Abstract
Gold colloids functionalized with amino acids provide a scaffold for effective DNA binding with subsequent condensation. Particles with lysine and lysine dendron functionality formed particularly compact complexes and provided highly efficient gene delivery without any observed cytotoxicity. Nanoparticles functionalized with first generation lysine dendrons (NP–LysG1) were ~28-fold superior to polylysine in reporter gene expression. These amino acid-based nanoparticles were responsive to intracellular glutathione levels, providing a tool for controlled release and concomitant expression of DNA.
Keywords: gold nanoparticles, transfection, glutathione, delivery, DNA
Gene therapy offers great promise for curing cancer and genetic disorders of both innate and acquired origin.1,2 Successful therapy requires the transport of nucleic acids into cells by delivery vehicles, as DNA is not efficiently translocated through the cell membrane.3 Recombinant viruses provide effective transfection vectors;4 however, issues of immunogenicity, carcinogenicity, and inflammation raise serious concerns for clinical applications.5–7 The challenges faced with viral vectors have inspired the parallel development of nonviral vectors based on polymers,8,9 dendrimers,10,11 and liposomes.12,13 These synthetic systems, however, are less efficient than viral systems.14,15 Therefore, efforts continue to focus on designing safe and efficient vectors.16
Inorganic nanoparticles including silica,17 iron oxide,18 and CdSe,19 have been exploited recently as alternate nonviral vectors. Gold nanoparticles provide particularly attractive scaffolds for the creation of transfection agents.20–22 Gold colloids are bioinert, nontoxic, and readily synthesized and functionalized.23,24 They also provide a multifunctional platform for both therapeutic and diagnostic purposes.25–28 Finally, through proper functionalization, these particles can be engineered to accumulate preferentially at tumor sites using targeting ligands, providing a powerful tool for cancer gene therapy.29,30
We report here the use of these nanoparticles for DNA transfection. There are several challenges to the design of polycations for gene transfection, including the need for effective complexation and condensation of the DNA, cellular uptake through endocytosis coupled with endosomal escape, protection from nuclease in cytoplasm, and finally delivery of the DNA to the nucleus.31 Nature provides insight into DNA packaging where DNA wraps around histone octamers; the nucleosome core proteins are ~6 nm in diameter featuring a large proportion of basic residues (lysine and arginine) that form salt bridges with the phosphate backbone of DNA.32 Using this structure as a starting point we designed DNA packaging agents using small spherical gold nanoparticles (core diameter, ~2 nm, overall diameter, 6 nm) functionalized with amino acids (Figure 1). These particles resemble histones in shape, size, and surface functionality. In these studies we find that compaction of DNA can be improved by increasing the density of ammonium groups on the nanoparticle surface, which facilitate delivery. Conjugation of lysines on particles in a dendritic fashion yielded efficient vectors, ~28-fold higher than polylysine for in vitro transfections. Amino acid-coated particles showed no cytotoxicity. Finally, we show that the gold-thiolate binding of ligands to particles allows the transfection ability of these materials to be regulated by manipulation of intracellular glutathione levels.
Figure 1.
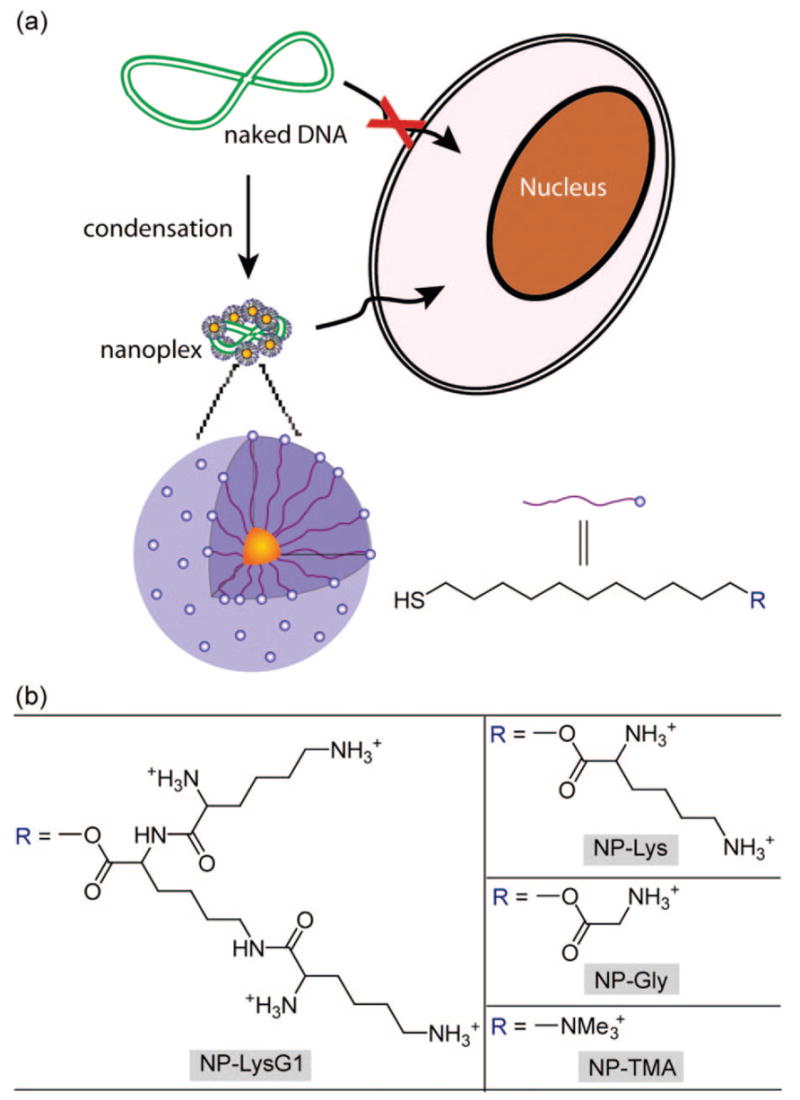
(a) Schematic illustration of the monolayer protected gold nanoparticles used as transfection vectors in this study; (b) chemical structures of headgroups presented on the surface of the nanoparticles.
RESULTS AND DISCUSSION
Fabrication of Vectors
Amino acid-conjugated thiols were synthesized in a straightforward procedure with terminal ammonium group(s) (see Supporting Information). 1-Pentanethiol-protected gold clusters (Au–C5, core diameter ≈ 2 nm) were functionalized with these ligands via Murray place-exchange reaction providing water-soluble particles.33 To explore the structural effect of headgroups on tranfection efficiency, we fabricated three vectors (NP–LysG1, NP–Lys, and NP–Gly) featuring varying density of cationic sites (Figure 1).
Nanoparticle-DNA Complexation
The comparable size of DNA and small functionalized nanoparticles facilitates their interaction.34 In our previous studies, we have demonstrated that quaternary ammonium-functionalized gold colloids effectively recognize DNA strands.35 Nanoparticles bearing primary ammonium groups (p Ka ≈10) on the surface are also expected to bind with anionic DNA via ion-pairing at physiological pH (pH = 7.4).36 Gel electrophoresis was carried out initially to test the association of nanoparticles with DNA (gWiz β-gal plasmid). Nanoparticle-DNA mixtures at molar ratios (MR = molNP:molDNA, at a fixed amount of DNA) of 200 (MR200) and 2000 (MR2000) were electrophoresed on agarose gel and stained with ethidium bromide (EtBr). While a band corresponding to free DNA was observed at MR200 suggesting incomplete complexation, electrophoretic mobility of DNA toward the positive electrode was completely retarded at MR2000 (Figure 2). In white light image, we observed that complexes were retained in the wells (Figure S1, see Supporting Information). Also, these complexes do not form extended aggregates over longer incubation periods, as evident from absorbance spectra (Figure S2, see Supporting Information). Binding was further verified by EtBr exclusion assay, which indicated complexation between DNA and nanoparticles by quenching of EtBr-fluorescence (Figure S3; see Supporting Information).
Figure 2.
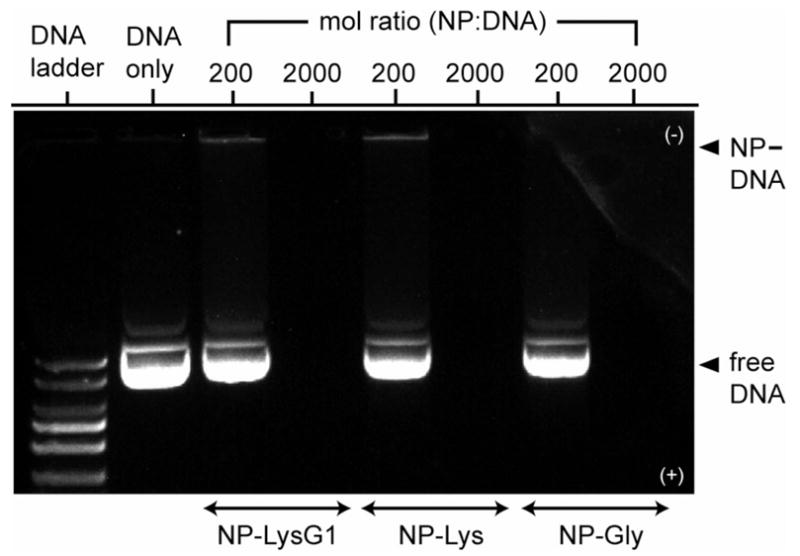
Gel retardation assay demonstrating DNA–nanoparticle complexation. A constant amount of DNA (333 ng/well) was complexed with nanoparticles at two different ratios in HEPES (10 mM, pH 7.4). No NP–DNA band is observed because of fluorescence quenching by NP complexation.
Characterization of Nanoplexes
Surface charge and size of DNA complexes are two important parameters in determining the efficiency of cellular uptake. ζ-Potential measurements revealed that complexes of each head-group (MR2000) bear positive surface potentials ranging from +25 to +34 mV (Table 1) that should promote initial adhesion on a negatively charged cell surface.37 In general, polycation–DNA complexes enter into mammalian cells via endocytosis, a process that is limited to particles smaller than ~150 nm in diameter.38 Dynamic light scattering (DLS) showed that NP–Gly formed large complexes (>150 nm). However, efficient condensation was achieved by increasing the density of ammonium groups on the particle surface. NP–LysG1 condensed DNA (<100 nm) most effectively (Table 1). In addition to enhanced cellular uptake, the tight packing with NP–LysG1 should prove useful for cancer gene therapy, where particles ~100 nm can more easily extravasate through open endothelial gaps in tumor tissues.39,40
TABLE 1.
Surface Charge and Size of Nanoparticle-DNA Complexes (mol Ratio 2000) in HEPES Buffer (10 mM, pH 7.4) at 25 °Ca
| nanoparticles | ζ-potential (mV) | size (nm) |
|---|---|---|
| NP–LysG1 | 30 ± 2 | 91 ± 2 |
| NP–Lys | 34 ± 1 | 112 ± 1 |
| NP–Gly | 25 ± 1 | 233 ± 40 |
| NP–TMA | 34 ± 2 | 118 ± 10 |
Size of DNA alone, 425 ± 51.
Transfection of Mammalian Cells
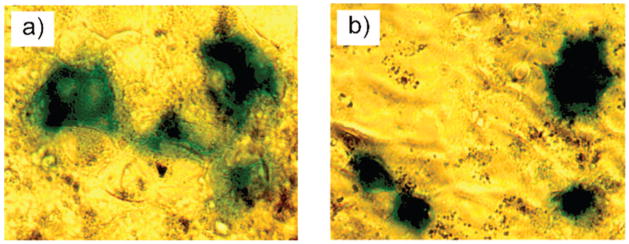
We have shown earlier that trimethyl ammonium-functionalized nanoparticles (NP–TMA) can protect DNA substantially from DNase digestion,41 and transfect 293T cells in the presence of serum and chloroquine.20 An X-gal staining assay was conducted to qualitatively assess the amino acid-coated nanoparticles mediated transfection of monkey kidney cells (Cos-1) with β-galactosidase (β-gal) reporter plasmid at MR2000. After 3 h of staining, blue spots were detected under an optical microscope from cells transfected with NP–LysG1 and NP–Lys, indicating that these particles serve as effective gene delivery agents (Figure 3).
Figure 3.
Optical micrographs showing transfected cells turned into blue after 3 h of X-gal staining. Cos-1 cells were transfected with β-gal reporter gene using (a) NP–LysG1 and (b) NP–Lys.
The expression of the β-gal reporter gene was monitored by enzyme activity assay to quantify the efficiency of DNA delivery. First, Cos-1 cells were treated with nanoparticle-DNA complexes at various ratios (MR1000, MR2000, MR4000, and MR6000) to determine the optimal ratio for transfection. Expression of β-gal was maximal at MR2000 (Figure S4, see Supporting Information), consistent with our previous studies.20 We next compared transfection efficiency of the synthesized nanoparticles at MR2000. As illustrated in Figure 4, lysine-coated particles NP–Lys and LP-LysG1 efficiently expressed the reporter gene, whereas NP–Gly showed negligible enzyme activity. Cells were also transfected with two other effective vectors as positive controls, previously reported NP–TMA,20 and extensively studied polylysine.42,43 Lysine and lysine dendrimer-coated particles were significantly better than both positive controls (Figure 4). In particular, NP–LysG1 and NP–Lys were superior to polylysine by ~28-fold and ~5-fold, respectively. As expected, no measurable β-gal activity was detected when cells were treated with naked DNA.
Figure 4.
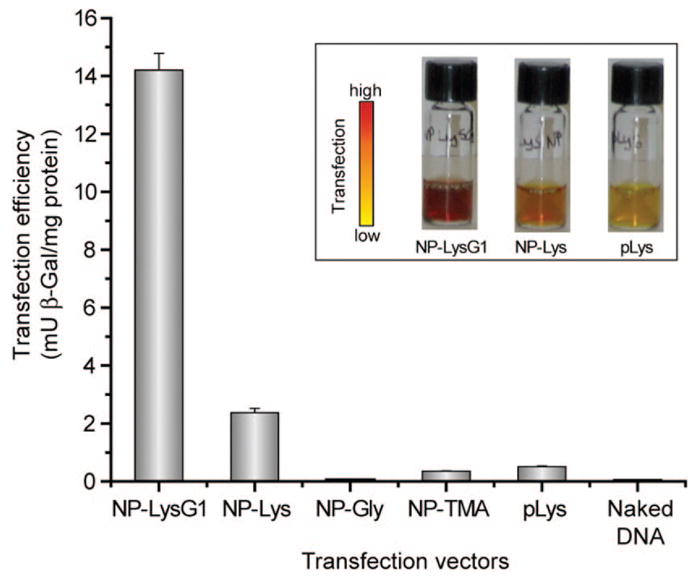
Enhanced transfection using NP–LysG1 and NP–Lys relative to positive controls, NP–TMA, and polylysine (pLys). No appreciable enzyme activity was observed in the absence of vectors. Inset shows solution colors during β-Gal activity assay performed after transfection. The color changes from yellow (substrate) to red (product) in the presence of active enzyme.
Cytotoxicity
Cellular metabolic activity was measured by alamar blue assay to evaluate possible toxicity that might arise from nanoparticles during transfection. As depicted in Figure 5, amino acid-coated particles displayed no decrease in viability. However, trimethyl ammonium-functionalized particles were moderately toxic, which probably originates from their strong interaction with the cell surface.44
Figure 5.
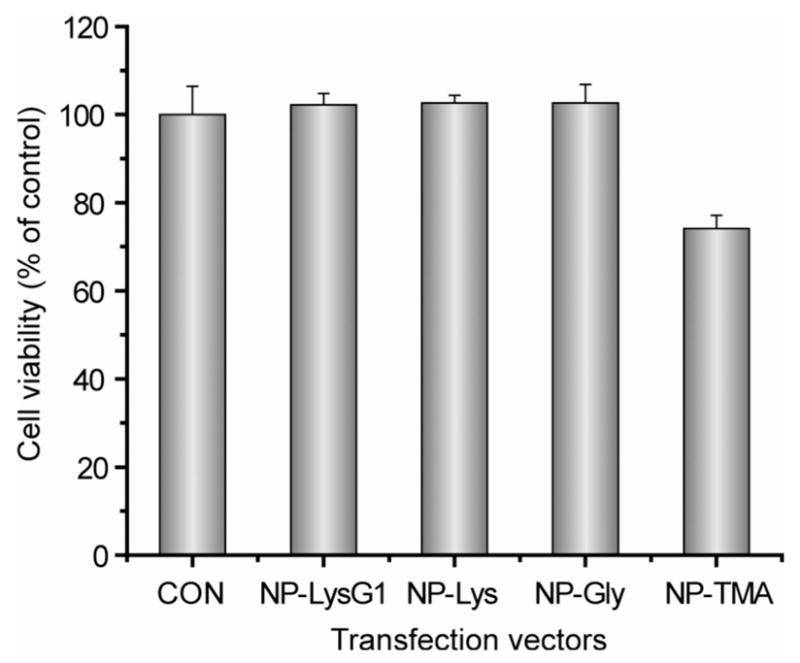
Cell viability determined by alamar blue assay at the end of transfection (total period 48 h) using the optimum molar ratio (MR2000).
Glutathione Regulation of Transfection Efficiency
The cationic ligands of Au nanoparticles can be displaced by glutathione (GSH), which would alter the surface charge and loosen the DNA–NP association (Figure 6a).45,46 This mode of release utilizes the dramatic differential between extra- and intracellular GSH levels. Moreover, manipulation of intracellular GSH levels provides a potential mechanism for external control of transfection. We investigated the regulation of transfection efficiency of nanoparticles, NP–Lys (MR2000), by both increasing and decreasing intracellular GSH concentration. The glutathione level was transiently increased by treating cells with glutathione monoester (GSH–OEt).26 Glutathione monoester is rapidly internalized by cells and processed into glutathione (GSH) by esterases.47 Transfection efficiency increased upon treatment of cells with GSH–OEt in a concentration-dependent fashion (Figure 6b). In a complementary study, baseline GSH production was suppressed by prolonged (24 h) treatment of cells with L-buthionine-[S,R]-sulfoximine (BSO), an inhibitor of γ-glutamylcysteine synthetase.48 As expected, BSO-treated cells showed lower transfection efficiency compared to untreated cells (Figure 6c).
Figure 6.
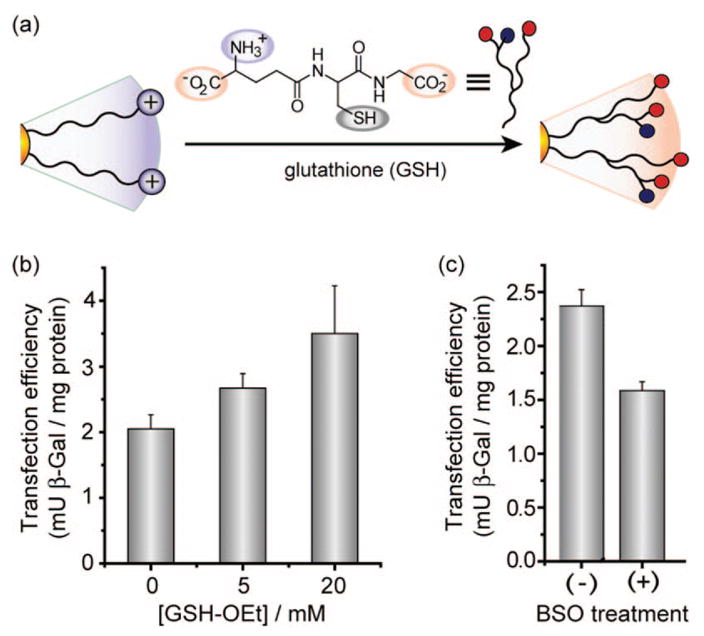
(a) Schematic depiction of place-exchange between native cationic ligands and cellular glutathione (GSH) on nanoparticle surface. (b) Elevation in transfection level depending on dose of glutathione monoester (GSH–OEt). Cells were preincubated with GSH–OEt for 1 h then washed prior to transfection. (c) Decrease in transfection efficiency upon BSO treatment. Cells were plated in BSO-containing (2 mM) media and incubated for 24 h.
CONCLUSION
In summary, we have demonstrated that coating gold nanoparticles with lysine-based headgroups produces effective transfection vectors. DNA delivery efficiency strongly depends on the structure of head-groups and their concomitant ability to condense DNA. The lysine dendron-functionalized nanoparticle NP–LysG1 was most effective at condensing DNA, and was the most potent vector, ~28 times more effective than polylysine. Importantly, these amino acid-functionalized particles showed no cytotoxicity when used as transfection agents. These materials were also responsive to cellular glutathione level during in vitro transfection, providing insight into their mode of activity as well as being a potential tool for orthogonal control of transfection.
EXPERIMENTAL SECTION
Materials
All chemicals were purchased from Aldrich unless otherwise stated. The organic solvents were bought from Pharmco-Aaper and used as received except dichloromethane and toluene which were distilled in the presence of calcium hydride. Flash column chromatography was carried out for purification using silica gel (SiO2, particle size, 40–63 μm). gWiz β-gal plasmid (8278 bp) was purchased from Aldevron (Fargo, ND). L-Glutathione reduced ethyl ester (GSH–OEt) was obtained from Fluka. Cell culture medium powder and phosphate buffer saline (PBS) were bought from Aldrich.
Gel Electrophoresis
To prepare each mixture (27 μL total volume), 400 ng of DNA was incubated with required amount of nanoparticles at room temperature in 10 mM HEPES buffer (pH = 7.4). After 10 min of incubation, 3 μL of gel loading dye (6x) was added into each mixture. A 25 μL aliquot from the resulting 30 μL solution was loaded into 0.6% agarose gel, prestained with EtBr. The samples were electrophoresed at 100 V for 60 min in TBE buffer (0.045 M Tris-borate; 0.001 M EDTA) and the bands were visualized on a UV trans-illuminator.
Cell Culture
Cos-1 cells were cultured in a humidified atmosphere (5% CO2) at 37 °C. The cells were grown in Dulbecco’s modified eagle’s medium (DMEM, 4.0 g/L glucose) supplemented with 10% fetal bovine serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). All transfection experiments were performed with complete growth medium without antibiotics and in the presence of 100 μM chloroquine. Cells (1 × 105 per well) were seeded on a 24-well plate 24 h prior the experiments. For GSH-OEt treatment, old medium was removed after 24 h of plating, and cells were incubated with fresh medium containing GSH-OEt for 1 h and washed three times with PBS before adding transfection medium.26
Transfection Protocol
All experiments were done in triplicate. Nanoplexes were first prepared at room temperature as follows (for 3 wells): (a) a 100 μL solution of 2.4 μg of plasmid and a 200 μL solution of the required concentration of nanoparticles were prepared separately in PBS. (b) After 5 min, particles were mixed with DNA and incubated together for 10 min. The nanoplexes were diluted with 1600 μL of prewarmed DMEM with 100 μM chloroquine. The resulting transfection medium was added into wells (600 μL/well, 3 wells) after washing cells once with PBS. Six hours later, the medium was removed, cells were washed 3× with PBS, and complete medium (600 μL/well) was added for another 42 h of incubation. Cells were transfected similarly with polylysine/DNA at a mass ratio of 2.5 as previously reported (Mw of pLys ≈ 50 K).42 Gene expression and cytotoxicity were tested after 48 h of total transfection period.
X-Gal Assay
Cells were stained following the assay kit (Genlantis, USA). After 3 h of staining, cells were washed twice with PBS and visualized on an optical microscope (Zeiss, 20×).
Reporter Gene Expression
β-Gal activity was assayed using chlorophenol red-β-D-galactopyranoside as a substrate (CPRG kit, Genlantis, USA). Absorbance (A570) was measured on a SpectroMax M5 microplate reader (Molecular Device), and the amount of expressed protein was calculated from a calibration curve constructed with pure β-gal. Total cellular protein was determined by bicinchoninic acid assay (Pierce, USA) according to the manufacturer’s protocol.
Cytotoxicity Assay
The alamar blue assay was performed according to the manufacturer’s protocol (Invitrogen Biosource, USA). After 48 h of transfection, cells were treated with 10% alamar blue solution and kept at 37 °C for another 2 h. Red fluorescence, resulting from the reduction of alamar blue, was monitored (excitation/emission: 535/590) on a SpectroMax M5 microplate reader (Molecular Device).
DLS and ζ-Potential
DLS experiments and ζ-potential measurements were carried out using a Malvern Zetasizer (Nano series, Malvern Instruments Inc., USA). Nanoplexes were prepared as mentioned above in transfection protocol using HEPES (10 mM, pH = 7.4) only, instead of PBS/media. After 10 min of incubation, data were collected and reported as an average of three measurements.
Supplementary Material
Nanoparticle and ligand synthesis, binding studies, and transfection efficiencies at varying particle/DNA ratios. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This research was support by the NIH (GM077173), the NSF-sponsored Center for Hierarchical Manufacturing (DMI-0531171), and MRSEC facilities.
REFERENCES AND NOTES
- 1.McCormick F. Cancer Gene Therapy: Fringe or Cutting Edge. Nat Rev Cancer. 2001;1:130–141. doi: 10.1038/35101008. [DOI] [PubMed] [Google Scholar]
- 2.Opalinska JB, Gewirtz AM. Nucleic-Acid Therapeutics: Basic Principles and Recent Applications. Nat Rev Drug Discovery. 2002;1:503–514. doi: 10.1038/nrd837. [DOI] [PubMed] [Google Scholar]
- 3.Sokolova V, Epple M. Inorganic Nanoparticles as Carriers of Nucleic Acids into Cells. Angew Chem, Int Ed. 2008;47:1382–1395. doi: 10.1002/anie.200703039. [DOI] [PubMed] [Google Scholar]
- 4.Kay MA, Glorioso JC, Naldini L. Viral Vectors for Gene Therapy: The Art of Turning Infectious Agents into Vehicles of Therapeutics. Nat Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 5.Check E. Gene Therapy Put on Hold as Third Child Develops Cancer. Nature. 2005;433:561. doi: 10.1038/433561a. [DOI] [PubMed] [Google Scholar]
- 6.Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao GP, Wilson JM, Batshaw ML. Fatal Systemic Inflammatory Response Syndrome in a Ornithine Transcarbamylase Deficient Patient Following Adenoviral Gene Transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, et al. A Serious Adverse Event after Successful Gene Therapy for X-linked Severe Combined Immunodeficiency. New Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 8.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and Development of Polymers for Gene Delivery. Nat Rev Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 9.Liu YM, Reineke TM. Hydroxyl Stereochemistry and Amine Number within Poly(glycoamidoamine)s Affect Intracellular DNA Delivery. J Am Chem Soc. 2005;127:3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- 10.Nishiyama N, Iriyama A, Jang WD, Miyata K, Itaka K, Inoue Y, Takahashi H, Yanagi Y, Tamaki Y, Koyama H, et al. Light-Induced Gene Transfer from Packaged DNA Enveloped in a Dendrimeric Photosensitizer. Nat Mater. 2005;4:934–941. doi: 10.1038/nmat1524. [DOI] [PubMed] [Google Scholar]
- 11.Dufes C, Uchegbu IF, Schatzlein AG. Dendrimers in Gene Delivery. Adv Drug Delivery Rev. 2005;57:2177–2202. doi: 10.1016/j.addr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 12.de Lima MCP, Neves S, Filipe A, Duzgunes N, Simoes S. Cationic Liposomes for Gene Delivery: From Biophysics to Biological Applications. Curr Med Chem. 2003;10:1221–1231. doi: 10.2174/0929867033457430. [DOI] [PubMed] [Google Scholar]
- 13.Mahato RI. Water Insoluble and Soluble Lipids for Gene Delivery. Adv Drug Delivery Rev. 2005;57:699–712. doi: 10.1016/j.addr.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Luo D, Saltzman WM. Synthetic DNA Delivery Systems. Nat Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 15.Kodama K, Katayama Y, Shoji Y, Nakashima H. The Features and Shortcomings for Gene Delivery of Current Non-Viral Carriers. Curr Med Chem. 2006;13:2155–2161. doi: 10.2174/092986706777935276. [DOI] [PubMed] [Google Scholar]
- 16.Cavazzana-Calvo M, Thrasher A, Mavilio F. The Future of Gene Therapy. Nature. 2004;427:779–781. doi: 10.1038/427779a. [DOI] [PubMed] [Google Scholar]
- 17.Torney F, Trewyn BG, Lin VSY, Wang K. Mesoporous Silica Nanoparticles Deliver DNA and Chemicals into Plants. Nat Nanotechnol. 2007;2:295–300. doi: 10.1038/nnano.2007.108. [DOI] [PubMed] [Google Scholar]
- 18.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In Vivo Imaging of siRNA Delivery and Silencing in Tumors. Nat Med. 2007;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 19.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Targeted Quantum Dot Conjugates for siRNA Delivery. Bioconjugate Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- 20.Sandhu KK, McIntosh CM, Simard JM, Smith SW, Rotello VM. Gold Nanoparticle-Mediated Transfection of Mammalian Cells. Bioconjugate Chem. 2002;13:3–6. doi: 10.1021/bc015545c. [DOI] [PubMed] [Google Scholar]
- 21.Thomas M, Klibanov AM. Conjugation to Gold Nanoparticles Enhances Polyethylenimine’s Transfer of Plasmid DNA into Mammalian Cells. Proc Natl Acad Sci US A. 2003;100:9138–9143. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 23.Daniel MC, Astruc D. Gold Nanoparticles: Assembly, Supramolecular Chemistry, Quantum-Size-Related Properties, and Applications toward Biology, Catalysis, and Nanotechnology. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 24.Connor EE, Mwamuka J, Gole A, Murphy CJ, Wyatt MD. Gold Nanoparticles Are Taken up by Human Cells but Do Not Cause Acute Cytotoxicity. Small. 2005;1:325–327. doi: 10.1002/smll.200400093. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharya R, Mukherjee P, Xiong Z, Atala A, Soker S, Mukhopadhyay D. Gold Nanoparticles Inhibit VEGF165-Induced Proliferation of HUVEC Cells. Nano Lett. 2004;4:2479–2481. [Google Scholar]
- 26.Hong R, Han G, Fernandez JM, Kim BJ, Forbes NS, Rotello VM. Glutathione-Mediated Delivery and Release Using Monolayer Protected Nanoparticle Carriers. J Am Chem Soc. 2006;128:1078–1079. doi: 10.1021/ja056726i. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh P, Han G, De M, Kim CK, Rotello VM. Gold Nanoparticles in Delivery Applications. Adv Drug Delivery Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Shi XG, Wang SH, Meshinchi S, Van Antwerp ME, Bi XD, Lee IH, Baker JR. Dendrimer-Entrapped Gold Nanoparticles as a Platform for Cancer-Cell Targeting and Imaging. Small. 2007;3:1245–1252. doi: 10.1002/smll.200700054. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharya R, Patra CR, Earl A, Wang SF, Katarya A, Lu LC, Kizhakkedathu JN, Yaszemski MJ, Greipp PR, Mukhopadhyay D, et al. Attaching Folic Acid on Gold Nanoparticles Using Noncovalent Interaction via Different Polyethylene Glycol Backbones and Targeting of Cancer Cells. Nanomedicine. 2007;3:224–238. [Google Scholar]
- 30.Paciotti GF, Kingston DGI, Tamarkin L. Colloidal Gold Nanoparticles: A Novel Nanoparticle Platform for Developing Multifunctional Tumor-Targeted Drug Delivery Vectors. Drug Dev Res. 2006;67:47–54. [Google Scholar]
- 31.Davis ME. Non-Viral Gene Delivery Systems. Curr Opin Biotechnol. 2002;13:128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 32.Voet D, Voet JG. Biochemistry. 2. John Wiley & Sons, Inc; New York: 1995. pp. 1124–1131. [Google Scholar]
- 33.Templeton AC, Wuelfing MP, Murray RW. Monolayer Protected Cluster Molecules. Acc Chem Res. 2000;33:27–36. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 34.Verma A, Rotello VM. Surface Recognition of Biomacromolecules Using Nanoparticle Receptors. Chem Commun. 2005:303–312. doi: 10.1039/b410889b. [DOI] [PubMed] [Google Scholar]
- 35.McIntosh CM, Esposito EA, Boal AK, Simard JM, Martin CT, Rotello VM. Inhibition of DNA Transcription Using Cationic Mixed Monolayer Protected Gold Clusters. J Am Chem Soc. 2001;123:7626–7629. doi: 10.1021/ja015556g. [DOI] [PubMed] [Google Scholar]
- 36.Andre I, Linse S, Mulder FAA. Residue-Specific pKa Determination of Lysine and Arginine Side Chains by Indirect N-15 and C-13 NMR Spectroscopy: Application to Apo Calmodulin. J Am Chem Soc. 2007;129:15805–15813. doi: 10.1021/ja0721824. [DOI] [PubMed] [Google Scholar]
- 37.Mansouri S, Cuie Y, Winnik F, Shi Q, Lavigne P, Benderdour M, Beaumont E, Fernandes JC. Characterization of Folate-Chitosan-DNA Nanoparticles for Gene Therapy. Biomaterials. 2006;27:2060–2065. doi: 10.1016/j.biomaterials.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 38.Je JY, Cho YS, Kim SK. Characterization of (Aminoethyl)chitin/DNA Nanoparticle for Gene Delivery. Biomacromolecules. 2006;7:3448–3451. doi: 10.1021/bm060373d. [DOI] [PubMed] [Google Scholar]
- 39.Hobbs SK, Monsky WL, Yuan F, Roberts WG, Griffith L, Torchilin VP, Jain RK. Regulation of Transport Pathways in Tumor Vessels: Role of Tumor Type and Microenvironment. Proc Natl Acad Sci US A. 1998;95:4607–4612. doi: 10.1073/pnas.95.8.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mozafari MR, Reed CJ, Rostron C. Prospects of Anionic Nanolipoplexes in Nanotherapy: Transmission Electron Microscopy and Light Scattering Studies. Micron. 2007;38:787–795. doi: 10.1016/j.micron.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Han G, Martin CT, Rotello VM. Stability of Gold Nanoparticle-Bound DNA toward Biological, Physical, and Chemical Agents. Chem Biol Drug Des. 2006;67:78–82. doi: 10.1111/j.1747-0285.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 42.Ramsay E, Hadgraft J, Birchall J, Gumbleton M. Examination of the Biophysical Interaction between Plasmid DNA and the Polycations, Polylysine and Polyornithine, as a Basis for Their Differential Gene Transfection in Vitro. Int JPharm. 2000;210:97–107. doi: 10.1016/s0378-5173(00)00571-8. [DOI] [PubMed] [Google Scholar]
- 43.Zauner W, Ogris M, Wagner E. Polylysine-Based Transfection Systems Utilizing Receptor-Mediated Delivery. Adv Drug Delivery Rev. 1998;30:97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 44.Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Toxicity of Gold Nanoparticles Functionalized with Cationic and Anionic Side Chains. Bioconjugate Chem. 2004;15:897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- 45.Han G, Chari NS, Verma A, Hong R, Martin CT, Rotello VM. Controlled Recovery of the Transcription of Nanoparticle-Bound DNA by Intracellular Concentrations of Glutathione. Bioconjugate Chem. 2005;16:1356–1359. doi: 10.1021/bc050173j. [DOI] [PubMed] [Google Scholar]
- 46.Li D, Li GP, Guo WW, Li PC, Wang EK, Wang J. Glutathione-Mediated Release of Functional Plasmid DNA from Positively Charged Quantum Dots. Biomaterials. 2008;29:2776–2782. doi: 10.1016/j.biomaterials.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Levy EJ, Anderson ME, Meister A. Transport of Glutathione Diethyl Ester into Human-Cells. Proc Natl Acad Sci US A. 1993;90:9171–9175. doi: 10.1073/pnas.90.19.9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balakirev M, Schoehn G, Chroboczek J. Lipoic Acid-Derived Amphiphiles for Redox-Controlled DNA Delivery. Chem Biol. 2000;7:813–819. doi: 10.1016/s1074-5521(00)00030-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nanoparticle and ligand synthesis, binding studies, and transfection efficiencies at varying particle/DNA ratios. This material is available free of charge via the Internet at http://pubs.acs.org.