Abstract
The recent structure and associated biochemical studies of the metazoan-specific p300/CBP and fungal-specific Rtt109 histone acetyltransferases (HATs) have provided new insights into the ancestrial relationship between HATs and their functions. These studies point to a common HAT ancester that has evolved around a common structural framework to form HATs with divergent catalytic and substrate binding properties. These studies also point to the importance of regulatory loops within HATs and autoacetylation in HAT function. Implications for furture studies are discussed.
Introduction
The genetic material present in the nucleus of eukaryotic cells is packaged into chromatin, which functions as a dynamic scaffold for the regulation of various nuclear processes including DNA transcription, replication, repair, chromosome segregation, and apoptosis. The basic, repetitive unit of chromatin is the nucleosome core particle, a nucleoprotein structure that contains 147 base pairs of DNA wrapped around a histone octomeric core of a histone H3/H4 heterotetramer and two histone H2A/H2B heterodimers. Together with the H1 linker histone, nucleosome core particles form arrays of compacted higher-order chromatin structures that allow the entire human genome with a linearized length of more than 1 meter to be packed into a nucleus with a micrometer-scale diameter. Compacted chromatin restricts the access of many DNA regulatory proteins, thus neccesitating the need for chromatin regulatory proteins to facilitate DNA regulatory activities. Of the proteins that regulate chromatin function, the enzymes that post-translationally modify the histone proteins, particularly on the N-terminal histone tail regions, form the largest family. Histone acetyltransferases (HATs) were the first family of histone modification enzymes characterized at the molecular, biochemical and structural levels.
HAT enzymes catalyze the transfer of an acetyl group from the co-factor acetyl-coenzyme A (acetyl-CoA) to the ε-amine of a substrate lysine side chain. Since the isolation and characterization of the Tetrahymena Gcn5 HAT in 1996 [1], a large number of HATs have now been identified and characterized. To date, nuclear HATs can be categorized into four major families based on primary sequence homology: Gcn5/PCAF (General control nonrepressed protein 5 and p300 and CBP associated factor); MYST (named for the founding members MOZ, Ybf2/Sas3, Sas2, and Tip60); p300/CBP (protein of 300 kDa and CREB Binding Protein); and Rtt109 (Regulator of Ty1 Transposition gene product 109). Other putative HATs that have been reported include ATF-2 [2], TAF250 [3], nuclear steroid receptor coactivators [4,5], and more recently the circadian CLOCK protein [6], but their enzymatic properties have not been rigorously characterized. In this review, we will describe the two HAT families that have been most recently characterized, p300/CBP and Rtt109, with a particular focus on their strucutres and chemistry with implications for HAT evolution and function.
A brief review of p300/CBP and Rtt109 biology
The p300 and CBP (CREB binding protein) paralogs were originally identified as binding partners of the adenovirus early-region 1A (E1A) protein [7,8], and the cAMP-regulated enhancer (CRE) binding proteins [9], respectively. These proteins were later shown to function as transcriptional adaptors, and, about a decade ago, they were shown to harbor HAT activity [10,11]. p300 and CBP HAT domains have >90% sequence identity and are conserved in metazoans with many overlapping functions and will henceforth be referred to as p300/CBP. p300/CBP has multiple domains and, in addition to the enzymatic HAT domain, contains other protein interaction domains including three cysteine-histidine rich domains (CH1, CH2 and CH3), a KIX domain, a bromodomain, and a steroid receptor coactivator interaction domain (SID, also the SRC-1 interaction domain). p300/CBP not only catalyzes the acetylation of all four core histones, but has been reported to acetylate over 70 other proteins and itself (Table S1). Therefore, p300/CBP appears to be a versatile, and perhaps rather general, transcriptional integrator.
Abberrent p300/CBP activity has also been observed in various diseases. For example, Rubinstein-Taybi syndrome (RTS) is a developmental disorder caused by heterozygous germline mutations in the CBP or p300 genes [12,13]. p300/CBP gene mutations have been detected in human tumors with loss-of-function point mutations found in colorectal, breast, ovarian, oral, gastric, lung, and pancreatic carcinomas [14–16]. p300/CBP involved chromosome translocations have been observed in acute myeloid leukemia (AML) and in hematological disorders such as myeloid/lymphoid or mixed lineage (MLL) [17]. p300/CBP also functions as a transcriptional cofactor for proteins involved in tumorigenic pathways, including oncoproteins (such as myb, jun, and fos), transforming viral proteins (such as E1A, E6 and large T antigen), and tumor-suppressor proteins (such as p53, E2F, Rb, Smads, and BRCA1) [17]. p300/CBP may also contribute to respiratory epithelial carcinogenesis via cyclin D1, cyclooxygenase-2 (COX-2), and activator protein-1 (AP-1) [18]. More recent studies have linked p300/CBP to other diseases like progressive neurodegenerative diseases (such as Huntington Disease) [19], Kennedy’s disease [20], Alzheimer’s disease [21], cardiac disease [22], and fibrosis [23].
In contrast to the exclusive presence of p300/CBP sequence homologues in metazoans, Rtt109 sequence homologues are only present in fungi. Rtt109 was first identified to be a budding yeast regulator of Ty1 transposition that also plays an important role in the DNA damage response to genotoxic agents [24,25]. Recent genomic and proteomic screens as well as a more directed effort led to the observation that Rtt109 promotes genome stability and resistance to a variety of DNA-damaging agents through the direct acetylation of histone Lys56 of histone H3 (H3K56), a residue positioned at the DNA entry and exit points of the nucleosome core particle, during S-phase [•26, •27, •28, •29]. Rtt109 was subsequently reported to also acetylate H3K9 [•30].
Similar to the p300/CBP family, Rtt109 has no detectable sequence homology to other known HATs. Unlike other HATs, Rtt109 does not contain additional protein interaction domains and requires the presence of one of two histone chaperone proteins, Asf1 or Vps75, for HAT activity [•27, •29, •30, 31, 32].
Structure and chemistry of p300/CBP
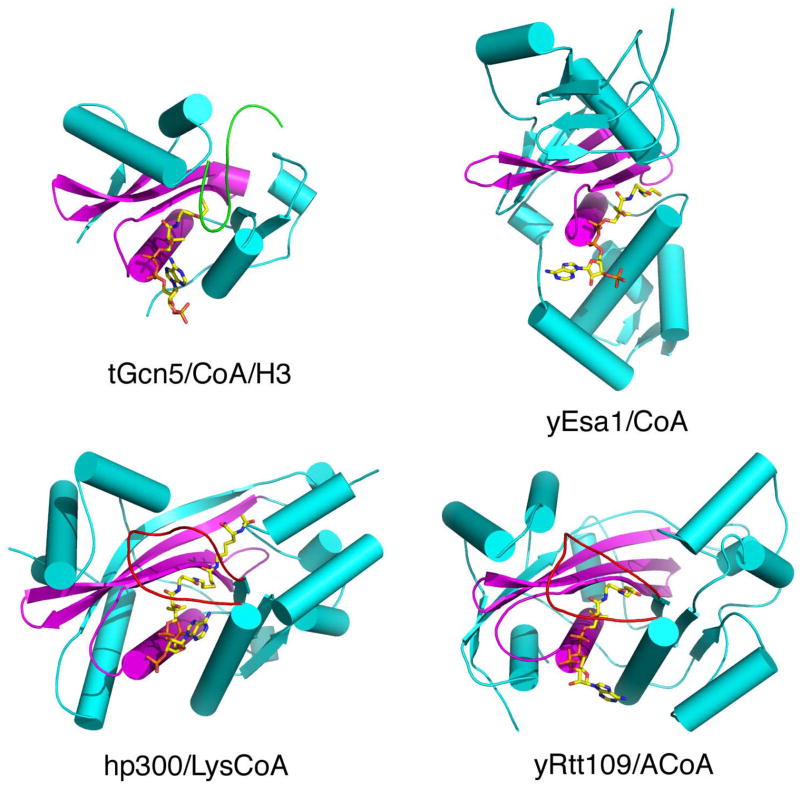
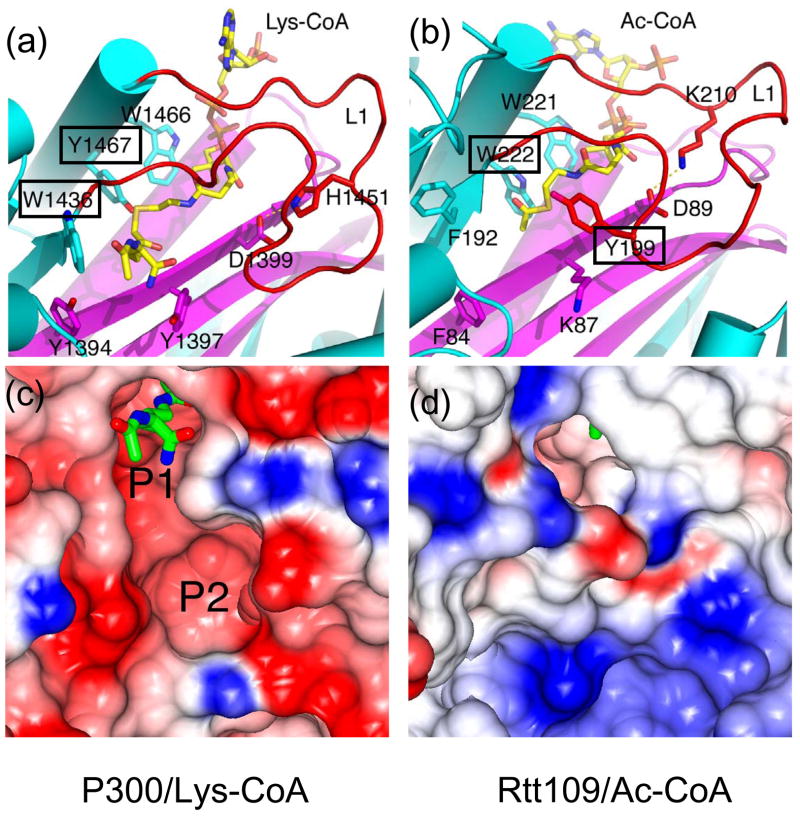
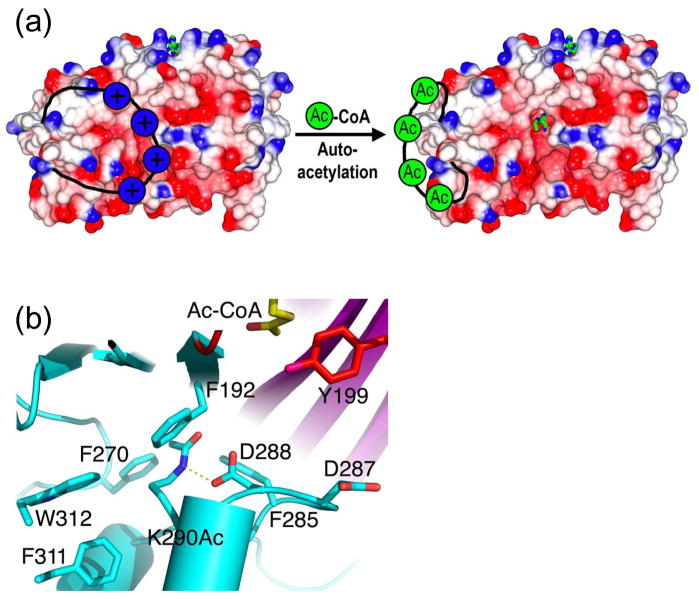
In early 2008, we reported on the crystal structure of the p300 HAT domain in complex with a Lys-CoA bisubstrate inhibitor and accompanying biochemical analysis of the enzyme [••33]. The overall fold of the p300 HAT domain consists of a central β-sheet composed of 7 β-strands surrounded by 9 α-helices and several loops (Figure 1c). Despite its sequence divergence from other HATs, a central core region of the p300 HAT domain, associated with acetyl-CoA cofactor binding, overlays well with the corresponding regions of the Gcn5/PCAF [34] and MYST [35] HATs (Figure 1a,b). This structurally conserved core region corresponds to the A, B and D sequence motifs of the GNAT (Gcn5 related N-acetyltransferase) superfamily of proteins reported by Neuwald and Landsman [36]. In contrast, the structural elements that flank the central core diverge significantly between the three HAT familes. Two other features also distinguish p300 from other HATs. First, the acetyl-CoA binding site of p300 is more buried than other HATs. This is largely due to the presence of a p300-specific L1 loop that covers one side of the cofactor and contributes to about one third of the protein-cofactor interactions (Figure 2a). This loop was also shown to be important for p300 catalysis. Second, the surface charge of the p300 HAT domain, particularly proximal to the lysine substrate binding site, is largely electronegative, compared to the more neutral character of the corresponding substrate binding sites of other HATs. Indeed, two shallow electronegative patches, one harboring the lysine moiety of the Lys-CoA inhibitor (P1) and another located about 10 Å away (P2) were shown to be mutationally sensistive for histone binding and catalysis (Figure 2c). Correlating with the importance of the P1 and P2 pockets, positively-charged residues are typically present within 3–4 amino acid residues (about 10 Å) upstream or downstream of the acetylated lysine residues of known p300/CBP substrates [37]. The promiscuous substrate specificity of p300 relative to other HATs can be accounted for by such a shallow and presumably versatile P2 pocket governing substrate recognition. An interesting property of the p300/CBP HATs is the presence of a highly basic autoacetylation loop that is hyperacetylated in the activated form of the p300/CBP HAT [•38] and that has been proteolytically cleaved in the current p300 HAT domain structure [••33]. We propose that this highly basic loop sits in the electronegative substrate binding site to block lysine substrate binding and is released from this site upon autoacetylation, as schemetically depicted in Figure 3a.
Figure 1. Overall structure of nuclear HATs.
(a) Tetrahymena Gcn5 (as cartoon) in complex with CoA (stick model in CPK coloring) and histone H3 (backbone trace colored in green) illustrates a representative Gcn5/PCAF HAT. The structurally conserved HAT core region is highlighted in magenta and the more structurally variable flanking regions are highlighted in cyan. (PDB ID 1PUA)
(b) Yeast Esa1 in complex with CoA illustrates a representative MYST HAT, with the coloring scheme as in (a). (PDB ID 1FY7)
(c) Human p300 in complex with the Lys-CoA bisubstrate inhibitor is the founding member of the p300/CBP HAT. In addition to the coloring scheme as in (a), the substrate binding L1 loop is highlighted in red. (PDB ID 3BIY)
(d) Yeast Rtt109 in complex with Ac-CoA. The coloring scheme is as in (c). (PDB ID 3D35)
Figure 2. p300 and Rtt109 active sites.
(a) The active site of p300 showing residues that are in position to play potential catalytic roles, highlighting Trp1436 and Tyr1467 (boxed).
(b) The active site of Rtt109 showing the corresponding residues shown in (a) and also highlighting mutationally sensitive residues Trp222 and Tyr199 (boxed) for HAT activity. In both (a) and (b), highlighted residues, together with Lys-CoA and acetyl-CoA, are shown as sticks with CPK coloring, with overall protein structures shown as cartoons colored as in Figure 1.
(c) Electrostatic surface of the p300 HAT substrate-binding pockets (P1 and P2) with blue, red and white representing electropositive, electronegative and neutral areas, respectively. Lys-CoA is shown in stick representation.
(d) The electrostatic surface of Rtt109 corresponding to that of p300 in (c).
Figure 3. p300 and Rtt109 autoacetylation regions.
(a) A model for the regulation of p300/CBP by autoacetylation is shown where it is proposed that the lysine-rich basic activation loop blocks the substrate-binding site when unacetylated but is displaced upon autoacetylation.
(b) Autoacetylated Lys290 in Rtt109 is shown hydrogen bonded to Asp288 and buried within a hydrophobic core.
Recent enzymatic studies on the p300 HAT domain is consistent with a Theorell-Chance (or “hit and run”) catalytic mechanism that is distinct from other HATs. This mechanism, as it applies to p300, invokes that during catalysis, there is no stable ternary complex formed during the reaction. Rather, the p300 protein first forms a stable complex with the acetyl-CoA cofactor and then the positively charged substrate lysine side chain from the protein substrate transiently binds to the negatively charged P1 pocket and departs immediately after acetyl transfer. Such a mechanism differs from the classical ordered Bi-Bi ternary complex mechansim employed by the Gcn5/PCAF [39] and the ternary [40] and ping-pong mechanisms [35] that have been proposed for the Esa1 member of the MYST family of HATs. The Theorell-Chance mechanism proposed for p300/CBP is consistent with the broad substrate specificity of p300 as it does not require a specific substrate binding pocket. Structure-based p300 mutagenesis studies also identify two critical catalytic residues: Tyr 1467 that likely funcitons as a general acid to protonate the CoA leaving group, and Trp1436 that likely helps steer the substrate lysine into its binding site (Figure 2a). Interestingly, p300 does not critically depend on a specific Asp/Glu general base for catalysis, similar to serotonin N-acetyltransferase [41], but unlike the Gcn5/PCAF [34,42] and MYST [35] HATs that employ glutamate residues for this purpose.
The structure of p300 allows for a rationalization of several p300/CBP-inactivating mutations that have been observed in various cancers [14–16]. For example, the catalytic Trp1436 residue is mutated to cysteine in lung cancer, as is Arg1410, which plays a key role in acetyl-CoA cofactor binding. In addition, Asp1399, a residue important for the appropriate conformation of the L1 loop (Figure 2a), is mutated to a tyrosine residue in colon cancer.
Structure and chemistry of Rtt109
Shortly after the structural determination of the p300 HAT domain, we reported on the crystal structure of the Rtt109 HAT domain bound to the acetyl-CoA cofactor [••43]. Despite its sequence divergence from other HATs, the Rtt109 structure reveals a strucutrally conserved core region for CoA binding. Unexpectedly, the rest of the Rtt109 domain also looks remarkably similar to p300 with an overall r.m.s.d of 3.5 Å for 236 out of 354 Ca atoms (Figure 1d). This similarity extends to the L1 loop where an Asp89-Lys210 salt-bridge (reminiscent of the Asp1399-His1451 salt-bridge in p300) is essential for holding the loop in position for acetyl-CoA cofactor binding (Figure 2b). Given that the Rtt109 and p300/CBP HAT family only exists in fungi and metazoans, respectively, these structural similarities suggest an evolutionary link between p300/CBP and Rtt109.
Despite the structural similarities between p300 and Rtt109, several biochemical properties of the two enzymes suggest that they diverge significantly at the functional level. Specifically, the surface charge of Rtt109 proximal to the lysine binding site (Figure 2d) is unlike the electronegative surface of p300 (Figure 2c) and, in fact, more like the apolar surfaces of the Gcn5/PCAF and MYST family of HATs. Indeed, unlike the broad substrate specificity of p300/CBP, Rtt109 appears to be quite selective for H3K56 [•27, •28, •29] and H3K9 [•30]. The potent p300 inhibitor, Lys-CoA [44] does not show detectable inhibition of Rtt109 [••43]. The mechanism of acetyl transfer catalyzed by Rtt109 also appears to be distinct from that of p300, as well as the Gcn5/PCAF and MYST HATs. Similar to p300, Rtt109 does not contain an appropriately positioned glutamate residue in the active site that might function as a general base for catalysis, as seen for the Gcn5/PCAF HATs. However, dissimilar to p300, the two important catalytic residues of p300, Trp1436 and Tyr1467, are not structurally conserved in Rtt109 (Figure 2a,b). Interestingly, although tyrosine and tryptophan residues of Rtt109 (Tyr199 and Trp222) have been shown to be important for catalysis, they are in different positions in the three-dimensional structure from the corresponding catalytic residues in p300 and appear to play different catalytic roles (Figure 2b). Kinetic analysis of Rtt109 also reveals an unusual sigmoidal histone H3 substrate concentration dependence at fixed acetyl-CoA that fits to a Hill equation with a Hill coefficient of 3, suggesting a high degree of cooperativity of the histone substrate in the acetylation reaction. Although the delineation of the precise catalytic mechanism employed by Rtt109 requires further study, it is already clear that Rtt109 employs a catalytic mechanism that differs from other HATs and is consistent with a more complex nature of protein substrate recognition.
Like p300, Rtt109 has been shown to acetylate itself [45]. Indeed the structure of the Rtt109/Ac-CoA complex reveals that a partially buried lysine residue (K290), located about 8 Å away from the active site of the enzyme, is acetylated (Figure 3b). Moreover, mass spectrometry data on bacterially produced recombinant and yeast produced Rtt109 protein reveals that this lysine is autoacetylated in vitro and in yeast cells [••43]. Although the functional consequence of this autoacetylation is still unclear, it is interesting to note that in addition to Rtt109 and p300/CBP, other HATs, including Esa1 [40] and PCAF [46] have been shown to undergo autoacetylation. These observations point to the possibility that the autoacetylation of HATs may play conserved functional roles in regulating HAT funciton.
A unique feature of Rtt109 that distinguishes it from p300 and other HATs, is that it requires the association of histone chaperones, Asf1 and/or Vps75 for catalysis [•27, •29, •30, 31]. We suggest that the binding of these chaperones may help present the H3 substrate to Rtt109 protein, which is consistent with the observation that the addition of excess Vps75 to a stoichiometric Rtt109/Vps75 complex inhibits histone H3 acetylation by Rtt109 [••43]. The histone chaperone requirement for Rtt109 HAT activity may be related to the fact that Rtt109, unlike other HATs, does not contain other associated domains to interact with other chromatin modifiers.
During the review of this manuscript, two additional manuscripts describing analogous crystal structures of Rtt109 were reported [••51, ••52]. Both studies utilized protein constructs with internal loop deletions similar to what we had described (delta 130–179) and reveal essentially the same Rtt109 structure. Moreover, the study by Stavropoulos et al. also confirmed that this particular loop region is essential for Vps75 histone chaperone binding [••43, ••51]. Also similiar to the initial Rtt109 structure report is the finding that K290 of Rtt109 is acetylated [••51, ••52]. Interestingly, while our study found that a K290R mutant is wild-type for both H3K56 acetylation and sensitivity to genotoxic stress in vivo, the more recent studies show that mutation of K290 reduces the ability of Rtt109 to acetylate H3K56 in vitro [••52] and in vivo [••51]. These apparently conflicting results may be related to the fact that we employed a K290R mutant in the context of an intact Rtt109 protein using a W303 yeast strain [••43], while Stavropoulos et al. employed K290 mutants in the context of a (delta 128–170) loop deletion using a different BY4741 yeast strain [••51]. Lin and Yuan also found that the K290R Rtt109 mutant is inactive in vitro towards an Asf1/H3-H4 substrate [••52]. Additional experiments are clearly needed to resolve the importance of K290 acetylation in Rtt109 function.
Concluding remarks
The recent structural analyses of the p300 and Rtt109 HATs further extend the notion that, despite the lack of sequence conservation, HATs may contain a “universal” conserved central core region for acetyl-CoA binding but divergent flanking regions (Figure 1). These more divergent flanking regions appear to be important for HAT-specific functions. For example, these flanking regions play a key role in histone substrate binding by Gcn5 [39] and play a role in nucleosome binding by the MOZ member of the MYST HAT [47]. The conserved L1 loop in p300 and Rtt109 plays an important role in acetyl-CoA cofactor binding and possibly also histone substrate binding and catalysis. With regard to catalysis by HATs, it is noteworthy that they employ different catalytic mechanisms despite the presence of a strucutrally conserved core region. This may reflect the fact that acetyl transfer between a thioester and an amine is kinetically and thermodynamically facile and can be readily catalyzed by positioning and orientation using various easily evolved strategies. The finding that different HATs employ different catalytic mechanisms argues favorably for the prospect of preparing small-molecule HAT-specific inhibitors, an important consideration given that HATs such as p300/CBP and MOZ [17,48] have been implicated in human disease. We would also note that the design of alternative therapeutics that rescue HAT-inactivating mutations [49] may also be aided by the structure of p300. Finally, the lack of sequence conservation among HATs and the relatively recent identification of Rtt109 as a HAT, suggest that other HATs are likely to be uncovered and tools such as novel proteomic reagents [50] are needed to aid in discovery.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
• • of outstanding interest
- 1.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki H, Schiltz L, Chiu R, Itakura K, Taira K, Nakatani Y, Yokoyama KK. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature. 2000;405:195–200. doi: 10.1038/35012097. [DOI] [PubMed] [Google Scholar]
- 3.Mizzen CA, Yang X-J, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 4.Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou JX, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 6.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Yee SP, Branton PE. Detection of cellular proteins associated with human adenovirus type 5 early region 1A polypeptides. Virology. 1985;147:142–153. doi: 10.1016/0042-6822(85)90234-x. [DOI] [PubMed] [Google Scholar]
- 8.Harlow E, Whyte P, Franza BR, Jr, Schley C. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 11.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 12.Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 13.Petrij F, Dorsman JC, Dauwerse HG, Giles RH, Peeters T, Hennekam RC, Breuning MH, Peters DJ. Rubinstein-Taybi syndrome caused by a De Novo reciprocal translocation t(2;16)(q36.3;p13.3) Am J Med Genet. 2000;92:47–52. doi: 10.1002/(sici)1096-8628(20000501)92:1<47::aid-ajmg8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 14.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong JM, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 15.Kishimoto M, Kohno T, Okudela K, Otsuka A, Sasaki H, Tanabe C, Sakiyama T, Hirama C, Kitabayashi I, Minna JD, et al. Mutations and deletions of the CBP gene in human lung cancer. Clin Cancer Res. 2005;11:512–519. [PubMed] [Google Scholar]
- 16.Bryan EJ, Jokubaitis VJ, Chamberlain NL, Baxter SW, Dawson E, Choong DY, Campbell IG. Mutation analysis of EP300 in colon, breast and ovarian carcinomas. Int J Cancer. 2002;102:137–141. doi: 10.1002/ijc.10682. [DOI] [PubMed] [Google Scholar]
- 17.Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 18.Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. Roles of CREB-binding protein (CBP)/p300 in respiratory epithelium tumorigenesis. Cell Res. 2007;17:324–332. doi: 10.1038/cr.2007.10. [DOI] [PubMed] [Google Scholar]
- 19.Cong SY, Pepers BA, Evert BO, Rubinsztein DC, Roos RA, van Ommen GJ, Dorsman JC. Mutant huntingtin represses CBP, but not p300, by binding and protein degradation. Mol Cell Neurosci. 2005;30:12–23. doi: 10.1016/j.mcn.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Lieberman AP, Harmison G, Strand AD, Olson JM, Fischbeck KH. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum Mol Genet. 2002;11:1967–1976. doi: 10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- 21.Francis YI, Diss JK, Kariti M, Stephanou A, Latchman DS. p300 activation by Presenilin 1 but not by its M146L mutant. Neurosci Lett. 2007;413:137–140. doi: 10.1016/j.neulet.2006.11.036. [DOI] [PubMed] [Google Scholar]
- 22.Yanazume T, Morimoto T, Wada H, Kawamura T, Hasegawa K. Biological role of p300 in cardiac myocytes. Mol Cell Biochem. 2003;248:115–119. doi: 10.1023/a:1024132217870. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh AK, Varga J. The transcriptional coactivator and acetyltransferase p300 in fibroblast biology and fibrosis. J Cell Physiol. 2007;213:663–671. doi: 10.1002/jcp.21162. [DOI] [PubMed] [Google Scholar]
- 24.Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 25.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. This manuscript was the first to correlate Rtt109 protein with the acetylation of lysine 56 of histone H3, although no direct HAT activity was demonstrated for Rtt109. [DOI] [PubMed] [Google Scholar]
- 27•.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. This manuscript showed that lysine 56 of histone H3 is acetylated by Rtt109 upon Asf1 stimulation to promote genome stability and DNA repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. This manuscript showed that lysine 56 of histone H3 is acetylated by Rtt109, an essential event for genone stability and DNA repair. [DOI] [PubMed] [Google Scholar]
- 29•.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. This manuscript highlighted the essential roles of histone chaperones, Asf1 and Vps75, in the acetylation of lysine 56 of histone H3 by Rtt109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. This manuscript was the first to show that Rtt109 has other substrates than lysine 56 of histone H3, showing that lysine 9 of histone H3 is also a target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem. 2007;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- 32.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 33••.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. This mansucript reported the structure of the HAT domain of p300 bound to the Lys-CoA bisubstrate inhibitor as well as in vitro biochemical and enzymatic studies of wild-type and mutant p300 proteins to provide insights into catalysis and histone substrate binding by p300. [DOI] [PubMed] [Google Scholar]
- 34.Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci U S A. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan Y, Harper S, Speicher DW, Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat Struct Biol. 2002;9:862–869. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 36.Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 37.Thompson PR, Kurooka H, Nakatani Y, Cole PA. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J Biol Chem. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- 38•.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. This manuscript presented biochemical studies to demonstrate that p300 is autoacetylated, predominantly on a “activation loop” to regulate its function. [DOI] [PubMed] [Google Scholar]
- 39.Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 40.Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007;46:623–629. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheibner KA, De Angelis J, Burley SK, Cole PA. Investigation of the roles of catalytic residues in serotonin N-acetyltransferase. J Biol Chem. 2002;277:18118–18126. doi: 10.1074/jbc.M200595200. [DOI] [PubMed] [Google Scholar]
- 42.Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 43••.Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verrault A, Cole PA, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol. 2008;15:738–745. doi: 10.1038/nsmb.1448. This mansucript reported the structure of Rtt109 bound to acetyl-CoA as well as in vitro and in vivo studies of wild-type and mutant Rtt109 proteins to provide insights into catalysis and Vps75 binding by Rtt109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, Wolffe AP, Nakatani Y, Roeder RG, Cole PA. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5:589–595. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 45.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 46.Santos-Rosa H, Valls E, Kouzarides T, Martinez-Balbas M. Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 2003;31:4285–4292. doi: 10.1093/nar/gkg655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holbert MA, Sikorski T, Carten J, Snowflack D, Hodawadekar S, Marmorstein R. The human monocytic leukemia zinc finger histone acetyltransferase domain contains DNA-binding activity implicated in chromatin targeting. J Biol Chem. 2007;282:36603–36613. doi: 10.1074/jbc.M705812200. [DOI] [PubMed] [Google Scholar]
- 48.Troke PJ, Kindle KB, Collins HM, Heery DM. MOZ fusion proteins in acute myeloid leukaemia. Biochem Soc Symp. 2006:23–39. doi: 10.1042/bss0730023. [DOI] [PubMed] [Google Scholar]
- 49.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 50.Hwang Y, Thompson PR, Wang L, Jiang L, Kelleher NL, Cole PA. A selective chemical probe for coenzyme A-requiring enzymes. Angew Chem Int Ed Engl. 2007;46:7621–7624. doi: 10.1002/anie.200702485. [DOI] [PubMed] [Google Scholar]
- 51••.Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc Natl Acad Sci U S A. 2008;105:12236–12241. doi: 10.1073/pnas.0805813105. This mansucript reported the structure of Rtt109 bound to acetyl-CoA as well as in vitro and in vivo studies of wild-type and mutant Rtt109 proteins to provide insights into catalysis and Vps75 binding by Rtt109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin C, Yuan YA. Structural Insights into Histone H3 Lysine 56 Acetylation by Rtt109. Structure. 2008 doi: 10.1016/j.str.2008.07.006. Published online Aug. 16 2008 as doi:10.1016/j.str.2008.07.006 ••This mansucript reported the structure of Rtt109 alone and bound to acetyl-CoA as well as in vitro studies of wild-type and mutant Rtt109 proteins to provide insights into catalysis by Rtt109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.