolates). They were identified by traditional methods: growth rate, colonial morphology, pigmentation, and biochemical profiles. The automated probe assay displayed an excellent correlation with the reference results. The four members of the Nocardia and Rhodococcus genera tested did not cross-hybridize. This flexible random-access and automated technology was shown to suit the routine context of the laboratory by rapidly delivering the results.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Böddinghaus B., Rogall T., Flohr T., Blöcker H., Böttger E. C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990 Aug;28(8):1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenach K. D., Cave M. D., Bates J. H., Crawford J. T. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990 May;161(5):977–981. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- Jablonski E., Moomaw E. W., Tullis R. H., Ruth J. L. Preparation of oligodeoxynucleotide-alkaline phosphatase conjugates and their use as hybridization probes. Nucleic Acids Res. 1986 Aug 11;14(15):6115–6128. doi: 10.1093/nar/14.15.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet F., Hebrard C., Brand D., Chapuis E., Cros P., Allibert P., Besnier J. M., Barin F., Mandrand B. Enzyme-linked oligosorbent assay for detection of polymerase chain reaction-amplified human immunodeficiency virus type 1. J Clin Microbiol. 1993 Jun;31(6):1444–1449. doi: 10.1128/jcm.31.6.1444-1449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Noordhoek G. T., Kolk A. H., Bjune G., Catty D., Dale J. W., Fine P. E., Godfrey-Faussett P., Cho S. N., Shinnick T., Svenson S. B. Sensitivity and specificity of PCR for detection of Mycobacterium tuberculosis: a blind comparison study among seven laboratories. J Clin Microbiol. 1994 Feb;32(2):277–284. doi: 10.1128/jcm.32.2.277-284.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Results of a World Health Organization-sponsored workshop to characterize antigens recognized by mycobacterium-specific monoclonal antibodies. Infect Immun. 1986 Feb;51(2):718–720. doi: 10.1128/iai.51.2.718-720.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Urbance J. W. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J Bacteriol. 1990 Jan;172(1):116–124. doi: 10.1128/jb.172.1.116-124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


