Abstract
The termination of protein synthesis occurs through specific recognition of a stop codon in the A site of the ribosome by a release factor (RF), which then catalyzes the hydrolysis of the nascent protein chain from the P-site tRNA. Here we present the crystal structure at 3.5 Å resolution of release factor RF2 in complex with its cognate UGA stop codon in the 70S ribosome. The structure provides insight into how RF2 specifically recognizes the stop codon and suggests a model for the role of a universally conserved GGQ motif in catalysis of peptide release.
In nearly all species, three stop codons, UGA, UAG and UAA, signal the end of the coding sequence in mRNA. These stop codons are decoded by a protein factor termed a class I release factor (RF) (1, 2). In bacteria, there are two such factors with overlapping specificity: RF1 recognizes UAG, RF2 recognizes UGA and both recognize UAA. In eukaryotes, a single release factor, eRF1, recognizes all three stop codons. The mechanism by which release factors specifically decode stop codons and catalyze peptidyl-tRNA hydrolysis is a fundamental problem in understanding translation.
Elements of release factors involved in catalysis and stop-codon recognition have been proposed using sequence analysis combined with biochemistry and genetics. A universally conserved tripeptide sequence, GGQ (3), has been implicated in hydrolysis of the peptide chain from tRNA (4). Exchanging a tripeptide motif between RF1 and RF2 (P[A/V]T in RF1; SPF in RF2) switches their respective specificities for UAG and UGA (5). Hydroxyl-radical probing suggested that the SPF and GGQ motifs were close to the decoding center and peptidyl transferase center (PTC) respectively (6).
The structure of the eukaryotic eRF1 suggested that the distance between its codon-recognition and GGQ motifs was compatible with the approximately 75 Å distance between the decoding center and the PTC (7). However, eRF1 has no sequence or structural homology to bacterial RFs. The crystal structure of a bacterial RF2 (8) showed that the distance between the SPF and GGQ motifs was about 23 Å, and thus incompatible with their simultaneous involvement in decoding and peptide release. This anomaly was resolved when low resolution structures (9-11), showed that the conformation of RF2 when bound to the ribosome was different, so that the GGQ and SPF/PVT motifs were localized to the PTC and decoding center respectively. However, the resolution of these structures was too low to reveal detailed interactions of release factors with the ribosome.
Two years ago, our laboratory discovered a crystal form of the Thermus thermophilus ribosome that diffracts to high resolution in the presence of both A- and P-site ligands (12), thus providing a way out of this impasse. Using this crystal form, we report a structure refined to 3.45 Å resolution of release factor RF2 bound to a 70S ribosome containing the RF2-specific UGA stop codon (whose nucleotides are named U1, G2, A3) in the A site (13). The structure shows details of stop-codon recognition by RF2, as well as interactions of RF2 with the PTC and other regions of the ribosome including helix 69 (H69) and the L11 region. This crystal form was also used in a recent structure of RF1 bound to the ribosome containing a UAA codon (14).
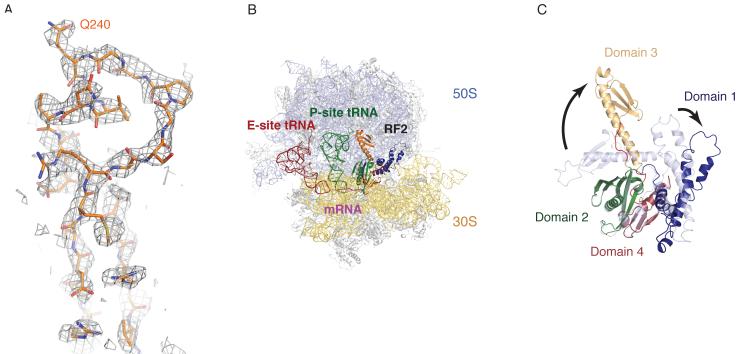
Initial refinement of the data by using a model of the empty ribosome showed clear difference density for the bound ligands (Fig. 1A). A complete model for the ribosome in complex with P- and E-site tRNAPhe, mRNA and RF2 was thus built and refined (13) (Fig. 1B). Compared to the crystal structure of isolated T. thermophilus RF2 (15), the factor in complex with the ribosome has undergone major conformational changes. As expected from earlier low-resolution studies (9-11), domain 3 of RF2 has peeled away from domains 2 and 4 (15) (Fig. 1C), placing the GGQ motif in the PTC, whereas domains 2 and 4 interact with the decoding center. Domain 1 has shifted slightly and interacts with the L11 region of the 50S subunit (see fig. S1 for details). These global conformational changes are accompanied by rearrangements of specific regions (Fig. 1C).
Fig. 1.
The structure of release factor RF2 in the ribosome. A. Unbiased difference Fourier maps showing density for RF2 in the peptidyl transferase center of the ribosome. B. Overview of the structure showing the 50S subunit (light blue), the 30S subunit (yellow) with E-site tRNA (red), P-site tRNA (green) and RF2 colored by domains as in C. The mRNA is shown in magenta. C. Conformational differences between the isolated crystal structure of RF2 (15) (shown in light blue), and the ribosome bound form (colored by domains as labeled) are indicated. Loop regions connecting domain 3 with domain 2 and 4, that undergo substantial conformational changes, are highlighted in red.
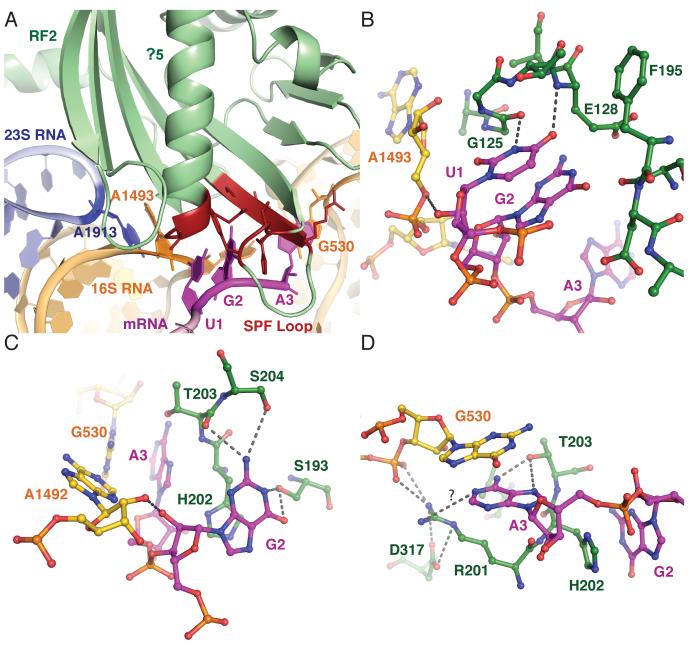
At the decoding center, RF2 induces changes that are markedly different from those induced by paromomycin or tRNA binding to sense codons (16, 17) (Fig. 2A). In standard decoding, three conserved nucleotides in the 30S ribosomal subunit, A1492, A1493 and G530 (Escherichia. coli numbering is used throughout), change conformation upon tRNA binding and closely interact with the minor groove of the codon-anticodon helix. These conformational changes would clash with RF2, explaining why paromomycin promotes tRNA binding but abolishes RF binding (18). In its new orientation, A1493 stacks on A1913 of 23S RNA, which likely plays a role in signal transduction from the decoding center to the PTC.
Fig. 2.
Interaction of RF2 with the stop codon in the decoding center. A. Overview of the decoding center showing the UGA stop codon (magenta). Domain 2 of RF2 is shown in green, except for parts interacting with the codon, which are shown in red. Key bases from 16S RNA and A1913 of 23S RNA are also shown. (B to D). Details of interactions at the first (B), second (C) and third (D) positions of the stop codon with elements of RF2 and the decoding center.
The structure sheds light on the basis of stop-codon recognition by RF2. At the first position, two conserved glycines at the tip of helix α5 in domain 2 make important backbone contacts with U1 of the stop codon (Fig. 2B). A purine at this position would result in a steric clash with the backbone of both G125 and G126. The N3 of U1 can make a hydrogen bond with the carbonyl group of G125, and the O4 is within hydrogen bonding distance of the backbone amide of E128. These interactions would not occur with a C. Together with the recent structure of RF1 bound to the ribosome (14), these observations explain why U is required at the first position of all stop codons.
Only the first amino acid in the SPF motif directly interacts with the second base of the stop codon, via hydrogen bonds between S193 and the O6 and N1 of G2 (Fig. 2C). Similar interactions with the hydroxyl group of S193 would be possible with the N6 and N1 of A, suggesting why an A would also be recognized at this position. The imidazole ring of H202 makes a stacking interaction with the second base. Though a pyrimidine would stack with H202 in a similar fashion, complete desolvation of the Watson Crick edge would be required without the formation of compensatory hydrogen bonds. Additional conserved elements are important for stop-codon recognition: both the hydroxyl of S204 and the carbonyl backbone of T203 can make hydrogen bonds with the N2 of G2.
Discrimination at the third position appears more complex. Although both release factors recognize an A at this position, RF2 miscodes a UGG codon with an error rate of 1 out of 2400, which is almost an order of magnitude worse than with UGU or UGC (19). The third base A3 does not stack on G2 but instead on G530 of 16S RNA (Fig. 2D). This stacking interaction would be far less favorable for a pyrimidine, which explains the discrimination against U or C at this position. The hydroxyl group of T203 can donate a proton to the N7 of A3 and accept one from its N6. With a G, similar hydrogen bonds could not be formed simultaneously to both its O6 and N7. However, T203 is also present in RF1, which can accept a G at the third position, suggesting that other interactions monitor this position. Interestingly, the terminal amino group of R201, which is conserved in RF2 but absent in RF1, is within hydrogen bonding distance of the phosphate of U531 and D317 in domain 4 of RF2. These interactions could position the arginine to act as a hydrogen bond donor for the N1 of A3, which would not be possible with a G. However, the arginine is poorly ordered in the structure.
The structure thus provides a rationale for specific recognition of UAA and UGA by RF2, and in conjunction with the recent structure of RF1, clarifies many of the elements involved in stop-codon recognition. However, it is clear that the tripeptide motif identified genetically (5) is only partially responsible for recognition. Other residues, some of which have been discussed, are likely involved in conferring specificity. Moreover, mutations in RF2 distant from the codon give rise to bacterial release factors with altered specificity (20). This suggests that recognition and miscoding by RFs is more complex and may involve the kind of tradeoff between binding energy and induced conformational changes that has been observed for tRNA recognition (21). The observation that the accuracy of termination arises from both a binding component and a catalytic rate component (19) is also fully consistent with the idea that codon recognition by RFs is followed by induced structural rearrangements that lead to catalysis.
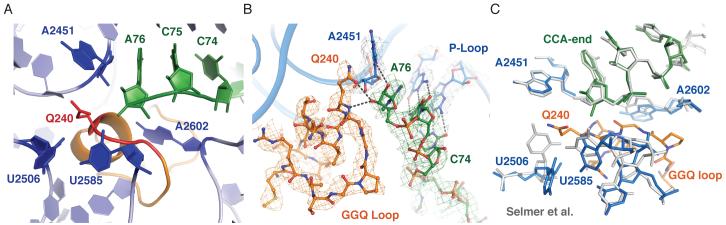
The GGQ motif essential for catalysis is positioned in the heart of the PTC (Fig. 3A). The loop within domain 3 containing this conserved tripeptide shifts considerably when bound to the ribosome compared to the isolated crystal structure (15). In the modeled loop, both glycines adopt a backbone conformation disallowed for any other amino acid, which explains their universal conservation and the drastic reduction in RF activity in vitro upon their mutation (22, 23). The glutamine side chain Q240 was modeled and refined in an orientation consistent with the initial unbiased difference Fourier maps (Fig 1A), which places its side-chain carbonyl within hydrogen bonding distance of the ribose of A76 of P-site tRNA (Fig. 3B). After refinement, difference Fourier maps also suggest the existence of a second conformation similar to that in the recent structure of RF1 bound to the ribosome, where it is pointed away from the ribose (14). The resolution is not high enough to distinguish between the C2′- and C3′-endo conformations of the ribose. In Fig. 3B, the ribose was modeled in a C3′-endo conformation, which in addition to a bond between the glutamine oxygen to the 3′-OH of A76, also allows a bond between the backbone amide of Q240 and the 3′-OH of A76. This backbone interaction was observed with RF1 (14), where it was suggested to be important for product stabilization. The alternative C2′-endo conformation seen in other structures (12, 24), would still allow the glutamine side-chain oxygen to hydrogen bond with the 2′-OH of A76 but would preclude formation of a hydrogen bond with the main-chain amide.
Fig 3.
Interaction of RF2 with the peptidyl transferase center. A. Overview of the peptidyl transferase center showing the terminal CCA of P-site tRNA (green), the conserved GGQ motif of RF2 (red) and key bases of 23S RNA (blue). B. Interaction of the GGQ loop of RF2 (orange) with the terminal ribose of P-site tRNA (green), showing sigma A weighted 3mFobs – 2DFcalc maps, where m is the figure of merit, D is the sigma A weight, and Fobs and Fcalc are the observed and calculated structure factors, respectively. Potential hydrogen bonds between the conserved Q240 and the ribose of P-site tRNA are shown. C. Changes in the peptidyl transferase center upon RF2 binding compared to the 70S structure with an empty A site (gray) (12). The structure shows that RF2 would clash with elements of 23S RNA and induces conformational changes, in particular in U2506 and U2585.
RF2 also induces changes in the PTC compared to the structure of the ribosome with an empty 50S A site (12). The GGQ loop would clash with U2506 and U2585 if they remained in the same conformation as when the A site is unoccupied. Thus both residues have moved as a result of RF2 binding (Fig. 3C). As suggested previously (24), the movement of U2585 away from the ester bond of peptidyl tRNA opens the bond to nucleophilic attack by water, whereas in the uninduced state it would be protected by U2585 in the absence of an A-site ligand. Clearly, the changes induced by the binding of the GGQ loop that expose the ester bond to nucleophilic attack must be a major component of the catalytic mechanism of release factors, and as suggested previously, show how similar changes induced by deacylated tRNA binding can catalyze peptide release (24).
The glutamine side chain was proposed to directly coordinate a water molecule during hydrolysis of the peptidyl-tRNA ester bond (7). Its mutation had a less drastic effect on activity than mutating either glycine (22, 23); however, the rate enhancement due to the glutamine must nevertheless be important because it is universally conserved, and RFs containing a glutamine mutation fail to complement defective RFs in vivo (25).
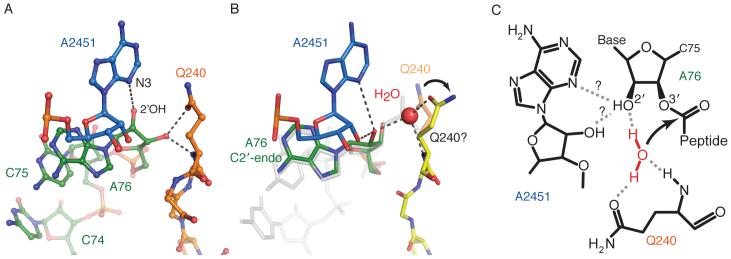
The structure, which is of the state after peptide release (Fig. 4A), makes it possible to propose a model for catalysis that rationalizes the available biochemical data using substrate and transition state analog structures. The superposition of a transition state analog of peptidyl transfer from a 50S subunit complex (24) shows that only a small change in the dihedral angles of the conserved Q240 would be sufficient to accommodate a water molecule positioned for nucleophilic attack on the peptidyl-tRNA ester bond (Fig. 4B). As in models presented earlier (7, 26), the side-chain oxygen of Q240 could be involved in hydrogen bonding with a water molecule (Fig. 4C). The glutamine is normally methylated in vivo, and the methylation is known to stimulate termination (27). Presumably the methylation would help direct the amine away from the ribose, thus positioning the oxygen for coordination with the water molecule. By acting as a hydrogen bond donor to N3 of A2451 (or its 2′-OH), the 2′-OH of A76 (modeled in the C2′-endo conformation, as seen in previous studies) is positioned to accept the second hydrogen of the attacking water, positioning the water oxygen for inline attack on the ester. Our proposed model also explains the minimal effect of A2451 mutants on peptide release (28) because all four bases contain a hydrogen-bond acceptor in the position of the N3 of A2451 and have a 2′-OH. Furthermore, the involvement of the 2′-OH of A76 agrees with data showing a significant effect on peptide hydrolysis of a 2′-deoxyadenosine mutant at this position (29). The specific coordination of a water by glutamine is consistent with studies showing that alternative nucleophiles are not affected by its mutation (23). A recent molecular dynamics study also proposes that the glutamine oxygen directly coordinates a water molecule, and suggests that mutation to alanine allows a second water molecule to compensate for the glutamine oxygen (26). Finally, the mechanism and the involvement of the 2′-OH of the peptidyl tRNA has similarities with the peptidyl transferase reaction (23).
Fig. 4.
A model of the substrate complex suggesting the basis for catalysis. A. The interaction of Q240 of RF2 and A2451 of 23S RNA in the current structure. B. A proposed structure showing how a minor change in the orientation of Q240 would allow it to coordinate a water molecule (transparent orange versus yellow). In gray is a transition-state analog superposed on this structure [1VQ7, taken from (24)] that was used to place the putative water that would take part in nucleophilic attack on the ester bond. C. A schematic representation of how a network of interactions among the conserved Q240, the coordinated water molecule that is the attacking nucleophile, the ribose of A76 of P-site tRNA (in C2′-endo conformation) and A2451 would act to facilitate catalysis.
Mutations of A2602 were observed to reduce the rate of peptide release (28), but other experiments reported that an abasic 2602 had no effect on the reaction rate (30). The structure shows that a direct involvement of A2602 in catalysis can be excluded, but it is possible that the nucleotide is involved in stabilizing the conformation of the GGQ loop, as also proposed for RF1 (14). Even so, the structure cannot explain why a mutation to G would result in a 90-fold reduced rate (28).
In the ribosome, the closed form of RF2 would clash with P-site tRNA and H69, consistent with the observation that E. coli RF1 is primarily in the open form in solution (31). While Thermus RF2 is mainly in the closed form at 20 °C, thermodynamic data suggested that it too exists predominantly in the open form at the physiological temperature of 75 °C (15). Thus it is unlikely that RF2 binds in the closed form to the ribosome and switches into the open form as a result of codon recognition. More likely the open form is conformationally variable and codon recognition imposes restraints on domain 3 that place the GGQ motif in the correct position at the PTC. As also noted for RF1 (14), local rearrangements of specific regions of RF2 are induced by ribosome binding (Fig. 1C).
Helix 69 of the 50S subunit is in a different orientation relative to the structure with a partially ordered tRNA in the A site (fig. S2)(12). Upon stop-codon recognition, A1913 of this helix stacks onto A1493 (Fig. 2A and fig. S2), thus establishing a connection between the decoding center and the 50S subunit. Also, Q121 of RF2 can make hydrogen bonds with both the 2′-OH of C1914 and the phosphate backbone of U1915. This is consistent with previous biochemical data showing that the pseudouridylation (32) or deletion (33) of H69 can affect translation termination. A similar change was also seen with RF1 (14) although the details of the A1913/A1493 stack appear different.
The high-resolution structure of RF2 bound to the ribosome with its UGA stop codon provides a structural basis for understanding its role in translational termination. Together with the recently published structure of the RF1-ribosome complex (14), these structures represent a major advance in understanding both stop-codon recognition and peptide release. Stop-codon recognition appears to have many components, and the remaining factor-codon combinations, along with mutagenesis studies will help clarify additional elements of recognition. We have proposed a model for catalysis that is consistent with the structure and biochemical data. However, both this and the RF1 structure represent the state after hydrolysis and peptide release, with a deacylated tRNA in the P site. Structures of substrate analog complexes at a resolution at which ordered water molecules would be visible, along with more directed biochemical experiments, will therefore be essential for understanding catalysis by these factors.
Supplementary Material
Materials and Methods, Figs. S1 to S2, Table S1, References
Acknowledgments
We thank Martin Schmeing for his useful comments and Martin Fuchs and Clemens Schulze-Briese for their help and advice with data collection at the Swiss Light Source. This work was supported by the Medical Research Council UK, a program grant from the Wellcome Trust and awards from the Agouron Institute and Louis-Jeantet Foundation. CN is supported by a Boehringer-Ingelheim fellowship and RMV is supported by a Gates-Cambridge fellowship. Coordinates for the structure have been deposited in the PDB with accession codes 2jl5-2jl8.
Footnotes
One sentence summary
The crystal structure of release factor RF2 bound to its cognate UGA codon in the ribosome sheds light on how the factor recognizes its stop codon as well as on the role played by elements involved in catalysis of peptide release.
References
- 1.Capecchi MR. Proc Natl Acad Sci U S A. 1967;58:1144. doi: 10.1073/pnas.58.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youngman EM, McDonald ME, Green R. Annu Rev Microbiol. 2008;62:353. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- 3.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 4.Frolova LY, et al. RNA. 1999;5:1014. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito K, Uno M, Nakamura Y. Nature. 2000;403:680. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 6.Wilson KS, Ito K, Noller HF, Nakamura Y. Nat Struct Biol. 2000;7:866. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 7.Song H, et al. Cell. 2000;100:311. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 8.Vestergaard B, et al. Mol Cell. 2001;8:1375. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 9.Rawat UB, et al. Nature. 2003;421:87. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 10.Klaholz BP, et al. Nature. 2003;421:90. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 11.Petry S, et al. Cell. 2005;123:1255. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 12.Selmer M, et al. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 13.Materials and methods are described in Supporting Online Materials.
- 14.Laurberg M, et al. Nature. 2008;454:852. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 15.Zoldak G, et al. Nucleic Acids Res. 2007;35:1343. doi: 10.1093/nar/gkl696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogle JM, et al. Science. 2001;292:897. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 17.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Cell. 2002;111:721. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 18.Youngman EM, He SL, Nikstad LJ, Green R. Mol Cell. 2007;28:533. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. Proc Natl Acad Sci U S A. 2000;97:2046. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito K, Uno M, Nakamura Y. Proc Natl Acad Sci U S A. 1998;95:8165. doi: 10.1073/pnas.95.14.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogle JM, Ramakrishnan V. Ann Rev Biochem. 2005;74:129. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 22.Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Mol Cell. 2002;10:789. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 23.Shaw JJ, Green R. Mol Cell. 2007;28:458. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmeing TM, Huang KS, Strobel SA, Steitz TA. Nature. 2005;438:520. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 25.Mora L, et al. Mol Microbiol. 2003;47:267. doi: 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- 26.Trobro S, Aqvist J. Mol Cell. 2007;27:758. doi: 10.1016/j.molcel.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 27.Dincbas-Renqvist V, et al. Embo J. 2000;19:6900. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Youngman EM, Brunelle JL, Kochaniak AB, Green R. Cell. 2004;117:589. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 29.Brunelle JL, Shaw JJ, Youngman EM, Green R. Rna. 2008;14:1526. doi: 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amort M, et al. Nucleic Acids Res. 2007;35:5130. doi: 10.1093/nar/gkm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vestergaard B, et al. Mol Cell. 2005;20:929. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Ejby M, Sorensen MA, Pedersen S. Proc Natl Acad Sci U S A. 2007;104:19410. doi: 10.1073/pnas.0706558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Mol Cell. 2006;23:865. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods, Figs. S1 to S2, Table S1, References