Abstract
Nanopores are increasingly utilized as tools for single-molecule detection in biotechnology. Many nanopores are fabricated through procedures that require special materials, expensive facilities and experienced operators, which limiting their usefulness on a wide-scale. We have developed a simple method of fabricating a robust, low-noise nanopore by externally penetrating a nanocavity enclosed in the terminal of a capillary pipette. The nanocavity was shown to have a pore size on the scale of a single molecule, verified by translocation of molecules of known sizes, including double-stranded DNA (2 nm), gold nanoparticles (10 nm) and ring-shaped cyclodextrin (1.5 nm). The small pore size allows entrapment of a single cyclodextrin molecule. Cyclodextrin in the nanopore may prove useful as a molecular adapter for chiral enantiomer discrimination.
INTRODUCTION
Nanopores have proven useful as single-molecule detectors for various biotechnological applications such as biosensing,1-5 detecting nucleic acids (which has potential for DNA sequencing),6-15 regulating membrane transportation,16;17 and constructing supramolecule18, as well as for basic research in single-molecule chemistry.19 The protein pore has unique capacities developed through structure-directed genetic engineering and chemical modification.20 However, it is limited by a fixed pore size and by the fragile lipid bilayer in which the pore is embedded. The stability of the membrane is being improved through new techniques, such as the creation of biochips that integrate the sandwich membrane structure21;22 and a glass nanopore membrane-supported lipid bilayer.23
Nanotechnology provides sophisticated tools for constructing a variety of molecular-scale pores in solid substrates.12;15;24-28 For example, a solid nanopore can be “sculpted” with a focused ion beam in the Si3N4 membrane12. A nanopore can also be lithographed with a high-energy electron beam from a transmission electron microscope.15;26-28 Such nanopores can be microscopically observed after fabrication. Another well-established method for creating a single nanopore involves ion-track etching on a polymer film.24 5;24;25;29 The dimension of this internally-etched pore can be controlled by controlling the etching conditions. However, these methods of pore fabrications require expensive instruments, well-trained operators or special materials (such as a polymer film with low-density ion tracks). Glass capillaries can be easily manipulated with a programmable puller to form nanopipettes that have the potential to be used for controlled delivery, ion conductance microscopes, and nanosensor and DNA sequencing.30-36 The technique is inexpensive. Pore sizes as small as 5 nm have been achieved.36 However, it is difficult to control the pore size at this scale. Recently White and his colleagues have created a glass nanopore electrode from a glass membrane-coated platinum nanoelectrode for a variety of nano-electrochemistry studies.37;38
In this report, we discovered a much more efficient process for creating a low-noise nanopore at the molecular scale on the terminal of a glass capillary. We were able to calibrate the pore size by controlling the level of pore conductance, and demonstrated the capability of this nanopore for single-molecule detection.
EXPERIMANTAL SECTION
Chemicals and materials
Au-NP with a diameter of 10 nm was purchased from Ted Pella Inc. According to the company, the sample contained 5.7×1012 particles/ml (∼9.5 nM) with a coefficient of size variation < 8%. The Au-NP was diluted to 4.75 nM in 15 mM NaCl for translocation experiments. A fresh nanoparticle preparation was used for each test. The 4.75 nM Au-NP in a 15 mM NaCl solution and in pure water (control) have been analyzed using a DynaPro99 Molecular Size Instrument (Dynamic Light Scattering) to confirm no aggregation in the low salt concentration (Fig.S1). The 1.0 kb dsDNA was from the 4.45 kb plasmid DNA pT7-αHL, which was digested with HindIII and NdeI restrictive endonucleases overnight, separated on an agarose gel and purified with the DNA purification kit (QiaGen). The purified 1.0 kb ds-DNA was 22 ng/μl and diluted to 10 nM in a 1 M NaCl solution. PCR was performed with the Polymer Chain Reaction (PCR) kit (Roche Corporation) using a Robocycler PCR instrument (Stratagene Inc). The primers for DNA amplification were synthesized and purified by Integrated DNA Technologies Inc. The sequences of the forward and backward primers were 5′-AGTGGTTTAGCCTGGCCTTC-3′ and 5′-TCTTGGAACCCGGTATATGG-3′. The PCR product was 350 base pairs and was examined on the agarose gel. (R)-(+)-ibuprofen was purchased from Toronto Research Company (Canada); cyclodextrins and other chiral enantiomers were purchased from Sigma-Aldrich.
A borosilicate capillary with an inner filament (1.5 mm o.d. and 0.86 mm i.d.) was purchased from A-M System Inc. This filament can facilitate backfilling of the pipette. Before use, the capillary was washed with ethanol and double-deionized water (ddH2O), followed by drying in nitrogen air. The cleaned tube was heated in a furnace to around 400 °C for 1 hr to improve surface smoothness. The P-2000 laser puller is an instrument from Sutter Inc. The micro-forge setup was self-made. Orthodontic wax was purchased from Sunstar Americas, the photoresist SU-8 Resists from MicroChem, and silicone elastomer Sylgard 184 from Dow Corning.
Recording solutions
In the DNA translocation experiment, both internal and external solutions contained 1 M NaCl. 10 nM DNA was presented in the external solution. In the nanoparticle translocation experiment, both internal and external solutions contained 15 mM NaCl and 4.75 nM Au-NP was presented in the external solution. To trap βCD in the nanopore, 100 μM βCD was loaded into the pipette. Both internal and external solutions contained 1 M NaCl. Before loading the guest into the pipette, the βCD concentration in the pipette was diluted with 1 M NaCl so that it was much lower than the guest concentration, in order to accurately quantify the guest-cyclodextrin interaction in the nanopore. All solutions were buffered with 10 mM Tris and titrated to pH7.2.
Pico-ampere electrical recording and data analysis
Conductance was measured using an Axopatch 200B patch-clamp amplifier (Molecular Device Inc.), then low-pass filtered with a built-in 4-pole Bessel filter at 1-5 kHz and sampled at 5-10 kHz with a Digidata 1332 A/D converter (Molecular Device Inc.). For all measurements, the external solution was grounded as the convention for voltage polarity. Current amplitudes and durations of single molecule block events were obtained from histograms constructed using pClamp software (Molecular Device Inc.). The amplitudes were obtained by fitting the amplitude peaks to Gaussian functions or averaging the amplitudes of measured block events. The average block duration and block interval (the time between adjacent blocks) were obtained from the arithmetic mean of all blocks. The reciprocal of the average block interval gave the block occurrence. Data were given as the mean ±SD, based on at least three separate measurements.
RESULTS AND DISCUSSION
Creation and properties of the nanopore
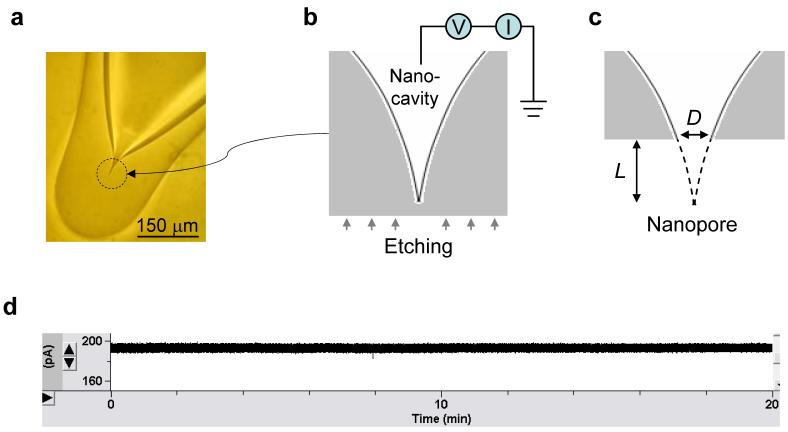
The capillary was first pulled into a micropipette. The narrow tip was melted with a micro-forge until the terminal was completely closed and a spherical terminal with a curvature radius of 100±10 μm was formed (Fig.1a). The terminal encloses an asymmetrical, gradually-narrowed cavity. The pipette was back-filled with the recording solution (1 M NaCl, 10 mM Tris and pH7.2) and examined under an optical microscope to ensure that no air bubble was isolated inside.
Figure 1.
Nanopore fabrication and electrical properties. a. Sealed glass ball (100 μm in radius) enclosing a nanocavity on the pipette terminal. b. External etching from bottom-up with electrical monitoring. c. Perforation on nanocavity terminal and nanopore formation. d. 20 min recording of nanopore current (+200 mV, 1 M NaCl).
The terminal was protected with a coat of material that resists the etchant, such as orthodontic wax or photoresist. The coating on the bottom of the terminal was carefully removed, allowing the local silica surface to be accessible to the chemical solution for bottom-up etching. The coating was used is to ensure the etching-through occurred at the end of the cavity and not on the sides of the tube. The pipette terminal was immersed in a solution of 40% NH4F/49% HF for the external etching and monitored through two Ag/AgCl electrodes spanning the solutions inside and outside the pipette (Fig.1b). The gradually increasing current should indicated the cavity enclosed in the terminal was ultimately perforated to form a nanopore (Fig.1c). Once etched through, the pipette terminal was washed with NaOH (50 mM) for 1-5 seconds, then washed with ddH2O to neutralize the surface etchant. The terminal was then transferred to a 1 M NaCl solution, the same as inside the pipette, to determine the pore conductance. This process effectively fixed the pore size and minimized contamination from byproducts, allowing accurate measurement of the conductance. The nanopore-terminated pipette was transferred back into the chemical solution for repeated etching until the desired conductance was achieved.
The glass nanopore created in this fashion has several advantages. The fabrication is greatly simplified: it requires no special and expensive equipments and can be accomplished with resources readily available in laboratories at all levels. Furthermore, the electrical noise of the nanopore is low (Fig.1d). Measurement over over a period of 20 mins (1 M NaCl, +200 mV) shows the current was consistently flat without drift and discrete change. The noise was 1.8 pA (IRMS value displayed on the amplifier panel), comparable to the 1.2-1.8 pA of αHL, the standard low-noise protein pore. The stable conductance and the low level of electrical noise make this nanopore ideal for electrically addressing single-molecule block events. Most importantly, this method of fabrication makes it possible to control the pore size. We were able to achieve a success rate of 80% in fabricating a nanopore with the desired conductance within an error range of ±0.3 nS. The method of nanopore fabrication is also programmable, since the conditions of etchant concentration and temperature can be adjusted. It may also be possible in the future to automate the control of pore conductance with a robotic device.
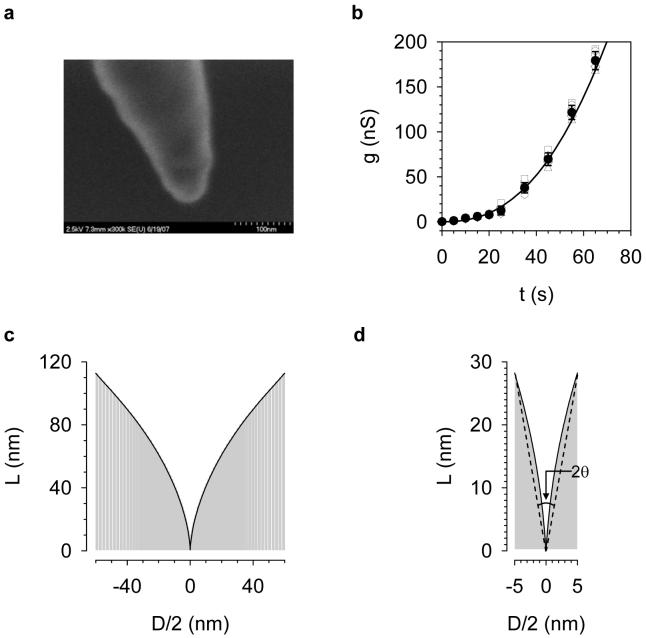
The profile of the cavity enclosed in the terminal of the pipette determines the pore size. The cavity profile can be described by L, the length begining from the narrow end of the cavity, and D, the diameter of the cavity cross-section at length L (Fig.1b and c). We used the cavity as the template to form silicone elastomer nanowire. The nanowire and the cavity should adopt the same shape. The image from the scanning electron microscope showed a conical-shaped nanowire with an aperture of ∼30° and terminal diameter of ∼40 nm (Fig.2). However, the image did not clearly reveal the geometry at the very end of the nanowire. Given this limitation in assessing the cavity profile, we proposed an electrical measurement, based on the relationship of the pore conductance (g) and etching time (t), i.e. the etching curve (g—t curve). Fig.2b shows the measures of etching curves for six different cavities fabricated following the procedures described above. Every etching curve showed increasing conductance with increasing the etching time. Each etching time showed similar conductance, with a confidence interval of g(1±0.17) at a confidence level of 95%. The mean etching curve (black line) was obtained by the polynomial fitting of averaged conductance. From the etching curve, L and D (geometrical parameters of cavity profile) can be determined using the following expression (derivation shown in Supplementary Materials),
| (1) |
In this expression, both the conductance g(t) and its derivative dg(t)/dt were obtained from the measured etching curve. v is the etching speed. The v determined for the borosilicate capillary in 40% NH4F: 49% HF (30:1 v/v) was 0.63 nm·s-1.
Figure 2.
Detection of nanocavity profile. a. SEM image of a nanopore-templated polydimethylsiloxane (PDMS) nanowire coated with a 3-nm platinum layer. The liquid PDMS, which has been vacuumed to remove the bubbles, was injected into the micropipette, followed by spinning in a microcentrifuge at 10,000 rpm for 2 minutes to deliver the polymer to the cavity end. After curing at 90 °C for 4 hours, the terminal glass was carefully removed manually to form a cavity-templated PDMS. b. Etching curve (g—t curve). The pipette was filled with 1 M NaCl (pH7.0) and etched in a bath containing 40% NH4F: 49% HF (30:1 v/v). When perforated, each pipette was transferred to a 1 M NaCl (pH7.0) solution every 5 to 10 s to determine the conductance at +20 mV. The unfilled symbols represent conductance of pores made from six independent nanocavities under standard fabrication processes (see text). Filled circles were mean conductance. Error bars represent standard deviation. Polynomial fitting of mean conductance yielded a continuous mean etching curve (black curve). c. The profile of the nanocavity (D—h curve) calculated from the etching curve (panel b), using Eq.1 in the text. The conductivity (K) of 1 M NaCl at 22 °C was 7.8 Sm-1. d. Magnified profile of the narrow end of the nanocavity. This section resembled a conical shape with an aperture of 20°, as marked by the dashed line.
The nanopore conductivity in Eq.1, K, is a key parameter that varies with the surface charges, ion concentration and pore shape. Both experimental27;39 and theoretical40 studies have revealed that, in low ion concentrations (such as 1 mM), the surface charges of the nanopore produce higher conductivity than in the bulk solution. The surface charges in asymmetrical nanopores also determine the spatial variation and voltage dependence of the pore conductivity40;41. In high ion concentrations (such as 1 M), however, the surface charges are effectively shielded, making the conductivity of the nanopore comparable to that in the bulk solution27;39;40. These properties have been studied for different pore materials such as SiO2,39 glass,40 Si3N4.27Thus in high ion concentrations, the conductivities of our borosilicate pore and in bulk solution are presumably similar. In the calculation, we used the K value of 1 M NaCl as the pore conductivity.
Fig.2c shows the cavity profile evaluated from the etching curve in Fig.2b. Because all independent cavities generated similar etching curves, we expected them to feature a uniform profile, making it possible to achieve a desired pore size D by etching the cavity for a corresponding length L. Fig.2d shows a magnified profile at the terminal of the cavity. Once perforated, this 10 nm×30 nm (D×L) domain was able to form a nanometer scale conical-shaped pore with an aperture 2θ of about 20° (dashed line). The conductance of the conical pore (in high ion concentrations) has been determined in several previous studies.24;37;38;42
Molecular translocation to assess the pore size
The etching curve (Fig.2b) and the profile of the nanocavity (Fig.2c, d) suggested that a nanometer-scale pore can be formed at the terminal of the terminal. However, it was difficult to observe the pore size at this level under the electron microscope, due to the much smaller pore size (∼102 nm2 in cross area) compared with the 108-fold larger terminal ball surface (104 μm2). Among other pore sizing methods, the stead-state voltammetric measurement of ion transfer has been used to evaluate the nanometer-size pipette.36 The translocation of analytes of known dimension has been shown to be a valid method of assessing the in many instances of nanopore detection.2;43;44 In the absence of direct microscopic observation, we employed this method to reveal the pore dimension.
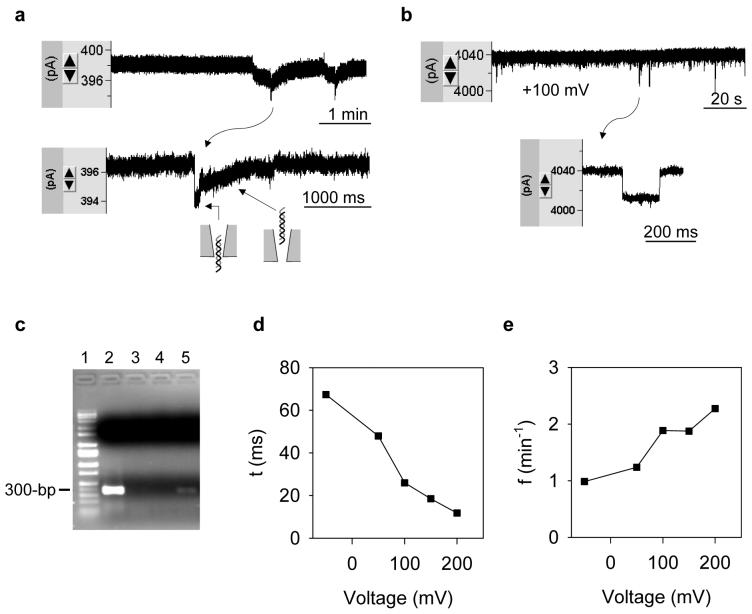
Fig.3 shows the translocation of 1 kb dsDNA (10 nM) from the external solution. The DNA block was not detected until the pore conductance was raised to 4.9±1.0 nS (n=3) (Fig.3a). At this conductance level (the threshold), the average block duration (τ ) was 1.1±0.6 s and block occurrence ( f ) was 0.28±0.10 min-1 (+100 mV). At a higher pore conductance such as 39 nS, τ was shortened to 26 ms, and f was increased to 1.9 min-1 (Fig.2b). The DNA translocation can be verified by the polymer chain reaction (PCR). We monitored DNA translocation in a 5.3 nS pore for 24 hrs, and collected 10 μl of the internal solution (around the nanopore) for PCR. The DNA amplification revealed by the agarose gel is shown in Fig.2c. The thick band in Lane-2 in the 350 bps position was for the external solution containing DNA; no band was detected in Lane-3 and Lane-4 for the water and internal solution (1 M NaCl) before the DNA was applied. Lane-5 shows the internal solution after DNA translocation. The weaker band at the same position as in Lane-2 shows the amplified DNA. The DNA amplification by PCR at various template levels has been calibrated in Fig.S2. Due to the negative charge, DNA translocation was also verified by the voltage-dependence of block duration and occurrence. Fig.3d shows the decrease of block duration; Fig.3e shows the increase of block occurrence as the voltage increased from -50 mV to +200 mV. The DNA translocation at a low negative voltage (-50 mV) could be an effect of the diffusion, the weak electrical force that prevents DNA from entering the pore, and the electroosmotic force that is reverse to the electrical force to drive molecules into the pore.
Figure 3.
Translocation of dsDNA in nanopores. a. Current blocks by 1 kb DNA (10 nM) in the external solution (1 M NaCl) through a 3.8 nS nanopore (+100 mV). b. Current blocks with DNA through a 39 nS nanopore (+100 mV). c. Reveals DNA translocation by PCR. Lane-1, marker; Lane-2, 10 μl bath solution (containing DNA); Lane-3, 10 μl dd-H2O; Lane-4, 10 μl internal solution without DNA (control); Lane-5, 10 μl internal solution near nanopore after the DNA translocation. d. Voltage-dependent duration of DNA translocation. e. Voltage-dependent occurrence of DNA translocation.
Earlier studies have shown that the duration of DNA translocation varies from ∼100 μs to ∼100 ms, depending on both the properties of nanopore and DNA. For example, the block for a 0.5 kb dsDNA molecule traversing a 5 nm Si3N4 pore (10 nm in length) at +120 mV12 was 5 ms, and the block for a 7.5 kb dsDNA molecule through a 40 nm polymer conical pore at +700 mV was 100 ms.45 The 26 ms block duration in our 39 nS pore (Fig.3b) is consistent with this time scale. However, the 1.1 s duration in the 4.9 nS pore (Fig.3a) is longer than shown in other studies. One possible explanation for this result was that the pore size at the threshold conductance is comparable to the dimension of the dsDNA molecule (2 nm). Thus, the traversing speed is slowed down due to stronger DNA-pore interaction. The block profile at the threshold conductance (Fig.3a) was also different than the typical rectangular block in a larger nanopore (Fig.3b), suggesting sequential steps involved in a translocation event (model in Fig.3a): the initial current drop marks the moment when a DNA molecule is entering the pore. The ensuing recovering current corresponds to the DNA threading the conical-shaped lumen and escaping from the wide opening of the pore into the pipette.
The current amplitude of the DNA block in the threshold conductance pore (∼2 pA, Fig.3a) is very small compared with that of the wider pore (∼30 pA, Fig.3b). The variation of DNA block conductance has been studied previously. The Bashir group first discovered that DNA translocation can increase, rather than decrease the pore conductance at 100 mM KCl.46 The Dekker group further found that the conductance variation due to DNA translocation (in a 10 nm pore) changed gradually from blockades to enhancements, as the salt concentration was lowered from 1 M, and the conductance amplitude was reduced to zero at 400 mM KCl.39 To better understand this phenomenon, an interfacial cation flow (Iint) between the DNA and nanopore was introduced that could compensate for the current loss from the DNA block.39;46 This Iint mechanism could be used to interpret the small conductance change for DNA translocation in our glass pore. However, due to the difference in the surface charge density, pore size and pore shape, the specific property of conductance change such as the ion concentration, may be different.
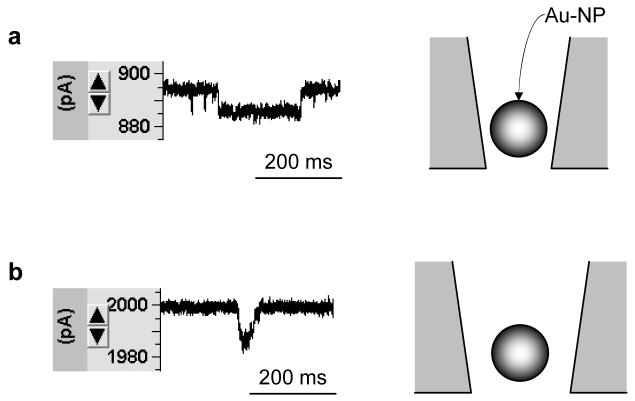
The translocation of gold nanoparticles (Au-NP, 10 nm) was detected in 15 mM NaCl. As with DNA molecules, the current block observed with Au-NP translocation can only be detected at positive voltages, due to the negative charge of Au-NP. We found that the nanoparticle block appeared only after the pore conductance was increased to the level of 6.5±0.7 nS (n=5) (Fig.4a). The block duration (+150 mV) was 110±30 ms and the block occurrence was 0.26±0.1 min-1. This conductance is the threshold for Au-NP translocation. At a higher conductance level, such as 13 nS, the block duration was 24 ms, and the occurrence was 0.65 min-1 (Fig.4b), indicating a decrease in translocation duration and increase in occurrence as the pore size increases.35 Karhanek and co-workers has reported a nanoparticle translocation in a 50-60 nm nanopipette.35 The translocation they observed had a duration of 20 ms and a current block amplitude of ∼10 pA (180 mV). This result is similar to the 24 ms duration and 15 pA current block amplitude (150 mV) in our 13 nS pore. Both results showed large residual current with a nanoparticle block, a phenomenon similar to that of the DNA molecule in nanopores as described above.
Figure 4.
Translocation of Au-NP through nanopore. Recording solutions inside and outside nanopore contained 15 mM NaCl. Au-NP of 10 nm in diameter (4.75 nM) was presented in external solution. Current was monitored at +150 mV. a, Current blocks in a 5.9 nS nanopore and b, a 13 nS nanopore.
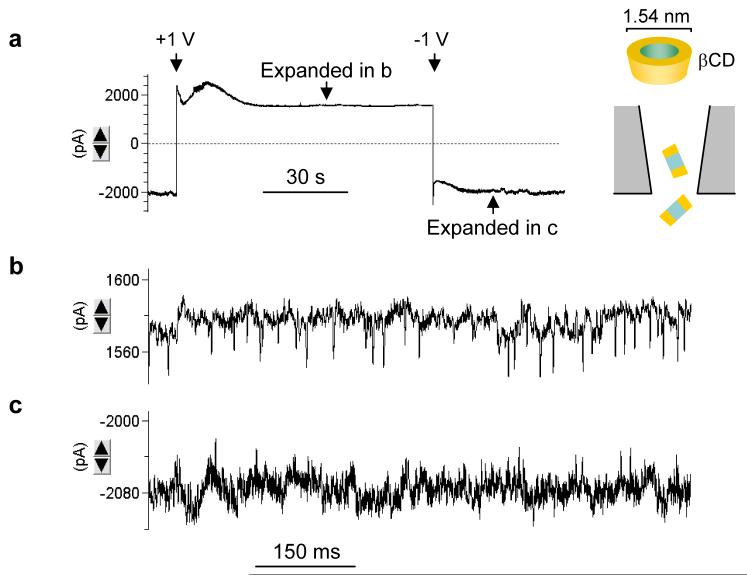
The translocation of β-cyclodextrin (βCD, 1.5 nm) from the internal solution was detected in 1 M NaCl. We found that βCD (100 μM) in the pipette produced detectable blocks as the pore conductance was increased to 2.9±0.4 nS (n=4) (Fig.5). Changing the voltage to +1 V, the conductance gradually decayed and stabilized at 1.6 nS (Fig.5a). The expanded current trace at this conductance level (Fig.5b) demonstrates a series of current blocks with a duration of 1.5±0.5 ms and occurrence of 26±7 s-1. As the voltage jumped to -1 V, the reduced conductance gradually recovered to the level of 2.9 nS without cyclodextrin (Fig.5a). The expanded trace at -1 V is shown in Fig.5c. Using the block current at +1 V (42 pA), we analyzed the blocks at -1 V. The block occurrence was 3.2±0.5 s-1, much lower than that at +1 V. This voltage-dependent current profile repeatedly occurred as the voltage switched between +1 V and -1 V. The current blocks are attributed to the translocation of individual βCD molecules. The voltage-dependence of βCD blocks in the solid nanopore is consistent with those observed in the α-hemolysin protein pore,17 where the voltage-dependence was attributed to the electroosmotic effect. The negative charge on glass surface induces a cation flow that can accumulate βCD at positive voltage to enhance the translocation; the cation flow is reversed at negative voltages, driving βCD dispersion into the pipette to reduce translocation events.
Figure 5.
Translocation of βCDs in nanopore. a. Voltage-dependent current profile in the presence of 100 μM βCD in a 2.4 nS nanopore. b. Expanded current trace from a +1 V domain in a, showing stochastic translocation of individual βCD molecules. c. Expanded current trace from a -1 V domain in a, showing fewer translocation events compared with +1 V in b.
The threshold conductance (g) for molecular translocation detected above was used in Eq.1 to evaluate the pore sizes. The pore size evaluated from DNA translocation was 1.9 nm, comparable to the 2 nm dimension of dsDNA. The pore size of βCD translocation was 1.4 nm, similar to the 1.5 nm diameter of βCD. The nanoparticle translocation was detected in 15 mM NaCl. The conductance of the same pores at 1 M NaCl was also determined for the calculation. The result shows that the pore size detected by the nanoparticles was 14 nm, similar but slightly larger than the 10 nm diameter of the nanoparticle. The molecular translocation results verified the molecular scale of the pore size and revealed the capability of the nanopore for single-molecule detection.
Trapping a single cyclodextrin in the nanopore and chiral-discrimination
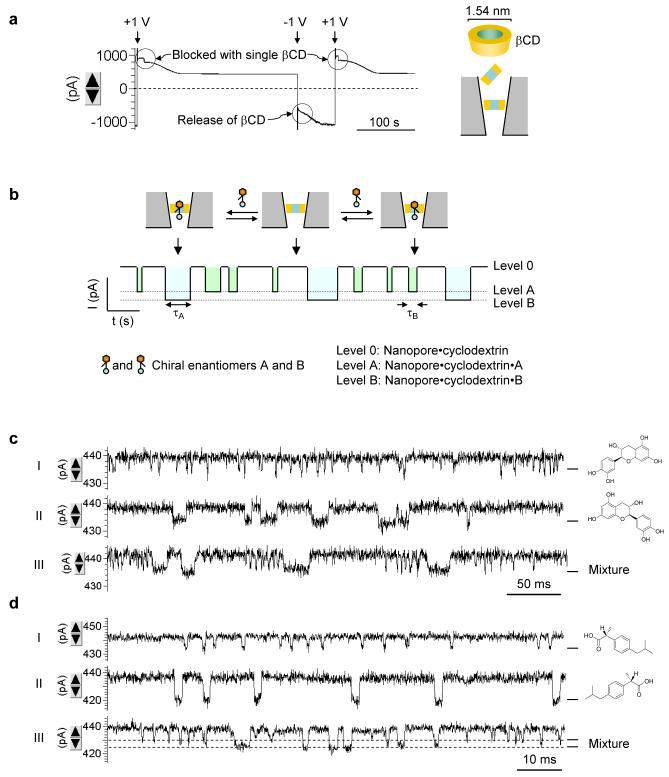
By achieving conductance lower than the threshold level, it is possible to create pores smaller than the probing molecule, therefore making the pore useful in trapping single molecules. Fig.6a shows the current profile for a 1 nS pore with 100 μM βCD presented in the pipette. Compared with the pore at the threshold conductance 2.9 nS (Fig.4a), the current profile of the 1 nS pore was characterized by a discrete current drop at +1 V, which repeatedly occurred as the voltage switched between -1 V and +1 V. We attribute the current step-down observed at +1 V to the block of a single cyclodextrin molecule in this narrower nanopore, driven by the electroosmotic force; βCD at -1 V is released from the nanopore into the pipette by a reversed cation flow.
Figure 6.
Chiral-discrimination by cyclodextrin trapped in nanopore. a. Current trace showing trapping and release of single βCD molecules (100 μM) in a 1 nS nanopore. b. Diagram showing single-molecule discrimination of chiral enantiomers with the cyclodextrin trapped in nanopore. c. Current profiles showing the binding of individual enantiomers of catechin to the βCD. The three traces were recorded for 50 μM 2S,3R-(-)-catechin (top), 50 μM 2R,3S-(+)-catechin (middle) and a mixture of 50 μM (-)-catechin and 50 μM (+)-catechin (bottom). D. Current profiles showing the binding of individual enantiomers of ibuprofen to the trapped βCD. The three traces were for 100 μM R-(-)-ibuprofen (top), 100 μM S-(+)-ibuprofen (middle) and a mixture 100 μM R-(-)-ibuprofen and 100 μM S-(+)-ibuprofen (bottom). The current block levels by R- and S-ibuprofen were marked with dash lines.
βCD has been reported to lodge in the αHL protein pore, and has been used as a molecular adapter for sensing of organic compounds.3 The sequential binding-release of individual guest molecules from the trapped cyclodextrin results in a series of discrete current blocks useful for identifying and quantifying guest compounds in the mixture (Fig.6b). Recently the βCD·αHL system has been used for identify chiral molecules.47 Here we demonstrated the enantiomer discrimination with βCD in the solid nanopore. Fig.6c-d shows the identification of catechin enantiomers.48 The presence of (+)-catechin (Fig.6c, top trace) or (-)-catechin (Fig.6c, middle trace) in the βCD solution inside the 1 nS nanopore discretely and stochastically altered the current. Their block durations were 14±3 ms (Fig.6c) and 1.5±0.4 ms (Fig.6d) respectively. In control tests, no such event was detectable in the absence of the guest or when presenting the guest in a βCD-free nanopore. As (+)- and (-)-catechin were simultaneously presented, we observed two types of single-molecule blocks distinguished by the duration (Fig.6c, bottom trace). One was 14 ms for (+)-catechin and the other was 1.5 ms for (-)-catechin. In another example, Fig.6d shows the separation of ibuprofen.49 The duration of blocks with (S)-ibuprofen was 2.0±0.3 ms (Fig.6d, top trace), compared with 0.79±0.21 ms for the (R)-ibuprofen blocks (Fig.6d, middle trace). Both block durations are in agreement with the 1.5 ms and 0.83 ms for the two ibuprofen in βCD·αHL reported previously.47 When the mixture of (S)- and (R)-ibuprofen is applied, we can not only separate the two molecules from their block durations, but also the amplitude of block current: (S)-ibuprofen (∼17 pA) reduced more current than (R)-ibuprofen (11 pA) (Fig.6d, bottom trace). The same trend of current block amplitude was also identified for βCD·αHL47. These consistent results confirm the binding of guests to the trapped cyclodextrin. Because only a single-level current block was identified for each guest, it was expected that a single molecule of βCD would be trapped in the nanopore, since the binding of this guest with multiple cyclodextrins would have produced addition block levels of conductance. It should be noted that the residual current at the point of the guest binding was still large compared with that of protein pore with a cyclodextrin.47 Possibly, the orientation of the cyclodextrin in the solid pore is not parallel to the axis of the pore lumen, as it did in the αHL pore. Instead, the cyclodextrin may be vertically anchored into the pore, yielding a large “leaking” of the current that bypasses the cyclodextrin. The guest molecule binding to the cavity of cyclodextrin in this orientation will block little bypass current. We used nanopores to separate not only catechin and ibuprofen, but camphanic acid enantiomers and adamantanes (non-chiral compounds), described in Supplementary Materials.
The current signatures yielded by the guest binding should be useful in studying host-guest kinetics. The reciprocal of the block duration represents the “off” rate constant, and the block occurrence divided by the guest concentration yields the “on” rate constant for guest-host interaction. At present, the guest concentration near the nanopore in the pipette terminal is unknown. Previous studies of entrapping biomolecules have shown optically higher analyte concentration in the nanopore than in the bulk solution under high voltages,50;51 through a complex process determined by electroosmosis, electrophoresis and dielectrophoresis.50;51 The guest concentration in nanopore needs further investigation, to obtain the true kinetic constants for host-guest interactions.
The findings of this research are significant both for the practice of fabricating nanopores and for understanding physical and chemical properties in nano-confinement. Because the pore can be made smaller than a molecule, it is possible to implant a single molecule into the pore to reveal host-guest interaction. It could be used to screen synthesized libraries of cyclic peptides52 or exploring bio-processes, such as DNA digestion by the enzymatic ring-shaped motors.53 Nanopores could also be made of special materials to achieve new properties. For example, we might create a pore with a hydrophobic lumen. Once filled with conductors such as gold, for example, the nanopore could become a molecular-scaled electrode for in vivo detection. Nanopores could also be used as atomic-scaled pens for the delivery of single molecules;54 and used to track single molecule processes.55
Supplementary Material
Acknowledgements
We thank Gabor Forgacs for the suggestion of the DNA translocation experiment and for his help the electron microscopic observation of the nanopore. We also thank Rainer Glaser and Kevin Gillis for invaluable comments on the manuscript. This investigation was supported by grants from the National Science Foundation, the National Institutes of Health, the University of Missouri Research Board and the University of Missouri Research Council. This investigation was conducted in a facility constructed with support from a Research Facilities Improvement Program Grant (C06-RR-016489) from the National Center for Research Resources at the National Institutes of Health.
Reference List
- 1.Bayley H, Cremer PS. Nature. 2001;413:226–30. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 2.Bezrukov SM, Vodyanoy I, Parsegian VA. Nature. 1994;370:279–81. doi: 10.1038/370279a0. [DOI] [PubMed] [Google Scholar]
- 3.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Nature. 1999;398:686–90. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 4.Movileanu L, Howorka S, Braha O, Bayley H. Nature Biotechnology. 2000;18:1091–95. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 5.Siwy Z, Trofin L, Kohli P, Baker LA, Trautmann C, Martin CR. Journal of the American Chemical Society. 2005;127:5000–01. doi: 10.1021/ja043910f. [DOI] [PubMed] [Google Scholar]
- 6.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13770–73. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1079–84. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Nature Biotechnology. 2001;19:248–52. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 9.Howorka S, Cheley S, Bayley H. Nature Biotechnology. 2001;19:636–39. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 10.Mathe J, Visram H, Viasnoff V, Rabin Y, Meller A. Biophysical Journal. 2004;87:3205–12. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Quesada J, Saghatelian A, Cheley S, Bayley H, Ghadiri MR. Angewandte Chemie-International Edition. 2004;43:3063–67. doi: 10.1002/anie.200453907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Nature. 2001;412:166–69. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 13.Li JL, Gershow M, Stein D, Brandin E, Golovchenko JA. Nature Materials. 2003;2:611–15. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 14.Kohli P, Harrell CC, Cao ZH, Gasparac R, Tan WH, Martin CR. Science. 2004;305:984–86. doi: 10.1126/science.1100024. [DOI] [PubMed] [Google Scholar]
- 15.Iqbal SM, Akin D, Bashir R. Nature Nanotechnology. 2007;2:243–48. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- 16.Gu LQ, Dalla Serra M, Vincent JB, Vigh G, Cheley S, Braha O, Bayley H. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3959–64. doi: 10.1073/pnas.97.8.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu LQ, Cheley S, Bayley H. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15498–503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu LQ, Cheley S, Bayley H. Science. 2001;291:636–40. doi: 10.1126/science.291.5504.636. [DOI] [PubMed] [Google Scholar]
- 19.Luchian T, Shin SH, Bayley H. Angewandte Chemie-International Edition. 2003;42:3766–71. doi: 10.1002/anie.200351313. [DOI] [PubMed] [Google Scholar]
- 20.Bayley H, Jayasinghe L. Molecular Membrane Biology. 2004;21:209–20. doi: 10.1080/09687680410001716853. [DOI] [PubMed] [Google Scholar]
- 21.Shim JW, Gu LQ. Analytical Chemistry. 2007;79:2207–13. doi: 10.1021/ac0614285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang XF, Cheley S, Rice-Ficht AC, Bayley H. Journal of the American Chemical Society. 2007;129:4701–05. doi: 10.1021/ja068654g. [DOI] [PubMed] [Google Scholar]
- 23.White RJ, Ervin EN, Yang T, Chen X, Daniel S, Cremer PS, White HS. J.Am.Chem.Soc. 2007;129:11766–75. doi: 10.1021/ja073174q. [DOI] [PubMed] [Google Scholar]
- 24.Siwy Z, Gu Y, Spohr HA, Baur D, Wolf-Reber A, Spohr R, Apel P, Korchev YE. Europhysics Letters. 2002;60:349–55. [Google Scholar]
- 25.Harrell CC, Lee SB, Martin CR. Analytical Chemistry. 2003;75:6861–67. doi: 10.1021/ac034602n. [DOI] [PubMed] [Google Scholar]
- 26.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Nature Materials. 2003;2:537–40. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 27.Ho C, Qiao R, Heng JB, Chatterje A, Timp RJ, Aluru NR, Timp G. Proc.Natl.Acad.Sci.U.S A. 2005;102:10445–50. doi: 10.1073/pnas.0500796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MJ, Wanunu M, Bell DC, Meller A. Adv Mater. 2006;18:3149. [Google Scholar]
- 29.Powell MR, Sullivan M, Vlassiouk I, Constantin D, Sudre O, Martens CC, Eisenberg RS, Siwy ZS. Nature Nanotechnology. 2008;3:51–57. doi: 10.1038/nnano.2007.420. [DOI] [PubMed] [Google Scholar]
- 30.Gorelik J, Gu YC, Spohr HA, Shevchuk AI, Lab MJ, Harding SE, Edwards CRW, Whitaker M, Moss GWJ, Benton DCH, Sanchez D, Darszon A, Vodyanoy I, Klenerman D, Korchev YE. Biophysical Journal. 2002;83:3296–303. doi: 10.1016/S0006-3495(02)75330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klenerman D, Korchev Y. Nanomedicine. 2006;1:107–14. doi: 10.2217/17435889.1.1.107. [DOI] [PubMed] [Google Scholar]
- 32.Piper JD, Clarke RW, Korchev YE, Ying LM, Klenerman D. Journal of the American Chemical Society. 2006;128:16462–63. doi: 10.1021/ja0650899. [DOI] [PubMed] [Google Scholar]
- 33.Ying LM, Bruckbauer A, Rothery AM, Korchev YE, Klenerman D. Analytical Chemistry. 2002;74:1380–85. doi: 10.1021/ac015674m. [DOI] [PubMed] [Google Scholar]
- 34.Wei C, Bard AJ, Feldberg SW. Analytical Chemistry. 1997;69:4627–33. [Google Scholar]
- 35.Karhanek M, Kemp JT, Pourmand N, Davis RW, Webb CD. Nano Lett. 2005;5:403–07. doi: 10.1021/nl0480464. [DOI] [PubMed] [Google Scholar]
- 36.Shao Y, Mirkin MV. Journal of the American Chemical Society. 1997;119:8103–04. [Google Scholar]
- 37.White RJ, Zhang B, Daniel S, Tang JM, Ervin EN, Cremer PS, White HS. Langmuir. 2006;22:10777–83. doi: 10.1021/la061457a. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, Zhang YH, White HS. Analytical Chemistry. 2004;76:6229–38. doi: 10.1021/ac049288r. [DOI] [PubMed] [Google Scholar]
- 39.Smeets RMM, Keyser UF, Krapf D, Wu MY, Dekker NH, Dekker C. Nano Letters. 2006;6:89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 40.White HS, Bund A. Langmuir. 2008;24:2212–18. doi: 10.1021/la702955k. [DOI] [PubMed] [Google Scholar]
- 41.Kalman EB, Vlassiouk I, Siwy ZS. Adv Mater. 2008;20:293–97. [Google Scholar]
- 42.Apel PY, Korchev YE, Siwy Z, Spohr R, Yoshida M. Nuclear Instruments & Methods in Physics Research Section B-Beam Interactions with Materials and Atoms. 2001;184:337–46. doi: 10.1016/s0168-583x(01)00693-0. [DOI] [PubMed] [Google Scholar]
- 43.Ito T, Sun L, Crooks RM. Anal.Chem. 2003;75:2399–406. doi: 10.1021/ac034072v. [DOI] [PubMed] [Google Scholar]
- 44.Merzlyak PG, Yuldasheva LN, Rodrigues CG, Carneiro CM, Krasilnikov OV, Bezrukov SM. Biophys.J. 1999;77:3023–33. doi: 10.1016/S0006-3495(99)77133-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrell CC, Choi Y, Horne LP, Baker LA, Siwy ZS, Martin CR. Langmuir. 2006;22:10837–43. doi: 10.1021/la061234k. [DOI] [PubMed] [Google Scholar]
- 46.Chang H, Kosari F, Andreadakis G, Alam MA, Vasmatzis G, Bashir R. Nano Letters. 2004;4:1551–56. [Google Scholar]
- 47.Kang XF, Cheley S, Guan XY, Bayley H. Journal of the American Chemical Society. 2006;128:10684–85. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 48.Smith VK, Ndou TT, Warner IM. Journal of Physical Chemistry. 1994;98:8627–31. [Google Scholar]
- 49.Reijenga JC, Ingelse BA, Everaerts FM. Journal of Chromatography A. 1997;792:371–78. [Google Scholar]
- 50.Clarke RW, White SS, Zhou DJ, Ying LM, Klenerman D. Angewandte Chemie-International Edition. 2005;44:3747–50. doi: 10.1002/anie.200500196. [DOI] [PubMed] [Google Scholar]
- 51.Ying LM, White SS, Bruckbauer A, Meadows L, Korchev YE, Klenerman D. Biophysical Journal. 2004;86:1018–27. doi: 10.1016/S0006-3495(04)74177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghadiri MR, Granja JR, Buehler LK. Nature. 1994;369:301–04. doi: 10.1038/369301a0. [DOI] [PubMed] [Google Scholar]
- 53.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Science. 2003;301:1235–38. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 54.Bruckbauer A, Ying L, Rothery AM, Zhou D, Shevchuk AI, Abell C, Korchev YE, Klenerman D. J.Am.Chem.Soc. 2002;124:8810–11. doi: 10.1021/ja026816c. [DOI] [PubMed] [Google Scholar]
- 55.Harms GS, Orr G, Montal M, Thrall BD, Colson SD, Lu HP. Biophysical Journal. 2003;85:1826–38. doi: 10.1016/S0006-3495(03)74611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.