Abstract
Enzymes of the Rnf family are believed to be bacterial redox-driven ion pumps, coupling an oxidoreduction process to the translocation of Na+ across the cell membrane. Here we show for the first time that Rnf is a flavoprotein, with FMN covalently bound to threonine-175 in RnfG and a second flavin bound to threonine-187 in RnfD. Rnf subunits D and G are homologous to subunits B and C of Na+-NQR, respectively. Each of these Na+-NQR subunits includes a conserved S(T)GAT motif, with FMN covalently bound to the final threonine. RnfD and RnfG both contain the same motif, suggesting that they bind flavins in a similar way. In order to investigate this, the genes for RnfD and RnfG from Vibrio cholerae were cloned and expressed individually in that organism. In both cases the produced protein fluoresced under UV illumination on an SDS gel, further indicating the presence of flavin. However, analysis of the mutants RnfG-T175L, RnfD-T278L, and RnfD-T187V showed that RnfG-T175 and RnfD-T187 are the likely flavin ligands. This indicates that, in the case of RnfD, the flavin is bound, not to the SGAT sequence but to the final residues of a TMAT sequence, a novel variant of the flavin binding motif. In the case of RnfG, flavin analysis, followed by MALDI-TOF-TOF mass spectrometry, showed that an FMN is covalently attached to threonine-175, the final threonine of the S(T)GAT sequence. Studies by visible, EPR, and ENDOR spectroscopy showed that, upon partial reduction, the isolated RnfG produces a neutral semiquinone intermediate. The semiquinone species disappeared upon full reduction and was not observed in the denatured protein. A topological analysis combining reporter protein fusion and computer predictions indicated that the flavins in RnfG and RnfD are localized in the periplasmic space. In contrast, in NqrC and NqrB the flavins are located in a cytoplasmic loop. This topological analysis suggests that there may be mechanistic differences between the Rnf and Na+-NQR complexes.
Rnf1 is a little understood family of membrane proteins that are believed to be involved in energetic coupling of redox reactions to Na+ translocation. Rnf was originally described in Rhodobacter capsulatus, where it was found to be essential for nitrogen fixation and hence named Rhodobacter nitrogen fixation protein (1-4). Recently, Rnf has been identified in the genomes of a number of other bacteria and archae, including Vibrio cholerae, Porphyromonas gingivalis, Azotobacter vinelandii, Escherichia coli, Clostridium tetanomorphum, Acetobacter woodii, and Methanosarcina acetivorans (5-10). In nitrogen fixing bacteria, Rnf is believed to transport electrons from NADH to ferredoxin, with this uphill redox reaction driven by inward (downhill) transport of sodium ions across the cell membrane (6). In the anaerobic bacterium, C. tetanomorphum, the same enzyme apparently runs in the reverse direction: electron transfer from ferredoxin to NAD+ drives electrogenic pumping of Na+ out of the cell (8, 11). In some other bacteria, Rnf apparently uses different substrates: in the marine archaeon M. acetivorans, ferredoxin is still the electron donor but the electron acceptor is methanophenazine (10, 12).
Rnf shows significant homology to the Na+-pumping NADH:quinone oxidoreductase (Na+-NQR), a bacterial respiratory enzyme in which electron transfer from NADH to quinone drives the electrogenic transport of Na+ outward across the cell membrane (13-16). However, the role of NAD+/NADH in Rnf is not entirely analogous to that in Na+-NQR; in Na+-NQR, electron donation from NADH is coupled to outward Na+ pumping, while in all known Rnf's electron donation from NADH would correspond to inward flow of Na+. Thus, the Rnf and Na+-NQR superfamily of enzymes exhibits a high degree of functional variability.
The Rnf complex from V. cholerae is an assembly of six subunits, RnfA, RnfB, RnfC, RnfD, RnfG, and RnfE, that include several conserved sequence motifs indicating the binding of redox cofactors: RnfC has an NADH binding motif similar to that of NuoF in NDHI (complex I) and also similar to the binding site found in NqrF of Na+-NQR. RnfB and RnfC each contain the sequence motif for an iron—sulfur center. EPR signals consistent with the presence of [4Fe-4S] centers have been reported in subunits B and C of Rnf from R. capsulatus when expressed individually (2, 3).
In addition to the NADH and iron—sulfur binding sequences, similarities also include flavin binding motifs. The B and C subunits of Na+-NQR each contain an FMN, covalently attached to a threonine residue. In both subunits the threonine is part of a conserved -serine- (or threonine-) glycine-alanine-threonine (S(T)GAT) motif (17-19), and similar sequences are found in both RnfG and RnfD.
RnfG includes a conserved SGAT sequence that coincides with the FMN binding motif of NqrC when the sequences are aligned. In this paper we present the biochemical identification and characterization of the flavin attached to subunit G. We show RnfG binds FMN through a phosphoester bond between the phosphate group of FMN and threonine-175, the final threonine of the conserved S(T)GAT sequence. This is the first direct demonstration that Rnf is a flavoprotein.
RnfD includes two partially conserved sequences similar to the SGAT flavin binding motif of NqrB. At positions 274-278 there is a complete SGAT sequence which is partially conserved in RnfD but not in NqrB. At position 183-187 there is a TMAT sequence which aligns with the S(T)GAT FMN binding motif of NqrB and is at least partially conserved in all known RnfD sequences, with threonine or serine at the ligand binding position. Here, we present spectroscopic and mutational evidence that there is a flavin covalently attached to threonine-187 and that it is unlikely that there is a flavin attached to threonine-278. The TMAT sequence represents a new version of the S(T)GAT flavin binding motif suggesting additional variability in this motif.
MATERIALS AND METHODS
Strains and Growth Conditions
V. cholerae O395N-1, V. cholerae Δnqr, RnfG-pBAD in V. cholerae Δnqr, and RnfD-pBAD in V. cholerae Δnqr were grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/mL) and/or streptomycin (50 μg/mL). E. coli Smart-Cells (Gene Therapy Systems, Inc.) were grown in LB medium supplemented with ampicillin (100 μg/mL). All cells were grown at 37 °C with vigorous agitation (200 rpm).
Cloning of rnfD and rnfG Genes
The rnfD and rnfG genes were amplified by PCR using Platinum PCR SuperMix (Invitrogen). The following primers were used: VcRnfD-forward, GAGGAATAATAAATGGCCTTCTTTATTGCTAGCTTCC; VcRnfD-reverse, GTGTCCGTAAGTTCTCGGTTTGGTGTAGTAG; VcRnfG-forward, GAGGAATAATAAATGCTGACAGCAATTCGAAAAAATGG; VcRnfG-reverse, TTGACCCTCACACGGATTCGGCTG. Genomic DNA from V. cholerae (0395N1) was used as a template for the reactions. The amplification reactions were run in a Minicycler (MJ Research) with the cycle program of 95 °C for 5 min, 45 °C for 30 s, 68 °C for 1 min, for 20 cycles, and then 72 °C for 7 min. The PCR products were purified (MinElute PCR purification kit, Qiagen) and cloned into the pBAD-TOPO vector (Invitrogen) which appends code for a 6×-histidine tag at the C-terminus. The cloning reactions were transformed into Smart Cells high-efficiency chemically competent E. coli cells (Gene Therapy Systems). Cells from the transformation reaction were plated on LB plates containing ampicillin and grown for at least 8 h at 37 °C. Several colonies from each cloning reaction were grown in liquid medium in order to prepare plasmid DNA. DNA was analyzed by restriction digestion and sequencing.
Plasmid DNA from the correct clones, containing the rnfD or rnfG genes, was electroporated into V. cholerae Δnqr competent cells, prepared as reported previously (20). Transformed cells were plated on LB agar containing ampicillin and streptomycin and incubated for 12 h at 37 °C. DNA from transformants was checked by restriction digest analysis and DNA sequencing.
The production of RnfD and RnfG subunits was verified by Western blotting using antibodies against the 6×-histidine tags, located in the C-terminus of the proteins. Cells were grown in two 30 L (working volume) fermenters (BioFlo-5000; New Brunswick Scientific) in LB medium containing ampicillin and streptomycin with constant agitation of 300 rpm at 37 °C. The expression of the recombinant proteins was achieved by the addition of 0.2% (w/v) arabinose to the fermenter during the early log phase of growth. Cells were harvested at the end of the log phase using a continuous flow centrifuge (Cepa Z41; New Brunswick Scientific), and membrane fractions were prepared as reported previously (13).
Mutagenesis of RnfG To Obtain the T175L Mutant and of RnfD To Obtain the T278L and T187V Mutants
All mutagenesis reactions were carried out using the QuikChange II XL site-directed mutagenesis kit (Stratagene) using the appropriate plasmid as a template. Sequences of the (forward) primers were as follows: RnfG-T175L, GGCGGTCAGTTTGACCAATTTACGGGAGCTCTCATTACTCCTCGCGCT; RnfD-T278L, CATGATCCATCTACTCTCGGGAGCTCTCATGCTGGGCGCATTTTTTATCG; RnfD-T187V, CATCGATGGCATTACCATGGCGGTACCGCTCGATGCGTTCAAGAC. The resulting mutants were verified by restriction enzyme digestion analysis and DNA sequencing.
Purification of RnfG
Membranes from the V. cholerae cells expressing the recombinant RnfG protein were washed with 50 mM NaPi buffer (pH 7.5), containing 300 mM NaCl, 5% (w/v) glycerol, and 10 mM imidazole, and then solubilized in the same buffer by the addition of 1% (w/v) n-dodecyl β-maltoside (DM) (Anatrace). Solubilized membranes were mixed with Ni-NTA resin (Qiagen) and incubated at 4 °C for 1 h in the presence of a protease inhibitor cocktail (Sigma). The mixture of solubilized membranes and resin was loaded into a column and washed with buffers with increasing concentration of imidazole, in the presence of 0.1% (w/v) DM (from 10 to 200 mM imidazole). RnfG eluted from the column at 100 mM imidazole. The protein was concentrated using centrifugal concentrators, and the buffer was exchanged to 50 mM Tris-HCl, pH 8, 1 mM EDTA, 5% glycerol, and 0.1% DM (Tris buffer) or 10 mM NH4HCO3, pH 7.6, and 0.1% DM, depending on the experiment. Aliquots were frozen in liquid nitrogen until needed.
The purified protein was run on a precast SDS gel (4-12%) (Invitrogen) and examined under UV light prior to staining with Coomassie. Visible spectra of the purified protein in its air-oxidized form and during anaerobic titrations with dithionite were recorded using a diode array spectrophotometer (Agilent). Protein concentration was measured using the BCA method (Pierce).
Purification of RnfD
Membranes from the V. cholerae cells expressing the recombinant RnfD protein were washed with 50 mM NaPi buffer (pH 8), containing 300 mM NaCl, 5% (w/v) glycerol, and 5 mM imidazole, and then solubilized in the same buffer by the addition of 1% (w/v) SDS. Solubilized membranes were mixed with Ni-NTA resin (Qiagen) and incubated at room temperature for 1 h in the presence of a protease inhibitor cocktail (Sigma). The mixture of solubilized membranes and resin was loaded into a column and washed with buffers with 10 and 20 mM imidazole, in the presence of 0.1% DM (w/v). RnfD eluted from the column at 40 mM imidazole. The protein was concentrated using centrifugal concentrators, and the buffer was exchanged to 50 mM Tris-HCl, pH 8, 1 mM EDTA, 5% glycerol, and 0.1% DM. The concentrated protein was further purified by gel filtration chromatography (HiLoad 16/60 Superdex 200 column). Aliquots of the purified protein were frozen in liquid nitrogen until used.
Redox Titration of RnfG for Visible, EPR, and ENDOR Spectroscopies
RnfG was titrated in anaerobic conditions, following the visible spectra. The sample and a 2 mM dithionite solution in Tris buffer were each maintained under a flowing atmosphere of water-saturated argon. The enzyme sample was stirred continuously with a magnetic stirrer. Small aliquots of the dithionite solution were added to the enzyme sample using “gastight” syringes (Hamilton). A similar titration was performed directly in EPR tubes. The EPR were placed in the sample holder of the spectrophotometer using a block the size of a standard cuvette that masked the EPR tube, blocking all light not passing through the contents of the tube. In this way, the optical signal of the flavin radical could be observed before freezing the tubes for EPR analysis. EPR samples were frozen using a bath of n-pentane over dry ice. The titration for EPR was performed at the following pH values: 6.3, 8.0, 8.6, and 9.3. EPR Spectra were recorded at X-band (ν = 9.05 GHz) in a Varian E-122 spectrometer. The data were acquired from frozen samples at 70 K using an Air Product Helitran cryostat with liquid helium. The magnetic field was calibrated with a Varian NMR Gauss meter, and the microwave frequency was measured with an EIP frequency meter.
Pulsed ENDOR spectra were collected on a Bruker E580-10 Elexsys spectrometer. Samples were maintained at 85 K using liquid helium in an Oxford CF935 cryostat controlled by an Oxford ITC-4 temperature controller. Pulsed ENDOR experiments were carried out using a Davies three-pulse scheme (π-T-π/2-τ-π-τ-echo, with the RF pulse applied during time T). The magnetic fields were calibrated with a Varian NMR Gauss meter. All instruments were maintained by the Illinois EPR Research Center (IERC), University of Illinois at Urbana-Champaign. EPR spectra were simulated with the SIMPOW6. ENDOR spectra were simulated using SIMEND as described earlier (21), except that a term accounting directly for the hyperfine selection was added (22).
Mass Spectrometry
MALDI mass spectra were recorder using an Ultraflex III MALDI-TOF-TOF mass spectrometer (Bruker Daltonics). Ionization was performed by an Nd:YAG “smart” laser beam. Sinapinic acid was used as the matrix for analyses of intact proteins, and α-cyano-4-hydroxycinnamic acid was used for analyses of digests. Calibration of the mass range in linear mode was performed using Protein Standard Mixture I (Bruker Daltonics) and with lysozyme as an internal standard. Peptide Standard Mixture (Bruker Daltonics) was employed for calibration in reflector and in MS/MS (PSD) experiments. Accurate mass measurements were performed using the same mixture of peptides as an internal standard. The mass accuracy was better than 3 ppm.
Sample Preparation for Tandem Mass Spectrometry
RnfG was digested with trypsin using trypsin-spin columns (Sigma) or/and DigestTip trypsin (ProteoGen Bio) following the protocol of the manufacturer. The digested protein was purified and fractionated by HPLC (Shimadzu) using an Altima C18 5× column, length 250 mm, OD 4.6 mm, with the following protocol. Solvents: A, 0.2% formic acid in water; B, 0.2% formic acid in acetonitrile. Gradients: T = 0 (min), 2% B; T = 150-155, 80% B; flow rate, 0.5 mL/min. The detector was set at 280 and 450 nm. Fractions containing flavins were collected for mass spectrometry analysis.
Topology Mapping of RnfG and RnfD
The following computer-based topology prediction algorithms were used: HMMTOP (23), TMPRED (http://www.ch.embnet.org/software/TMPRED_form.html), TMHMM (24), MEMSAT3 (25), TOPORED II (26), and ConPred II (27, 28).
For experimental verification an alkaline phosphatase reporter gene fusion method was used; alkaline phosphatase activity was measured as reported before (29, 34).
The pBAD-phoA-rnfG, full-length C-terminal fusion was constructed using the protocol reported previously (29) with the following primers: ARGNC-for, GGTACCGCTGACAGCAATTCGAAAAAATG; ARGNC-rev, GGTACCTTTTGACCCTCACACGGATTC.
For RnfD, several fusion constructs were created using the following primers (Table 5 and Figure 6B): ARD-for, GGTACCGGCCTTCTTTATTGCTAGCTC; ARDN92-rev, GGTACCAAGAGGGGCGGGATAG; ARDN266-rev, GGTACCAAGGTTGTTCCCGGTGC; ARDNC-rev, GGTACCTTGTGTCCGTAAGTTCTCGG.
Table 5.
Alkaline Phosphatase (PhoA) Activities of Constructed Fusions of RnfG and RnfD
| subunit | fusion | PhoA activity (units)a |
|---|---|---|
| RnfG | ARGC | 525 |
| RnfD | ARDN92 | 559 |
| ARDN266 | 746 | |
| ARDNC | no activity |
Activity is measured as the pNPP hydrolyzed per minute per optical density at 600 nm of cells.
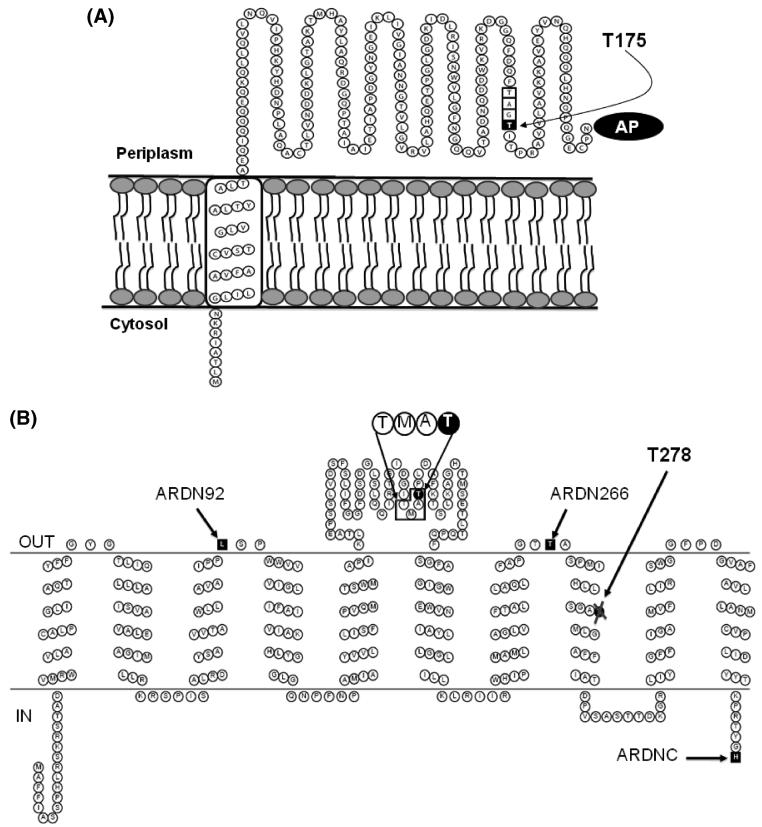
Figure 6.
(A) Schematic showing membrane topology of RnfG. The figure highlights the FMN binding motif, TGAT, where last threonine of the motif, Thr-175, is the ligand of the flavin. (B) Schematic showing the most probable topology of RnfD. The figure highlights the new motif for binding of flavin, TMAT, where Thr-187 is the ligand. The alkaline phosphatase fusion sites are labeled as follows: ARDN92, alkaline phosphatase fused to the C-terminus of a subset of RnfD from the N-terminus to residue 92; ARDN266, alkaline phosphatase fused to the C-terminus of a subset of RnfD from the N-terminus to residue 266; ARDNC, alkaline phosphatase fused to the C-terminus of the complete RnfD. Thr-278 is part of the conserved SGAT motif labeled in the figure.
RESULTS
It has been previously noted that the Rnf complexes from R. capsulatus and other photosynthetic bacteria share homology with Na+-NQR, suggesting that these two groups of proteins may make up a family of enzymes that carry out electron transfer reactions coupled to the translocation of sodium ions in a variety of bacteria (3, 6).
RnfG shows strong homology to subunit C of Na+-NQR, with 15.7% protein sequence identity and 39.5% similarity (Figure 1A), while RnfD shows homology to NqrB, with 12% identity and 35.6% similarity (Figure 1B). Significantly, RnfG and RnfD each contain a flavin binding motif also found in NqrC and NqrB. In both Na+-NQR subunits this conserved motif, S(T)GAT, is a binding site for FMN in which the final threonine forms a phosphoester bond to the flavin (17-19) (Figure 1A, B). This suggested that RnfG and RnfD might also be a flavoprotein.
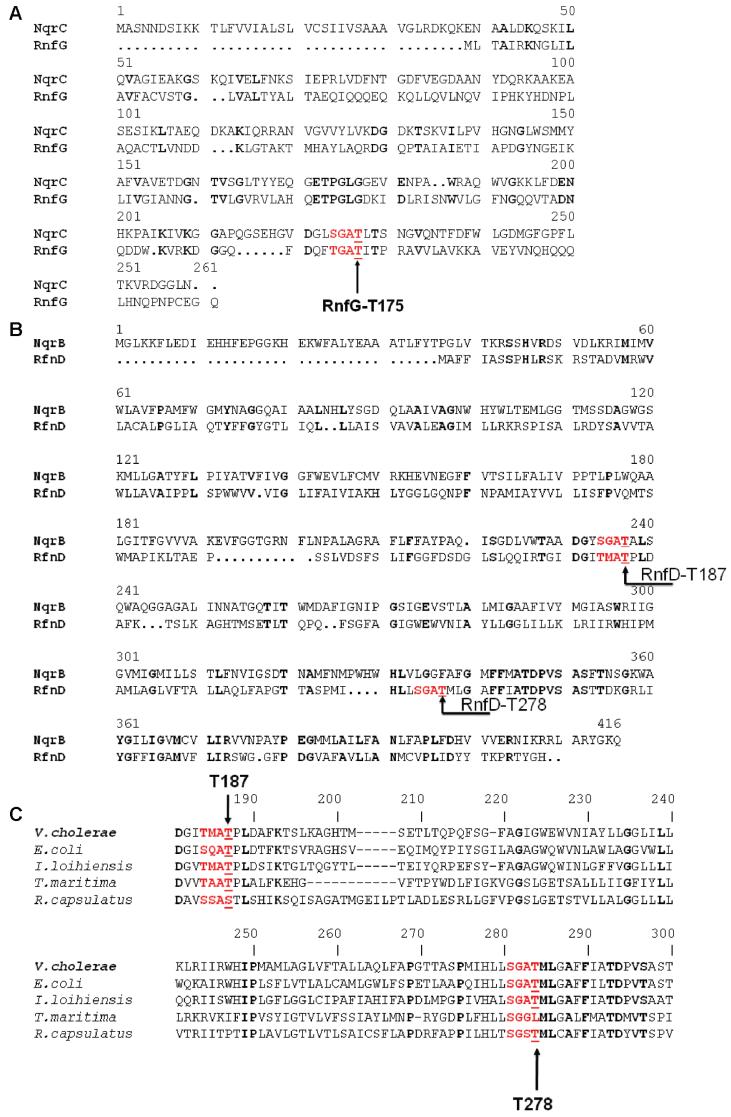
Figure 1.
(A) Alignment between RnfG and NqrC from V. cholerae showing the conserved FMN binding motif, S(T)GAT. (B) Alignment between RnfD and NqrB from V. cholerae showing the two possible flavin binding sequences: the SGAT sequence containing Thr-278 and the TMAT sequence containing Thr-187. (C) Alignment between RnfD's from different sources showing that the two possible flavin binding sequences are both conserved.
In our current work, isolating and characterizing the Rnf complex from V. cholerae, we found that when a semipurified preparation is run on an SDS-PAGE gel, two fluorescent bands are observed, corresponding to the molecular weights of subunits D and G (data not shown), again suggesting the presence of flavin in these two subunits. Given these two independent lines of evidence, we decided to express RnfG and RnfD individually, in order to look for covalently bound flavins and, if present, to determine the type of flavin, as well as the precise location and chemical character of their attachment to the protein.
Production and Purification of RnfG and RnfD
RnfG and RnfD subunits were produced individually using a strain of V. cholerae from which the genes for Na+-NQR had been deleted. Gene expression was controlled by the arabinose promoter system, p-BAD, which has been shown to work well in V. cholerae cells (13, 18). A 6×-histidine tag was genetically engineered into the carboxyl terminal of each subunit. Production of each protein was confirmed by Western blot analysis using anti-His tag antibodies. The predicted molecular masses for RnfG and RnfD are around 26 and 38 kDa, respectively, and the Western blot showed bands at the predicted molecular weights, when extracts of cells producing the proteins were run on SDS gels and then transferred to PVDF membranes.
RnfG was purified by affinity chromatography using Ni-NTA resin and gel filtration, and the purified protein was analyzed by SDS-PAGE. When the gel was examined under UV illumination, prior to staining, a fluorescent band was observed (Figure 2B). The corresponding band, which appeared after Coomassie staining, had an apparent molecular mass of 26 kDa, the same as predicted for RnfG (Figure 2A). The band, when excised from an unstained gel, gave a fluorescence spectrum typical of flavins (not shown). The fact that this fluorescence is still present after electrophoretic separation in denaturing conditions shows that the fluorescent cofactor is covalently attached to the protein, while the character of the fluorescence suggests that this cofactor is a flavin and that it becomes incorporated in the individually expressed subunit. A visible spectrum of the purified RnfG in the oxidized (and nondenatured) form showed features typical of flavins: a peak at 390 nm and a peak at 450 nm with a shoulder at about 479 nm (Figure 3A). Based on the absorbance, the ratio of protein to flavin was between 0.8 and 0.9.
Figure 2.
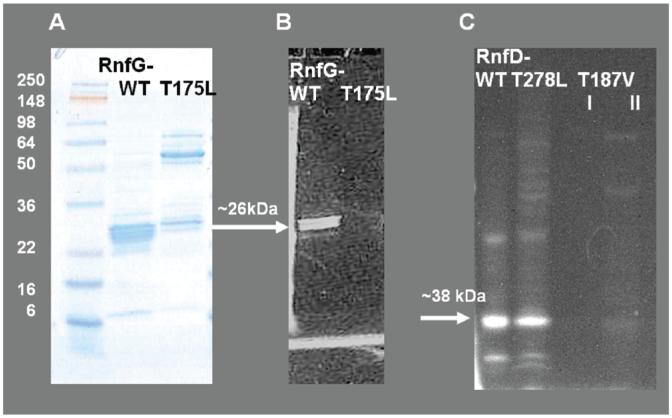
SDS-PAGE comparing the wild-type RnfG to the RnfGT175L mutant and wild-type RnfD to the RnfD-T278L mutant. Panel A: RfnG and RnfG-T175L, gel stained with Coomassie. Panel B: The same gel under UV illumination prior to staining, showing flavin fluorescence. Panel C: RfnD, RnfD-T278L, and RnfD-T187V (I, after Ni-NTA; II, after gel filtration), gel under UV illumination prior to staining, showing flavin fluorescence. Approximately 20 μg of protein was loaded in each lane.
Figure 3.
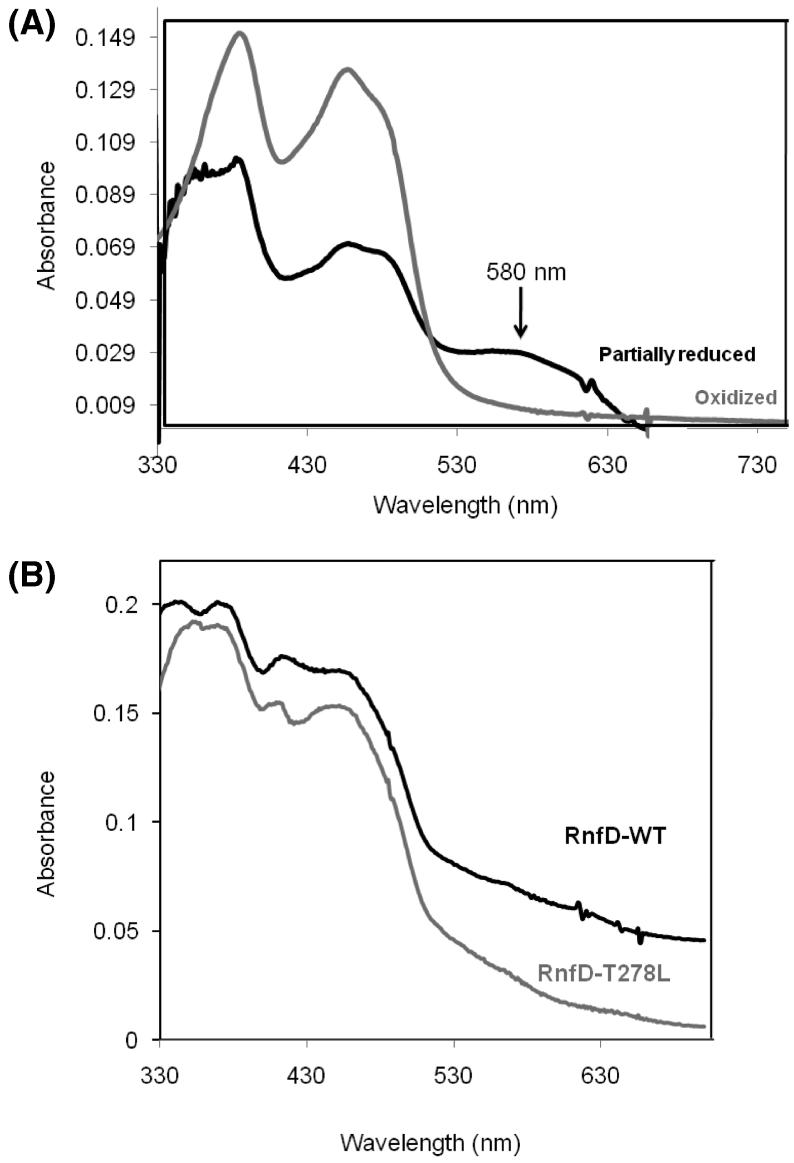
(A) Visible spectra of oxidized and partially reduced RnfG. Approximately 30 μg of protein was used. Gray spectrum: oxidized (as isolated) RnfG. Black spectrum: partially reduced RnfG. The partially reduced spectrum was acquired under anaerobic conditions using dithionite as reductant. (B) Visible spectrum of oxidized RnfD wild type (black line) and the RnfD-T278L mutant (gray line). Approximately 30 μg of protein was used.
Similarly, RnfD was purified by affinity chromatography, but the yield and purity are not as good as for RnfG. However, when run on an SDS gel and examined under UV illumination, a fluorescent band around 38 kDa was observed (Figure 2C). This band, when excised from the gel, also gave a spectrum typical of flavins (not shown). As in the case of RnfG, a visible spectrum of the purified RnfD indicated the presence of flavin (Figure 3B).
RnfG-T175L, RnfD-278L, and RnfD-T187V Mutants. RnfG-T175L
In order to determine if the binding motif Thr-172, Gly-173, Ala-174, Thr-175, in RnfG, is involved in the binding of the flavin, and specifically if threonine-175 is the site of covalent attachment, a site-directed mutant was prepared. Threonine-175 was mutated to leucine, a residue that is not able to form a phosphoester bond with the phosphate of the flavin. The production of the RnfG-T175L protein was confirmed by Western blotting with both whole cells and cell membranes, and the protein was then purified by affinity chromatography, as described above. The wild-type and mutant proteins were run in adjacent lanes on an SDS gel. Figure 2A shows the gel after Coomassie staining, while the same gel, under UV illumination prior to staining, is shown in Figure 2B. The fluorescent band, corresponding to RnfG, is clearly seen in the wild-type protein, whereas no fluorescent band can be seen in the RnfG-T175L mutant. It should be noted that RnfG-T175L appeared to form high molecular mass aggregates, which appear after staining. These high molecular mass bands react with the anti-His tag antibodies, confirming that they contain the correct protein, but they are not fluorescent, indicating that they do not contain flavin. This suggests that the absence of the flavin cofactor may lead to a protein conformation that is susceptible to aggregation. The fact that the mutation abolishes flavin binding indicates that threonine-175 is the ligand for the covalently bound flavin, confirming the similarity between Rnf and Na+-NQR. However, the type flavin involved, FMN or FAD, remained to be shown.
RnfD-278L and RnfD-T187V
As described above, RnfD includes two sequences that are candidates for covalent FMN binding sites. We initially focused on the Ser-275, Gly-276, Ala-277, Thr-278 sequence because it replicated the known S(T)GAT binding motif exactly and made a mutant in which Thr-278 was replaced by leucine. The production of the RnfD-278 L protein was confirmed by Western blotting with cell membranes. The protein was purified using the same protocol as for the wild-type RnfD. However, the mutant protein still had a flavin-like visible absorbance spectrum (Figure 3B) and fluoresced when an SDS gel was examined under UV illumination (Figure 2C), indicating that a covalently attached flavin was still present.
We therefore, turned our attention to the second sequence. When RnfD is aligned with NqrB (Figure 1B), the SGAT FMN binding site in NqrB aligns to Thr-184, Met-185, Ala-186, Thr-187 in RnfD. This TMAT sequence is partially conserved in all RnfDs (Figure 1C). To study this site, we mutated Thr-187 to valine. Whereas wild-type RnfD and the RnfD-T278L mutant both showed strong fluorescence on an SDS gel, no such clear band is observed in the RnfD-T187V mutant (Figure 3B). Similarly, this mutant shows no clear flavin-like features in the visible absorbance or fluorescence spectra. The results from these two mutants suggest that Thr-187 is attached covalently to a flavin, whereas Thr-278 is probably not a flavin binding site.
Mass Spectrometry Analysis of RnfG
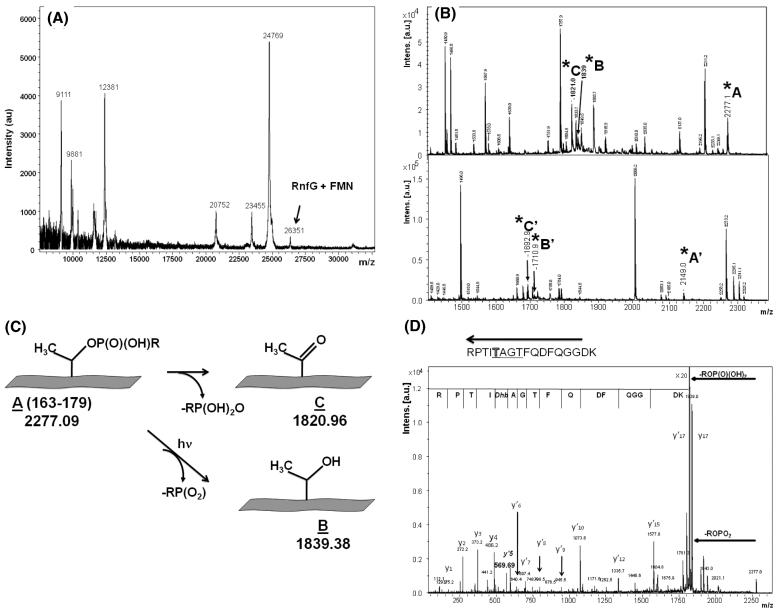
To further investigate the nature of the flavin and the character of its binding, the isolated RfnG subunit was studied by MALDI-TOF mass spectrometry. The resulting spectrum (Figure 4A) includes a peak at m/z = 26351, consistent with the expected mass of the RnfG protein plus one FMN (26350) (Table 1). The appearance of this spectrum, including the other peaks, was consistent for several preparations. In each case, the m/z = 26351 peak was relatively small, in spite of the fact that biochemical analysis of the samples consistently gave a protein-to-flavin ratio close to 1, indicating that the polypep-tide undergoes partial chemical degradation as part of the mass spectrometry sample preparation.
Figure 4.
(A) MALDI mass spectrum of RnfG. The peak corresponding to RnfG with the covalently attached FMN is shown. (B) MALDI mass spectra of two fluorescent FMN-containing peptide fractions in m/z range 1200-2400. Peaks corresponding to modified threonine are marked as A, A′, B, B′, C, and C′. (C) Scheme illustrating the two mechanisms of phosphoester bond breaking leading to the results shown in (B). (D) MS/MS (postsource decay) mass spectra of the MS peak at m/z 2277, showing the protein sequence in the vicinity of T175 including the TGAT flavin binding motif. The dephosphorylated state of Thr-175 is evident.
Table 1.
Predicted and Experimental Masses of RnfG and Tryptic Peptides
| species | theoretical mass | experimental mass |
|---|---|---|
| RnfG + FMN | 26350 | 26351 |
| tryptic peptide A' + FMN | 2276 | 2277.09 |
| B': tryptic peptide A - FMN | 1838 | 1839.38 |
| C': tryptic peptide A - FMN - water | 1819 | 1820.96 |
| tryptic peptide A′ + FMN | 2148 | 2148.90 |
| B′: tryptic peptide A - FMN | 1709 | 1710.81 |
| C′: tryptic peptide A - FMN - water | 1691 | 1692.9 |
To analyze the flavin binding in more detail, the RnfG was digested with trypsin to obtain fragments small enough for tandem mass spectrometry. The tryptic fragments were separated by HPLC, and fractions containing flavin were detected by absorbance at 450 nm. Two fractions with significant absorbance at 450 nm were collected, and their mass spectra are shown in the two panels of Figure 4B. In each case it is possible to identify a fragment corresponding to a peptide with a covalently bound flavin and two additional fragments corresponding to the same peptide, having lost the flavin by two different chemical mechanisms, as described in ref 30 and illustrated in Figure 4C. In the upper panel of Figure 4B, fragment A (2277.09 Da) corresponds to a predicted tryptic peptide, which includes the putative flavin binding site (TGAT) from RnfG, plus the mass of FMN. Fragment C (1820.96) corresponds to the same tryptic peptide, after the loss of the FMN and a water molecule, while fragment B (1839.38 Da) corresponds to the fragment after the loss of FMN without dehydration (Figure 4C). The lower panel of Figure 4B shows a similar pattern of peaks for a slightly smaller peptide (Table 1). Accurate mass measurements were performed for fragments A′, B′, and C′, shown in Figure 4C (m/z measured 2148.9022, 1710.8105, and 1692.7988, and m/z calculated 2148.9008, 1710.8082, and 1692.7976, respectively). These results show that the covalently bound flavin in RnfG is FMN and that it is attached by a phosphoester bond to a threonine: Thr-172, Thr-175, or Thr-177.
In order to determine which of the three threonines is the actual site of FMN attachment, PSD (postsource decay) mass spectra of the fragments at m/z 2277 (Figure 4D) and m/z 2149 were recorded. In both cases, very strong signals corresponding to the loss of the FMN were observed, with formation of dehydrated threonine (Dhb). By analysis of y-ions it was possible to confirm the identity, and position in the amino acid sequence, of the group, which binds FMN. A well-defined breakup of the peptide chain clearly indicated that the modified threonine residue is the fifth amino acid from the C-terminus in these products (Figure 4D and Table 2). This corresponds to threonine-175 in the RnfG protein. These results were supported by the analysis of the fragmentation of the corresponding dephosphorylated peptides (not shown).
Table 2.
Theoretical and Experimental Masses of Postsource Decay Peaksa
| peak | theoretical mass | experimental mass | sequence |
|---|---|---|---|
| y1 | 175.1190 | 175.2 | R |
| y2 | 272.1717 | 272.1 | RP |
| y3 | 373.2194I | 373.1 | RPT |
| y4 | 486.3035 | 486.2 | RPTI |
| y′5 | 569.3406 | 569.2 | RPTIDhb |
| y′6 | 640.3777 | 640.4 | RPTIDhbA |
| y′7 | 697.991 | 697.4 | RPTIDhbAG |
| y′8 | 798.4468 | 798.5 | RPTIDhbAGT |
| y′9 | 945.5152 | 945.5 | RPTIDhbAGTF |
| y′10 | 1073.5738 | 1073.6 | RPTIDhbAGTFQ |
| y′12 | 1335.6692 | 1335.7 | RPTIDhbAGTFQDF |
| y′15 | 1577.7707 | 1577.8 | RPTIDhbAGTFQDFQGG |
Dhb: dephosphorylated and dehydrated threonine.
Redox Properties of the Bound Flavin Using Visible and EPR Spectroscopy
Na+-NQR is known to exhibit spectra from neutral and anionic flavin radicals under different redox conditions (31, 32). We were interested in studying whether the FMN in RnfG is able to form a similar one-electron-reduced intermediate. FMN, free in solution, would not be expected to form a stable semiquinone species, but if the protein environment stabilizes the one-electron-reduced form, a semiquinone species may be observed during a reductive titration. In order to observe the possible formation of a flavosemiquinone species, a reductive titration of the purified RnfG, by dithionite, was carried out, monitored by UV-vis absorbance. As shown in Figure 3, the formation of a neutral semiquinone flavin is observed as an absorbance increase around 600 nm. This was only observed in the native protein; when RnfG was denatured by the addition of 6 M guanidine, the titration showed only the oxidized and reduced forms of the flavin. Similarly, when the titration was carried out using FMN, no one-electron-reduced intermediates were detected. These results indicate that the RnfG protein folds around the covalently bound FMN in a way that stabilizes the one-electron-reduced state of the flavin.
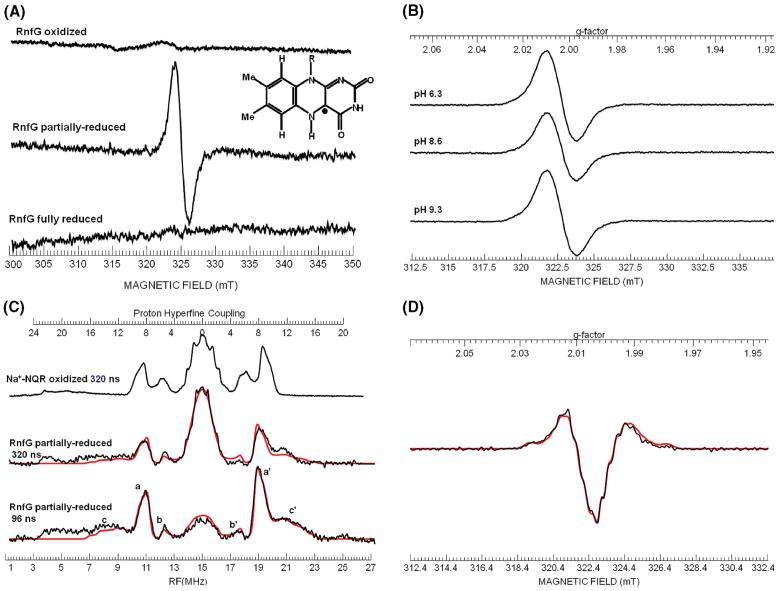
In order to confirm the presence of the semiquinone form of the flavin, and determine its character, we repeated the titration in an EPR tube, first observing the changes by visible spectroscopy. As soon as a significant population of the semiquinone form was observed, the sample was rapidly frozen in a bath of n-pentane and dry ice. The freeze-trapped sample was subsequently analyzed by EPR spectrometry. Figure 5A shows the EPR spectra of this partially reduced sample, as well as oxidized, fully reduced controls. The partially reduced spectrum shows a flavin semiquinone radical with a line width of 20 G (2 mT). This is consistent with the neutral semiquinone flavin observed in Na+-NQR which has a line width of 20 G (2 mT), whereas the anionic semiquinone has a line width of 15 G (1.5 mT). However, some of the RnfG sample preparations gave EPR signals with line widths of 17-18 G (1.7-1.8 mT). The second derivative CW X-band spectra and the ENDOR spectra of the latter samples exhibited poor resolution of hyperfine structure. This would be consistent with the presence of a small population of an anionic radical, in the presence of a large population of a neutral flavosemiquinone radical. In order to check whether the presence of this putative anionic radical is due to a protonation equilibrium, the redox titration of RnfG was repeated at a range of pH values (6.4, 8.3, and 9.3, in addition to the pH 7.5 experiment above). As shown in Figure 5B there were no significant changes in the EPR spectra, and all spectra had line widths of 20 G. Thus, if the flavin semiquinone in RnfG can form a mixture of neutral and anionic forms, this is not due to a protonation equilibrium with the solvent.
Figure 5.
(A) X-band EPR spectra of oxidized, partially reduced, and fully reduced RnfG. Inset: Structure of the neutral flavin radical showing the location of the unpaired electron spin on the isoalloxazine ring. (B) X-band EPR spectra of partially reduced RnfG samples at different pH values. (C) ENDOR spectra of the partially reduced RnfG at π pulse lengths of 320 and 96 ns. The spectrum of the neutral flavin semiquinone radical of Na+-NQR for a π pulse length of 320 ns is shown for comparison. The peaks marked a and a′ are due to the C(8α) methyl protons; peaks marked b and b′ are due to the C(6) proton, and peaks marked c and c′ are due to the C(1′) β-methylene protons. The simulation of the partially reduced RnfG is presented in red overlaying the actual spectrum. (D) Second derivative X-band EPR spectrum of partially reduced RnfG. Simulated spectrum is shown in red.
ENDOR Analysis
In order to confirm the nature of the flavosemiquinone species present in the partially reduced RnfG, we analyzed the radical signal by ENDOR spectroscopy where the features of a neutral flavin radical can be clearly discerned (31, 33). Figure 5C shows the ENDOR spectrum of RnfG for a typical preparation that had a CW line width of 20 G (2 mT), together with the neutral flavin radical spectrum from Na+-NQR obtained with the same value of 320 ns for the π pulse in the Davies pulse sequence. The ENDOR spectrum clearly shows peaks at 11 and 19 MHz which are readily assignable to the C(8α) methyl protons. The average splitting of 8.2 MHz is typical for a neutral radical (31). Also seen are a set of peaks at 12 and 18 MHz which can be assigned to the C(6) ring proton. The splittings for the C(8α) methyl protons and the C(6) proton are similar to those observed for the neutral radical in Na+-NQR, but the splittings are slightly smaller (~7%). The “matrix” peak is narrower in the RnfG spectrum than the Na+-NQR spectrum, suggesting that the splittings for C(7α) methyl protons and the C(9) ring proton may also be smaller. This implies that the spin density on the aromatic ring is smaller for the RnfG neutral radical than the Na+-NQR neutral radical.
Also observed in the RnfG ENDOR spectrum is a broad peak at 21 MHz corresponding to a proton splitting of 12-14 MHz. The corresponding low frequency peak expected at 8.5 MHz is obscured by noise/baseline. However, by recording spectra with shorter π pulses, one can improve the signal-to-noise ratio, and for a π pulse of 96 ns one can observe these sets of peaks better. We believe these broad peaks are due to two slightly inequivalent C(1′) protons. The second derivative spectrum of a RnfG sample with a 20 G line width showed the presence of weak hyperfine structure with a spacing of ~13 MHz. Simulation of both ENDOR and second derivative CW spectra confirmed the presence of two protons with average hyperfine couplings in the range of 11-13 MHz (Table 6). For the neutral radical in Na+-NQR both C(1′) proton couplings are much smaller, and their peaks lie underneath the C(6) and C(8α) methyl proton peaks (21). The peaks for the C(6) protons of RnfG appear weaker than those in Na+-NQR, and this can be explained because the underlying C(1′) proton peaks in Na+-NQR are now located outside the C(8α) methyl protons peaks in RnfG. In the case of DNA photolyase (33), one C(1′) proton splitting is small (2.7 MHz) while the other splitting is large (9.7 MHz). Our ENDOR results imply that the orientation of the ribityl side chain relative to the isoalloxazine ring is considerably different than that of either Na+-NQR or DNA photolyase. The values of the hyperfine coupling for the two β-methylene C(1′) protons are determined by the dihedral angle between the pπ orbital on the N(10) and the Cβ -Hβ bond (21). The large hyperfine values for both C(1′) protons are consistent with dihedral angles of ~30° and ~150° for the C(1′) protons and a dihedral angle of ~270° for the ribityl side chain.
Table 6.
Spin Hamiltonian Parameters for the Neutral Semiquinone Observed in RnfG Obtained by Simulation of ENDOR and Second Derivative CW Spectraa,b
| Ax | Ay | Az | Aiso | |
|---|---|---|---|---|
| 14N(5)c | 2 | 2 | 50.0 | 18.0 |
| 14N(10)c | 1 | 1 | 28.8 | 10.3 |
| N(5)Hc | 5 | 42.4 | 27.7 | 25.0 |
| C(8α)Hd | 9.35 | 7.62 | 7.62 | 8.20 |
| C(6)Hd | -2.5 | -5.8 | -5.4 | -4.6 |
| (C(1′)H1)c,d | 9.4 | 16.3 | 12.0 | 12.6 |
| (C(1′)H2)c,d | 8.3 | 14.9 | 11.2 | 11.5 |
Isotropic and principal hyperfine values in MHz.
Principal values of the electronic Zeeman: 2.00390, 2.00312, 2.00227.
Values determined using second derivative CW spectra.
Values determined using ENDOR.
Topological Studies of RnfG and RnfD by Protein Fusion Analysis. RnfG
In order to localize the FMN cofactor of RnfG with respect to the membrane, we analyzed the topology of the protein in the membrane, first using prediction algorithms (Table 3), and then experimentally, by genetically fusing rnfG to alkaline phosphatase. All of the prediction methods used found one main transmembrane helix, from amino acids 8-10 to amino acids 25-31. The predictions also agree that the N-terminus of RnfG is located in the cytoplasm and therefore the C-terminus is located in the periplasm (Figure 6A). These models locate T125, where FMN binds, in the periplasmic space. To corroborate these predictions, we made a C-terminal fusion of the complete rnfG gene to alkaline phosphatase. The expression of the chimeric fusion protein was confirmed by Western blotting, using anti-alkaline phosphatase antibodies (not shown). The alkaline phosphatase activity of the fusion protein was 525 units, a high value compared to controls (29, 34). This result confirms the predicted topology, suggesting that RnfG has one transmembrane helix, and locating the flavin binding site in the periplasmic space. This result is in contrast with the topological map obtained for the homologous protein in Na+-NQR, subunit C, where the flavin cofactor is located in the cytoplasmic side of the membrane.
Table 3.
Membrane Topology Predictions for RnfG
| method | N-terminal/C-terminal locations | no. of TMHs | TMH segments |
|---|---|---|---|
| HMMTOP | in/out | 1 | 9-31 |
| TMPRED | in/in | 2 | 9-25, 105-124a |
| TMHMM | in/out | 1 | 9-31 |
| MEMSAT | in/out | 1 | 10-27 |
| TOPPRED | in/out | 1 | 9-29 |
| ConPred II | in/out | 1 | 9-29 |
Low probability.
RnfD
We also analyzed the topology of RnfD by prediction algorithms (Table 4) and constructed three reporter group fusions using alkaline phosphatase, as shown in Figure 6B and Table 5. The experimental results locate the flavin binding site, threonine-187, in the periplasmic space. In contrast, the corresponding flavin binding site in NqrB is located on the cytoplasmic side of the membrane. Interestingly, the complete SGAT motif, containing Thr-278, which apparently does not bind flavin, is located within the membrane in helix VIII (Figure 6B).
Table 4.
Membrane Topology Predictions for RnfD
| method | N-terminal/C-terminal locations | no. of TMHs | TMH segments |
|---|---|---|---|
| HMMTOP | in/in | 10 | 25-42 |
| 47-64 | |||
| 77-94 | |||
| 99-116 | |||
| 125-144 | |||
| 214-233 | |||
| 246-265 | |||
| 270-287 | |||
| 300-317 | |||
| 322-339 | |||
| TMPRED | in/out | 7 | 43-64 |
| 88-109 | |||
| 120-143 | |||
| 216-236 | |||
| 243-262 | |||
| 269-285 | |||
| 304-337 | |||
| TMHMM | in/in | 10 | 20-39 |
| 43-65 | |||
| 72-91 | |||
| 95-117 | |||
| 124-146 | |||
| 212-234 | |||
| 241-263 | |||
| 268-287 | |||
| 300-317 | |||
| 322-341 | |||
| MEMSAT | in/in | 8 | 23-29 |
| 47-64 | |||
| 84-108 | |||
| 211-234 | |||
| 243-262 | |||
| 270-286 | |||
| 300-316 | |||
| 323-340 | |||
| TOPPRED | 7 | 37-57 | |
| 89-109 | |||
| 124-144 | |||
| 214-234 | |||
| 242-262 | |||
| 267-287 | |||
| 300-320 | |||
| ConPred II | in/in | 8 | 42-62 |
| 89-109 | |||
| 122-142 | |||
| 217-237 | |||
| 243-263 | |||
| 268-288 | |||
| 299-319 | |||
| 321-341 |
DISCUSSION
In this paper we present the first direct confirmation that Rnf is a flavoprotein. We have shown by biochemical and mass spectrometry analysis that RnfG contains FMN covalently attached to threonine-175. This was further confirmed when no flavin was found in the RnfG-T-175L mutant. Additionally, the RfnD subunit appears to have a flavin covalently attached to threonine-187; the subunit fluoresces on a gel, and the fluorescence is lost when the Thr-187 is replaced by valine, a nonbinding residue.
These results are further evidence of the relatedness of Rnf to Na+-NQR and of the importance of the S(T)GAT motif, or variations thereof, for binding FMN. The original prediction that RnfG and RnfD would contain covalently bound flavins was based on sequence homology between these subunits and NqrC and NqrB, respectively. It has now been confirmed that the SGAT sequence in NqrB, NqrC, RnfG, and NosR (N2O reductase) is the flavin binding site (18, 19, 35). The case of RnfD is slightly different. This subunit contains an SGAT sequence, but the part of the protein that aligns to the SGAT of NqrB has the sequence TMAT, and it appears to be the later sequence that binds flavin. This suggests that methionine is a viable option for the second amino acid in the motif. Alignments of available RnfD sequences suggest that even more variation is possible, including substitution of the threonine ligand itself by serine. In fact, a mutant in which the threonine of NqrB was replaced by serine appears to have the same properties as the wild-type enzyme, confirming the viability of serine as an alternative to threonine (21).
Although the SGAT site in RnfD, which includes Thr-278, does not appear to have a covalently attached flavin, this motif is conserved in RnfD sequences, though not in NqrB. The fact that the sequence is conserved suggests that it may have a function as yet to be discovered.
In another apparent similarity between Rnf and Na+-NQR, the FMN in RnfG could be observed during redox titrations as a one-electron-reduced neutral flavosemiquinone. The presence of a neutral semiquinone form was evident from the spectrum in the visible region; a broad peak appeared between 509 and 650 nm, with a maximum at 580 nm (Figure 3, black line). The flavosemiquinone was only present in the native protein and could not be observed in titrations of the denatured protein, suggesting a role for the protein structure in the stabilization of the radical. The presence of the radical was confirmed by X-band EPR and ENDOR. ENDOR spectra confirmed the presence of a neutral flavosemiquinone radical. The radical is somewhat different from the neutral radical in Na+-NQR in that the aromatic ring methyl and proton splittings are slightly smaller, while the splittings of the C(1′) β-methylene protons are significantly larger. The signal had a line width in the range 17-20 G (1.7-2 mT (29)), suggesting that the primary radical formed is a neutral semiquinone (Figure 5A, B). Some samples gave line widths smaller than 20 G (2 mT), and in these cases there could be a mixture of neutral and anionic flavosemiquinone species. However, the radical spectrum is consistent over a pH range of at least 6.3-9.3, indicating that the putative population of anionic radical does not arise through a protonation equilibrium with the solvent. Caution must be used in applying these conclusions to the complete Rnf complex since the stabilization of the radical in the whole complex may involve interactions between RnfG and the other subunits, and possibly even between other subunits and the FMN moiety itself. Thus, these results must await confirmation with the complete complex.
The results of the gene fusion experiments are in agreement with the computer-based topology analysis. On this basis we can propose topological models for RnfG and RnfD. In the topology model of RnfG (Figure 6A) the N-terminus is located in the periplasmic space, as is the FMN attachment site, Thr-175. In the topology model of RnfD (Figure 6B) the N-terminal in the cytosol but the predicted flavin binding site, Thr-187, is located in the periplasmic space. In the models of both subunits, the location of the flavin is opposite to that predicted for the corresponding subunit in Na+-NQR; in both NqrC and NqrB the FMN is predicted to be in the cytosol. This could be indicative of a difference between the two enzymes in the details of the mechanism that couples electron transfer to ion translocation.
It is also possible that these differences reflect differences in the physiological roles of Rnf and Na+-NQR. In nitrogen fixing bacteria, Rnf provides a supply of low-potential reducing equivalents for nitrogenase. The enzyme oxidizes NADH and reduces ferredoxin, the energy for this uphill process coming from the sodium motive force across the cell membrane (6, 36). It has been suggested that in E. coli Rnf has a similar function, providing low-potential reducing equivalents, which maintain the oxygen sensor, SoxR, in its reduced state (9, 37). In both cases, the redox reaction is driven by downhill flow of Na+ into the cell. In contrast, in Na+-NQR the redox reaction drives uphill pumping of Na+ across the cell membrane (38). In Na+-NQR all of the redox cofactors are located on the cytoplasmic side of the membrane (29). The results presented above indicate that in Rnf from V. cholerae the flavin cofactors are located on the periplasmic side. This could suggest a relationship between the location of the cofactors and whether Na+ translocation is the driving or driven reaction, but experimental evidence from many more examples from both classes of enzymes is needed.
ACKNOWLEDGMENT
We thank Dr. Joel Morgan for help and advice in the anaerobic titrations, for many discussions, and for critical reading of the manuscript, Dr. Sarah Brooks for help with the HPLC separation, and Irina Barash for skillful technical assistance.
This work was supported by National Institutes of Health Grant R01 GM 069936.
Footnotes
Abbreviations: BCA, bicinchoninic acid; ENDOR, electron nuclear double resonance; EPR, electron paramagnetic resonance; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; HPLC, high-pressure liquid chromatography; kDa, kilodaltons; MALDI, matrix-assisted laser desorption/ionization; Na+-NQR, Na+-pumping NADH: quinone oxidoreductase; NDHI, H+-pumping NADH:quinone oxidoreductase; PCR, polymerase chain reaction; pNPP, p-nitrophenyl phosphate; PSD, postsource decay; PVDF, polyvinylidene fluoride; Rnf, Rhodobacter nitrogen fixing protein; rpm, revolutions per minute; SDS, sodiumdodecylsulfate; SDS-PAGE,sodiumdodecylsulfate-polyacrylamide gel electrophoresis; TMH, transmembrane helices; TOF, time of flight; Tris, tris(hydroxymethyl)aminomethane; w/v, weight /volume.
REFERENCES
- 1.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 2.Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus-characterization of two membrane-bound iron-sulfur proteins. Eur. J. Biochem. 1998;251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 3.Kumagai H, Fujiwara T, Matsubara H, Saeki K. Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry. 1997;36:5509–5521. doi: 10.1021/bi970014q. [DOI] [PubMed] [Google Scholar]
- 4.Jeong HS, Jouanneau Y. Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes. J. Bacteriol. 2000;182:1208–1214. doi: 10.1128/jb.182.5.1208-1214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duran-Pinedo AENK, Duncan MJ. The RprY response regulator of Porphyromonas gingivalis. Mol. Microbiol. 2007;64:1061–1074. doi: 10.1111/j.1365-2958.2007.05717.x. [DOI] [PubMed] [Google Scholar]
- 6.Curatti L, Brown CS, Ludden PW, Rubio LM. Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6291–6296. doi: 10.1073/pnas.0501216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Hetzel M, Boiangiu CD, Buckel W. Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of alpha-amino acids by anaerobic bacteria. FEMS Microbiol. Rev. 2004;28:455–468. doi: 10.1016/j.femsre.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Muller V, Imkamp F, Biegel E, Schmidt S, Dilling S. Discovery of a ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium woodii: a novel potential coupling site in acetogens. Ann. N.Y. Acad. Sci. 2008;1125:137–146. doi: 10.1196/annals.1419.011. [DOI] [PubMed] [Google Scholar]
- 9.Koo MS, Lee JH, Rah SY, Yeo WS, Lee KL, Koh YS, Kang SO, Roe JH. A reducing system of the superoxide sensor SoxR. Escherichia coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Li L, Rejtar T, Lessner DJ, Karger BL, Ferry JG. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 2006;188:702–710. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imkamp F, Biegel E, Jayamani E, Buckel W, Muller V. Dissection of the caffeate respiratory chain in the acetogen Acetobacterium woodii: identification of an Rnf-type NADH dehydrogenase as a potential coupling site. J. Bacteriol. 2007;189:8145–8153. doi: 10.1128/JB.01017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beifuss U, Tietze M, Baumer S, Deppenmeier U. Methanophenazine: structure, total synthesis, and function of a new cofactor from methnogenic archaea. Angew. Chem., Int. Ed. 2000;39:2470–2473. doi: 10.1002/1521-3773(20000717)39:14<2470::aid-anie2470>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 13.Barquera B, Hellwig P, Zhou W, Morgan JE, Hase CC, Gosink KK, Nilges M, Bruesehoff PJ, Roth A, Lancaster CR, Gennis RB. Purification and characterization of the recombinant Na(+)-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry. 2002;41:3781–3789. doi: 10.1021/bi011873o. [DOI] [PubMed] [Google Scholar]
- 14.Bogachev AV, Bertsova YV, Aitio O, Permi P, Verkhovsky MI. Redox-dependent sodium binding by the Na(+)-translocating NADH:quinone oxidoreductase from Vibrio harveyi. Biochemistry. 2007;46:10186–10191. doi: 10.1021/bi700440w. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi M, Nakayama Y, Unemoto T. Recent progress in the Na(+)-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biochim. Biophys. Acta. 2001;1505:37–44. doi: 10.1016/s0005-2728(00)00275-9. [DOI] [PubMed] [Google Scholar]
- 16.Steuber J. Na(+) translocation by bacterial NADH:quinone oxidoreductases: an extension to the complex-I family of primary redox pumps. Biochim. Biophys. Acta. 2001;1505:45–56. doi: 10.1016/s0005-2728(00)00276-0. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi M, Nakayama Y, Yasui M, Maeda M, Furuishi K, Unemoto T. FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na(+)-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 2001;488:5–8. doi: 10.1016/s0014-5793(00)02404-2. [DOI] [PubMed] [Google Scholar]
- 18.Barquera B, Hase CC, Gennis RB. Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na(+) pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 2001;492:45–49. doi: 10.1016/s0014-5793(01)02224-4. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama Y, Yasui M, Sugahara K, Hayashi M, Unemoto T. Covalently bound flavin in the NqrB and NqrC subunits of Na(+)-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 2000;474:165–168. doi: 10.1016/s0014-5793(00)01595-7. [DOI] [PubMed] [Google Scholar]
- 20.Barquera B, Nilges MJ, Morgan JE, Ramirez-Silva L, Zhou W, Gennis RB. Mutagenesis study of the 2Fe-2S center and the FAD binding site of the Na(+)-translocating NADH: ubiquinone oxidoreductase from Vibrio cholerae. Biochemistry. 2004;43:12322–12330. doi: 10.1021/bi048689y. [DOI] [PubMed] [Google Scholar]
- 21.Barquera B, Ramirez-Silva L, Morgan JE, Nilges MJ. A new flavin radical signal in the Na+-pumping NADH: quinone oxidoreductase from Vibrio cholerae: An EPR/ENDOR investigation of the role of the covalently bound flavins in subunits B and C. J. Biol. Chem. 2006;281:36482–36491. doi: 10.1074/jbc.M605765200. [DOI] [PubMed] [Google Scholar]
- 22.Fan C, Doan PE, Davoust CE, Hoffman BE. J. Magn. Reson. 1992;98:62–72. [Google Scholar]
- 23.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 24.Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- 25.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 26.Claros MG, von Heijne G. ToPRed II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 27.Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda M, Arai M, Lao DM, Shimizu T. Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a data set of experimentally-characterized transmembrane topologies. In Silico Biol. 2002;2:19–33. [PubMed] [Google Scholar]
- 29.Duffy EB, Barquera B. Membrane topology mapping of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis. J. Bacteriol. 2006;188:8343–8351. doi: 10.1128/JB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazurkiewicz P, Thomas J, Thompson JA, Liu M, Arbibe L, Sansonetti P, Holden DW. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol. Microbiol. 2008;67:1371–1383. doi: 10.1111/j.1365-2958.2008.06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barquera B, Morgan JE, Lukoyanov D, Scholes CP, Gennis RB, Nilges MJ. X- and W-band EPR and Q-band ENDOR studies of the flavin radical in the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. J. Am. Chem. Soc. 2003;125:265–275. doi: 10.1021/ja0207201. [DOI] [PubMed] [Google Scholar]
- 32.Bogachev AV, Bertsova YV, Ruuge EK, Wikstrom M, Verkhovsky MI. Kinetics of the spectral changes during reduction of the Na+-motive NADH:quinone oxidoreductase from Vibrio harveyi. Biochim. Biophys. Acta. 2002;1556:113–120. doi: 10.1016/s0005-2728(02)00342-0. [DOI] [PubMed] [Google Scholar]
- 33.Kay CWM, Feicht R, Schulz K, Sadewater P, Sancar A, Bacher A, Mobius K, Richter G, Weber S. EPR, ENDOR, and TRIPLE resonance spectroscopy on the neutral flavin radical in Escherichia coli DNA photolyase. Biochemistry. 1999;38:16740–16748. doi: 10.1021/bi991442u. [DOI] [PubMed] [Google Scholar]
- 34.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 35.Wunsch P, Zumft WG. Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration. J. Bacteriol. 2005;187:1992–2001. doi: 10.1128/JB.187.6.1992-2001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeki K, Kumagai H. The rnf gene products in Rhodobacter capsulatus play an essential role in nitrogen fixation during anaerobic DMSO-dependent growth in the dark. Arch. Microbiol. 1998;169:464–467. doi: 10.1007/s002030050598. [DOI] [PubMed] [Google Scholar]
- 37.Ding H, Demple B. In vivo kinetics of a redox-regulated transcriptional switch. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8445–8449. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogachev AV, Verkhovsky MI. Na+- translocating NADH:quinone oxidoreductase: progress achieved and prospects of investigations. Biochemistry (Moscow) 2005;70:143–149. doi: 10.1007/s10541-005-0093-4. [DOI] [PubMed] [Google Scholar]