Abstract
Muscle activity contributes to formation of the neuromuscular junction and affects muscle metabolism and contractile properties through regulated gene expression. However, the mechanisms coordinating these diverse activity-regulated processes remain poorly characterized. Recently, it was reported that histone deacetylase 4 (HDAC4) can mediate denervation-induced myogenin and nicotinic acetylcholine receptor gene expression. Here, we report that HDAC4 is not only necessary for denervation-dependent induction of genes involved in synaptogenesis (nicotinic acetylcholine receptor and muscle-specific receptor tyrosine kinase) but also for denervation-dependent suppression of genes involved in glycolysis (muscle-specific enolase and phosphofructokinase). In addition, HDAC4 differentially regulates genes involved in muscle fiber type specification by inducing myosin heavy chain IIA and suppressing myosin heavy chain IIB. Consistent with these regulated gene profiles, HDAC4 is enriched in fast oxidative fibers of innervated tibialis anterior muscle and HDAC4 knockdown enhances glycolysis in cultured myotubes. HDAC4 mediates gene induction indirectly by suppressing the expression of Dach2 and MITR that function as myogenin gene corepressors. In contrast, HDAC4 is directly recruited to myocyte enhancer factor 2 sites within target promoters to mediate gene suppression. Finally, we discovered an HDAC4/myogenin positive feedback loop that coordinates gene induction and repression underlying muscle phenotypic changes after muscle denervation.
INTRODUCTION
Skeletal muscle is composed of a spectrum of fiber types with different structural and functional properties. The composition of myosin heavy chain (MHC) isoforms specifies muscle contractile properties. In rodents, MHC IIa, IIb, or both are enriched in fast twitch fibers, whereas MHC I is enriched in slow twitch fibers. These fibers also express different enzymes that result in fiber type-specific metabolic properties. Type IIb fibers are rich in glycolytic enzymes, whereas type I and IIa fibers are rich in enzymes necessary for oxidative metabolism. The extent to which a fiber is glycolytic or oxidative is largely determined by the activity pattern of its innervating motor neuron (Gundersen et al., 1988). Nerve activity also plays a crucial role in maintaining muscle mass (Jackman and Kandarian, 2004) and in shaping the formation of the neuromuscular junction (NMJ) (Sunesen and Changeux, 2003; Kummer et al., 2006).
Although many of the genes contributing to muscle fiber phenotype are regulated by muscle activity, not all are regulated in the same direction. For example, in the fast tibialis anterior (TA) muscle some genes, such as those encoding the MyoD family of basic helix-loop-helix transcription factors, and synaptic components such as nicotinic acetylcholine receptors (nAChRs) and muscle-specific kinase (MuSK) are induced (Goldman et al., 1985, 1986; Eftimie et al., 1991; Dutton et al., 1993; Tang et al., 2006), whereas other genes, such as those encoding muscle-specific enolase (MSE), phosphofructose kinase (PFK), and MHC IIb, are suppressed after muscle denervation (Huey and Bodine, 1998; Nozais et al., 1999; Tang et al., 2000; Raffaello et al., 2006). Interestingly, in contrast to denervation-dependent MHC IIb mRNA suppression in fast TA muscle (Huey and Bodine, 1998), muscle inactivity induces its expression in the slow soleus muscle (Pandorf et al., 2006), suggesting muscle fiber-type–specific regulation.
A key molecule mediating denervation-dependent nAChR and MuSK gene regulation is myogenin (Mgn), a member of the basic helix-loop-helix family of transcription factors, whose expression is regulated by muscle depolarization (Tang et al., 2006). Muscle denervation induces Mgn gene expression, and this induction requires histone deacetylase (HDAC) activity (Tang and Goldman, 2006). HDAC activity serves to relieve Mgn gene inhibition by suppressing expression of Dach2, a Mgn gene corepressor whose levels are high in innervated muscle and low in denervated muscle (Tang and Goldman, 2006). Dach2 repression after muscle denervation seems to be mediated, in part, via HDAC4 expression (Cohen et al., 2007). In addition, the HDAC9 splice variant MITR is also induced in innervated muscle and contributes to activity-dependent Mgn suppression by functioning as a corepressor in a complex with the myocyte enhancer factor (MEF) 2 transcription factor (Méjat et al., 2005). These two deacetylases (HDAC4 and -9) belong to a subgroup of class II HDACs that include HDAC5 and -7 (Khochbin et al., 2001; Verdin et al., 2003; Yang and Seto, 2008). Although the mechanisms underlying activity-dependent gene regulation in muscle are emerging (Chin et al., 1998; Liu et al., 2005; Méjat et al., 2005; Tang and Goldman, 2006), relatively little is known about how the muscle is able to coordinate activity-dependent gene induction and gene suppression.
Here, we report that HDAC4 is induced in denervated TA muscle and is not only necessary for the denervation-dependent induction of Mgn and synaptic protein gene expression but also for the denervation-dependent suppression of genes involved in glycolytic metabolism and fiber-type specification. Investigation of the mechanisms underlying HDAC4-dependent gene regulation showed that HDAC4 mediates gene repression by recruitment to MEF2 sites in the promoters of repressed genes, whereas HDAC4 indirectly contributes to gene induction by coordinately inhibiting the expression of the Mgn gene corepressors Dach2 and MITR. Consistent with its role in regulating the expression of genes involved in muscle metabolism, we found that HDAC4 is preferentially enriched in myonuclei of fast oxidative fibers in innervated muscle and its knockdown in myotubes enhances glycolysis. Finally, we found that HDAC4 and Mgn positively regulate each other's expression to allow maintenance and coordination of gene activation and repression after muscle denervation.
MATERIALS AND METHODS
Plasmids, Cells, and Reporter Gene Assay
Plasmids pCS2:green fluorescent protein (GFP) and pCS2:β-galactosidase (β-gal) were gifts from D. Turner (University of Michigan, Ann Arbor, MI) and pmU6:MgnshRNA (myogenin-targeting short hairpin RNA [shRNA] under control of the mouse U6 promoter) were described previously (Tang et al., 2004). pcDNA3:HDAC4-GFP was a gift from T. Kouzarides (Gurdon Institute, Cambridge, United Kingdom), pcDNA3.1-HDAC4:3SA was described previously (Wang et al., 2000), pEGFPN1-HDAC6-GFP was a gift from F. Sánchez-Madrid (Universidad Autónoma de Madrid, Madrid, Spain). The human 1.6-kb HDAC4 promoter (NT 022173) was cloned into the HindIII/XhoI sites of pXP2 luciferase reporter vector by using genomic DNA from human embryonic kidney (HEK) 239 cell line with primers 5′-CTGCCAAGCTTACAGGTGTTCCAGAATACCTAG and 5′-ATCCGCTCGAGTGCTAGCAGCGTCAGTGCCTT. The mouse 1.7-kb MSE promoter (NT 096135) was cloned into HindIII/BamH I sites of pXP2 by using genomic DNA from C2C12 cells with primers 5′-TTTTTACAAGCTTGCGCTCGAGAGCCGGCC and 5′-GAGGAAAGATCTGGCTGCACCACAGGAGATGTG. The MEF2 mutation in the MSE promoter was generated by polymerase chain reaction (PCR)-directed mutagenesis with primer 5′-ATGGGGTATGTTTAGCTCCAGCA (single base [T to G] change in underlined MEF2 site). C2C12 cells were cultured in DMEM, 10% fetal bovine serum with 5% CO2. Primary muscle cultures were prepared as described previously (Tang et al., 2004) and cultured on collagen-coated dishes in DMEM, 10% horse serum, 10% fetal bovine serum, and 5% CO2. Muscle cells were induced to differentiate at ∼80% confluence by dropping the serum concentration to 5% horse serum. HEK293 cells were cultured in DMEM, 10% fetal bovine serum, and 5% CO2. Cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cultured muscle cells were observed using an inverted DMIL fluorescent microscope (Leica Microsystems, Deerfield, IL) equipped with a 40 × 0.5 numerical aperture (NA) objective and a MagnFIRE SP digital camera.
Animals and Muscle Denervation
All animal studies were approved by the University of Michigan Committee on Use and Care of Animals. Dach2 null and myogenin (Mgn) conditional knockout mice were described previously (Davis et al., 2006; Knapp et al., 2006). To induce recombination in Mgn conditional knockout mice, 1 mg of tamoxifen suspended in sunflower oil was injected intraperitoneally into adult mice for five consecutive days followed by a 2-d rest and then repeated. For all muscle denervation experiments, mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). For TA and soleus muscle denervation, fur covering the lower back to proximal thigh was removed, the region was swabbed with Betadine, and a small posterolateral cutaneous incision (∼1–2 cm) was made beginning at the approximate region of the sciatic notch. The superficial facia was cut and the hamstring, and gluteal muscles were separated bluntly. With the sciatic nerve exposed, a 1-cm-long section was removed and the incision closed with wound clips. For experiments investigating the in vivo effects of HDAC inhibitors and mithramycin on gene expression, we used innervated and denervated sternomastoid muscle because it sits in a bed of tissue that allows one to apply drugs that pool over the muscle and not readily escape into the surrounding tissue. To denervate the sternomastoid muscle, anesthetized mice were shaved around the neck, and the skin was swabbed with Betadine. A midline incision was made from the apex of the mandible to the sternal notch. The ventral surface of the sternomastoid muscle was exposed and unilaterally denervated by removing a 2-mm section of the sternomastoid nerve while viewing under a dissecting microscope. Animals were kept warm on a heating pad to maintain body temperature, and the anesthetic plane was maintained, over a period of 12 h, by intraperitoneal injection of one third of the initial dose of anesthetic every 2 h. HDAC inhibitor trichostatin A (TSA; 5 μM), mithromycin (100 μM), or vehicle (0.9% NaCl, 0.1% dimethyl sulfoxide [DMSO]) were applied directly to the innervated and denervated muscles in 0.5-ml volume every 2 h to maintain constant drug exposure. Muscles were then harvested for quantification of gene expression by using real-time PCR and primers listed in Table 1.
Table 1.
PCR primers
| Primer namea | Primer sequence (5′ to 3′) forward | Primer sequence (5′ to 3′) reverse |
|---|---|---|
| AChRα | CGT-CTG-GTG-GCA-AAG-CT | CCG-CTC-TCC-ATG-AAG-TT |
| Actinγ | ACC-CAG-GCA-TTG-CTG-ACA-GGA-TGC | CCA-TCT-AGA-AGC-ATT-TGC-GGT-GGA-CG |
| CPT-1 | ACC-TGG-GCT-ACA-CGG-AGA-CA | CCT-TGG-CTA-CTT-GGT-ACG-AGT |
| CS | TGG-ACA-ATT-TTC-CAA-CCA-ATC-T | ATT-GTG-TGA-CCA-GTC-TAG-CC |
| Cyto B | CTT-CTA-GGA-GTC-TGC-CTA-AT | GGC-ACC-TCA-GAA-TGA-TAT-TTG |
| Dach2 | ACT-GAA-AGT-GGC-TTT-GGA-TAA | TTC-AGA-CGC-TTT-TGC-ATT-GTA |
| FBP1 | CTG-ATA-TTC-ACC-GCA-CTC-TG | TCC-TGC-ACA-TCT-TCA-GAG-GA |
| GLUT4 | GCA-GCG-AGT-GAC-TGG-AAC-A | CCA-GCC-ACG-TTG-CAT-TGT-AG |
| GS1 | CTT-CCA-CAA-GCC-CTC-TCC-T | CTG-AGG-GAT-CTG-CGA-TGT-GT |
| GS2 | AGA-GTT-TGT-CCG-AGG-TTG-TCA | TAT-GCA-GTA-GGG-TCA-GCC-ACA-T |
| HDAC1 | TCA-ATG-TTG-GTG-AGG-ACT-GT | CTT-AAA-GAT-GGC-TTC-ATA-GGA |
| HDAC2 | GCC-TCA-TAA-AGC-CAC-TGC-TG | GTA-ATT-CGA-GGA-TGG-CAA-GC |
| HDAC3 | ATG-TGT-TGA-ATA-TGT-CAA-GAG-T | GCT-GTA-GTT-CTC-CTC-GGG-A |
| HDAC4 | CAG-GAG-ATG-CTG-GCC-ATG-AA | GCA-CTC-TCT-TTG-CCC-TTC-TC |
| HDAC5 | GCT-TCT-TTG-GAC-CAG-AGT-TCC | CAT-CTC-AGT-GGG-GAT-GTT-GG |
| HDAC6 | TCT-CAA-CTG-ATC-TCT-CCA-GG | ACG-CTG-ACT-ACA-TTG-CTG-CT |
| HDAC7 | TCA-CCA-TCA-GCC-TCT-GAG | AGC-TGG-CTG-AAG-TGA-TCC |
| HDAC8 | AAC-ACG-GCT-CGT-TGC-TGG | CCA-GCT-GCC-ACT-TGG-TGC |
| HDAC9 (MITR) | TCA-GAG-GTT-CCT-ATG-GGC-CTG | TGG-AGA-CGT-TCC-ACT-GAG-GG |
| Mdh2 | ATC-CAG-CTC-GAG-TCA-ACG-T | CTT-CAC-GAC-TTC-TGT-GCC-A |
| MHC-Iβ | AGA-TGG-CTG-CAT-TTG-GGG-CTG | GTC-CTC-GAT-CTT-GTC-GAA-CTT |
| MHCIIA | ACA-AAT-CTA-TCC-AAG-TTC-CG | TTC-GGT-CAT-TCC-ACA-GCA-TC |
| MHCIIB | CCG-TGA-TAT-ACA-GGA-CAG-TG | GTT-CCG-TAA-GAT-CCA-GCA-CG |
| MSE | AAA-AGT-TGA-CAA-ATT-TAT-GAT-TGA | AAT-CAT-GAA-CTC-CTG-CAT-GG |
| MuSK | CTC-GTC-CTC-CCA-TTA-ATG-TAA-AAA | TCC-AGC-TTC-ACC-AGT-TTG-GAG-TAA |
| Mgn | CTC-AGC-TTA-GCA-CCG-GAA-GCC-CGA | ATT-GCC-CCA-CTC-CGG-AGC-GCA-GGA-G |
| PFKm | TGC-CAC-TAA-GAT-GGG-TGC-TA | GTC-TGG-TCC-TTC-AGC-TCA-GT |
| PGC-1α | CGC-ACA-ACT-CAG-CAA-GTC-CTC | CTT-GCT-GGC-CTC-CAA-AGT-CTC |
| SDHa | GGA-GCA-AAT-TCT-CTC-TTG-GA | CAA-ATC-TCA-ACT-TGT-CAA-GAT-TC |
| SDHb | AGC-CTT-ATC-TGA-AGA-AGA-AGG | TAC-TTG-TCT-CCG-TTC-CAC-CAG |
| SDHc | GAA-GAA-CAC-GAG-TTC-AAA-CCG | AAA-GTT-CCC-AGG-AAG-CAG-CA |
a CPT-1, carnitine palmitoyltransferase-1; CytoB, cytochrome b; CS, citrate synthase; FBP, fructose 2,6-bisphosphatase; GS, glycogen synthase; GLUT4, glucose transporter 4; PGC-1α, peroxisome proliferators-activated receptor γ coactivator 1α.
Tissue Staining and Western Blots
Dissected muscle was embedded in OCT, quickly frozen in dry ice-isopentane, and sectioned to 10 μm in thickness by using a cryostat. Immunohistochemistry was performed using standard conditions. Antibody dilutions were as follows: anti-HDAC4, 1:200 dilution for immunocytochemistry and 1:1000 for Western blot; anti-sarcomere myosin (MF20, hybridoma supernatant), 1:100 for Western blot; anti-cytochrome c oxidase subunit 1 (COX1; a gift from Dr. Steve Lentz, University of Michigan), 1:100; anti-laminin (hybridoma supernatant), 1:50; anti-myc (hybridoma supernatant), 1:50; and anti-myogenin antibody (ascites), 1:1000. The latter three antibodies were purchased from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). Secondary antibodies included anti-rabbit cyanine 3 (1:500; Invitrogen) and anti-rabbit biotin (1:200; Sigma-Aldrich, St. Louis, MO). Neuromuscular junctions were visualized by staining nAChRs with α-bungarotoxin (Sigma-Aldrich). Succinate dehydrogenase (SDH) staining was performed by incubating fresh muscle tissue sections in 0.1 M phosphate buffer, pH 7.6, 5 mM EDTA, pH 8.0, 1 mM KCN, 21.8 mg/ml sodium succinate, and 1.24 mg/ml nitroblue tetrazolium for 20 min at room temperature. Sections were then rinsed, dehydrated, and mounted before microscopic visualization. Sections were observed using an Axiophot microscope (Carl Zeiss, Thornwood, NY) equipped with fluorescence optics and a 10 × 0.3 NA objective (Figure 5A), a 40 × 0.85 NA objective (Figures 5, B–D, and 6A), or a 100 × 1.3 NA oil immersion objective. A Sony digital camera was used to capture images with Digital Acquire software (Optronics, Goleta, CA). Neuromuscular junctions were sometimes observed using a FluoView FV1000 confocal imaging system (Olympus, Tokyo, Japan) by using 60 × 1.2 NA water immersion objective (Figure 5D). Confocal images were acquired with FV10-ASW 1.7 software (Olympus). Standard procedures were used for Western blotting.
Figure 5.
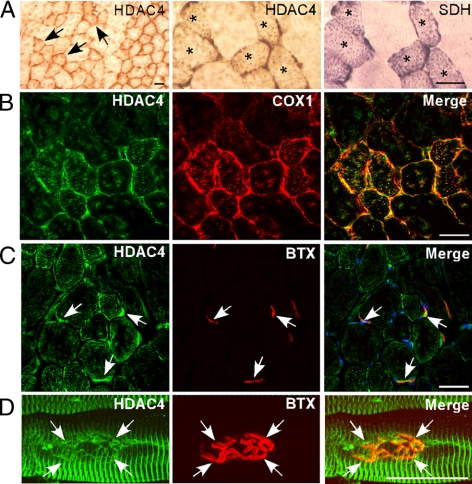
HDAC4 is enriched in TA muscle small oxidative fibers and localizes to the subsarcolemma region and NMJ. (A) Innervated TA muscle was immunostained with an anti-HDAC4 antibody (HDAC4) or assayed for SDH activity. Arrows in the low-magnification (100×) panel (left) identify representative HDAC4-positive small oxidative fibers. Asterisks in higher magnification (400×) panels (middle and right) identify small oxidative fibers expressing both HDAC4 and SDH. (B) Immunofluorescence showing HDAC4 immunoreactivity colocalizes with COX1 (400× magnification). (C and D) HDAC4 immunoreactivity colocalizes with BTX-stained NMJs (in merge [C], blue color is 4,6-diamidino-2-phenylindole-stained nuclei). Bars, 40 μm.
Figure 6.
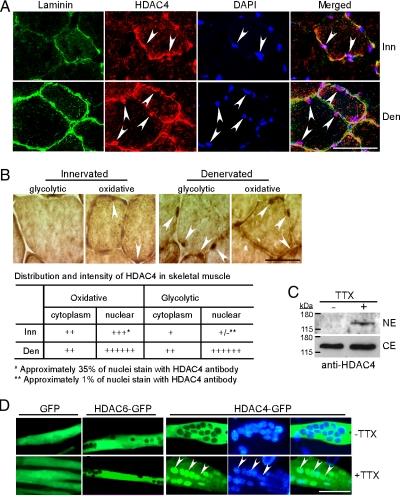
Muscle inactivity stimulates nuclear accumulation of HDAC4. (A) Triple fluorescence immunostaining for laminin, HDAC4, and nuclei on innervated and denervated TA muscle sections. Arrowheads point to HDAC4+ myonuclei located within the muscle fibers basal lamina; 600× magnification. (B) HDAC4 immunoreactivity shows low-level nuclear accumulation of HDAC4 in innervated small oxidative fibers and high level nuclear accumulation in denervated small oxidative and larger glycolytic fibers. Arrowheads point to nuclei with weak HDAC4 immunoreactivity in oxidative fibers of innervated muscle and high HDAC4 immunoreactivity in both oxidative and glycolytic denervated muscle fibers; 1000× magnification. Qualitative assessment of HDAC4 expression in cytoplasm and nuclei is presented in the table. (C) Western blot analysis of HDAC4 nuclear levels (NE) and cytoplasmic levels (CE) in active (−TTX) and inactive (+TTX) primary mouse myotube cultures. (D) Inactivity induced nuclear accumulation of HDAC4 in cultured primary mouse myotubes. Primary myotubes were transfected with indicated expression vectors and either kept inactive with TTX or allowed to spontaneously contract (−TTX); 400× magnification. Bars, 40 μm. Inn, innervated; Den, denervated.
Muscle Electroporation and RNA Analysis
Stealth siRNA (Invitrogen) or plasmid DNA was delivered into TA muscle by electroporation as described previously (Tang et al., 2006). Briefly, TA or soleus muscle was injected with 20 μl of solution containing 2–5 μg of pCS2EGFP plasmid for identifying electroporated fibers and stealth siRNA (Invitrogen) (160 pM) targeting either HDAC4, MITR, Mgn, or a control small interfering RNA (siRNA). At least two different siRNAs targeting different sequences were used for each gene-specific knockdown to ensure results were not due to off-target effects of siRNAs. Control siRNAs have a similar GC content as the experimental siRNA but do not target any known gene. The molar ratio of stealth siRNA to enhanced green fluorescent protein (EGFP) plasmid was >500-fold so as to ensure all GFP+ fibers harbor siRNA. Nucleic acid uptake was facilitated by placing electrodes, coated with ultrasound transmission gel, on either side of the leg and using a BTX square wave electroporator to deliver six pulses of 140 V/cm of 60-ms duration with an interval of 100 ms. Animals were allowed to recover 12 d before denervation or isolation of electroporated/GFP+ fibers by using a stereomicroscope equipped with fluorescence optics. A 12-d postelectroporation recovery ensures muscle fibers had recovered from any electroporation-induced damage as determined by anatomy and gene expression (Bertrand et al., 2003; Golzio et al., 2005; Méjat et al., 2005; Sadasivam et al., 2005; Tang et al., 2006). In some experiments, we first denervated the lower hindlimb muscles and then 9 d later we electroporated the TA muscle with siRNAs before harvesting electroporated muscle 14–15 d after denervation. Both denervation/electroporation regimens gave essentially identical results. GFP+ fibers were dissected in ice-cold phosphate-buffered saline (PBS) and then immediately lysed in TRIzol (Invitrogen) for RNA isolation. This process takes ∼10–20 min. RNA was reverse transcribed with oligo(dT) primer and SuperScript II (Invitrogen), and 1/20 of the cDNA mixture served as template for PCR reactions. Real-time PCR was performed on a iCycler (Bio-Rad, Hercules, CA) by using SYBR Green SuperMix (Bio-Rad). Radioactive PCR was performed using [α-32P]dCTP to spike the reaction mix. Radiolabeled products were separated on 6% polyacrylamide gels and visualized by exposure of dried gels to x-ray film. Stealth siRNAs targeted the following sequences: HDAC4 2#, CCACACACUCCUCUACGGCACAAAU; HDAC4 3#, CCGUGUAAACCACUCAACUCAUCUU; HDAC6 2#, AAAGUUGGCACCUUCACGGUGCAGA; HDAC6 3#, ACUAGGGUCAUCCUCAAAUAUGUGC; MITR 1#, CAACUUGAAGGUGCGGUCCAGGUUA; and MITR 3#, GGAAAUACAGCUUGUUCAUUCUGAA; siRNAs targeting Mgn were described previously (Macpherson et al., 2006). Primers for amplifying specific RNAs are shown in Table 1.
Chromatin Immunoprecipitation (ChIP)
ChIP assay kit (Millipore, Billerica, MA) was used for ChIP analysis according to manufacturer's directions with the following modifications. Denervated muscle, electroporated with either a myc-tagged HDAC4 expression vector or an empty myc-tag expression vector, were dissected, frozen in liquid nitrogen, and finely ground with a mortar and pestle. Samples were thawed on ice and fixed in 1% paraformaldehyde/PBS for 20 min and then quenched with 0.125 M glycine. After three washes with ice-cold PBS, cells were lysed using an SDS lysis buffer, and the chromatin was sheared with a sonicator to generate chromatin fragments of ∼400–800 base pairs. Antibody binding and washes were based on methods recommended by Millipore ChIP protocol. Anti-HDAC4 or anti-myc antibodies were used to immunoprecipitate the HDAC4-bound chromatin fragments and immunoprecipitates harboring the muscle-specific enolase promoter's MEF2 site were quantified using primers flanking the MEF2 site in real-time PCR. Primers used for amplifying the MSE promoter's MEF2 region were 5′-AACCCTCGATTCTCTTGATTG and 5′-AGAGTGGACAGTTGATCCCTT. Primers used for amplifying the myogenin promoter's MEF2 region were 5′-AAAAGGCTTGTTCCTGCCACT and 5′-ACTGGAAACGTCTTGATGTGCA.
Pyruvate Assay
Pyruvate assay kit (Biovision, Mountain View, CA) was used to measure the concentration of intracellular pyruvate following the manufacturer's protocol. Briefly, cultured C2C12 cells were transfected with either control or HDAC4 siRNAs (60 nM) by using Lipofectamine 2000. Transfected cells were then switched to pyruvate-free differentiation medium (5% horse serum, high glucose DMEM), and 2 d later cells were collected and lysed with 0.5 ml of pyruvate assay buffer. Pyruvate is oxidized by pyruvate oxidase, to generate a fluorogenic product that can be measured at excitation/emission 535/590 nm. Products were measured on a FluroStar OPTIMA 96-well plate reader (BMG Labtech, Chicago, IL). Values were normalized to total protein.
RESULTS
HDAC Activity Contributes to Gene Induction and Repression in Developing Myotubes and Denervated Muscle
We demonstrated previously that HDAC inhibitors sodium butyrate (NaB) or TSA, block muscle differentiation- and denervation-dependent myogenin (Mgn) gene induction (Tang and Goldman, 2006). Here, we investigated whether HDAC activity was also necessary for differentiation- and denervation-dependent gene suppression. We chose the muscle-specific enolase (MSE) gene for this analysis because it is robustly suppressed after muscle denervation (Nozais et al., 1999).
As shown in Figure 1A, inhibition of HDAC activity with NaB resulted in increased MSE mRNA levels in differentiated C2C12 cells and the protein synthesis inhibitor cycloheximide had little effect on this induction. In contrast, NaB suppressed Mgn mRNA levels in C2C12 myotubes, and this repression required new protein synthesis as reported previously (Tang and Goldman, 2006). Similar results were obtained with the HDAC inhibitor TSA.
Figure 1.
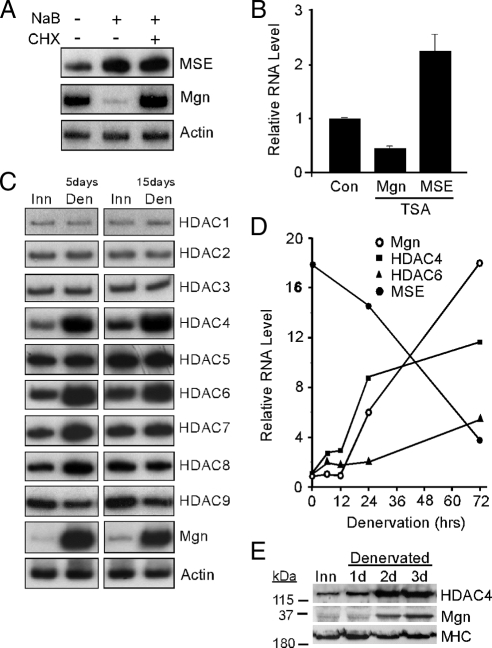
Regulation of Mgn, MSE, and HDAC levels by HDAC inhibitors and muscle denervation. (A) Radioactive PCR showing MSE, Mgn, and γ-actin mRNA levels after treatment of differentiated C2C12 myotubes with the HDAC inhibitor NaB (5 mM) or vehicle (0.9% NaCl, 0.1% DMSO) for 6 h in the presence and absence of the protein synthesis inhibitor cycloheximide (CHX; 50 μg/ml). (B) Real-time PCR quantification of Mgn and MSE mRNA levels in denervated sternomastoid muscle that was bathed, in vivo, in the HDAC inhibitor TSA (5 μM) or vehicle (Con; 0.9% NaCl, 0.1% DMSO) for 12 h. Error bars are standard deviations, n = 3. (C) Radioactive PCR showing HDAC1-9, Mgn, and γ-actin mRNA levels in innervated (Inn), and 5- and 15-d denervated (Den) TA muscle. (D) Real-time PCR was used to quantify the temporal induction of Mgn, HDAC4, HDAC6, and MSE mRNAs after TA muscle denervation. Values are normalized to γ-actin and reported relative to innervated muscle RNA levels (0-h denervation). (E) Western blot analysis of denervation-dependent HDAC4 and Mgn protein induction. Proteins from innervated (Inn) or denervated TA muscles were assayed by Western blot analysis as described in Materials and Methods. Note the increased expression of HDAC4 and Mgn shortly after muscle denervation, whereas the MHC expression remains relatively constant.
We next investigated whether HDAC inhibition also prevented MSE suppression in denervated TA muscle. For these experiments, we used the sternomastoid muscle because it sits in a bed of tissue that allows one to apply drugs that pool over the muscle and do not readily escape into the surrounding tissue (Tang and Goldman, 2006). We chose TSA for these experiments because it is specific for class II HDACs, and it was more potent than NaB in blocking HDAC activity in vivo in preliminary studies. These experiments revealed that HDAC inhibition blocks denervation-dependent Mgn induction and MSE suppression (Figure 1B).
HDAC4 and HDAC6 Are Induced in Denervated Muscle
The above-mentioned data suggest specific HDACs may be necessary for both denervation-dependent gene induction and suppression. We suspected that the relevant HDACs may themselves be regulated by muscle activity. To identify these candidate deacetylases, we assayed HDAC1–9 mRNA levels in innervated and denervated TA muscle. Interestingly, like Mgn, HDAC4 and HDAC6 mRNA levels increased in 5- and 15-d denervated muscle (Figure 1C), whereas HDAC7 and HDAC8 mRNA levels increased transiently at 5 d after denervation, but returned to basal levels at 15 d after denervation (Figure 1C). Real-time PCR analysis of denervation-dependent gene expression at early times after muscle denervation showed temporal changes in HDAC4 mRNA levels but not HDAC6 most closely reflects the temporal changes in Mgn and MSE mRNA levels (Figure 1D). This time course of denervation-dependent Mgn and HDAC4 gene expression was also reflected in protein levels with HDAC4 induction preceding Mgn induction (Figure 1E).
HDAC4 Regulates Genes Involved in Synapse Formation, Glycolytic Metabolism, and Fiber-Type Specification
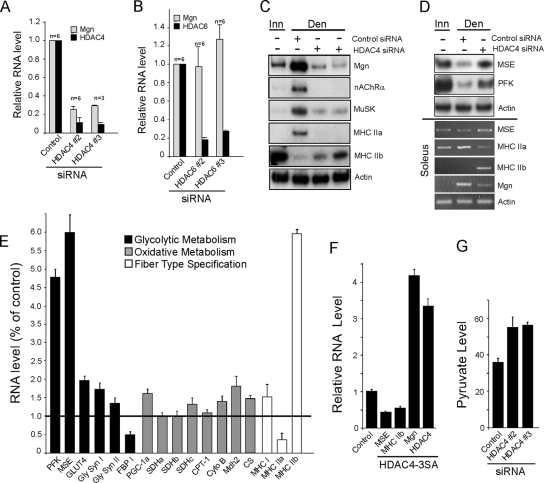
Although the temporal pattern of HDAC4 gene expression suggests it is a better candidate than HDAC6 in mediating denervation-dependent Mgn gene regulation, they are both induced in denervated muscle and they are both likely to contribute to activity-dependent gene regulation. To determine whether these HDACs are necessary for denervation-dependent Mgn gene regulation, we knocked down their expression by using siRNAs. Consistent with the temporal pattern of their expression, HDAC4 but not HDAC6 knockdown in denervated TA muscle dramatically suppressed Mgn mRNA induction (Figure 2, A and B). As expected, the denervation-dependent increase in nAChR α-subunit and MuSK mRNA expression, shown previously to be induced by Mgn (Tang et al., 2006), was also blocked after HDAC4 knockdown (Figure 2C). We also investigated the effect that HDAC4 knockdown had on the expression of genes involved in muscle contraction and metabolism. Interestingly, HDAC4 knockdown reciprocally regulated the expression of MHC isoforms by blocking denervation-dependent induction of MHC IIa and relieving denervation-dependent suppression of MHC IIb (Figure 2, C and E). In addition, HDAC4 knockdown relieved suppression of MSE and PFK that normally occurs in denervated TA muscle (Figure 2D, top; and E). Similar results were observed in the denervated slow soleus muscle where denervation-dependent induction of Mgn and MHC IIa mRNAs was suppressed and denervation-dependent suppression of MSE and MHC IIb mRNAs was relieved by HDAC4 knockdown (Figure 2D, bottom). The extremely low expression of MHC IIb mRNA in soleus is consistent with the preponderance of slow-type MHC I and IIa fibers in this muscle. Quantification of the effects of HDAC4 knockdown on a variety of glycolytic, oxidative, and contractile protein encoding mRNAs in TA muscle showed that those involved in glycolysis (PFK and MSE) and muscle contraction (MHC IIa and IIb) were the most influenced, whereas genes related to oxidative metabolism were less influenced (Figure 2E).
Figure 2.
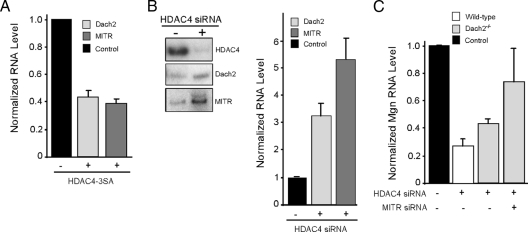
HDAC4 is necessary for denervation-dependent gene induction and suppression. Experiments were performed on adult innervated and denervated TA muscle except bottom panel in D, in which soleus muscle was used. Denervated muscles were electroporated with the indicated siRNAs (E; data from siRNAs targeting HDAC4) as described in Materials and Methods. (A and B) Quantitative real-time PCR showing that knockdown of HDAC4, but not HDAC6, prevents denervation-dependent Mgn mRNA induction. (C and D) Radioactive and qualitative PCR analysis showing HDAC4 regulates denervation-dependent gene expression in TA and soleus muscle. (E) Quantitative real-time PCR analysis of the effects of HDAC4 knockdown on the expression of genes involved in glycolytic and oxidative metabolism, and fiber-type specification in denervated TA muscle. Reported is the -fold change relative to the control siRNA electroporated muscle, n = 3. (F) Expression of nuclear localized HDAC4-3SA in innervated TA muscle represses MSE and MHC IIb mRNA levels and induces Mgn mRNA levels. Adult innervated TA muscle was electroporated with an HDAC4-3SA expression vector or an empty expression vector (control) and 12 d later the TA muscle was isolated for RNA analysis by real-time PCR. (G) HDAC4 knockdown in C2C12 muscle cells increases pyruvate production. Error bars are SDs, n = 6. In all studies using real-time PCR, gene expression values were normalized to γ-actin expression. FBP, fructose 2,6 bisphosphatase; GLUT4, glucose transporter 4; PGC-1α, peroxisome proliferators-activated receptorγ coactivator 1α; CPT-1, carnitine palmitoyltransferase-1; CytoB, cytochrome b; CS, citrate synthase.
The above-mentioned data suggest HDAC4 is necessary for both denervation-dependent gene induction and suppression. To determine whether HDAC4 is sufficient for mediating these changes in gene expression in response to muscle denervation, we expressed the mutant HDAC4-3SA in innervated TA muscle. This mutant harbors three mutations, S246A, S467A, and S632A, conferring its resistance to nuclear export (Grozinger and Schreiber, 2000; Wang et al., 2000). Quantitative real-time PCR revealed ∼3.5-fold increase in the HDAC4 mRNA level after electroporation of TA muscle with this expression vector (Figure 2F). This increased expression of nuclear localized HDAC4 in innervated muscle mimicked the effect of muscle denervation by suppressing MSE and MHC IIb mRNA levels and increasing Mgn mRNA levels (Figure 2F).
Because HDAC4 suppresses both MSE and PFK gene expression and because PFK is a rate-limiting enzyme in glycolysis, we investigated whether HDAC4 influences glycolysis. For these experiments, we transfected HDAC4 siRNA into C2C12 differentiated myotubes and measured pyruvate production. After HDAC4 knockdown, intracellular pyruvate increased by ∼50%, indicating HDAC4 does inhibit glycolysis in muscle cells (Figure 2G).
HDAC4 Coordinates the Suppression of Multiple Repressors to Allow for Denervation-dependent Mgn Gene Induction
The Mgn corepressors Dach2 and MITR are induced in innervated muscle and suppressed in denervated muscle (Méjat et al., 2005; Tang and Goldman, 2006). To test whether HDAC4 contributes to this regulation, we expressed HDAC4-3SA in innervated TA muscle (Figure 3A) and knocked down HDAC4 in denervated TA muscle (Figure 3B). Consistent with the idea that HDAC4 contributes to denervation-dependent Dach2 and MITR gene repression, we found HDAC4-3SA suppressed the expression of both of these corepressors in innervated muscle and HDAC4 knockdown partially relieved their suppression in denervated muscle.
Figure 3.
HDAC4-dependent Mgn induction in denervated muscle requires Dach2 and MITR suppression. (A) Overexpression of nuclear-localized HDAC4-3SA in innervated muscle results in Dach2 and MITR mRNA suppression. Adult innervated TA muscle was electroporated with an HDAC4-3SA expression vector or an empty vector (−) and 12 d later electroporated fibers were harvested for real-time PCR analysis of Dach2 and MITR mRNA levels. (B) HDAC4 knockdown in denervated muscle relieves Dach2 and MITR suppression. Adult innervated TA muscle was electroporated with control or HDAC4-targeting siRNA and 12 d later muscles were denervated for 5 d before harvesting tissue for radioactive PCR analysis of the indicated mRNAs (left). Quantification of the radioactive PCR data by signal density is shown (right). mRNA values were normalized to γ-actin mRNA and are reported as -fold difference from control siRNA, which is assigned a value of 1 (black bar). (C) HDAC4-knockdown-dependent Mgn suppression in denervated muscle is mediated by both Dach2 and MITR. Wild-type and Dach2−/− innervated TA muscle was electroporated with the indicated siRNAs and 12 d later denervated for 5 d before harvesting tissue for real-time PCR analysis of Mgn and γ-actin mRNA expression. Mgn mRNA levels were normalized to γ-actin mRNA levels and reported as a -fold difference from control siRNA electroporated muscle that is assigned a value of 1 (black bar). Error bars are SDs, n = 3.
Because HDAC4 expression is necessary for denervation-dependent Mgn gene induction and knockdown of HDAC4 in denervated muscle returned it to a more innervated pattern of gene expression (Figure 2), we investigated whether the effect of HDAC4 knockdown on Mgn gene expression is due to increased Dach2 and MITR expression. For this, we evaluated Mgn mRNA levels in HDAC4-siRNA electroporated denervated TA muscle from wild-type mice, Dach2−/− mice (Davis et al., 2006), and Dach2−/− mice that also had MITR levels reduced by RNA interference (Figure 3C). Consistent with the idea that HDAC4 mediates its effects on Mgn gene expression by suppressing both Dach2 and MITR expression, we found that HDAC4-knockdown in Dach2 null animals partially relieved Mgn suppression, whereas the addition of MITR knockdown (∼65% knockdown) in Dach2 null animals abrogated Mgn suppression more dramatically (Figure 3C). Therefore, HDAC4 seems to coordinately suppress Dach2 and MITR expression after muscle denervation, and suppression of both these genes is required for robust denervation-dependent Mgn gene induction.
HDAC4 Directly Suppresses MSE Gene Expression
HDAC4 represses genes involved in glycolytic metabolism (Figure 2, D and E), and we suspected this may result by recruitment of HDAC4 to MEF2 sites residing in glycolytic enzyme gene promoters (McKinsey et al., 2002). To investigate this, we focused our analysis on the MSE promoter because it is robustly regulated in an HDAC4-dependent manner. As expected, expression of nuclear localized HDAC4-3SA suppressed MSE promoter activity in transfected C2C12 cells (Figure 4A). Conversely, knockdown of endogenous HDAC4 expression resulted in a dramatic induction of MSE promoter activity (Figure 4B). This regulated expression required a putative MEF2 binding site (TATTTTTA) at position −113 (relative to the initiator ATG), because when mutated no induction was observed (Figure 4B). Consistent with these studies, MEF2 expressed in 293 cells specifically bound an oligonucleotide harboring the MSE promoter's putative MEF2 element (Figure 4C).
Figure 4.
HDAC4 suppresses MSE promoter activity via recruitment to the MSE promoter's MEF2 element. (A) Nuclear-localized HDAC4-3SA suppresses expression from a cotransfected MSE:Luciferase expression vector in C2C12 myotubes. Confluent C2C12 cells were transfected with a CMV:HDAC4-3SA expression vector, MSE:luciferase expression vector, and a CMV:β-gal expression vector that was used for normalization. Cells were differentiated in low serum, and 48 h later they were harvested for luciferase and β-gal assays. Reported MSE promoter activity is luciferase activity normalized to β-gal activity. Error bars are standard deviations, n = 3. (B) The MSE promoter's MEF2 site is required for HDAC4-dependent regulation. C2C12 cells were transfected with wild-type or a MEF2 mutant MSE:luciferase expression vector along with either a control or HDAC4-targeting siRNA and a normalizing CMV:β-gal vector as described above. Samples were analyzed as described in A. (C) Autoradiogram showing that overexpression of MEF2 in HEK293 cells increases binding to the MSE promoter's MEF2 element. HEK293 cells were transfected with increasing amounts of CMV:MEF2 or an empty vector (−) and 48 h later they were harvested and nuclear extracts were prepared. Nuclear extracts with (+) and without (−) overexpressed MEF2 were incubated with a radiolabeled oligonucleotide containing the MSE promoter's MEF2 site in the absence (−) or presence (+) of a 50-fold excess of unlabeled MEF2 oligonucleotide (competitor). Samples were resolved on a native polyacrylamide gel that was then dried and exposed to x-ray film. Shown is a representative autoradiogram. (D) ChIP assay shows that after muscle denervation HDAC4 or overexpressed myc-HDAC4 binds to the MSE promoter's MEF2 site but not the Mgn promoter's MEF2 site. Samples were either 5-d denervated TA muscle or 5-d denervated TA muscle that was electroporated with a myc-HDAC4 expression vector 12 d before denervation. Real-time PCR with primers flanking either the Mgn or MSE promoter's MEF2 site was used to quantify the amount of immunoprecipitated DNA.
We also examined whether HDAC4 is recruited to the MSE promoter's MEF2 site in vivo. For this analysis, we performed an in vivo ChIP assay by using an anti-HDAC4 or anti-myc antibody to immunoprecipitate sheared chromatin from either normal denervated TA muscle or denervated TA muscle overexpressing myc-HDAC4, respectively. Interestingly, quantitative real-time PCR of immunoprecipitated DNA by using primers flanking the MEF2 site of either the MSE or the Mgn promoter showed that HDAC4 is preferentially associated with the MSE promoter's MEF2 site (Figure 4D). We were unable to detect HDAC4 at the Mgn promoter's MEF2 site.
HDAC4 Is Enriched in Oxidative Skeletal Muscle Fibers and Nuclear Import Is Stimulated by Muscle Denervation
We next analyzed HDAC4 expression in innervated TA muscle that contains predominantly type II fibers (99%) that can be further subdivided into a mixture of small oxidative fibers and larger glycolytic fibers (Gorza, 1990). Interestingly, immunohistochemistry revealed a mosaic pattern of HDAC4 expression, with small oxidative fibers tending to have more intense staining than the relatively larger glycolytic fibers (Figure 5A). We confirmed that HDAC4 expression was enriched in oxidative fibers by colocalizing HDAC4 with the oxidative fiber-specific mitochondrial markers SDH and cytochrome c oxidase subunit 1 (COX1) (Figure 5, A and B). We also found that HDAC4 was present at NMJs by colocalization of HDAC4 immunoreactivity with α-bungarotoxin (BTX) staining (Figure 5, C and D). HDAC4 staining in extrasynaptic regions of the muscle fiber is consistent with its colocalization with mitochondria (Figure 5, B and C) that are enriched in the I-band region of muscle fibers (Ogata and Yamasaki, 1985).
To investigate whether HDAC4 is enriched in denervated muscle myonuclei, we triply stained denervated TA muscle cross-sections for laminin, HDAC4, and nuclei (Figure 6A). This analysis clearly demonstrated nuclear HDAC4 immunoreactivity within the confines of the muscle fibers basal lamina (pink nuclei in merged image). Under high magnification, we quantified the distribution and intensity of HDAC4 staining (Figure 6B). In innervated muscle, HDAC4 staining was higher in oxidative fibers compared with glycolytic fibers. Within oxidative fibers, HDAC4 was found in the cytoplasm and could be faintly detected in ∼35% of the myonuclei. In denervated muscle, cytoplasmic staining remained relatively constant, but we now observed pronounced HDAC4 nuclear accumulation in both oxidative and glycolytic fibers (Figure 6B). Consistent with the above-mentioned results, Western blot analysis showed nuclear enrichment of HDAC4 in cultured primary myotubes treated with sodium channel blocker tetrodotoxin (TTX) compared with cells that were allowed to spontaneously contract in the absence of TTX (Figure 6C). In addition, we observed TTX-dependent nuclear import of GFP-tagged HDAC4, but not GFP-tagged HDAC6 or GFP (Figure 6D). Therefore, muscle activity stimulates HDAC4 nuclear export.
A Positive HDAC4/Mgn Feedback Loop Coordinates and Maintains Gene Expression Changes in Denervated Muscle
HDAC4 is induced in denervated muscle, and this induction is necessary for denervation-dependent changes in gene expression (Figures 1 and 2). The molecular mechanisms mediating HDAC4 induction in denervated muscle have not been investigated. A previous study suggested that the HDAC4 promoter harbors GC-rich regulatory elements that bind Sp proteins and that mithramycin treatment of cultured cells, which impedes Sp protein binding to GC-rich sequences (Nehls et al., 1993), down-regulates HDAC4 gene expression (Liu et al., 2006). Therefore, we investigated whether mithramycin affected denervation-dependent induction of HDAC4 in vivo. For these experiments, we bathed innervated and denervated sternomastoid muscle in 100 μM mithramycin for 12 h, and we found mithramycin suppressed HDAC4 and Mgn gene expression in innervated but not denervated muscle (Figure 7A). Therefore, GC-binding proteins seem to play a role in maintaining the basal expression of HDAC4 and Mgn genes in innervated muscle, but they are not required for maintaining high levels of expression in denervated muscle.
Figure 7.
HDAC4 and Mgn positively regulate each others expression. (A) Mithramycin inhibits HDAC4 and Mgn mRNA expression in innervated (Inn) but not denervated (Den) sternomastoid muscle. Inn and Den sternomastoid muscle was bathed in mithramycin (100 mM) or vehicle (−), in vivo, for 12 h before harvesting tissue for assaying HDAC4, Mgn, and γ-actin mRNA levels by using real-time PCR. HDAC4 and Mgn RNA levels are normalized to γ-actin RNA levels. Experiments were performed as described in the legend of Figure 1B and in Materials and Methods. (B) Mgn overexpression induces HDAC4 promoter activity in C2C12 myotubes. C2C12 myotubes were transfected with an HDAC4:luciferase and CMV:β-gal expression vector, +/− a CMV:Mgn expression vector. Forty-eight hours later, cells were harvested for luciferase and β-gal activities. Reported HDAC4 promoter activity is luciferase activity normalized to β-gal activity. (C) Mgn knockdown reduces HDAC4 promoter activity in C2C12 myotubes. C2C12 myotubes were transfected with an HDAC4:luciferase and CMV:β-gal expression vector, +/− a U6:MgnshRNA expression vector for knocking down Mgn expression. Forty-eight hours later, cells were harvested for luciferase and β-gal activities. Reported HDAC4 promoter activity is luciferase activity normalized to β-gal activity. (D) Mgn knockdown in denervated TA muscle blocks denervation-dependent induction of HDAC4. Innervated TA muscle was electroporated with control or Mgn-targeting siRNAs; and 12 d later, muscles either remained innervated (Inn) or were denervated for 5 d (Den) before harvesting electroporated muscle fibers for analysis of mRNA expression. Two different Mgn-targeting siRNAs were used in this experiment, and radioactive PCR was used to visualize Mgn, HDAC4 and γ-actin mRNA levels. (E) Conditional deletion of the Mgn gene in adult mice blocks denervation-dependent induction of HDAC4 mRNA. Adult mice harboring a conditionally expressed Mgn gene were either treated with tamoxifen (Mgn−/−) or remained untreated (Wt). The left hindlimb muscles were denervated for 5 d, whereas the right hindlimb remained innervated. Five days after denervation, innervated and denervated TA muscles were isolated and Mgn, HDAC4, and γ-actin mRNA levels were assayed by real-time PCR. Mgn and HDAC4 mRNA levels were normalized to γ-actin mRNA. –Fold difference from innervated RNA level is reported. (F) Denervation-dependent suppression of PFK, MSE, and MHC IIb mRNA levels are abrogated in mice where the Mgn gene is deleted in adult animals. Experimental protocol was as described in E. Error bars are SDs, n = 3.
Because E-box sequences (CANNTG) are known to mediate denervation-dependent induction of nAChR and MuSK promoters via their interaction with Mgn (Tang et al., 2004, 2006), we looked for similar elements residing in the HDAC4 promoter. Indeed, three E-boxes (−572, −924, and −1383 relative to the translation initiation codon) were found upstream of the HDAC4 coding sequence. Interestingly, these E-boxes conform to the CACCTG or CACGTG sequences that seem to be required for denervation-dependent activation of human, rat, mouse, and chicken nAChR and MuSK promoters (Tang et al., 2006), suggesting that Mgn may also mediate denervation-dependent HDAC4 gene induction. Indeed, we found Mgn overexpression in C2C12 cells increased HDAC4 promoter activity (Figure 7B) and that shRNA-mediated Mgn knockdown reduced HDAC4 promoter activity (Figure 7C).
The above-mentioned data suggest Mgn activates the HDAC4 promoter; therefore, we investigated whether Mgn induction after muscle denervation influences HDAC4 gene expression. For this analysis, we measured endogenous HDAC4 mRNA before and after siRNA-mediated Mgn knockdown in denervated TA muscle. Interestingly, Mgn knockdown blocked denervation-dependent HDAC4 gene induction (Figure 7D). As shown in Figure 7E, similar results were obtained when we assayed denervation-dependent HDAC4 induction in conditional Mgn null mice in which the Mgn gene was deleted in adult muscle (Knapp et al., 2006). These data suggest that Mgn, via its effect on HDAC4 expression, may also be necessary for denervation-dependent suppression of MSE, PFK, and MHCIIb gene expression. To test this idea, we used Mgn null mice and compared denervation-dependent regulation of MSE, PFK, and MHCIIb mRNA expression with that found in normal mice (Figure 7F). Consistent with the idea that Mgn and HDAC4 regulate each others expression, denervation-dependent suppression of MSE, PFK, and MHCIIb mRNAs was impaired in the absence of Mgn (Figure 7F). Thus, Mgn positively regulates HDAC4 expression and HDAC4 induction negatively regulates its target genes.
DISCUSSION
Nerve–muscle communication influences muscle structure and function via regulated gene expression. Loss of nerve activity by injury or disease impacts the expression of genes involved in synapse formation, metabolism, and muscle fiber-type specification (Buonanno et al., 1998). The ability of muscle cells to regulate the expression of these genes in response to muscle activity allows them to adapt to the changing demands of their environment. A major goal of muscle biologists is to understand the mechanisms coupling muscle activity to muscle phenotype. The data described herein identify an HDAC4/Mgn positive feedback loop that coordinates the opposing patterns of muscle gene expression in response to denervation.
HDAC4 Coordinates Gene Induction and Suppression after Muscle Denervation
Although muscle denervation is known to cause activation and suppression of gene expression (Buonanno et al., 1998), the molecular mechanisms underlying this regulation have remained elusive. We found that HDAC4 is necessary for both denervation-dependent gene induction and suppression (Figure 2). HDAC4 nuclear localization is regulated by calcium and calcium-regulated protein kinases (McKinsey et al., 2002; Liu et al., 2005) and denervation-dependent accumulation of HDAC4 in the nucleus seems to initiate changes in gene expression (McKinsey et al., 2002). We demonstrated that HDAC4 contributes indirectly to Mgn induction by coordinately inhibiting the expression of the Mgn gene corepressors Dach2 and MITR (Figure 3). HDAC4 contributes to gene suppression via its interaction with MEF2 transcription factors (Lu et al., 2000; Miska et al., 1999), and we identified putative MEF2 elements ∼980 base pairs upstream of the Dach2 coding sequence and 742 base pairs, 1568 base pairs, 2558 base pairs, and 2771 base pairs upstream of the MITR coding sequence. We also found HDAC4 inhibits MSE, PFK, and MHC IIb gene expression. Consistent with the idea that HDAC4 mediates inhibition by interaction with MEF2, we found HDAC4 is recruited to the MSE promoter's MEF2 site. The MHC IIb and PFK promoters also harbor MEF2 sites at positions −172 and −1780 relative to the translation initiation codon, respectively, which likely serve as HDAC4 docking sites.
Our observation that HDAC4 is recruited to the MSE, but not the Mgn promoter's MEF2 site is intriguing and may suggest that HDAC4 recruitment to MEF2 sites is context dependent. Whether the MEF2 site and flanking sequence or the constellation of transcription factors and cofactors that bind these promoters affect HDAC4 recruitment is not currently known.
Subcellular Localization of HDAC4 in Innervated and Denervated Muscle
We observed enrichment of HDAC4 in small oxidative fibers of innervated TA muscle that is composed predominantly of fast oxidative and glycolytic fiber types (Figure 5). Because HDAC4 can suppress glycolytic enzyme gene expression and induce Mgn expression (Figure 2) and because Mgn expression is highest in oxidative fibers (Hughes et al., 1993) and can increase the expression of oxidative enzymes (Hughes et al., 1999; Ekmark et al., 2003), it seems reasonable to propose that HDAC4-regulated gene expression contributes to the oxidative phenotype of innervated fast muscle fibers.
After muscle denervation, HDAC4 is enriched in nuclei of both fast oxidative and glycolytic fibers (Figure 6). Muscle denervation is also accompanied by an inhibition of glycolytic enzyme expression (Gundersen et al., 1988), and we showed that this is mediated by an HDAC4-dependent mechanism (Figure 2). Although Mgn is reported to increase expression of oxidative enzymes in innervated muscle, we did not find HDAC4/Mgn-dependent induction of oxidative enzymes in denervated muscle. This is consistent with previous reports showing muscle denervation is not correlated with increased oxidative metabolism (Gundersen et al., 1988) and may suggest Mgn differentially regulates oxidative enzyme gene expression in innervated and denervated muscle.
In addition to its nuclear localization, we found HDAC4 concentrated beneath the sarcolemma and also at the NMJ (Figure 5). Interestingly, mitochondria are found in both these regions (Ogata and Yamasaki, 1985; Perkins et al., 2001) and colocalize with HDAC4 (Figure 5). We suspect that mitochondria, or other cellular components distributed in a similar manner as mitochondria, serve as docking sites for HDAC4 and function either to sequester HDAC4 from nuclear entry and/or provide novel substrates for HDAC4-dependent modification. Of relevance, HDAC7 has also been found to localize to mitochondria (Bakin and Jung, 2004). More interestingly, HDAC4 interacts with the sarcolemma Z-disk–associated muscle LIM protein (MLP) in cardiac sarcomeres and impacts muscle contractile activity (Gupta et al., 2008). Whether MLP also associates with HDAC4 in skeletal muscle is not known. These unexpected findings suggest that HDAC4 localizes to the cytoplasm not just simply as a passive regulatory mechanism but may indicate that novel regulatory mechanisms remain to be elucidated.
An HDAC4/Mgn Positive Feedback Loop Coordinates and Maintains Denervation-dependent Transcriptional Programs
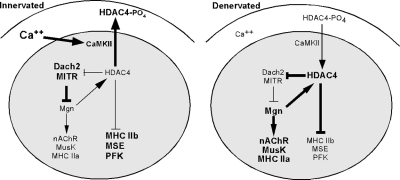
Our results reveal a novel positive feedback loop coordinating the induction and repression of genes after muscle denervation (Figure 8). We found that in denervated muscle HDAC4 expression is necessary for Mgn gene induction and that Mgn induction is necessary for increased HDAC4 gene expression (Figures 2 and 7). This signaling loop seems to be initiated upon muscle denervation by reduced intracellular calcium levels that stimulate nuclear retention of HDAC4. Increased nuclear levels of HDAC4 repress expression of the Mgn transcriptional repressors Dach2 and MITR. Relief of Mgn transcriptional repression, along with activating factors, results in increased Mgn levels that can then stimulate transcription of the HDAC4 gene.
Figure 8.
Diagram depicting the HDAC4/Mgn positive feedback loop and its regulation of downstream genes in innervated and denervated muscle. Bold is used to represent increased expression, levels, or activity. See text for details.
We have shown that the HDAC4/Mgn regulatory loop is capable of coordinating gene induction and suppression in denervated muscle. Muscle fibers are defined by the type of MHC protein expressed and their unique program of gene expression. In rodents, slow muscle fibers express MHC I and are rich in oxidative metabolism, whereas fast muscle fibers express a variety of MHC II isoforms (IIa, IIx, and IIb) to various extents. At one extreme, MHC IIa fibers are more like slow muscle in that they are rich in oxidative enzymes and exhibit a high resistance to fatigue; at the other extreme are MHC IIb fibers that have high glycolytic metabolism and fatigue easily. Endurance training will often result in a shift of fiber type to the more fatigue-resistant MHC I and MHC IIa type fibers. Similarly denervation of fast muscle is often accompanied by a shift to the more fatigue resistant type IIa phenotype (Gundersen et al., 1988). It seems that the HDAC4/Mgn signaling pathway contributes to this shift in muscle phenotype by suppressing expression of the fast MHC IIb contractile protein along with a variety of glycolytic enzymes and inducing expression of the more fatigue-resistant MHC IIa contractile protein (Figure 2).
Because HDAC4 nuclear localization is regulated by muscle activity and calcium levels (Liu et al., 2005), it is uniquely poised to respond to the changing demands put on muscle. In this respect, it is similar to the calcineurin/nuclear factor of activated T cells (NFAT) signaling complex that seems to drive expression of genes that are involved in determining a slow fiber phenotype (Chin et al., 1998). In this case, calcineurin responds to increased muscle calcium by dephosphorylating cytoplasmic NFAT, inducing its nuclear translocation where it can act on genes involved in determining the slow fiber phenotype. Thus, these data suggest that the HDAC4/Mgn signaling pathway, along with the calcineurin/NFAT pathway, represent master controls of muscle phenotype in response to the changing demands put on muscle by neural input.
ACKNOWLEDGMENTS
We are grateful to Drs. Steve Lentz and David Turner, Tony Kouzarides, Stuart Schreiber (Harvard, Cambridge, MA), and Francisco Sánchez-Madrid for sharing reagents. We thank Dr. Graeme Mardon (Baylor College of Medicine, Houston, TX) for providing Dach2 null mice; Boris Vidri, Pritish Iyer, and Gabriel Sotomayor for technical assistance; Drs. Laurent Schaeffer (Centre National de la Recherche Scientifique), Jack Parent, Audrey Seasholtz, Ted Ueda, Hisashi Umemori, and John Faulkner (University of Michigan) for advice, protocols, and shared equipment; and James Beal for assistance in microscopic imaging. We also thank members in the Goldman laboratory for experimental suggestions and helpful discussions. D. G. was supported by a grant from the Muscular Dystrophy Association, W.H.K. was supported by a grant from the Muscular Dystrophy Association and Robert A. Welch Foundation, and X.J.Y was supported by a grant from the Canadian Institute of Health Research.
Abbreviations used:
- BTX
α-bungarotoxin
- COX1
cytochrome c oxidase subunit 1
- HDAC
histone deacetylase
- Mdh2
malate dehydrogenase 2
- Mgn
myogenin
- MHC
myosin heavy chain
- MSE
muscle-specific enolase
- MuSK
muscle-specific receptor tyrosine kinase
- NaB
sodium butyrate
- nAChRα
nicotinic acetylcholine receptor α-subunit
- NMJ
neuromuscular junction
- PFK
phosphofructokinase
- SDH
succinate dehydrogenase
- TA
tibialis anterior
- TSA
trichostatin A
- TTX
tetrodotoxin.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0759) on December 24, 2008.
REFERENCES
- Bakin R. E., Jung M. O. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J. Biol. Chem. 2004;279:51218–51225. doi: 10.1074/jbc.M409271200. [DOI] [PubMed] [Google Scholar]
- Bertrand A., Ngo-Muller V., Hentzen D., Concordet J. -P., Daegelen D., Tuil D. Muscle electrotransfer as a tool for studying muscle fiber-specific and nerve-dependent activity of promoters. Am. J. Physiol. Cell Physiol. 2003;285:C1071–C1081. doi: 10.1152/ajpcell.00104.2003. [DOI] [PubMed] [Google Scholar]
- Buonanno A., Cheng J., Venepally P., Weis J., Calvo J. Activity-dependent regulation of muscle genes: repressive and stimulatory effects of innervation. Acta Physiol. Scand. 1998;163:S17–S26. doi: 10.1046/j.1365-201X.1998.1630s3S17.x. [DOI] [PubMed] [Google Scholar]
- Chin E. R., Olson E. N., Richardson J. A., Yang Q., Humphries C., Shelton J. M., Wu H., Zhu W., Bassel-Durby R., Williams R. S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:573–588. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T. J., Waddell D. S., Barrientos T., Lu Z., Feng G., Cox G. A., Bodine C, S., Yao T. -P. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J. Biol. Chem. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- Davis R. J., Pesah Y. I., Harding M., Paylor R., Mardon G. Mouse Dach2 mutants do not exhibit gross defects in eye development or brain function. Genesis. 2006;44:84–92. doi: 10.1002/gene.20188. [DOI] [PubMed] [Google Scholar]
- Dutton E., Simon A., Burden S. Electrical activity-dependent regulation of the acetylcholine receptor delta-subunit gene, MyoD, and myogenin in primary myotubes. Proc. Natl. Acad. Sci. USA. 1993;90:2040–2044. doi: 10.1073/pnas.90.5.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftimie R., Brenner H., Buonanno A. Myogenin and MyoD join a family of skeletal muscle genes regulated by electrical activity. Proc. Natl. Acad. Sci. USA. 1991;88:1349–1353. doi: 10.1073/pnas.88.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekmark M., Gronevik E., Schjerling P., Gundersen K. Myogenin induces higher oxidative capacity in pre-existing mouse muscle fibers after somatic DNA transfer. J. Physiol. 2003;548:259–269. doi: 10.1113/jphysiol.2002.036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D., Boulter, Heinemann J., S., Patrick J. Muscle denervation increases the levels of two mRNAs coding for the acetylcholine receptor α-subunit. J. Neurosci. 1985;5:2553–2558. doi: 10.1523/JNEUROSCI.05-09-02553.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D., Brenner H. R., Heinemann S. Acetylcholine receptor α-, β-, γ-, and δ-subunit mRNA levels are regulated by muscle activity. Neuron. 1986;1:329–333. doi: 10.1016/0896-6273(88)90081-5. [DOI] [PubMed] [Google Scholar]
- Golzio M., Mazzolini L., Moller P., Rols M. P., Theissie J. Inhibition of gene expression in mice muscle by in vivo electrically mediated siRNA delivery. Gene Therapy. 2005;12:246–251. doi: 10.1038/sj.gt.3302405. [DOI] [PubMed] [Google Scholar]
- Gorza L. Identification of a novel type 2 fiber population in mammalian skeletal muscle by combined use of histochemical myosin ATPase and anti-myosin monoclonal antibodies. J. Histochem. Cytochem. 1990;38:257–265. doi: 10.1177/38.2.2137154. [DOI] [PubMed] [Google Scholar]
- Grozinger C. M., Schreiber S. I. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K., Leberer E., Lomo T., Pette D., Staron R. S. Fibre types, calcium-sequestrating proteins and metabolic enzymes in denervated and chronically stimulated muscles of the rat. J. Physiol. 1988;398:177–189. doi: 10.1113/jphysiol.1988.sp017037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. P, Samant S., Smith S. H., Shroff S. G. HDAC4 and PcAF bind to cardiac sarcomeres and play a role in regulating the myofilament contractile activity. J. Biol. Chem. 2008;283:10135–10146. doi: 10.1074/jbc.M710277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey K., Bodine S. Changes in myosin mRNA and protein expression in denervated rat soleus and tibialis anterior. Eur. J. Biochem. 1998;256:45–50. doi: 10.1046/j.1432-1327.1998.2560045.x. [DOI] [PubMed] [Google Scholar]
- Hughes S. M., Taylor J. M., Tapscott S. J., Gurley C. M., Carter W. J., Peterson C. A. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- Hughes S., Chi M., Lowry O., Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. J. Cell Biol. 1999;145:633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman R. W., Kandarian S. C. The molecular basis of skeletal muscle atrophy. Am. J. Cell Physiol. 2004;287:C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- Khochbin S., Verdel A., Lemerecier C., Seigneurin-Berny D. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 2001;11:162–166. doi: 10.1016/s0959-437x(00)00174-x. [DOI] [PubMed] [Google Scholar]
- Knapp J., Davie J., Myer A., Meadows E., Olson E., Klein W. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development. 2006;133:601–610. doi: 10.1242/dev.02249. [DOI] [PubMed] [Google Scholar]
- Kummer T. T., Misgeld T., Sanes J. R. Assembly of the postsynaptic membrane at the neuromuscular junction: paradigm lost. Curr. Opin. Neurobiol. 2006;16:74–82. doi: 10.1016/j.conb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Liu F., Pore N., Kim M., Voong K., Dowling M., Maity A., Kao G. Regulation of histone deacetylase 4 expression by the SP family of transcription factors. Mol. Biol. Cell. 2006;17:585–597. doi: 10.1091/mbc.E05-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Randall W., Schneider M. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J. Cell Biol. 2005;168:887–897. doi: 10.1083/jcb.200408128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., McKinsey T., Zhang C., Olson E. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- Macpherson P.C.D., Cieslak D., Goldman D. Myogenin-dependent nAChR clustering in aneural myotubes. Mol. Cell Neurosci. 2006;31:649–660. doi: 10.1016/j.mcn.2005.12.005. [DOI] [PubMed] [Google Scholar]
- McKinsey T., Zhang C., Olson E. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- Méjat A., Ramond F., Bassel-Duby R., Khochbin S., Olson E., Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- Miska E., Karlsson C., Langley E., Nielsen S., Pines J., Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls M., Brenner D., Gruss H., Dierbach H., Mertelsmann R., Herrmann F. Mithramycin selectively inhibits collagen-alpha 1(I) gene expression in human fibroblast. J. Clin. Invest. 1993;92:2916–2921. doi: 10.1172/JCI116914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nozais M., Merkulova T., Keller A., Janmot C., Lompré A., D'Albis A., Lucas M. Denervation of rabbit gastrocnemius and soleus muscles: effect on muscle-specific enolase. Eur. J. Biochem. 1999;263:195–201. doi: 10.1046/j.1432-1327.1999.00486.x. [DOI] [PubMed] [Google Scholar]
- Ogata T., Yamasaki Y. Scanning electron-microscopic studies on the three-dimensional structure of mitochondria in the mammalian red, white and intermediate muscle fibers. Cell Tissue Res. 1985;241:251–256. doi: 10.1007/BF00217168. [DOI] [PubMed] [Google Scholar]
- Pandorf C. E., Haddad F., Roy R. R., Qin A. X., Edgerton V. R., Baldwin K. M. Dynamics of myosin heavy chain gene regulation in slow skeletal muscle. J. Biol. Chem. 2006;281:38330–38342. doi: 10.1074/jbc.M607249200. [DOI] [PubMed] [Google Scholar]
- Raffaello A., Laveder P., Romualdi C., Bean C., Toniolo L., Germinario E., Megighian A., Danieli-Betto D., Reggiani C., Lanfranchi G. Denervation in murine fast-twitch muscle: short-term physiological changes and temporal expression profiling. Physiol. Genomics. 2006;25:60–74. doi: 10.1152/physiolgenomics.00051.2005. [DOI] [PubMed] [Google Scholar]
- Perkins G. A., et al. PKA, PKC, and AKAP localization in and around the neuromuscular junction. BMC Neurosci. 2001;2:17. doi: 10.1186/1471-2202-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam G., Willmann R., Lin S., Erb-Vogtli S., Kong X. C., Ruegg M. A., Fuhrer C. Src-family kinases stabilize the neuromuscular synapse in vivo via protein interactions, phosphorylation, and cytoskeletal linkage of acetylcholine receptors. J. Neurosci. 2005;25:10479–10493. doi: 10.1523/JNEUROSCI.2103-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunesen M., Changeux J. Transcription in neuromuscle junction formation: who turns on whom? J. Neurocytol. 2003;32:677–684. doi: 10.1023/B:NEUR.0000020616.53664.80. [DOI] [PubMed] [Google Scholar]
- Tang H., Cheung W., Ip F., Ip N. Identification and characterization of differentially expressed genes in denervated muscle. Mol. Cell Neurosci. 2000;16:127–140. doi: 10.1006/mcne.2000.0864. [DOI] [PubMed] [Google Scholar]
- Tang H., Macpherson P., Argetsinger L. S., Cieslak D., Suhr S. T., Carter-Su C., Goldman D. CaM kinase II-dependent phosphorylation of myogenin contributes to activity-dependent suppression of nAChR gene expression in developing rat myotubes. Cell Signal. 2004;16:551–563. doi: 10.1016/j.cellsig.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Tang H., Veldman M. B., Goldman D. Characterization of a muscle-specific enhancer in human MuSK promoter reveals the essential role of myogenin in controlling activity-dependent gene regulation. J. Biol. Chem. 2006;281:3943–3953. doi: 10.1074/jbc.M511317200. [DOI] [PubMed] [Google Scholar]
- Tang H., Goldman D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc. Natl. Acad. Sci. USA. 2006;103:16977–16982. doi: 10.1073/pnas.0601565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Dequiedt F., Kasler H. G. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Kruhlak M. J., Wu J., Bertos N. R., Vezmar M., Posner B. I., Bazett-Jones D. P., Yang X. J. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. -J., Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]