Abstract
Transformations of 2-hydroxybenzoate and fluorobenzoate isomers were investigated in the strictly anaerobic Syntrophus aciditrophicus to gain insight into the initial steps of the metabolism of aromatic acids. 2-Hydroxybenzoate was metabolized to methane and acetate by S. aciditrophicus and Methanospirillum hungatei cocultures and reduced to cyclohexane carboxylate by pure cultures of S. aciditrophicus when grown in the presence of crotonate. Under both conditions, transient accumulation of benzoate but not phenol was observed, indicating that dehydroxylation occurred prior to ring reduction. Pure cultures of S. aciditrophicus reductively dehalogenated 3-fluorobenzoate with the stoichiometric accumulation of benzoate and fluorine. 3-Fluorobenzoate-degrading cultures produced a metabolite that had a fragmentation pattern almost identical to that of the trimethylsilyl (TMS) derivative of 3-fluorobenzoate but with a mass increase of 2 units. When cells were incubated with deuterated water, this metabolite had a mass increase of 3 or 4 units relative to the TMS derivative of 3-fluorobenzoate. 19F nuclear magnetic resonance spectroscopy (19F NMR) detected a metabolite in fluorobenzoate-degrading cultures with two double bonds, either 1-carboxyl-3-fluoro-2,6-cyclohexadiene or 1-carboxyl-3-fluoro-3,6-cyclohexadiene. The mass spectral and NMR data are consistent with the addition of two hydrogen or deuterium atoms to 3-fluorobenzoate, forming a 3-fluorocyclohexadiene metabolite. The production of a diene metabolite provides evidence that S. aciditrophicus contains dearomatizing reductase that uses two electrons to dearomatize the aromatic ring.
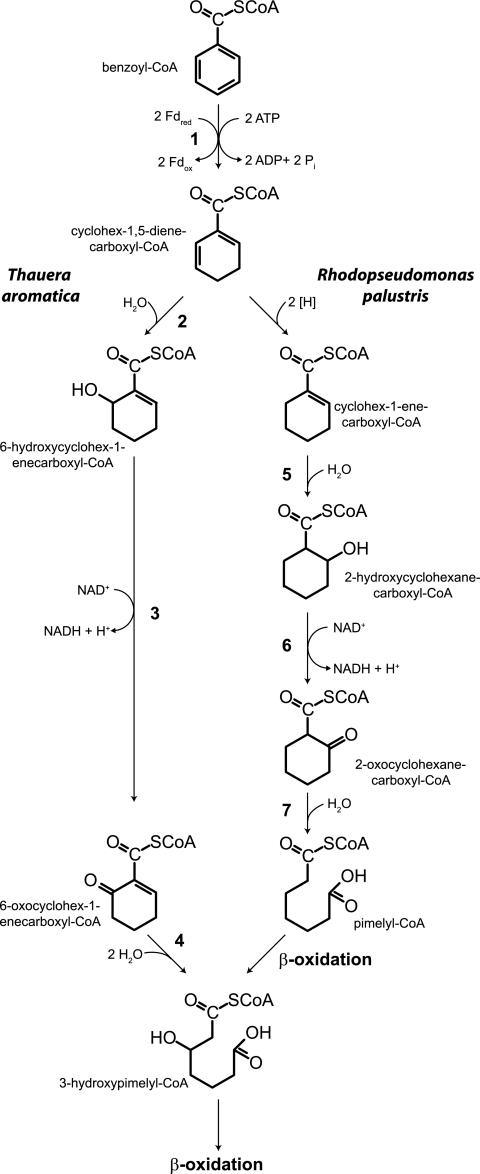
In anaerobic environments, the biodegradation of aromatic compounds plays an important role in the cycling of carbon and the bioremediation of environmental contaminants (7, 12, 13, 19, 20). The benzene ring is the main structural component of lignin, which comprises about 30% of plant material and is the second most abundant polymer in nature next to cellulose. Initial anaerobic transformations of aromatic compounds generally lead to the conversion of diverse aromatic compounds into benzoyl-coenzyme A (benzoyl-CoA) (13, 17, 20, 25, 44). Two pathways are known for the anaerobic metabolism of benzoyl-CoA (Fig. 1) (3, 17, 19, 20). The first step in both pathways is the ATP-dependent reduction of benzoyl-CoA to cyclohexa-1,5-diene-1-carboxyl-CoA (2-4, 8, 18). The reduction of benzoyl-CoA represents a considerable energy barrier for anaerobic microorganisms because of the high resonance energy that stabilizes the aromatic ring (2-4, 20). In Thauera aromatica, a benzoyl-CoA reductase that hydrolyzes two ATP molecules per two electrons is used to overcome this barrier (2). Rhodopseudomonas palustris contains badDEFG genes whose deduced amino acid sequences have high degree of sequence identity to the BcrCBAD subunits of the benzoyl-CoA reductase of T. aromatica (8), which suggests that a similar ATP-requiring reaction is involved in benzoyl-CoA reduction. Geobacter metallireducens (a benzoate-degrading iron reducer) and Desulfococcus multivorans (a benzoate-degrading sulfate reducer) require molybdenum and selenium for growth with benzoate (40, 48). These data coupled with the results of proteomic studies with radioactive selenium indicate that obligate anaerobes have a completely different benzoyl-CoA reductase that contains molybdenum and selenium (40, 48).
FIG. 1.
Pathways for anaerobic metabolism of benzoate. Pathways: 1, benzoyl-CoA reductase; 2, cyclohexa-1,5-diene-1-carboxyl-CoA hydratase; 3, 6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase; 4, 6-oxocyclohex-1-ene-1-carboxyl-CoA hydrolase; 5, cyclohex-1-ene-1-carboxyl-CoA hydratase; 6, 2-hydroxycyclohexane-1-carboxyl-CoA dehydrogenase; 7, 2-oxocyclohexane-1-carboxyl-CoA hydrolase. Fdred, reduced ferredoxin; Fdox, oxidized ferredoxin; [H], reducing equivalents.
Denitrifiers use three enzymatic steps to convert cyclohexa-1,5-diene-1-carboxyl-CoA to 3-hydroxypimelyl-CoA; the enzymes involved in these steps are cyclohexa-1,5-diene-1-carboxyl-CoA hydratase, 6-hydroxyclohex-1-ene-1-carboxyl-CoA dehydrogenase, and 6-oxocyclohex-1-ene-1-carboxyl-CoA hydrolase (5, 27, 28) (Fig. 1). Amino acid sequence analyses of the deduced gene products indicate that facultative anaerobes in the genera Thauera, Azoarcus, and Magnetospiriullum have enzymes that are highly similar to the above three T. aromatica enzymes (17, 30, 31). The genomes of Geobacter metallireducens (GenBank accession no. CP000148) and the syntrophic benzoate degrader Syntrophus aciditrophicus (GenBank accession no. CP000252) (32) each contain homologs with similarity at the amino acid level to the above three enzymes in T. aromatica or Azoarcus sp. strain EbN1 (30, 41, 48). The bamRGeo gene (gi 78223357) and bamAGeo gene (gi 78223295) of Geobacter metallireducens and the bamRSyn gene (gi 85860872) and bamASyn (gi 85860871) of Syntrophus aciditrophicus were each heterologously expressed in Escherichia coli. The expressed BamR gene products from both organisms catalyzed the hydration of cyclohexa-1,5-diene-1-carboxyl-CoA to 6-hydroxycyclohex-1-ene-1-carboxyl-CoA (reaction 2 [Fig. 1]) (41), and the expressed BamA gene products of both organisms catalyzed the hydrolysis of 6-oxocyclohex-1-ene-1-carboxyl-CoA to 3-hydroxylpimelyl-CoA (reaction 4 [Fig. 1]) (26). The presence of a gene that encodes cyclohexa-1,5-diene-1-carboxyl-CoA hydratase in S. aciditrophicus and G. metallireducens and the detection of this activity in cell extracts of these two organisms and of D. multivorans provide evidence that the product of ring dearomatization in facultative and obligate anaerobes is cyclohexa-1,5-diene-1-carboxyl-CoA even though obligate anaerobes have a different type of benzoyl-CoA reductase. From the above studies, it appears that, once cyclohexa-1,5-diene-1-carboxyl-CoA is formed, aromatic acid-degrading facultative and obligate anaerobes use the benzoyl-CoA pathway found in T. aromatica (Fig. 1). The exception is Rhodopseudomonas palustris which uses a different pathway for cyclohexa-1,5-diene-1-carboxyl-CoA metabolism; cyclohexa-1,5-diene-1-carboxyl-CoA is reduced to cyclohex-1-ene-1-carboxyl-CoA and then metabolized to pimelyl-CoA by a series of β-oxidation-like reactions (Fig. 1) (8, 19, 33, 38, 39).
In methanogenic environments, syntrophic associations of a benzoate-degrading microorganism and hydrogen- and/or formate-using methanogens catalyze the reduction and cleavage of the aromatic ring (14, 34, 46). The degradation of benzoate to acetate, carbon dioxide, and hydrogen (equation 1) is unfavorable unless the hydrogen partial pressure is kept low by the methanogen (21, 34, 46) (equation 1):
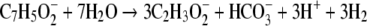 |
(1) |
(ΔG°′ = +70.1 kJ mol−1 of benzoate [47]).
The amounts of energy available during syntrophic benzoate metabolism are small (ΔG′ of −20 to −45 kJ mol−1 of benzoate) (21, 22, 43), which raised the question of whether there is sufficient energy to support an ATP-dependent, two-electron reduction of benzoyl-CoA to a dienoyl-CoA intermediate (40, 48). For this reason, Schöcke and Schink (45) proposed that benzoate may be reduced by a four- or six-electron reduction reaction. The reduction of benzoate to cyclohex-1-ene-1-carboxylate (ΔG°′ of −71.3 kJ/mol) or to cyclohexane carboxylate (ΔG°′ of −94.5 kJ/mol) is favorable (10, 45). However, regardless of the number of electrons involved in the reaction, the activation barrier should still be the same. S. aciditrophicus transiently accumulates cyclohex-1-ene-1-carboxylate and up to 260 μM of cyclohexane carboxylate during syntrophic benzoate metabolism (9), ferments benzoate to acetate and cyclohexane carboxylate (10), and reduces benzoate to cyclohexane carboxylate with crotonate as the electron donor (35). Cyclohex-1-ene-1-carboxylate and cyclohexane carboxylate could be formed by a four-electron or a six-electron reduction, respectively, of benzoyl-CoA. However, the presence of a cyclohexa-1,5-diene-1-carboxyl-CoA hydratase suggests that these compounds may be formed by a series of two-electron reduction steps (41). Hydroxylation of benzoyl-CoA prior to its reduction has been proposed as an alternative mechanism of dearomatization; the hydroxyl moiety would weaken the aromatic character of the ring and make reductive dearomatization by an electron donor, such as ferredoxin, favorable without ATP input (24, 42). The reduction of 2-hydroxybenzoyl-CoA may yield 6-oxocyclohex-1-ene-1-carboxyl-CoA after keto/enol tautomerizations.
Fluorine analogs have been used to detect metabolites derived from the metabolism of diverse aromatic compounds (11, 15, 29). The fluorine atom is only 20% larger than the hydrogen atom, which allows fluorinated analogs to be metabolized to a point where the strong electronegativity of the fluorine atom slows or prevents further degradation of the metabolite (11, 23). Recently, we showed that pure cultures of S. aciditrophicus use benzoate as an electron acceptor with crotonate as the electron donor (35). Here, we use fluorinated and hydroxylated compounds to investigate the steps involved benzoate reduction by S. aciditrophicus with crotonate as the electron donor.
MATERIALS AND METHODS
Media and cultivation conditions.
Pure cultures of Syntrophus aciditrophicus strain SB (ATCC 700169) and Methanospirillum hungatei (ATCC 27890) and cocultures of these two organisms were grown anaerobically as described previously (9). The media and stock solutions were prepared according to the anaerobic techniques described by Balch and Wolfe (1). All cultures were incubated at 37°C without shaking. The culture purity was checked daily by microscopic examination and by periodic inoculation of thioglycolate-containing medium.
To determine whether pure cultures of S. aciditrophicus metabolize aromatic compounds other than benzoate with crotonate as the electron donor, triplicate 10-ml cultures with 10 mM crotonate and 2 mM of the aromatic compound were each inoculated with 1 ml of a pure culture of S. aciditrophicus grown with 20 mM crotonate. The following aromatic compounds were tested: o-phthalate, 2-fluorobenzoate, 3-fluorobenzoate, 4-fluorobenzoate, 4-aminobenzoate, 2-chlorobenzoate, 3-chlorobenzoate, 4-chlorobenzoate, 2-hydroxybenzoate, 3-hydroxybenzoate, 4-hydroxybenzoate, and o-toluate. Growth was monitored and compared to growth with crotonate alone or crotonate with benzoate. Additionally, samples were taken immediately after inoculation and after 16 days of incubation to measure substrate depletion.
The degradation of hydroxybenzoate isomers by pure cultures and cocultures of S. aciditrophicus was also monitored in triplicate 50-ml cultures with 10 mM crotonate and 2 mM of the hydroxybenzoate. Each culture was inoculated with 5 ml of a pure culture of S. aciditrophicus grown with 20 mM crotonate. Cocultures were established by coinoculation of the serum bottles with 5 ml of a pure culture of M. hungatei. Heat-killed controls were included for each condition. One-milliliter samples were taken daily to measure substrate depletion and product formation. Methane formation by cocultures was measured by daily headspace analysis. Pure cultures of S. aciditrophicus were grown in duplicate 500-ml cultures with 10 mM crotonate and 2 mM 2-hydroxybenzoate, and samples (50 ml) were taken daily for gas chromatography-mass spectroscopy (GC-MS) analyses as described previously (36).
The metabolism of fluorinated benzoate isomers by pure cultures of S. aciditrophicus was monitored as described above. Each serum bottle contained 50 ml of medium with 10 mM crotonate and 0.5 mM of 2-fluorobenzoate, 3-fluorobenzoate, or 4-fluorobenzoate. Heat-killed controls were included for each condition and served as negative controls. Growth in medium with 10 mM crotonate or with 10 mM crotonate and 3 mM benzoate served as positive controls. One-milliliter samples were taken daily to monitor growth, substrate depletion, and product formation. Hydrogen formation was measured in the headspace. Larger-volume cultures (300 ml) were used to detect the formation of metabolites. Fifty-milliliter samples were taken initially and after 12, 26, and 90 days of incubation for analysis by GC-MS and 19F nuclear magnetic resonance (19F NMR) spectroscopy (36).
Metabolite formation by washed cell suspensions of S. aciditrophicus was also monitored in the presence of deuterated water. The pure culture of S. aciditrophicus was grown in 1 liter of medium with 20 mM crotonate. At the mid-log phase of growth (optical density at 600 nm of 0.2), cells of S. aciditrophicus were harvested by centrifugation at 14,300 × g for 20 min at 4°C and washed twice with 50 mM phosphate buffer (pH 7.0) by resuspending the pellet and centrifuging the cell suspension as described above. The final cell pellet was resuspended in 10 ml of anoxic, phosphate buffer (pH 7.0), and 5 ml was used as an inoculum. Duplicate washed cell suspensions (final volume of 50 ml) each contained 45 ml of deuterated water, 2.5 ml each of Pfennig I (50 mg/liter of K2HPO4), Pfennig II (33 mg/liter of MgCl2, 40 mg/liter of NaCl, 40 mg/liter of NH4Cl, and 5 mg/liter CaCl2), resazurin (10 mg/liter), cysteine-HCl (0.5 g/liter), Na2S (0.5 g/liter), 0.5 mM of 3-fluorobenzoate, and 10 mM crotonate. Twenty-five-milliliter samples were taken initially and after 2 days of incubation at 37°C for GC-MS analyses (36).
Analytical procedures.
Growth was monitored by measuring optical density at 600 nm. Acetate was analyzed with a GC apparatus equipped with a flame ionization detector and a glass column (2 m by 2 mm) packed with 80/120 Carbopack B-DA-4% Carbowax 20M. The carrier gas was nitrogen with a flow rate set at 24 ml/min. The isothermal column temperature was 155°C. The injector and detector temperatures were 200°C. Each sample contained 30 mM oxalic acid. Fluoride was analyzed by ion-exchange chromatography with a Dionex system (Dionex Corporation, Sunnyvale, CA) equipped with an AS-4A column (4-mm particle size) and bicarbonate buffer as the mobile phase at a flow rate of 2 ml/min. The concentrations of the other nongaseous products and the substrates were analyzed by high-performance liquid chromatography as described previously (36).
Hydrogen was quantified by using a gas chromatograph equipped with a mercury vapor detector (RGA3 reduction gas analyzer; Trace Analytical, Menlo Park, CA). Methane was measured by using a gas chromatograph with a flame ionization detector equipped with a Poropak Q 80/100 column (6 feet by 1/8 in.) (Supelco, Bellefonte, PA). The injector temperature was set at 100°C, the column was set at 100°C, and the detector was set at 125°C. Helium was used as a carrier gas.
Samples were derivatized and analyzed by GC-MS as described previously (9, 36). Each sample (cells and culture fluid) was adjusted to a pH of 12 with 1 N NaOH to hydrolyze possible thioester bonds, then acidified to a pH of <2 with 12 N HCl, extracted with ethyl acetate three times, and filtered through anhydrous sodium sulfate to remove water. The sample was then concentrated under vacuum at 47°C to a volume of 1 to 2 ml. The sample was split into two subsamples at this time if NMR analysis was to be done, and the samples and subsamples were evaporated to dryness under a stream of nitrogen gas. For GC-MS analysis, the sample was resuspended in 200 μl ethyl acetate and derivatized with 100 μl of N,O-bis-(trimethylsilyl)trifluoroacetamide (BSTFA). The subsample for NMR analysis was resuspended in 1 ml of deuterated chloroform. 19Fluorine NMR was carried out by using a Unity Inova 400-MHz spectrometer at a frequency of 376.110 MHz. A standard sample of 0.05% α,α,α-trifluorotoluene was used to determine the 90° pulse width and for chemical shift calibration. Spectra were collected at 23.5°C using a single pulse experiment with a 50° pulse width (14.5 μs), a delay time of 30 s, an acquisition time of 0.600 s, a spectral width of 50,000.0, and 400 acquisitions.
RESULTS
Transformation of aromatic compounds by S. aciditrophicus.
Previously, we found that the growth rate of crotonate-grown pure cultures of S. aciditrophicus decreased when benzoate was used as an electron acceptor (32). We used this approach to test whether other aromatic compounds could be metabolized by pure cultures of S. aciditrophicus grown with crotonate (Table 1). The growth of S. aciditrophicus in pure culture with crotonate was not affected by the addition of the following aromatic compounds: o-phthalate, 4-fluorobenzoate, 4-aminobenzoate, 2-chlorobenzoate, 3-chlorobenzoate, 4-chlorobenzoate, 2-hydroxybenzoate, 3-hydroxybenzoate, 4-hydroxybenzoate, and o-toluate (Table 1). In each case, S. aciditrophicus reached a maximum absorbance at 600 nm between 0.22 and 0.26 units within 10 days of incubation. However, when 2-fluorobenzoate or 3-fluorobenzoate was present, the growth of S. aciditrophicus with crotonate was inhibited (Table 1). Substrate depletion was observed after 16 days of incubation in cultures grown with 2-fluorobenzoate, 3-fluorobenzoate, or 2-hydroxybenzoate (see below), but not with any of the other aromatic compounds tested (data not show).
TABLE 1.
Growth of S. aciditrophicus with 10 mM crotonate and different cosubstrates
| Cosubstrate | Maximum absorbance reached after 10 days of incubationa |
|---|---|
| None | 0.26 ± 0.02 |
| Benzoate | 0.10 ± 0.01 |
| o-Phthalate | 0.24 ± 0.02 |
| 2-Fluorobenzoate | 0.04 ± 0.01 |
| 3-Fluorobenzoate | 0.04 ± 0.01 |
| 4-Fluorobenzoate | 0.22 ± 0.02 |
| p-Aminobenzoate | 0.24 ± 0.01 |
| 2-Chlorobenzoate | 0.25 ± 0.02 |
| 3-Chlorobenzoate | 0.20 ± 0.01 |
| 4-Chlorobenzoate | 0.24 ± 0.02 |
| 2-Hydroxybenzoate | 0.23 ± 0.01 |
| 3-Hydroxybenzoate | 0.25 ± 0.01 |
| 4-Hydroxybenzoate | 0.26 ± 0.01 |
| o-Toluate | 0.26 ± 0.01 |
Data are means ± standard deviations of triplicate cultures.
Metabolism of 2-hydroxybenzoate.
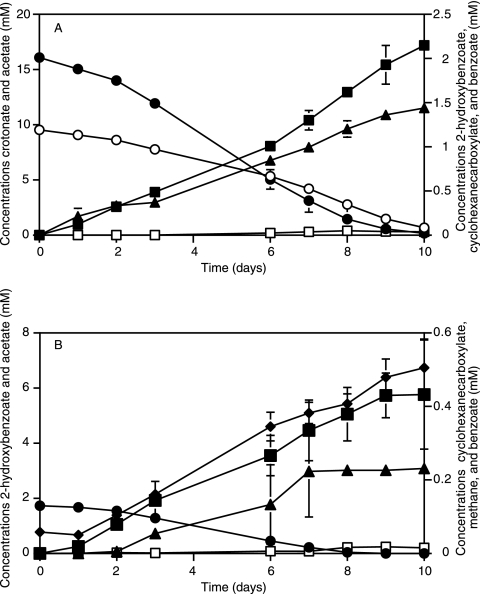
Pure cultures of S. aciditrophicus degraded crotonate and 2-hydroxybenzoate simultaneously (Fig. 2). After 10 days of incubation, 8.9 ± 0.1 mM crotonate and 2.0 ± 0.1 mM 2-hydroxybenzoate were degraded and 17.2 ± 0.5 mM acetate and 1.4 ± 0.1 mM cyclohexane carboxylate were formed. Benzoate transiently accumulated to a concentration of 0.05 ± 0.02 mM. The carbon recovery was 90%, and the hydrogen recovery was 88%. No degradation of the two substrates occurred in the heat-killed controls.
FIG. 2.
Metabolism of 2-hydroxybenzoate by S. aciditrophicus in pure culture with crototate as the cosubstrate (A) and in coculture with M. hungatei (B). The data are averages ± standard deviations (error bars) of triplicate cultures. Symbols: •, 2-hydroxybenzoate; ○, crotonate; ▴, cyclohexane carboxylate; ▪, acetate; □, benzoate; ⧫, methane.
The above stoichiometry is consistent with a coupled process where crotonate acts as the electron donor and 2-hydroxybenzoate acts as the electron acceptor (equation 2) (35):
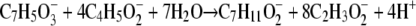 |
(2) |
(ΔG°′ = −183.9 kJ per mol of 2-hydrobenzoate) [47]).
The ratio of cyclohexane carboxylate to 2-hydroxybenzoate was 0.7, which indicates that some of the benzoate made from 2-hydroxybenzoate was further metabolized rather than being reduced to cyclohexane carboxylate.
Acetate, benzoate, and cyclohexane carboxylate were the only metabolites detected during crotonate and 2-hydroxybenzoate degradation. Peaks indicating the presence of phenol or 2-hydroxycyclohexane carboxylate were not detected by GC-MS analysis. In the absence of crotonate, no degradation of 2-hydroxybenzoate by pure cultures of S. aciditrophicus was observed during the 3-week incubation period (data not shown).
S. aciditrophicus and M. hungatei cocultures formed methane and acetate from 2-hydroxybenzoate (Fig. 2). After 10 days of incubation, 1.7 ± 0.1 mM 2-hydroxybenzoate was consumed and 5.8 ± 2 mM acetate and 0.5 ± 0.1 mmol liter−1 methane accumulated. Benzoate and cyclohexane carboxylate transiently accumulated up to 0.02 ± 0.01 mM. The carbon recovery was 101%, and the hydrogen recovery was 106%.
3-Hydroxybenzoate and 4-hydroxybenzoate were not degraded by S. aciditrophicus in pure culture with or without the presence of crotonate or by cocultures of S. aciditrophicus with M. hungatei (data not shown).
Reductive defluorination of 3-fluorobenzoate.
The addition of 0.5 mM 2-fluorobenzoate or 3-fluorobenzoate inhibited the growth of pure cultures of S. aciditrophicus in the presence of crotonate (Table 2). After 10 days of incubation, pure cultures of S. aciditrophicus metabolized about 26% of the added 3-fluorobenzoate (0.13 ± 0.02 mM) and formed 0.15 ± 0.02 mM fluoride and 0.17 ± 0.01 mM benzoate, indicating that dehalogenation of 3-fluorobenzoate occurred. Some dehalogenation of 2-fluorobenzoate was also observed. A loss of 0.06 ± 0.03 mM 2-fluorobenzoate and the formation of 0.06 ± 0.01 mM fluoride and 0.04 ± 0.01 mM benzoate was observed. The addition of 4-fluorobenzoate had no effect on growth or crotonate metabolism, and little, if any, 4-fluorobenzoate degradation was observed after 17 days of incubation (Table 2). The small amount of benzoate made in cultures with 4-fluorobenzoate could be due to crotonate metabolism (36).
TABLE 2.
Metabolism of benzoate and fluorobenzoate isomers by pure cultures of S. aciditrophicus grown with crotonatea
| Substrate(s) | Maximum OD600b | Crotonate used (mM) | Cosubstrate used (mM) | Acetate formed (mM) | CHAC formed (mM) | Fluoride formed (mM) | Benzoate formed (mM) | Final H2 (Pa) |
|---|---|---|---|---|---|---|---|---|
| Crotonate | 0.24 ± 0.01 | 7.1 ± 0.2 | NA | 10.4 ± 2.1 | 1.16 ± 0.08 | NA | 0.04 ± 0.01 | 9.4 ± 1.2 |
| Crotonate + benzoate | 0.25 ± 0.01 | 7.7 ± 0.4 | 1.79 ± 0.12 | 15.1 ± 0.9 | 1.67 ± 0.26 | NA | NA | 8.9 ± 2.3 |
| Crotonate + 2-fluorobenzoate | 0.09 ± 0.01 | 2.2 ± 0.5 | 0.06 ± 0.03 | 2.2 ± 2.0 | 0.17 ± 0.01 | 0.06 ± 0.01 | 0.04 ± 0.01 | 87.4 ± 7.8 |
| Crotonate + 3-fluorobenzoate | 0.08 ± 0.01 | 3.7 ± 0.6 | 0.13 ± 0.02 | 5.7 ± 0.3 | 0.53 ± 0.14 | 0.15 ± 0.02 | 0.17 ± 0.01 | 116.6 ± 13.9 |
| Crotonate + 4-fluorobenzoate | 0.25 ± 0.01 | 6.7 ± 1.0 | 0.01 ± 0.02 | 9.9 ± 1.8 | 1.3 ± 0.20 | 0.005 ± 0.14 | 0.04 ± 0.01 | 7.8 ± 0.8 |
Abbreviations: OD600, optical density at 600 nm; CHAC, cyclohexane carboxylate; NA, not applicable. The values are the averages ± standard deviations of triplicate cultures measured after 10 days of incubation.
The maximum absorbance was reached within 6 days for the cultures grown in the presence of crotonate alone or crotonate and 4-fluorobenzoate; it was reached within 10 days for the cultures grown with crotonate and benzoate, 2-fluorobenzoate, or 3-fluorobenzoate.
The final partial pressure of hydrogen was 87.4 ± 7.8 Pa and 116.6 ± 13.9 Pa when 2- or 3-fluorobenzoate was present, respectively, compared to final partial pressures of 7.8 ± 0.8 Pa, 9.4 ± 1.2 Pa, and 8.9 ± 2.3 Pa when crotonate and 4-fluorobenzoate, crotonate alone, and crotonate and benzoate, respectively, were used as substrates (Table 2).
Detection of a diene intermediate during fluorobenzoate metabolism.
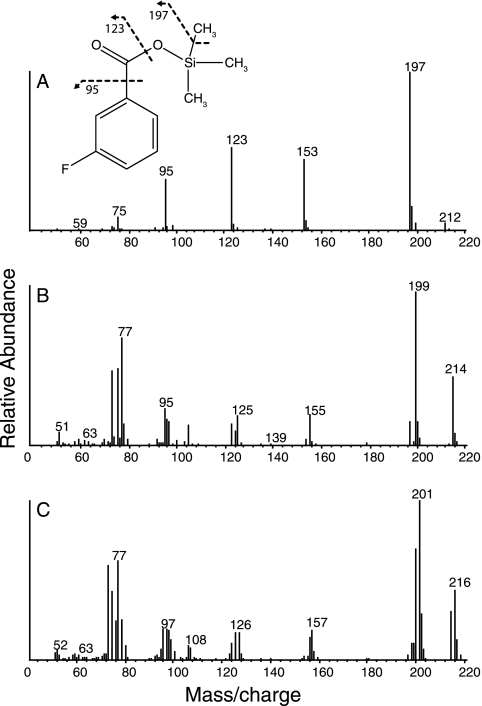
Because 2-fluorobenzoate and 3-fluorobenzoate affected the growth and metabolism of S. aciditrophicus, pure cultures of S. aciditrophicus were grown in large volumes with crotonate and these inhibitors to detect metabolites. Initially, peaks with the same mass spectrum profiles and retention times as the TMS derivatives of crotonate and 2-, 3-, or 4-fluorobenzoate, cyclohexane carboxylate, benzoate, cyclohex-1-ene-1-carboxylate, glutarate, and pimelate, were detected. The presence of compounds other than crotonate and the fluorinated benzoates was due to their presence in the inoculum of S. aciditrophicus grown with crotonate. After 12 and 26 days of incubation, no additional peaks were detected by GC-MS in cultures with either 2-fluorobenzoate or 4-fluorobenzoate. However, in cultures with 3-fluorobenzoate and crotonate, a new peak with a retention time of 25.07 min with major m/z ion fragments of 77, 95, 105, 125, 155, 199 (M −15), and 214 (total mass ion) mass units (Fig. 3B) was observed after 12 days of incubation. After 26 days, the area of the metabolite peak decreased as did the concentration of 3-fluorobenzoate. The relative amounts of 3-fluorobenzoate and the new metabolite in the 90-day sample were 56% to 44% based on the peak area of 19F NMR spectra. The GC retention time of the TMS derivative of 3-fluorobenzoate was 22.8 min, and the masses of its m/z ion fragments were 75, 95, 123, 153, 197 (M −15), and 212 (total mass ion) (Fig. 3A). The fragmentation pattern of the metabolite was almost identical to that of the 3-fluorobenzoate TMS derivative, but with a mass increase of 2 units. This is consistent with the addition of two hydrogen atoms to 3-fluorobenzoate, probably forming a 3-fluorocyclohexadiene-1-carboxyl TMS derivative. However, no chemical standard is available for this compound.
FIG. 3.
Mass spectrum of the TMS derivative of 3-fluorobenzoate (A), the metabolite with a retention time of 25.07 min detected in cultures grown with 3-fluorobenzoate and crotonate (B), and the metabolite detected in washed cell suspensions incubated in the presence of deuterated water with 3-fluorobenzoate and crotonate (C).
To confirm the formation of a diene intermediate, samples from the cultures grown with 3-fluorobenzoate and crotonate for 90 days were analyzed by 19F NMR. Initially, the 19F NMR spectrum contained a signal with six peaks centered at −113.97 ppm and displaying coupling constants of 3JF-H = 8.8 Hz (t) and 4JF-H = 5.1 Hz (d) corresponding to 3-fluorobenzoate. The 19F NMR spectrum contained two signals, one from the 3-fluorobenzoate and a second signal with nine peaks of a metabolite centered at −100.7 ppm. The chemical shift value demonstrates that the fluorine atom is bound to an sp2 carbon. The coupling pattern results from the overlap of a doublet and two triplets corresponding to 3JF-H = 13.1 Hz (d), 3JF-H = 3.7 Hz (t), and 4JF-H = 2.9 Hz (t). A doublet indicates that the fluorine atom is coupled to a single proton on an sp2 or sp3 carbon, and a triplet results from coupling between the fluorine nucleus and the two protons on an sp3 carbon; the greater the distance between the fluorine and the coupled proton, the smaller the coupling constant. Based on the coupling pattern, the fluorine atom is vicinal to only one proton and separated by three to four bonds from two sp3 carbon atoms, each containing two protons. Of the six possible molecular structures with fluorine bonded to an sp2 carbon, the only two structures that fit the data are 1-carboxyl-3-fluoro-3,6-cyclohexadiene or 1-carboxyl-3-fluoro-2,6-cyclohexadiene. The latter has the double bond arrangement found in cyclohexa-1,5-diene-1-carboxyl-CoA.
Further evidence for the formation of a diene intermediate was obtained during the metabolism of crotonate and 3-fluorobenzoate by washed cell suspensions of S. aciditrophicus in the presence of deuterated water. Initially, two peaks were observed by GC-MS, one corresponding to the TMS derivative of crotonate and the second one corresponding to the TMS derivative of 3-fluorobenzoate. After 2 days of incubation, several new peaks were detected by GC-MS with the same retention times and mass spectrum profiles as the TMS derivatives of metabolites of crotonate and benzoate metabolism, including pimelate, glutarate, cyclohexane carboxylate, cyclohex-1-ene carboxylate, 3-hydroxybutyrate, and benzoate, with benzoate being the major peak. One additional peak was present with a retention time of 25.07 min, the same as the TMS derivative of the putative diene metabolite. The major m/z ion fragments were 77, 96, 106, 126, 156, 200, and 201 (M −15), and 215 and 216 (total mass ion) mass units (Fig. 3C). This corresponds to an increase of 3 or 4 mass units compared to the TMS derivative of 3-fluorobenzoate, consistent with the addition of one or two deuterium atoms.
DISCUSSION
S. aciditrophicus can syntrophically degrade benzoate in coculture with H2-using microorganisms (21) and ferment benzoate to acetate and cyclohexane carboxylate (10) and reduce benzoate to cyclohexane carboxylate with crotonate as the electron donor in pure culture (35). The pathway for benzoyl-CoA metabolism by S. aciditrophicus is not completely understood. The formation of cyclohex-1-ene-1-carboxylate and cyclohexane carboxylate from benzoate could be the result of a four- or six-electron reduction reaction. However, the presence of a cyclohexa-1,5-diene-1-carboxyl-CoA hydratase in S. aciditrophicus argues that cyclohexa-1,5-diene-1-carboxyl-CoA is the product of benzoyl-CoA reduction and that S. aciditrophicus uses a two-electron reduction mechanism to reduce benzoyl-CoA (41). Here, we showed that a diene metabolite is formed by S. aciditrophicus when it grows in the presence of 3-fluorobenzoate. The detection of this intermediate shows that S. aciditrophicus contains a dearomatizing reductase that reduces the aromatic ring by adding two electrons. The genome of S. aciditrophicus (32) contains two sets of genes homologous to the putative benzoyl-CoA reductase genes in G. metallireducens (48), either of which could act as the benzoyl-CoA reductase. However, caution should be exercised in extrapolating this result to the mechanism of benzoyl-CoA metabolism, as the enzyme involved in 3-fluorobenzoate reduction may not be the same one as that used for benzoyl-CoA reduction. The formation of cyclohex-1-ene-1-carboxylate and cyclohexane carboxylate during syntrophic benzoate metabolism (9) may indicate diverging pathways once cyclohexa-1,5-diene-1-carboxyl-CoA is formed. Some of the cyclohexa-1,5-diene-1-carboxyl-CoA could be oxidized to acetate and CO2, while the remainder is reduced to cyclohexane carboxylate (10). When crotonate is present, the latter pathway may predominate.
It appears that S. aciditrophicus metabolizes 2-hydroxybenzoate by first reductively eliminating the hydroxyl moiety prior to ring reduction. Data in support of this mechanism are the transient accumulation of small amounts of benzoate during the reduction of 2-hydroxybenzoate (Fig. 2) and a ratio of crotonate oxidized to 2-hydroxybenzoate reduced of 4.5. This ratio would be close to 3 if a reductive dehydroxylation step were not involved (35). Since S. aciditrophicus is able to form small amounts of benzoate during crotonate fermentation, we cannot exclude the possibility that some of the benzoate was derived from crotonate. However, we previously found that cyclohexane carboxylate and benzoate formation from crotonate occurred after benzoate reduction was complete (35). 2-Hydroxybenzoate is probably activated to its CoA thioester prior to reductive dehydroxylation as shown for the degradation of other hydroxybenzoate isomers (16, 37). Bonting and Fuchs (6) showed that Pseudomonas sp. strain S100 metabolized 2-hydroxybenzoate under denitrifying conditions in two steps. First, 2-hydroxybenzoate was activated to 2-hydroxybenzoyl-CoA by a CoA ligase, followed by reductive dehydroxylation of 2-hydroxybenzoyl-CoA to form benzoyl-CoA.
We previously found that the growth rate of S. aciditrophicus in crotonate and benzoate was lower than the growth rate of S. aciditrophicus in crotonate alone, although the final cell densities were similar for the two growth conditions (36). The low cell density of cultures grown in crotonate and benzoate compared to the culture grown in crotonate recorded in Table 1 was probably because the culture grown in crotonate and benzoate had an unexpectedly long lag period. The metabolism of fluorinated benzoates by S. aciditrophicus was associated with high partial pressures of hydrogen (87 to 116 Pa) (Table 2). The partial pressure of hydrogen was much lower (about 8 Pa) when S. aciditrophicus was grown with crotonate and 4-fluorobenzoate; the latter was not metabolized by S. aciditrophicus (Table 2). These data show that the metabolism of 2- and 3-fluorobenzoatae affected the physiology of S. aciditrophicus, leading to an accumulation of hydrogen. The high partial pressure of hydrogen might explain why benzoate degradation was not observed in cultures containing 3-fluorobenzoate (and 2-fluorobenzoate) because high levels of hydrogen have been shown to inhibit benzoate degradation (10). The inhibitory activity of fluorine or the fluorinated diene could also be reasons why benzoate was not metabolized by these cultures.
Acknowledgments
We thank N. Q. Wofford for technical assistance and artwork.
We gratefully acknowledge the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (contract DE-FG02-96ER20214) for financial support of this work.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boll, M., S. S. Albracht, and G. Fuchs. 1997. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. A study of adenosine triphosphate activity, ATP stoichiometry of the reaction and EPR properties of the enzyme. Eur. J. Biochem. 244:840-851. [DOI] [PubMed] [Google Scholar]
- 3.Boll, M., and G. Fuchs. 2002. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Chem. Biol. 6:604-611. [DOI] [PubMed] [Google Scholar]
- 4.Boll, M., and G. Fuchs. 1995. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 234:921-933. [DOI] [PubMed] [Google Scholar]
- 5.Boll, M., D. Laempe, W. Eisenreich, A. Bacher, T. Mittelberger, J. Heinze, and G. Fuchs. 2000. Nonaromatic products from anoxic conversion of benzoyl-CoA with benzoyl-CoA reductase and cyclohexa-1,5-diene-1-carbonyl-CoA hydratase. J. Biol. Chem. 275:21889-21895. [DOI] [PubMed] [Google Scholar]
- 6.Bonting, C. F., and G. Fuchs. 1996. Anaerobic metabolism of 2-hydroxybenzoic acid (salicylic acid) by a denitrifying bacterium. Arch. Microbiol. 165:402-408. [DOI] [PubMed] [Google Scholar]
- 7.Diaz, F. 2004. Bacterial degradation of aromatic pollutants: a paradigm of metabolic versatility. Microbiology 7:173-180. [PubMed] [Google Scholar]
- 8.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 94:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elshahed, M. S., V. K. Bhupathiraju, N. Q. Wofford, M. A. Nanny, and M. J. McInerney. 2001. Metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by Syntrophus aciditrophicus strain SB in syntrophic association with H2-using microorganisms. Appl. Environ. Microbiol. 67:1728-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elshahed, M. S., and M. J. McInerney. 2001. Benzoate fermentation by the anaerobic bacterium Syntrophus aciditrophicus in the absence of hydrogen-using microorganisms. Appl. Environ. Microbiol. 67:5520-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engesser, K. H., G. Auling, J. Busse, and H. J. Knackmuss. 1990. Fluorobenzoate enriched bacterial strain FLB 300 degrades benzoate and all three isomeric monofluorobenzoates. Arch. Microbiol. 153:193-199. [Google Scholar]
- 12.Evans, W. C. 1977. Biochemistry of the bacterial catabolism of aromatic compounds in anaerobic environments. Nature 270:17-22. [DOI] [PubMed] [Google Scholar]
- 13.Evans, W. C., and G. Fuchs. 1988. Anaerobic degradation of aromatic compounds. Annu. Rev. Microbiol. 42:289-317. [DOI] [PubMed] [Google Scholar]
- 14.Ferry, J. G., and R. S. Wolfe. 1976. Anaerobic degradation of benzoate to methane by a microbial consortium. Arch. Microbiol. 107:33-40. [DOI] [PubMed] [Google Scholar]
- 15.Genthner, B. R., G. T. Townsend, and P. J. Chapman. 1989. Anaerobic transformation of phenol to benzoate via para-carboxylation: use of fluorinated analogues to elucidate the mechanism of transformation. Biochem. Biophys. Res. Commun. 162:945-951. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, J., M. Dispensa, and C. S. Harwood. 1997. 4-Hydroxybenzoyl-coenzyme A reductase (dehydroxylating) is required for anaerobic degradation of 4-hydroxybenzoate by Rhodopseudomonas palustris and shares features with molybdenum-containing hydroxylases. J. Bacteriol. 179:634-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, K. J., and J. J. Gibson. 1992. Potential early intermediates in anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl. Environ. Microbiol. 58:696-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harwood, C. S., G. Burchhardt, H. Herrmann, and G. Fuchs. 1999. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439-458. [Google Scholar]
- 20.Heider, J., and G. Fuchs. 1997. Anaerobic metabolism of aromatic compounds. Eur. J. Biochem. 243:577-596. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, B. E., V. K. Bhupathiraju, R. S. Tanner, C. R. Woese, and M. J. McInerney. 1999. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch. Microbiol. 171:107-114. [DOI] [PubMed] [Google Scholar]
- 22.Jackson, B. E., and M. J. McInerney. 2002. Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415:454-456. [DOI] [PubMed] [Google Scholar]
- 23.Key, B. D., and R. D. Howell. 1997. Fluorinated organics in the biosphere. Environ. Sci. Technol. 31:2445-2454. [Google Scholar]
- 24.Kluge, C., A. Tschech, and G. Fuchs. 1990. Anaerobic metabolism of resorcyclic acids (m-dihydroxybenzoic acids) and resorcinol (1,3-benzendiol) in a fermenting and in a denitrifying bacterium. Arch. Microbiol. 155:68-74. [Google Scholar]
- 25.Krumholz, L. R., M. E. Caldwell, and J. M. Suflita. 1996. Biodegradation of “BETX” hydrocarbons under anaerobic conditions, p. 61-99. In R. L. Crawford and D. L. Crawford (ed.), Bioremediation: principles and applications. Cambridge University Press, Cambridge, United Kingdom.
- 26.Kuntze, K., Y. Shinoda, H. Moutakki, M. J. McInerney, C. Vogt, H.-H. Richnow, and M. Boll. 2008. 6-Oxocyclohex-1-ene-1-carbonyl-coenzyme A hydrolases from obligately anaerobic bacteria: characterization and identification of its gene as a functional marker for aromatic compounds degrading anaerobes. Environ. Microbiol. 10:1547-1556. [DOI] [PubMed] [Google Scholar]
- 27.Laempe, D., W. Eisenreich, A. Bacher, and G. Fuchs. 1998. Cyclohexa-1, 5-diene-1-carboxyl-CoA hydratase, an enzyme involved in anaerobic metabolism of benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 255:618-627. [DOI] [PubMed] [Google Scholar]
- 28.Laempe, D., M. Jahn, and G. Fuchs. 1999. 6-Hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase and 6-oxocyclohex-1-ene-1-carboxyl-CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 262:420-429. [DOI] [PubMed] [Google Scholar]
- 29.Londry, K. L., and P. M. Fedorak. 1993. Fluorophenols and 3-fluorobenzoate in phenol-degrading methanogenic cultures. Arch. Microbiol. 160:137-143. [Google Scholar]
- 30.López Barragán, M. J., M. Carmona, M. T. Zamarro, B. Thiele, M. Boll, G. Fuchs, J. L. García, and E. Díaz. 2004. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 186:5762-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López Barragán, M. J., E. Díaz, J. L. García, and M. Carmona. 2004. Genetic clues on the evolution of anaerobic catabolism of aromatic compounds. Microbiology 150:2018-2021. [DOI] [PubMed] [Google Scholar]
- 32.McInerney, M. J., L. Rohlin, H. Mouttaki, U. Kim, R. S. Krupp, L. Rios-Hernandez, J. Sieber, C. G. Struchtemeyer, A. Bhattacharyya, J. W. Campbell, and R. P. Gunsalus. 2007. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc. Natl. Acad. Sci. USA 104:7600-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merkel, S. M., A. E. Eberhard, J. Gibson, and C. S. Harwood. 1989. Involvement of coenzyme A thioesters in anaerobic metabolism of 4-hydroxybenzoate by Rhodopseudomonas palustris. J. Bacteriol. 171:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mountfort, D. O., W. J. Brulla, L. R. Krumholz, and M. P. Bryant. 1984. Syntrophus buswellii gen. nov., sp. nov.: a benzoate catabolizer from methanogenic ecosystems. Int. J. Syst. Bacteriol. 34:216-217. [Google Scholar]
- 35.Mouttaki, H., M. J. McInerney, and M. A. Nanny. 2008. Use of benzoate as an electron acceptor for growth of Syntrophus aciditrophicus grown in pure culture with crotonate. Environ. Microbiol. 10:3265-3274. [DOI] [PubMed] [Google Scholar]
- 36.Mouttaki, H., M. A. Nanny, and M. J. McInerney. 2007. Cyclohexane carboxylate and benzoate formation from crotonate in Syntrophus aciditrophicus. Appl. Environ. Microbiol. 73:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller, J. A., and B. Schink. 2000. Initial steps in the fermentation of 3-hydroxybenzoate by Sporotomaculum hydrobenzoicum. Arch. Microbiol. 173:288-295. [DOI] [PubMed] [Google Scholar]
- 38.Pelletier, D. A., and C. S. Harwood. 2000. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J. Bacteriol. 182:2753-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelletier, D. A., and C. S. Harwood. 1998. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 180:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters, F., M. Rother, and M. Boll. 2004. Selenocysteine-containing proteins in anaerobic benzoate metabolism in Desulfococcus multivorans. J. Bacteriol. 186:2156-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters, F., Y. Shinoda, M. J. McInerney, and M. Boll. 2007. Cyclohexa-1,5-diene-1-carbonyl-coenzyme A hydratases of Geobacter metallireducens and Syntrophus aciditrophicus: evidence for a common benzoyl-CoA pathway in facultative and obligate anaerobes. J. Bacteriol. 189:1055-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichenbecher, W., and B. Philipp. 2000. Hydroxyhydroquione reductase, the initial enzyme involved in the degradation of hydroxyhydroquinone (1,2,4-trihydroxybenene) by Desulfovibrio inopinatus. Arch. Microbiol. 173:206-212. [DOI] [PubMed] [Google Scholar]
- 43.Schink, B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schink, B., B. Philipp, and J. Müller. 2000. Anaerobic degradation of phenolic compounds. Naturwissenschaften 87:12-23. [DOI] [PubMed] [Google Scholar]
- 45.Schöcke, L., and B. Schink. 1999. Energetics and biochemistry of fermentative benzoate degradation by Syntrophus gentianae. Arch. Microbiol. 171:331-337. [DOI] [PubMed] [Google Scholar]
- 46.Szewzyk, U., and B. Schink. 1989. Degradation of hydroquinone, gentisate, and benzoate by a fermenting bacterium in pure or defined mixed culture. Arch. Microbiol. 151:541-545. [Google Scholar]
- 47.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wischgoll, S., D. Heintz, F. Peters, A. Erxleben, E. Sarnighausen, R. Reski, A. Van Dorsselaer, and M. Boll. 2005. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 58:1238-1252. [DOI] [PubMed] [Google Scholar]