Abstract
Epithelial–stromal cell interactions have an important role in breast tumor progression, but the molecular mechanisms underlying these effects are just beginning to be understood. We previously described that fibroblasts promote, whereas normal myoepithelial cells inhibit, the progression of ductal carcinoma in situ (DCIS) to invasive breast carcinomas by using a xenograft model of human DCIS. Here, we report that the tumor growth and progression-promoting effects of fibroblasts are at least in part due to increased COX-2 expression in tumor epithelial cells provoked by their interaction with fibroblasts. Up-regulation of COX-2 in DCIS xenografts resulted in increased VEGF and MMP14 expression, which may contribute to the larger weight and invasive histology of COX-2-expressing tumors. Administration of celecoxib, a selective COX-2 inhibitor, to tumor-bearing mice decreased xenograft tumor weight and inhibited progression to invasion. Coculture of fibroblasts with DCIS epithelial cells enhanced their motility and invasion, and this change was associated with increased MMP14 expression and MMP9 protease activity. We identified the NF-κB pathway as one of the mediators of stromal fibroblast-derived signals regulating COX-2 expression in tumor epithelial cells. Inhibition of NF-κB and COX-2 activity and down-regulation of MMP9 expression attenuated the invasion-promoting effects of fibroblasts. These findings support a role for COX-2 in promoting the progression of DCIS to invasive breast carcinomas, and suggest that therapeutic targeting of the NF-κB and prostaglandin signaling pathways might be used for the treatment and prevention of breast cancer.
Keywords: breast cancer, stroma, tumor progression, DCIS, celecoxib
Increasing evidence supports the importance of epithelial–stromal cell interactions in tumor growth, progression, angiogenesis, and therapeutic resistance (1–4). Despite the importance of these interactions in tumorigenesis, the underlying molecular mechanisms are poorly characterized, in part because of their complexity and redundancy. Multiple factors secreted by multiple cell types can exert the same effects, creating a challenge for approaches aiming at their therapeutic targeting. Most studies analyzing epithelial–stromal cell interactions have focused on cytokines and chemokines as potential mediators of this cross-talk (5, 6), whereas the involvement of lipid signaling including various prostaglandins (PGs) has not been extensively investigated.
The prostaglandin synthetase complex of enzymes has a key role in converting membrane arachidonic acid to PGs that subsequently mediate their effects by means of G protein-coupled receptors (GPCRs) (7). One of the components of this complex is the COX-2 cyclooxygenase enzyme that is expressed in a cell type-specific manner and is inducible by various mitogenic and inflammatory stimuli (8). COX-2 has been implicated to have a role in breast tumorigenesis based on its increased expression in a significant fraction of breast carcinomas and the protective effects of nonsteroidal antiinflammatory drugs (NSAIDs) against breast cancer (9). Immunohistochemical analysis of COX-2 expression in human breast carcinomas have revealed variable and sometimes contradictory results both with regards of the fraction of tumors expressing COX-2 and the associations between its expression and clinicopathologic characteristics of the tumors (9). Variability of antibodies, heterogeneous expression of COX-2 within tumors, small sample size, and differences in data interpretation are potential explanations of the conflicting results. However, the general agreement is that a significant subset of breast tumors express COX-2, and that its expression is associated with unfavorable clinical behavior, including poorly differentiated histology, higher risk of distant metastasis, and shorter survival (10).
The potential role of COX-2 in the preinvasive stages of breast tumorigenesis has especially been intensely investigated due to human epidemiological studies linking the use of NSAIDs to decreased risk of breast cancer (11), and to our relative inability to predict the clinical course of these early stage lesions. Analysis of COX-2 expression in combination with that of other markers (p16 and Ki67) in DCIS was found to be associated with subsequent tumor events, although it did not predict the risk of invasive recurrence potentially due to small sample size (12). In line with this observation, a recent study reported that COX-2 expression in atypical epithelial hyperplasia correlated with the development of subsequent breast cancer, identifying COX-2 as a potential biomarker for risk prediction and selection of patients for chemopreventive studies (13).
We have previously analyzed the progression of in situ to invasive breast carcinomas by molecular profiling of human tumors and experiments in DCIS models (14–16). Using these approaches, we identified dramatic changes in the gene expression and epigenetic profiles of each cell type composing the microenvironment, and demonstrated a role for these alterations in the progression of DCIS tumors. In the present study, we investigated whether COX-2 is a potential mediator of tumor epithelial–stromal cell interactions that promote the growth and progression of in situ tumors using a xenograft model of human DCIS.
Results and Discussion
Expression of COX-2 in the MCFDCIS Xenograft Model.
Our prior studies demonstrated that a xenograft model based on the MCF10DCIS.com (referred to as MCFDCIS) breast cancer cell line reproduces main aspects of human basal-like breast tumor progression (15), and, thus, can be used for investigating molecular mechanisms underlying the transition of in situ to invasive breast carcinomas. We have also illustrated that coinjection of primary cultured stromal fibroblasts derived from normal breast tissue (PBS), breast tumors (PBTS), or rheumatoid arthritis synovium (RASF) promoted invasive progression, and this effect was inhibited by normal myoepithelial (HME) cells (15).
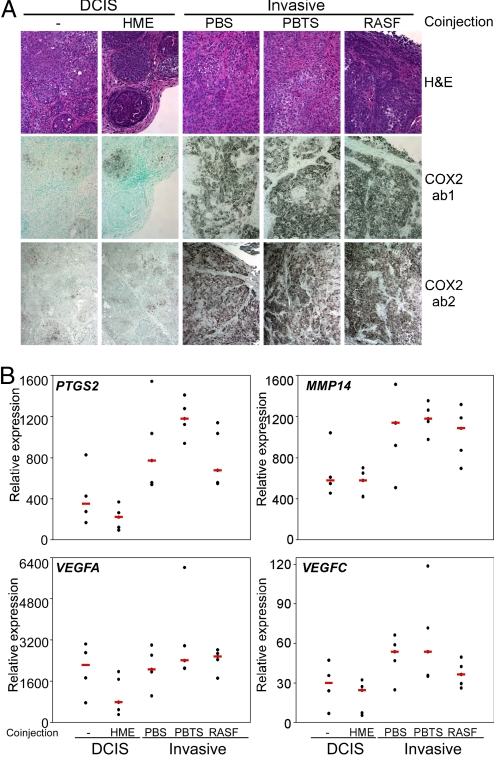
To investigate the role of COX-2 in the progression of DCIS to invasive carcinomas, we evaluated the expression of COX-2 in MCFDCIS xenografts with DCIS and invasive histology by immunohistochemical analysis of representative tumors from each experimental group. The level of COX-2 was low in MCFDCIS and MCFDCIS+HME tumors, only a few COX-2-positive cells could be detected, whereas coinjection of fibroblasts led to its dramatic up-regulation in tumor epithelial cells (Fig. 1A). This observation was consistent with findings in pancreatic cancer, where COX-2 expression was markedly augmented in tumor cells in response to coculture with fibroblasts, and down-regulation of COX-2 decreased the invasive properties of cancer cells acquired through epithelial-mesenchymal interactions (17). However, in contrast to this in vitro pancreatic cancer study, we did not see up-regulation of COX-2 in the stromal cells of any of the xenografts (Fig. 1A). The MCFDCIS cell alone xenografts displayed low COX-2 expression even at later time points (7–8 weeks) after injection, when all of the tumors acquired invasive morphology. Thus, the up-regulation of COX-2 in tumor epithelial cells might not be a consequence of invasive progression, but it is likely due to the coinjected human fibroblasts.
Fig. 1.
The expression of COX-2 and angiogenic factors in MCFDCIS xenografts. (A) Immunohistochemical analysis of COX-2 expression in MCFDCIS xenografts using an antibody specific for human COX-2 (ab1) and one that also recognizes murine COX-2 (ab2). Low expression was detected in MCFDCIS cells injected alone (−) or coinjected with HME cells, whereas coinjection of any fibroblast (e.g., PBS, PBTS, and RASF) up-regulated COX-2 expression in tumor epithelial cells. No COX-2-positive cells were detected in the stroma. (B) Quantitative RT-PCR analysis of human-specific COX-2 (PTGS2), MMP14, VEGFA, and VEGFC gene expression in MCFDCIS xenografts. Decreased and increased expression of these genes was detected in MCFDCIS cells coinjected with HME cells and fibroblasts (PBS, PBTS, and RASF), respectively, relative to human-specific hypoxanthine phosphoribosyltransferase 1 (HPRT1) used as loading control.
COX-2 is known to have a role in promoting tumor cell invasion and angiogenesis by means of its up-regulation of extracellular matrix-degrading proteases and angiogenic factors (18, 19). To investigate the association between COX-2 expression and markers of invasion and angiogenesis in our DCIS xenograft model, we analyzed the mRNA levels of human COX-2 (PTGS2), MMP9, MMP13, MMP14, MMP15, MMP16, VEGFA, VEGFC, and mouse CXCL12 by quantitative RT-PCR in representative tumors from each experimental group using species-specific primers. Coinjection of HME cells decreased, whereas that of fibroblasts increased the expression of PGTS2, MMP14, VEGFA, and VEGFC, but not all of the observed differences were statistically significant (Fig. 1B). The expression of human MMP9, MMP13, MMP15, MMP16, and murine CXCL12 were not significantly different in any of the xenografts analyzed. The up-regulation of MMP14 and VEGF by COX-2 might contribute to the increased growth and invasive histology of xenografts derived from MCFDCIS cells coinjected with fibroblasts, but the involvement of other genes and pathways cannot be excluded.
Effect of a Selective COX-2 Inhibitor on Tumor Growth and Progression to Invasion.
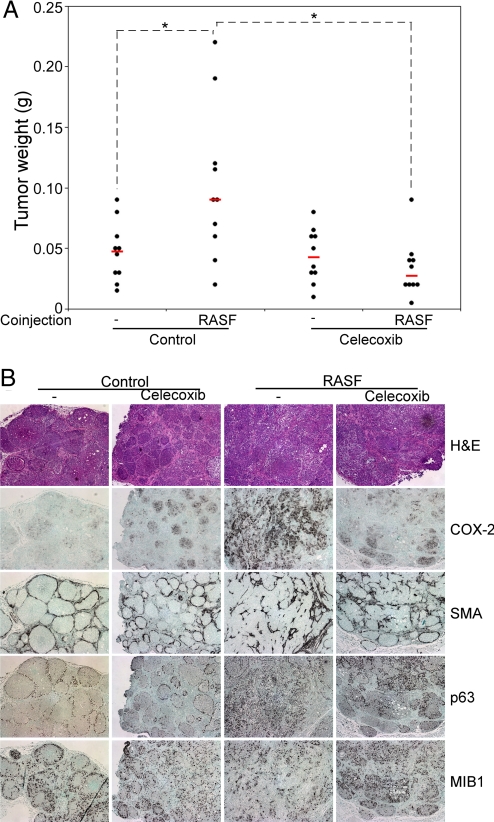
Based on these observations, we hypothesized that up-regulation of COX-2 in tumor epithelial cells by coinjected fibroblasts might be responsible for their tumor growth and progression promoting effects. Thus, these effects may be abolished or decreased by inhibiting COX-2 activity. To test this hypothesis, we analyzed the consequences of treatment of mice with celecoxib, a selective COX-2 inhibitor, on the weight and histology of xenografts derived from MCFDCIS cells injected alone or together with RASF inflammatory fibroblasts. We chose RASF coinjections for these studies, because, based on our previous results, the coinjection of RASFs produced the most consistent and significant increase in tumor weight and promotion to invasion (15). Feeding the mice with celecoxib containing diet had no significant effect on the growth of MCFDCIS cells-alone xenografts, but it completely eliminated the tumor growth-promoting effects of the coinjected RASFs (Fig. 2A), and partially inhibited DCIS progression to invasive tumors (Fig. 2B). Similar, although less significant, results were obtained using sulindac, another NSAID [supporting information (SI) Fig. S1]. Also, the increase in COX-2 expression induced in MCFDCIS cells by RASF coinjection was eliminated by celecoxib treatment (Fig. 2B), potentially due to the attenuation of factors secreted by RASFs (e.g., cytokines, chemokines, and PGs) that led to the up-regulation of COX-2 by means of paracrine interactions, whereas the mRNA levels of MMP14 were not significantly altered after celecoxib treatment (Fig. S2). These effects of celecoxib were not likely to be due to its inhibition of inflammatory leukocytes, because we used immunodeficient (NCRNU nude) mice and did not detect infiltrating inflammatory cells (CD45+ cells) in any of the xenografts at the time points we analyzed (Fig. S3). It is also unlikely that the results were due to the selective killing of RASFs by celecoxib, because these cells do not survive long term in mice (15), and coinjection of lethally irradiated cells had even more pronounced tumor-promoting effects. The absence of neutrophils and macrophages that were supposed to be functional even in NCRNU mice in the xenografts is interesting and may reflect lack of inflammatory reaction in this model, which could potentially influence the histology of the resulting tumors. However, the potential inhibition of macrophages and leukocytes by celecoxib cannot be completely excluded.
Fig. 2.
The effect of celecoxib on MCFDCIS xenografts. The effect of a selective COX-2 inhibitor (celecoxib) on the weight (A) and histology (B) of MCFDCIS xenografts derived from cells injected alone (−) or coinjected with RASFs on control or celecoxib-containing diet. Xenografts from MCFDCIS cells alone (−) had DCIS histology and low COX-2 expression, regardless of diet. Tumors from MCFDCIS cells coinjected with RASF showed invasive phenotype and high COX-2 levels in the control diet group. Celecoxib abolished the tumor growth-stimulating effects of RASF (P = 0.0001), partially inhibited the progression to invasive tumors, and decreased COX-2 protein levels. No significant difference in the number of cycling (MIB1+) cells was detected after celecoxib treatment, but this observation does not exclude the possibility of decreased proliferation due to increased cell cycle length or increased apoptosis after celecoxib treatment. Statistically significant (P < 0.05) differences in tumor weight are marked with dashed lines and asterisks.
Epithelial–Stromal Cell Interactions in Cell Culture Models.
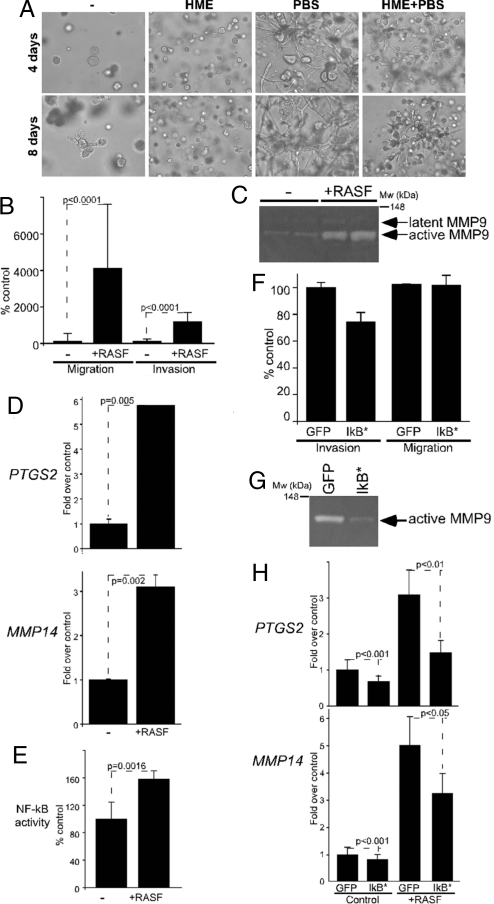
To begin delineating molecular mechanisms underlying interactions between MCFDCIS tumor epithelial cells and fibroblasts, and the role of these interactions in invasive progression, we developed 2D and 3D coculture models including both cell types. We previously described that coculture of MCFDCIS cells with breast stromal fibroblasts in Matrigel resulted in invasive branching growth, compared with spheroids formed by MCFDCIS cells cultured alone (20). We extended these studies further and determined that coculture of MCFDCIS cells with fibroblasts in Matrigel led to the invasive branching growth of MCFDCIS cells, and this behavior was suppressed by the inclusion of HME cells (Fig. 3A). These results were consistent with our findings in MCFDCIS xenografts, where all fibroblasts promoted progression to invasion and HME cells were able to suppress this effect (15). Based on these observations, we concluded that at least some aspects of invasive progression and cell–cell interactions that occur in MCFDCIS xenografts can be reproduced in our cell culture models; thus, these cultures can be used for the dissection of signaling pathways involved in these processes.
Fig. 3.
The effect of RASF coculture on MCFDCIS cells. (A) Morphology of MCFDCIS cells cultured alone (−) or together with the indicated cell types for 4 and 8 days after plating them into Matrigel. HME cells suppressed the growth of MCFDCIS cells and decreased the invasion-promoting effects of fibroblasts. (B) Coculture with RASFs statistically significantly (P < 0.0001) increased the migration and invasion of MCFDCIS cells. (C) MMP9 gelatinase activity detected in zymograms using conditioned media of MCFDCIS cells cultured alone or cocultured with RASFs. Latent and activate forms of MMP9 (determined based on molecular weight) are marked with arrows. Migration of the molecular weight marker is indicated. (D) Quantitative RT-PCR analysis of PTGS2 (COX-2) and MMP14 expression in MCFDCIS cells cultured alone (−) or with RASFs (+RASF); y axis indicates relative mRNA levels normalized to RPL39. (E) Activation of NF-κB in MCFDCIS cells by RASF coculture; y axis indicates NF-κB dependent firefly luciferase activity relative to Renilla luciferase. (F) The expression of IκB* in MCFDCIS cells attenuated their enhanced invasion (P = 0.049) induced by RASF cells without significant effect on their motility (P = 0.949). (G) The activity of MMP9 induced by RASF coculture was reduced by expressing IκB* in MCFDCIS cells. Cells infected with GFP expressing adenovirus were used as controls. (H) IκB* expression in MCFDCIS cells attenuated the increase in PTGS2 and MMP14 mRNA levels induced by RASF coculture. Infection with GFP expressing adenovirus had no effect; y axis indicates mRNA levels normalized relative to that of RPL39.
To quantitate the proinvasive effects of cocultured RASFs on MCFDCIS cells, we performed Boyden chamber assays using fluorescently labeled MCFDCIS cells to differentiate between the 2 cell types. These experiments showed that RASFs dramatically increased the motility of MCFDCIS cells and their invasion through Matrigel (Fig. 3B). The invasion-promoting effects of RASFs did not require direct cell–cell contact, because conditioned media of RASF cells produced essentially the same effects. Correlating with this finding, when conditioned media from these cocultures was analyzed by zymography, we detected increased MMP9 gelatinase activity, compared with conditioned media from epithelial cell only cultures (Fig. 3C).
To investigate gene expression alterations in MCFDCIS cells that accompanied changes in their phenotype provoked by RASFs, we performed quantitative RT-PCR (qPCR) analysis of selected genes. Using this approach, we determined that COX-2 and MMP14 mRNA levels were significantly higher in MCFDCIS cells recovered from 2D cocultures with RASFs correlating with results in xenografts (Fig. 3D). Despite increased MMP9 gelatinase activity in these cocultures, we were not able to detect changes in MMP9 mRNA and protein levels in MCFDCIS cells in any of the conditions analyzed. MMP14 (MT1-MMP) is an upstream activator of multiple MMPs (21); thus, increased MMP14 levels in cocultures might lead to increased MMP9 activity without altering MMP9 mRNA and protein levels. However, due to the complex regulation of MMPs and other ECM degrading proteases, alternative hypotheses cannot be excluded. Quantitative PCR analyses of MCFDCIS cells recovered from 3D colonies grown embedded in Matrigel essentially reproduced the findings obtained in 2D cultures with respect of COX-2, MMP14, and MMP9 expression. Thus, based on these results, the up-regulation of COX-2 and subsequent induction of MMP14 and MMP9 in MCFDCIS cells due to RASF coculture might be responsible for the invasion promoting effects of RASFs.
NF-κB Pathway As a Mediator of Epithelial–Stromal Cell Interactions in DCIS.
COX-2 activity and expression are regulated at multiple levels, including transcriptional and posttranscriptional mechanisms (22). One of the main transcriptional regulators of COX-2 expression is NF-κB (22), a pathway activated by many cytokines and chemokines secreted by inflammatory and mesenchymal cells (23). Gene expression profiling of invasion promoting RASFs and tumor-suppressing HMEs identified dramatic differences in the expression of numerous genes encoding for secreted proteins that might explain the opposing effects of these 2 cell types on in situ to invasive breast carcinoma progression (15). To extend these studies further, we assessed the levels of 120 cytokines in conditioned media of RASFs and HMEs, and found high levels of CCL2, IGFBP3, TIMP1, and TIMP2, specifically in RASFs (Fig. S4). Correlating with this data, conditioned media or coculture with RASFs led to increased NF-κB activity in MCFDCIS cells, determined by using a luciferase reporter assay (Fig. 3E).
To test the requirement for NF-κB activation in mediating the invasion-promoting effects of RASFs, we expressed a mutant “superrepressor” IκB in MCFDCIS cells using a recombinant adenovirus. The transcriptional activity of NF-κB is regulated at multiple levels, including its sequestration in the cytoplasm by the IκB repressor protein (24). Signals that activate the NF-κB pathway lead to the phosphorylation of IκB by IKKs and its subsequent ubiquitination and degradation by the proteasomal pathway, allowing for the translocation of NF-κB into the nucleus and activation of its target genes (24). The superrepressor IκB mutant (IκB*) cannot be phosphorylated and degraded; thus, it keeps NF-κB in its inactive cytoplasmic form (25).
MCFDCIS cells expressing this IκB* mutant demonstrated decreased invasion through Matrigel when cocultured with RASFs, but cellular motility was not altered by reduced NF-κB activity (Fig. 3F). The reduced invasiveness of MCFDCIS cells was accompanied with lower MMP9 gelatinase activity in zymograms using conditioned media from cocultures of IκB*-expressing MCFDCIS and RASFs (Fig. 3G). Expression of IκB* in MCFDCIS cells also decreased, but did not abolish the up-regulation of COX-2 and MMP14 expression induced by RASF coculture (Fig. 3H). Thus, inhibition of NF-κB signaling attenuated, but did not completely eliminate the effects of RASF cocultures on MCFDCIS cells. This incomplete inhibition could potentially be explained by the activation of multiple signaling networks induced by epithelial–stromal cell interactions. For example, numerous cytokines secreted by RASF cells (e.g., CCL2) activate ERK, AKT, and calcium signaling besides NF-κB that can also up-regulate the expression of COX-2 and MMPs and increase cell motility and invasion (26). Also, various PGs and other lipid mediators, and reactive oxygen species (ROS) produced by RASFs, can all exert tumor growth and invasion promoting effects. Correlating with this hypothesis, we were not able to reproduce the tumor growth and invasion-promoting effects of RASFs in cell culture and xenograft models by incubating MCFDCIS cells with any of the cytokines analyzed, suggesting that a combination of multiple factors are involved in this process.
The redundancy of secreted factors and networks mediating epithelial–stromal cell interactions highlight the difficulties associated with targeting alterations in the tumor microenvironment for cancer treatment. Multiple cell types (e.g., myofibroblasts, leukocytes, and endothelial cells) present in the tumor microenvironment can produce multiple cytokines and chemokines that can provoke very similar responses in tumor epithelial cells. Thus, inhibiting any of these pathways may not result in discernable therapeutic response. Similarly, this redundancy makes it difficult to envision how tumor epithelial cells could evolve dependency on any particular stromal cell type, and how this interaction could provide selective advantage for the expansion of a specific stromal cell clone.
Inhibition of COX-2 and NF-κB.
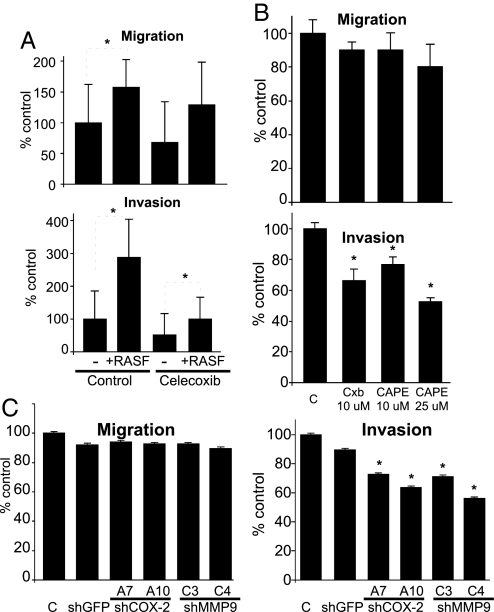
To further investigate the role of COX-2 and NF-κB in mediating the tumor invasion-promoting effects of RASFs in MCFDCIS cells, we analyzed the consequences of their pharmaceutical inhibition in cocultures. Celecoxib, a selective COX-2 inhibitor, significantly reduced the invasion of MCFDCIS cells in the presence of RASFs, but it had no effect on cellular motility (Fig. 4A). Similarly, an inhibitor of NF-κB activation, CAPE, dose-dependently inhibited the enhanced invasion of MCFDCIS cells induced by RASF coculture, whereas cell migration was not influenced to the same extent (Fig. 4B).
Fig. 4.
Inhibition of COX-2 or NF-κB in cocultures. Inhibition of COX-2 activity by using celecoxib (A) or via inhibition of NF-κB activation using CAPE (B) inhibited the increased invasive capacity of MCFDCIS cells induced by RASF coculture, whereas cell motility was not affected. Relative migration and invasion compared with MCFDCIS cells cultured alone are indicated on the y axis. Statistically significant (P < 0.05) differences are marked with dashed lines and asterisks. (C) Down-regulation of COX-2 and MMP9 expression in MCFDCIS cells using lentiviral shRNA decreased their enhanced invasion, but not migration induced by RASF coculture; y axis indicates migration and invasion relative to MCFDCIS cells cultured alone.
Because COX-2 inhibitors are known to have COX-2-dependent and COX-2 independent antitumorigenic effects (27), to better understand the role of COX-2 and MMP-9 in mediating epithelial–stromal cell interactions in our coculture model, we generated derivatives of MCFDCIS cells stably expressing shRNAs targeting these 2 genes using lentiviral shRNAs. Effective down-regulation of COX-2 and MMP9 was confirmed by qRT-PCR and immunoblot analyses (Fig. S5). When the migration and invasion of these shRNA clones were assess in cocultures, RASF-induced invasion of MCFDCIS cells was decreased after down-regulation of either COX-2 or MMP-9, but cell migration was not effected (Fig. 4C). Correlating with this finding, MMP9 gelatinase activity was slightly decreased in conditioned media derived from cocultures of RASFs with clones expressing COX-2 shRNA (Fig. S5). These results indicate that the invasion-promoting effects of RASFs in cocultures are at least in part mediated by the up-regulation of COX-2 and MMP9 in MCFDCIS breast cancer cells.
In summary, we identified COX-2 as a mediator of tumor epithelial–stromal cell interactions in breast cancer, and demonstrated that, by using a COX-2 inhibitor, we were able to eliminate the growth and invasive progression promoting effects of stromal fibroblasts in a human DCIS model. The use of NSAIDs and especially COX-2 inhibitors for the treatment and prevention of different human cancer types has been proposed and investigated (28). Similarly, results in several animal models support a cancer preventative role for COX-2 inhibitors (29). Our present study, using the MCFDCIS xenograft model of human DCIS, suggests that COX-2 inhibition may reduce breast tumor growth and progression to invasion. Thus, COX-2 inhibitors may decrease the incidence of invasive breast carcinomas by means of inhibiting the progression of premalignant and preinvasive tumors.
Materials and Methods
Cell Culture and Lentiviruses.
MCF10ADCIS.com and RASF cells were obtained and cultured as previously described (15). Cocultures of both cell types for RNA isolation were carried out as follows: 3 × 105 RASFs were plated in 100-mm dishes in DMEM supplemented with 10% FCS and allowed to grow for 24 h. We plated 1 × 105 MCFDCIS cells in the RASF-containing dishes, and cocultures were maintained for 48 h in MCF10A medium. MCFDCIS cells were isolated from cocultures by using CELLection Epithelial Enrich Dynabeads (Invitrogen), following protocols established in our lab (14). For conditioned media experiments, the same procedure was followed, except that 35-mm dishes were used. After 48 h of coculture, plates were washed thrice with PBS, and DMEM/F12 medium without additives was added. Plates were incubated for an additional 24 h; then conditioned media were collected, cleared by centrifugation, and kept at −80 °C; 3D Matrigel cultures were performed essentially as previously described (20), except multiple cell types were included in cocultures and different culture media were used. Adenoviral infection of MCFDCIS cells was carried out as previously described (30). Celecoxib and caffeic acid phenethyl ester (CAPE) were obtained from LKT Laboratories and Sigma–Aldrich, respectively. Generation of lentiviruses and infection of MCFDCIS cells were carried out as previously described (31, 32). The sequences of each of the clones used are listed in Table S1.
Mouse Xenograft Experiments and Statistical Analysis.
For xenograft studies, 100,000 MCFDCIS cells were injected s.c. into 6- to 9-week-old female nude mice alone or together with 2- to 3-fold excess of HME, PBS, PBTS, and RASF cells in 50% Matrigel (BD Biosciences). Tumors were allowed to grow for 3–8 weeks. Xenografts were weighed, then either snap frozen on dry ice and stored at −80 °C for DNA/RNA purification, or formalin fixed and paraffin embedded for histopathology. For sulindac and celecoxib experiments, mice were fed with control AIN-93G diet (Dyets) or AIN-93G diet with 0.9 g/kg celecoxib or 150 g/kg sulindac (LKT Laboratories), starting 7–10 days before injection and continued for the duration of the experiment. Xenograft weights and gene expression levels were evaluated by using statistical methods described in detail in our previous publication (15). RT-PCR and migration/invasion data (Figs. 3 and 4) were analyzed by using Student's t test and 1-way ANOVA with Bonferroni's posttest using the GraphPad Prism software. P <0.05 were considered significant. For Fig. 3H, 2-way ANOVA with Bonferroni's posttest was used.
Immunohistochemistry and Immunoblot Analyses, RNA isolation, and RT-PCR.
Immunohistochemistry and immunoblot analyses were performed essentially as previously described (15). COX2 antibodies were from Cayman Chemical, ab1 (anti-human, no. 160112) and ab2 (anti-human and anti-mouse, no. 160126). Total RNA was isolated and quantitative RT-PCR was performed as previously described (15). A list of primers used is available on request.
Luciferase Reporter Assays.
For luciferase assay experiments, 2.1 × 104 RASFs were seeded in 24-well plates. The same day, MCFDCIS cells were transfected by using Lipofectamine2000 (Invitrogen), with a reporter plasmid encoding for the firefly luciferase gene under the control of a promoter containing tandem NF-κB elements (33), and a Renilla luciferase encoding plasmid used as control for transfection efficiency. Next day, 7 × 103 of these transfected MCFDCIS cells were plated into the 24-well plates containing RASFs or empty wells. Cocultures were allowed to proceed for 24 h followed by measuring luciferase activity with Dual-Luciferase Reporter Assay System (Promega).
Gelatin Zymography and Cytokine Arrays.
For zymography, conditioned media standardized for cell numbers were mixed with equal volume of nonreducing sample buffer and resolved on 10% Novex Zymogram gels containing 0.1% gelatin (Invitrogen). Renaturation and detection were performed following the manufacturer's instructions. Clear bands corresponding to gelatinolytic activity were measured by densitometry. To analyze conditioned media on cytokine arrays, we cultured RASFs in DMEM supplemented with 0.5 g/mL BSA and 20 M Hepes for 4 days, followed by 5-fold concentration using Centricon filters and Western blotting of cytokine arrays (series C 1000; RayBiotech), as recommended by the manufacturer.
Cell Invasion and Migration Assays.
Cell invasiveness was assayed by using BD BioCoat Matrigel invasion chambers (BD Biosciences). Uncoated or Matrigel-coated filters (8-μm pore size, 6.5-mm diameter) were used for chemotaxis and chemoinvasion assays, respectively. MCFDCIS cells were prelabeled in vivo by using CellTracker Green CMF-DA (Invitrogen), following manufacturers instructions. We resuspended 1 × 104 MCFDCIS cells alone or mixed with 2 × 104 RASFs in 0.5 mL of serum-free DMEM/F12, and seeded in the upper compartment of the chamber. The lower compartment was filled with 0.75 mL of DMEM/F12 supplemented with 20% FCS as chemoattractant. After incubation at 37 °C in a humid atmosphere for 36 h, filters were rinsed with PBS. Cells on the upper surface of filters were wiped away with a wet cotton swab, and those on the lower surface were fixed with Carnoy fixative at room temperature), and stained with DAPI. Green cells (CMF-DA-stained MCFDCIS cells) were counted at 100× magnification using an inverted microscope. Each experiment was performed in triplicate and repeated at least twice.
Supplementary Material
Acknowledgments.
We thank Natasha Pliss for help with immunohistochemistry; Dr. Jerry Shay (University of Texas Southwestern Medical Center, Dallas, TX) for HME50 cells; Drs. John Mountz (University of Alabama, Birmingham, AL), and Steven Goldring (Beth-Israel Deaconess Medical Center, Boston, MA) for providing us with primary cultures derived from rheumatoid arthritis patients; Dr. Mark Ewen (Dana-Farber Cancer Institute, Boston, MA) for his generous gift of the IκB*-expressing adenoviral vector; and the Broad Institute TRC for shRNA constructs. This work was supported in part by National Institutes of Health Grants CA89393, CA94074, and CA116235, Department of Defense Grant W81XWH-07-1-0294, and American Cancer Society Grant RSG-05-154-01-MGO (to K.P.); the Susan G. Komen Breast Cancer Foundation Fellowship PDF042234 (to M.H.); and an International Union Against Cancer International Cancer Technology Transfer fellowship and National Institutes of Health Grant NO2-CO-41101 (to G.P.).
Footnotes
Conflict of interest statement: K.P. receives research support from and is a consultant to Novartis Pharmaceuticals, Inc., and is also a consultant to Aveo Pharmaceuticals, Inc., and Genego Inc.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813306106/DCSupplemental.
References
- 1.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinberg R, Mihich E. Eighteenth annual Pezcoller symposium: Tumor microenvironment and heterotypic interactions. Cancer Res. 2006;66:11550–11553. doi: 10.1158/0008-5472.CAN-06-3149. [DOI] [PubMed] [Google Scholar]
- 3.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 4.Hu M, Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008;18:27–34. doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, et al. Abrogation of TGFβ signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 8.Wang MT, Honn KV, Nie D. Cyclooxygenases, prostanoids, and tumor progression. Cancer Metastasis Rev. 2007;26:525–534. doi: 10.1007/s10555-007-9096-5. [DOI] [PubMed] [Google Scholar]
- 9.Singh-Ranger G, Salhab M, Mokbel K. The role of cyclooxygenase-2 in breast cancer: Review. Breast Cancer Res Treat. 2008;109:189–198. doi: 10.1007/s10549-007-9641-5. [DOI] [PubMed] [Google Scholar]
- 10.Ristimaki A, et al. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res. 2002;62:632–635. [PubMed] [Google Scholar]
- 11.Ulrich CM, Bigler J, Potter JD. Non-steroidal anti-inflammatory drugs for cancer prevention: Promise, perils and pharmacogenetics. Nat Rev Cancer. 2006;6:130–140. doi: 10.1038/nrc1801. [DOI] [PubMed] [Google Scholar]
- 12.Gauthier ML, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visscher DW, et al. Association between cyclooxygenase-2 expression in atypical hyperplasia and risk of breast cancer. J Natl Cancer Inst. 2008;100:421–427. doi: 10.1093/jnci/djn036. [DOI] [PubMed] [Google Scholar]
- 14.Allinen M, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Hu M, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu M, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 17.Sato N, Maehara N, Goggins M. Gene expression profiling of tumor-stromal interactions between pancreatic cancer cells and stromal fibroblasts. Cancer Res. 2004;64:6950–6956. doi: 10.1158/0008-5472.CAN-04-0677. [DOI] [PubMed] [Google Scholar]
- 18.Gately S, Li WW. Multiple roles of COX-2 in tumor angiogenesis: A target for antiangiogenic therapy. Semin Oncol. 2004;31:2–11. doi: 10.1053/j.seminoncol.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 19.Tsujii M, et al. Cyclooxygenase regulates angiogenesis induced by colon cancer cells. Cell. 1998;93:705–716. doi: 10.1016/s0092-8674(00)81433-6. [DOI] [PubMed] [Google Scholar]
- 20.Kleer CG, et al. Epithelial and stromal cathepsin K and CXCL14 expression in breast tumor progression. Clin Cancer Res. 2008;14:5357–5367. doi: 10.1158/1078-0432.CCR-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmbeck K, Bianco P, Yamada S, Birkedal-Hansen H. MT1-MMP: A tethered collagenase. J Cell Physiol. 2004;200:11–19. doi: 10.1002/jcp.20065. [DOI] [PubMed] [Google Scholar]
- 22.Howe LR, Subbaramaiah K, Brown AM, Dannenberg AJ. Cyclooxygenase-2: A target for the prevention and treatment of breast cancer. Endocr Relat Cancer. 2001;8:97–114. doi: 10.1677/erc.0.0080097. [DOI] [PubMed] [Google Scholar]
- 23.Shishodia S, Koul D, Aggarwal BB. Cyclooxygenase (COX)-2 inhibitor celecoxib abrogates TNF-induced NF-κB activation through inhibition of activation of IκBα kinase and Akt in human non-small cell lung carcinoma: Correlation with suppression of COX-2 synthesis. J Immunol. 2004;173:2011–2022. doi: 10.4049/jimmunol.173.3.2011. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 25.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 26.O'Hayre M, Salanga CL, Handel TM, Allen SJ. Chemokines and cancer: Migration, intracellular signalling and intercellular communication in the microenvironment. Biochem J. 2008;409:635–649. doi: 10.1042/BJ20071493. [DOI] [PubMed] [Google Scholar]
- 27.Grosch S, Maier TJ, Schiffmann S, Geisslinger G. Cyclooxygenase-2 (COX-2)-independent anticarcinogenic effects of selective COX-2 inhibitors. J Natl Cancer Inst. 2006;98:736–747. doi: 10.1093/jnci/djj206. [DOI] [PubMed] [Google Scholar]
- 28.Pereg D, Lishner M. Non-steroidal anti-inflammatory drugs for the prevention and treatment of cancer. J Intern Med. 2005;258:115–123. doi: 10.1111/j.1365-2796.2005.01519.x. [DOI] [PubMed] [Google Scholar]
- 29.Arun B, Goss P. The role of COX-2 inhibition in breast cancer treatment and prevention. Semin Oncol. 2004;31:22–29. doi: 10.1053/j.seminoncol.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 30.Seth P, et al. Cellular and molecular targets of estrogen in normal human breast tissue. Cancer Res. 2002;62:4540–4544. [PubMed] [Google Scholar]
- 31.Carroll DK, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 32.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 33.Li N, Karin M. Signaling pathways leading to nuclear factor-kappa B activation. Methods Enzymol. 2000;319:273–279. doi: 10.1016/s0076-6879(00)19027-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.