Abstract
Microarrays have been used extensively in gene expression profiling and genotyping studies. To reduce the high cost and enhance the consistency of microarray experiments it is often desirable to strip and reuse microarray slides. Our genome-wide analysis of microRNA expression involves the hybridization of fluorescently labeled nucleic acids to custom-made, spotted DNA microarrays based on the GAPSII coated slides. We describe here a simple and effective method to regenerate such custom microarrays, which uses a very low salt buffer to remove labeled, nucleic acids from microarrays. Slides can be stripped and reused multiple times without significantly compromising data quality. Moreover, our analyses of the performance of regenerated slides identified parameters that influence the attachment of oligonucleotide probes to GAPSII slides, which sheds light on the interactions between DNA and microarray surface and suggests ways to improve the design of oligonucleotide probes.
Keywords: custom microarray, GAPSII slide, microRNA, oligonucleotide probe
INTRODUCTION
microRNAs (miRNAs) are a class of approximately 22 nucleotides (nt) long, widely-expressed RNA molecules that predominantly inhibit gene expression at the post-transcriptional level in eukaryotes [1–3]. Because they regulate the expression of a large number of protein-encoding genes [4–7], miRNAs control a wide range of biological processes, such as metabolism, organogenesis, development, cell growth, cell death, and cell fate determination [8, 9]. Furthermore, under-expression or over-expression of miRNAs has been associated with human disease such as cancer [10, 11]. To investigate the important physiological functions of miRNAs, therefore, many laboratories have developed custom miRNA microarray techniques to detect and compare global miRNA expression in different cell types and tissues and under different conditions [e.g., 12–19].
To fabricate a typical miRNA microarray, DNA probes consisting of 20–70 nt long oligonucleotides are printed on a chemically modified, glass slide, similar to many other types of oligonucleotide-based microarrays. The slide is then incubated with labeled samples, and signals hybridized to the probes detected by a fluorophore-based method. Because the number of miRNA genes in any particular genome is currently below 1000, it is manageable and cost effective to produce miRNA microarrays and perform hybridizations in house. In addition, custom microarrays can be easily updated as miRNA databases evolve. Nevertheless, these microarray experiments can still be expensive because of the high cost of array fabrication and the large number of replicate hybridizations required to draw meaningful conclusions from the experiments. Complication further arises as the quality of custom slide printing usually varies from batch to batch. As a result, it is often advantageous to strip and subsequently reuse microarray slides that are of high quality to reduce cost as well as to enhance the consistency of miRNA and indeed other kinds of microarray experiments.
To date, various protocols for stripping slides have been reported [e.g., 20–26]. A common practice is to immerse slides in boiling or near-boiling sodium dodecyl sulfate solution, although high temperature is potentially detrimental to slide integrity. Since many expression profiling studies use labeled RNAs to hybridize to probe DNAs on the microarrays, one can also take advantage of the differential stability of DNA and RNA molecules at high pH to strip microarray slides under mild alkaline conditions. For example, Hu et al. [25] used a solution of 1 x standard saline citrate (SSC), 0.00025% Triton® X-102, 8 mM sodium hydroxide (NaOH), and 250 mM EDTA to treat high-density oligonucleotide microarrays from Agilent Technologies (Wilmington, DE, USA) at 60–62°C. It was believed that NaOH degraded RNAs while leaving the DNA probes and slide coating largely intact. While all these approaches yield overall reusable microarrays, whether and how individual probes were affected by the stripping procedures has never been examined closely. We introduce here a simple and effective method to regenerate custom miRNA microarrays and further evaluate factors that may contribute to probe retention on a microarray.
MATERIALS AND METHODS
Fabrication of custom miRNA microarrays
A miRNA probe set was purchased from Invitrogen™ (Carlsbad, CA, USA), which was designed based on the Sanger miRBase Sequence Database, Release 9.0 (October 2006). The set contains approximately 1,140 unmodified oligonucleotides of 34 to 44 nt long as probes. They are complementary to worm, fly, zebrafish, mouse, rat, and human miRNAs, and also include a collection of internal control probes. We further supplemented a number of probes as additional controls. All the oligonucleotides were dissolved in 3 x SSC and quadruply printed on Corning® GAPS™ II coated slides (Corning, NY, USA) using a BioRobotics Microgrid II spotter (Genomic Solutions, Ann Arbor, MI, USA) by the Biomedical Genomics Center Microarray Facility at the University of Minnesota, Minneapolis, MN. The slides were then irradiated by ultraviolet (UV) light and stored at 22°C.
Sample preparation for microarray experiments
Trizol® reagent (Invitrogen™) was used to extract total RNA from 293T, a human embryonic kidney cell line, and the quantity and quality of isolated RNA determined by A260nm and A280nm measurements. For RNA labeling, we ligated total RNA to a synthetic linker, 5′-pCU-DY547-3′ (Dharmacon, Lafayette, CO, USA), according to Thomson et al. [17]. To control for the hybridization process, reference DNA oligonucleotides (Invitrogen™) complementary to a subset of mammalian miRNAs were combined and labeled with a ULYSIS® Alexa Fluor® 647 kit (Invitrogen™).
Microarray hybridization, signal detection, and data analysis
A mixture of labeled RNA and reference DNA was hybridized to a miRNA microarray, according to Thomson et al. [17], except that hybridization was conducted inside a MAUI mixer in a MAUI hybridization system (BioMicro® System, Inc, Salt Lake City, UT, USA). After washing, the slide was scanned by a ScanArray 5000 machine (Perkin Elmer®, Waltham, MA, USA). BlueFuse (BlueGenome, Cambridge, UK) was used to quantify pixel intensities. Individual spots on microarrays were further inspected to exclude abnormal spots from further consideration. Typically, signals with intensities that were at least 150% above background (usually between 200 and 250) were deemed positive. As we were interested in the expression of mammalian miRNAs, we limited our analyses to the approximately 720 probes specific for the human, mouse, and rat miRNAs on our microarrays. GeneSpring GX 7.3.1 (Agilent Technologies) was used for data normalization to the internal control, 28S rRNA in channel (ch) 1. SPSS Windows Version 13.0 (SPSS Inc., Chicago, IL, USA) was used for calculating Pearson correlation coefficients and Intraclass Correlation Coefficients (ICC). While Pearson correlation coefficients are frequently used to assess the similarities between array results, ICC may be better at evaluating the reproducibility or agreement between the results [27].
Slide stripping procedure
Microarray slides were rinsed in distilled water for 1 min, immersed in approximately 250 ml of pre-warmed stripping buffer (0, 1, 2, 5, or 10 mM NaOH, in 0.1 x SSC) in a staining dish, and incubated at 62°C for 10 min. The incubation was repeated once with fresh buffer. The slides were washed three times in water for 15 min each time with gentle shaking at 22°C. Slides were spun dry and then scanned to verify the effect of stripping. Slides were stored in the dark at 22°C until use.
Evaluating factors that may influence the affinities between oligonucleotide probes and microarray slides
Hybridization data from slides that underwent stripping with 0.1 x SSC or with 1 mM NaOH, 0.1 x SSC for up to three times (described in Fig. 1 and Tables 1 and 2) were analyzed. We selected those probes that generated positive signals, at least in a new slide, and calculated their decay values (DVs) as in y = a*eDV*X, for raw signal intensities (y) after every stripping process (x = 0, 1, 2, 3), using Excel (Microsoft®, Redmond, Washington). Normalized signals gave similar results. There were maximally four DVs for each probe, as calculated under the above, two stripping conditions and using separate signals from the two channels, corresponding to hybridization by 293T RNA and the reference DNA, respectively. The actual number, however, was usually one to three, because RNA and DNA signals overlapped only partially (see Fig. 1), and weak signals might be detected in one microarray but not the other. We eliminated probes that yielded only one DV, although their inclusions did not affect the conclusions of our analysis. For each of the majority of the remaining, approximately 210 probes, its DVs computed under the different stripping conditions and in the two channels were quite similar, so we took the mean of the DVs as the final, representative DV for the probe. In the case of a small number of probes that had dissimilar DVs, we removed the clear outliners, or re-analyzed the raw data to select the ones with the stronger hybridization signals for subsequent calculations. A number of probes had small, positive DVs, which were included in our analysis as well. We then examined the correlation between DVs of the probes and physical attributes of the probes, such as probe length, nucleotide composition, and melting temperature (Tm), using Excel.
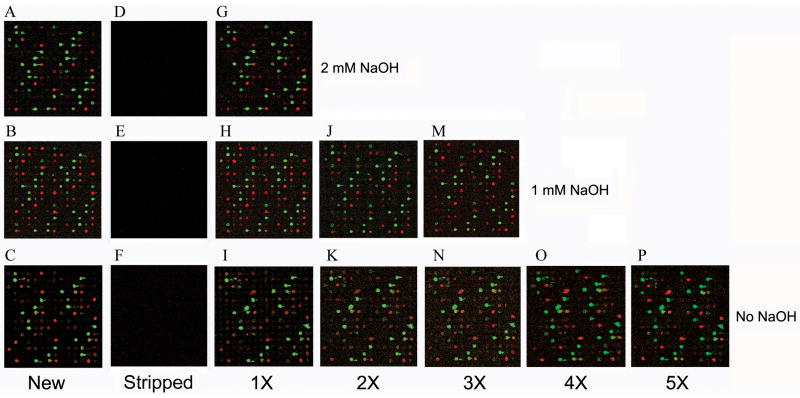
Fig. 1.
Scanned images of new slides and their stripped counterparts. (A–C) Composite images of the same area of three miRNA microarray slides hybridized with a mixture of labeled 293T RNAs and reference DNA oligonucleotides. RNA hybridization generated signals in the green channel, while reference DNA generated signals in the red channel. (D–F) Images of the same slides (A–C) after treated with 2 mM, 1 mM, or 0 mM NaOH in 0.1 x SSC. (G–I) Images of the stripped slides (D–F) after re-hybridized with the same sample of labeled 293T RNAs and reference DNAs as in (A–C). (J, K) Image of twice-stripped slides after re-hybridization. (M, N) Image of trice-stripped slides after re-hybridization. (O) Image of the slide stripped four times after re-hybridization. (P) Image of the slide stripped five times after re-hybridization.
Table 1.
Performance of a microarray slide after stripping in 0.1 x SSC.
| New slide | 1x | 2x | 3x | 4x | 5x | |
|---|---|---|---|---|---|---|
| New slide | 1 | |||||
| 1x | 0.988 (0.988) | 1 | ||||
| 2x | 0.970 (0.964) | 0.985 (0.980) | 1 | |||
| 3x | 0.964 (0.957) | 0.978 (0.973) | 0.993 (0.993) | 1 | ||
| 4x | 0.946 (0.946) | 0.967 (0.967) | 0.977 (0.973) | 0.984 (0.980) | 1 | |
| 5x | 0.927 (0.927) | 0.952 (0.952) | 0.964 (0.958) | 0.973 (0.967) | 0.994 (0.994) | 1 |
Normalized, ch1 RNA signals from hybridizations of the same RNA samples to a new slide, the same slide that was stripped once (1x), twice (2x), three time (3x), four times (4x), and five times (5x) are compared. Numbers given are Pearson correlation coefficients and ICC in parentheses.
Table 2.
Performance of a microarray slide after stripping in 1 mM NaOH, 0.1 x SSC.
| New slide | 1x | 2x | 3x | |
|---|---|---|---|---|
| New slide | 1 | |||
| 1x | 0.980 (0.948) | 1 | ||
| 2x | 0.968 (0.965) | 0.992 (0.975) | 1 | |
| 3x | 0.948 (0.930) | 0.980 (0.978) | 0.975 (0.968) | 1 |
Normalized, ch1 RNA signals from hybridizations of the same RNA samples to a new slide, the same slide that was stripped once (1x), twice (2x), and three time (3x) are compared. Numbers given are Pearson correlation coefficients and ICC in parentheses.
RESULTS
Establishing conditions for stripping custom miRNA microarrays
Our research entails the use of custom microarrays to profile global miRNA expression. The most vexing aspect of the experiments is that array fabrication is inconsistent, as different batches of printed slides have uneven probe deposits and print quality. This problem is not unusual of custom-made microarrays. We reasoned that reusing slides of good quality would reduce cost and waste as well as allow for better comparisons between different experimental samples.
Because our custom microarrays were hybridized with labeled RNA, which is labile under high pH, we first used different concentrations of NaOH to strip GAPSII slides. Newly printed slides were incubated with a mixture of labeled 293T total RNA and reference DNA oligonucleotides, and images acquired, three of which shown in Fig. 1, panels A–C. We next treated the slides with stripping buffers that contained 0–10 mM of NaOH in 0.1 x SSC at 62°C for 20 min, with a buffer change at the 10 min interval. We then washed and dried the slides, and scanned them to examine the effects of different solutions on the previously existing signals. As shown in Fig. 1, 0.1 x SSC, regardless of NaOH, was sufficient to strip the hybridization signals from the slides (compare panels D–F with panels A–C, respectively). Both RNAs (in the green channel, ch1) and reference DNAs (in the red channel, ch2), labeled by different chemical reactions, were effectively purged, indicating that the method removed fluorescently labeled nucleic acids irrespective of the ways labels were incorporated, and that stripping was largely due to the disruption of hydrogen bonds between probes and targets under low ionic strength conditions. Although NaOH was not required in most cases, we did notice that a low concentration of NaOH led to more efficient removal of saturating hybridization signals (data not shown). Such residual signals, however, usually do not pose a problem for slide reuse, because they are always in a tiny minority and easily identifiable. To strip slides that were relatively old, e.g., one to three months after hybridization, 10 mM NaOH in 0.1 x SSC was typically needed (data not shown). As 10 mM NaOH treatment led to more signal degradation (see below), we generally use 0.1 x SSC to strip slides within one week after hybridization.
Performance of the stripped microarrays
To determine if the stripped slides were reusable, we hybridized them with the same samples of labeled 293T RNAs and reference DNAs. Fig. 1, panels G–I show that the stripped slides yielded an almost identical hybridization pattern with similar signal intensities to new slides (panels A–C). To assess microarray reproducibility, we computed the correlation coefficients of results from the new and stripped slides. Tables 1 and 2 list the coefficient values based on normalized microarray data. Results generated using the new slide and results generated using the same slide stripped once by 0.1 x SSC had a Pearson correlation coefficient and an ICC value of approximately 0.99, indicating that the new slide and stripped slide had very similar properties (Table 1). Stripping with 1 mM NaOH, 0.1 x SSC gave almost identical correlation coefficients (Table 2) as in Table 1. Correlation coefficients determined based on the raw data (ch1 and ch2) or only the positive signals were essentially indistinguishable as those presented in Tables 1 and 2 (data not shown).
Hu et al. [25] applied 8 mM NaOH in a more complicated solution to strip microarray but did not recommend stripping arrays more than once, as it compromised slide coating and decreased hybridization intensities in the arrays. We investigated how well our current protocol preserved slides after multiple rounds of stripping. Fig. 1J and 1K show that the twice-stripped slides still produced strong signals after re-hybridization. Results from the twice-stripped slides mirrored those of the once-stripped slides, and their Pearson correlation coefficients are approximately 0.99, with an ICC of 0.98 (Tables 1 and 2). Results from the twice-stripped slides and the new slides also have a high Pearson correlation coefficient (0.97) and ICC (0.97). Indeed, we have routinely re-used 0.1 x SSC-stripped slides up to five times (six hybridizations in total) and still acquired satisfactory data at the end (Fig. 1, panels M–P, and Tables 1 and 2). The correlation coefficients between results of the first hybridization and the last did lower to 0.927 (Table 1), reflecting an eventual drop in slide performance due to wear and tear from repeated stripping and hybridizations.
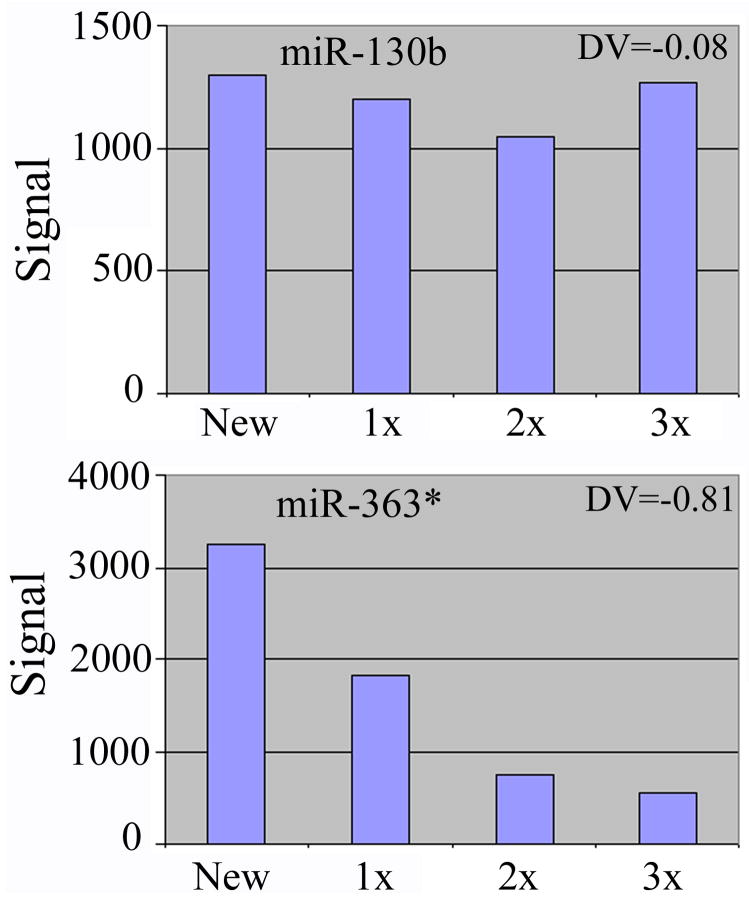
From the experiments described here, approximately 300 human, mouse, and rat miRNA probes exhibited positive signals in ch1 or ch2, or both. Most probes were minimally affected by stripping and continued to produce robust signals upon re-hybridizations, but a small number of probes, e.g., human miR-324-5p and miR-363*, had signals that decreased appreciably faster than the others. Two representative examples are shown in Fig. 2. miR-130b RNA hybridization signals were stable throughout the processes of stripping and hybridization, with a DV of −0.08. In contrast, miR-363* RNA signals were degraded much faster, with a DV of −0.81. Raw signal intensities for probes with diverse DVs were in similar ranges, and the degree of loss in signal strength was consistent in different print batches of slides, indicating that the phenomenon is probe-specific. Stripping with either 1 mM NaOH, 0.1 x SSC or 0.1 x SSC alone affected probes identically, although with 10 mM NaOH a larger number of probes lost signals faster (data not shown). Non-covalently linked, labeled nucleic acids should have been purged to free up the probes for a new hybridization, especially by a high concentration of NaOH. Because 10 mM NaOH impacted the same probes more severely than 0 or 1 mM NaOH, the failure to re-hybridize to certain probes was not due to incomplete stripping. A high concentration of NaOH may more effectively remove probes that are only weakly, e.g., non-covalently attached to the slides (see below).
Fig. 2.
Examples of changes in miRNA hybridization signals in fresh and stripped slides. The raw, ch1 signal intensities (y-axis) for miR-130b and miR-363* are shown. Slides were stripped three times (1x, 2x, 3x) in 0.1 x SSC. DVs were calculated as described in MATERIALS AND METHODS.
Examination of factors that may impact the responses of individual probes to stripping
Why DNA probes were differentially affected by the stripping process is unknown but an interesting issue. There are two plausible explanations. One is that prior hybridization, slide dehydration, and/or stripping makes certain probes on the slide less accessible for subsequent hybridization. Nonetheless, it is difficult at present to envision and test the underlying mechanisms. The other is that certain probes were preferentially washed off a slide during stripping. The attachment of unmodified DNAs to microarrays is mediated by complicated interactions that are poorly characterized. During a typical array printing, a solution containing an oligonucleotide probe is spotted onto the surface of a microarray slide. The probe probably adheres to the slide through hydrogen bonds and through electrostatic attraction between the negatively charged phosphate groups in the DNA and the positively charged glass surface, such as that in a GAPSII slide. Consequently, the longer the probe, the stronger it should bind to the slide. Probes are then immobilized to the slide by UV-crosslinking, which primarily induces covalent interactions involving thymine (T) bases between the DNAs and between the DNAs and glass [28]. As crosslinking efficiency is never 100%, all DNA is not crosslinked to a slide.
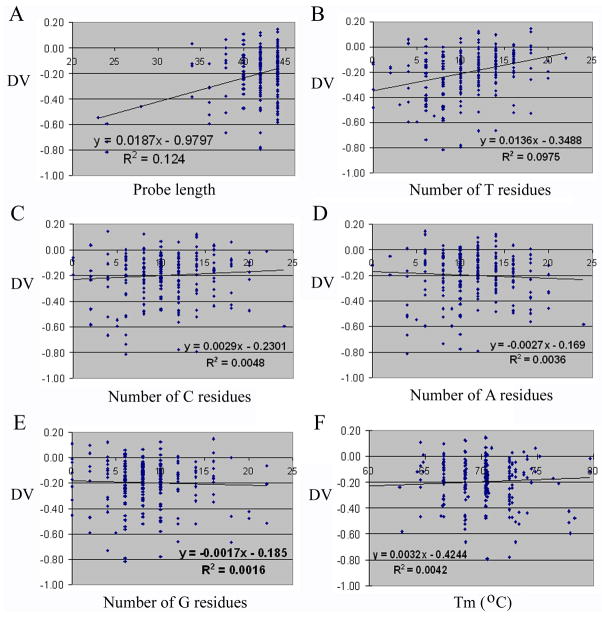
We analyzed our data to determine how the length and base or nucleotide composition of a probe affected its response to stripping (MATERIALS AND METHODS). Probe length played a clearly positive role in the retention of signals after re-hybridization, or, probably, the stability of probe attachment to a microarray, as Fig. 3A shows that the slope of the linear curve plotting DV against probe length is a positive number, 0.0187. The R2 value of 0.124 for the fitted curve is low, which is not unexpected, as many other parameters, e.g., nucleotide, sequence, and local structure information, likely contribute to glass:probe and probe:target interactions, and the relationship between DV and probe length is almost certainly nonlinear. When we examined the contributions of different bases, we found that plotting DV against the number of T residues generated a slope of 0.0136 (Fig. 3B), suggesting that the residue facilitates probe retention on microarray slides. For cytosines (C), the slope is much shallower, 0.0029 (Fig. 3C), while the slopes for adenines (A) and guanines (G) are small, negative values, −0.0027 and −0.0017, respectively (Figs. 3D and 3E). Plotting DV against the percentages of the individual nucleotides in the probes produced similar results (data not shown). Relationship between DV and Tm, calculated according to the GC content and length of a probe, has a slope of 0.032 (Fig. 3F).
Fig. 3.
Scatter plots of DVs versus various attributes of microarray probes. Spots in the graphs represent individual probes. DVs are shown on the y-axis. Trendlines, equations, and R2 values were generated using Excel. (A) Probe length on the x-axis. (B) The number of T residues in each probe on the x-axis. (C) The number of C residues in each probe on the x-axis. (D) The number of A residues in each probe on the x-axis. (E) The number of G residues in each probe on the x-axis. (F) Tm on the x-axis.
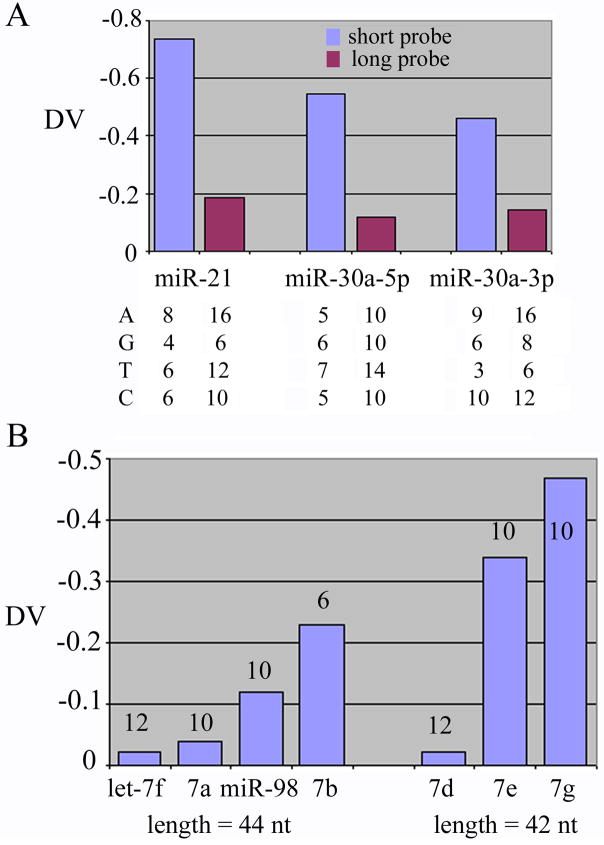
To compare the relative contributions by target sequence and by probe length and nucleotide contents, we evaluated probes that hybridized to identical or homologous targets. Probes in the Invitrogen probe set were designed so that each is a dimer of a sequence complementary to a miRNA. We added three oligonucleotides that were barely longer than the monomeric, complementary sequences as probes for miR-21, miR-30a-5p, and miR-30a-3p, and printed them along with the Invitrogen probe set. In all three cases, the longer, Invitrogen probes had the bigger, or less negative DVs than the shorter probes, even though both had similar nucleotide compositions and all recognized the same target sequences (Fig. 4A). Lastly, we examined probes for the human let-7 family of miRNAs, including let-7a, b, d, e, f, g, and miR-98. Again, probes that are longer and contain more T bases tend to have smaller negative DVs (Fig. 4B). Taken together, our analyses strongly suggested that the length and the T content of a probe positively contributed to the affinity between the probe and the slide surface.
Fig. 4.
Comparisons of probes that recognize identical or homologous target miRNAs. (A) DVs of short and long (Invitrogen) probes for miR-21, miR-30a-5p, and miR-30a-3p are shown. The numbers of A, G, C, and T residues in each probe are listed below the graph. (B) DVs of probes for various human let-7 family members are shown. Probes for let-7f, let-7a, miR-98, and let-7b are 44-nt-long. Probes for let-7d, let-7e, and let-7g are 42-nt-long. The number of T residues in each probe is indicated with the corresponding column.
DISCUSSIONS
Custom microarrays have been extensively used to quantify nucleic acids, e.g., miRNAs, a class of novel, important regulators of gene expression. While reusing microarray slides is a cost-saving measure by itself, we have an extra incentive to reuse well-printed slides because the quality of custom microarray fabrication varies among different print batches. Using the same print batch of slides for either technical or biological replicates can improve the consistency of custom microarray experiments in one-channel and probably in two-channel studies as well. For example, suppose samples A and B are hybridized to slides X and Y, respectively. Differences in the signals on X and Y could be due to intrinsic differences in A and B, or due to differences in the quantity and quality of probes printed on X and Y. If slides X and Y are stripped and then re-probed with the replicates of B and A, respectively, interference from the slide printing variations can be minimized.
We have demonstrated that it is possible to repeatedly strip and reuse custom-made, GAPSII coated microarray slides under a very mild condition, and that the quality of regenerated slides is very close to that of their predecessors. The fact that we use short, ≤ 44 nt oligonucleotide probes to recognize the even shorter, approximately 22 nt miRNAs may have permitted the use of a buffer with low ionic strength (0.1 x SSC) to strip our slides at a relatively low temperature, in contrast to published reports [20–26]. Other custom miRNA microarray platforms also use oligonucleotide probes to capture miRNAs [12–19], so our stripping method is potentially suitable for stripping those slides as well. Furthermore, GAPSII coated slides are widely used to produce custom DNA microarrays; for example, Cook et al. [29] recently printed unmodified oligonucleotide probes on GAPSII slides for yeast genome-wide, molecular barcode analysis. We expect that our stripping method is also applicable to arrays based on GAPSII slides or matrixes with similar chemistries that aim to detect nucleic acids other than miRNAs.
Although most our oligonucleotide probes resisted repeated stripping and produced robust signals after re-hybridization, we did find that certain probes lost signals more quickly. It seems probable that the same probes or probes with similar properties would also be adversely affected by other stripping protocols, as our slide-stripping condition is arguably the mildest among all the known methods. The number and percentage of such probes are small, and reference DNAs or RNAs complementary to these probes can be used to control for the reduction in sensitivity. Nevertheless, one must consider this apparent difference between new slides and regenerated slides when designing microarray experiments and analyzing microarray data. It is prudent to examine the most important or precious samples using fresh microarray slides, whereas to perform replicate hybridizations on stripped slides.
Our studies have also provided experimental evidence that the length and T content of oligonucleotide probes positively influence the retention of probes on microarray slides, which has important implications on the design of microarray probes and on our knowledge of the interactions between DNA and the surface of microarray support. Factors that regulate the attachment of unmodified DNA to microarray slides have not been dissected in any detail [30]. Although one could imagine that a long oligonucleotide would bind to slide surface more tightly than a shorter one, we showed here that probes that are 44 nt long with more than 25% T residues adequately maintained their attachment after stripping, while 20–40 nt long probes tended to lose potential signal intensities faster. Of the four bases, T appears to play a disproportionate and positive role in mediating probe retention. A tentative explanation is that DNA was immobilized by UV irradiation, which principally induced covalent crosslinking involving T residues. One should design microarray probes accordingly if slide reuse is being contemplated. Even if a slide will not be reused, insufficient probe deposit on a microarray can still pose a problem, as it may violate a common premise of microarray experiments that a probe is present in a vast excess over its targets. As the two aforementioned, simple parameters can predict, to a significant extent, the affinity between an oligonucleotide and a GAPSII slide, our results provide critical insights into the interactions between an oligonucleotide and a microarray slide. Other, more complicated factors, e.g., oligonucleotide sequence and local structure information, likely also determine how tightly DNA binds to glass surface, and the experimental system here can be further applied to dissect their potential contributions.
Acknowledgments
The work was supported in part by National Institute of Drug Abuse Center (DA 011806) and United States Army Department of Defense (W81XWH-07-1-0183).
Abbreviations
- A
adenine
- C
cytosine
- ch
channel
- DV
decay value
- G
guanine
- ICC
Intraclass Correlation Coefficients
- miRNA
microRNA
- NaOH
sodium hydroxide
- nt
nucleotides
- SSC
standard saline citrate
- T
thymine
- Tm
melting temperature
- UV
ultraviolet
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 2.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 3.Zeng Y. Principles of microRNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 4.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 11.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, Lobenhofer EK, Sharon E, Shiboleth YM, Shtutman M, Bentwich Z, Einat P. MicroRNA expression detected by oligonucleotide microarrays, system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 15.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dunmitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 18.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goff LA, Yang M, Bowers J, Getts RC, Padgett RW, Hart RP. Rational probe optimization and enhanced detection strategy for microRNAs using microarrays. RNA Biol. 2005;2:93–100. doi: 10.4161/rna.2.3.2059. [DOI] [PubMed] [Google Scholar]
- 20.Strother T, Hamers RJ, Smith LM. Covalent attachment of oligodeoxyribonucleotides to amine-modified Si (001) surfaces. Nucl Acids Res. 2000;28:3535–3541. doi: 10.1093/nar/28.18.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolan PL, Wu Y, Ista LK, Metzenberg RL, Nelson MA, Lopez GP. Robust and efficient synthetic method for forming DNA microarrays. Nucl Acids Res. 2001;29:E107–E117. doi: 10.1093/nar/29.21.e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consolandi C, Castiglioni B, Bordoni R, Busti E, Battaglia C, Berbardi LR, De Bellis G. Two efficient polymeric chemical platforms for oligonucleotide microarray preparation. Nucleosides Nucleotides Nucleic Acids. 2002;21:561–580. doi: 10.1081/NCN-120015069. [DOI] [PubMed] [Google Scholar]
- 23.Bao Z, Wenli W, Rong S, Ling L, Qiuye G, Wenling Z. Re-use of a stripped cDNA microarray. Br J Biomed Sci. 2002;59:118–119. [PubMed] [Google Scholar]
- 24.Benters R, Niemeyer CM, Drutschmann D, Blohm D, Wohrle D. DNA microarrays with PAMAM dendritic linker systems. Nucl Acids Res. 2002;30:E10. doi: 10.1093/nar/30.2.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z, Troester M, Perou CM. High reproducibility using sodium hydroxide-stripped long oligonucleotide DNA microarrays. BioTechniques. 2005;38:121–124. doi: 10.2144/05381MT02. [DOI] [PubMed] [Google Scholar]
- 26.Hahnke K, Jacobsen M, Gruetzkau A, Gruen JR, Koch M, Emoto M, Meyer TF, Walduck A, Kaufmann SHE, Mollenkopf H. Striptease on glass: Validation of an improved stripping procedure for in situ microarrays. Journal of Biotechnology. 2007;128:1–13. doi: 10.1016/j.jbiotec.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Pellis L, Franssen-van Hal NL, Burema J, Keijer J. The intraclass correlation coefficient applied for evaluation of data correction, labeling methods, and rectal biopsy sampling in DNA microarray experiments. Physiol Genomics. 2003;16:99–106. doi: 10.1152/physiolgenomics.00111.2003. [DOI] [PubMed] [Google Scholar]
- 28.Ehrenreich A. DNA microarray technology for the microbiologist: an overview. Appl Microbiol Biotechnol. 2006;73:255–273. doi: 10.1007/s00253-006-0584-2. [DOI] [PubMed] [Google Scholar]
- 29.Cook MA, Chan CK, Jorgensen P, Ketela T, So D, Tyers M, Ho CY. Systematic validation and atomic force microscopy of non-covalent short oligonucleotide barcode microarrays. PLoS ONE. 2008;3:e1546. doi: 10.1371/journal.pone.0001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Call DR, Chandler DP, Brockman F. Fabrication of DNA microarrays using unmodified oligonucleotide probes. Biotechniques. 2001;30:368–376. doi: 10.2144/01302tt06. [DOI] [PubMed] [Google Scholar]