Abstract
Lentiviruses are causal agents of severe pathologies of a variety of mammals, including cattle and humans (e.g., AIDS and different types of lymphoma). While endogenous forms of lentivirus do not occur in these species, A. Katzourakis and coworkers (A. Katzourakis, M. Tristem, O. G. Pybus, and R. J. Gifford, Proc. Natl. Acad. Sci. USA 104:6261-6265, 2007) recently reported the presence in the genome of the European rabbit (Oryctolagus cuniculus) of multiple sequences defining a lentiviral subgroup elegantly referred to as RELIK (rabbit endogenous lentivirus type K). Sequence comparisons indicated that the RELIK ancestor may have integrated into the rabbit lineage more than 7 million years ago. We have substantiated this by producing sequence data certifying the sharing of RELIK sequences among leporid lineages that diverged some 12 million years ago.
Lentiviruses are remotely related to known endogenous retroviruses (e.g., of the alpha- and betaretrovirus groups). Five subgroups of lentivirus have been described, each being restricted to a single mammalian family (1, 5, 14). Most intensively studied are the more recently discovered human immunodeficiency viruses. The origin of this virus group, however, remains obscure (11, 12), mainly because of the absence of endogenous forms, i.e., of virus that has integrated into the host genome. Compared to their exogenous counterparts, such germ line-imbedded copies are known to maintain much better their original sequence patterns and therefore provide valuable markers in evolutionary studies (3, 4). The discovery by Katzourakis and coworkers (6) of what appears to be the first reported endogenous lentivirus in any species therefore constitutes a crucial step toward our understanding of the history of lentivirus and associated diseases.
Interestingly, it was found in the European rabbit (Oryctolagus cuniculus), a species without known lentiviral affections. It was achieved by data mining of the WGS (Whole Genome Shotgun) archives released by the rabbit genome project (genome project 12819). By measuring the genetic distances between segmentally duplicated regions, assuming a divergence rate of 4 × 10−9 nucleotide substitutions per site per year for the rabbit genome (7, 8), Katzourakis and coworkers dated the oldest intragenomic segmental duplication at 7 million years (My) and estimated at 11 My the maximal time depth of the RELIK evolutionary tree. The authors suggested that a more exact estimate of RELIK persistence times might be obtained by searching for the occurrence of related lentiviral sequences in other lagomorph species.
We have done this for several lagomorph genera by targeting the RELIK gag gene sequence, which is among the more conserved gene regions of lentivirus (6). The gag sequence of the RELIK consensus region was at first subjected to intensive BLAST research on WGS data banks, both in the trace file archives (http://www.ncbi.nlm.nih.gov/blast/mmtrace.shtml) and in the assembled WGS reads (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The most closely related nonrabbit sequences were parts of the gag genes of equine infective anemia virus and of puma feline lentivirus (d > 0.75). Except for Oryctolagus cuniculus, no vertebrate sequence even remotely similar to the query was found in any of the available databases, including that of Ochotona princeps (pikas, or whistling hares), the only other lagomorph species for which WGS sequences are available. The order Lagomorpha is indeed composed of two families, which diverged more than 35 My ago (7, 8): Ochotonidae, with the genus Ochotona (pikas), and Leporidae (rabbits and hares), with 11 extant genera, Brachylagus, Caprolagus, Nesolagus, Pentalagus, Pronolagus, Poelagus, Romerolagus, Sylvilagus, Oryctolagus, Bunolagus, and Lepus (2, 7).
Genomic DNA from tissue samples of Orytcolagus cuniculus algirus (OCA), Bunolagus monticularis (BM), Lepus granatensis (LG), and Sylvilagus brasiliensis (SB) was prepared using a Qiagen extraction kit (Qiagen, Vienna, Austria). Taxonomic verification for DNA samples was carried out using mitochondrial and nuclear markers. (Genomic DNA of Ochotona princeps was kindly made available by Dennis Lanning [Loyola University Chicago]. The specimen of Oryctolagus was collected in the aboriginal range of the species [Mértola, SE Portugal] [7].) Amplification of a 0.7-kb fragment of the RELIK gag gene (below GagC) was performed using the primer pair Gag_1F (5′-GGACGTCCCAGTCAAAAGAA-3′) and Gag_1R (5′-AGGGTTCTGGCATCAGCAAA-3′), designed according to the RELIK consensus sequence described in reference 6. The PCR conditions and procedures were standard and are available upon request. PCR products with the expected fragment size were cloned into the pGEM-T Easy vector (Promega, Madison, WI). Sequencing of cloned and uncloned products was performed on an ABI PRISM 310 genetic analyzer. Sequence data were analyzed using the software packages MEGA4 (13, 15) and DnaSP4 (10).
PCR amplification proved to be successful with the four leporid species but not with pikas. The sequencing of the uncloned PCR products revealed sequence patterns undeniably characteristic of GagC although often obscured by multiple polymorphic positions and frameshift mutations due to insertions and deletions (data not shown). Sequencing of cloned products was limited to two clones per individual. For Sylvilagus (SB), only one sequence was obtained, while the two sequences of one Portuguese wild rabbit (OCA) turned out to be identical, although the electrochromatographs of the crude PCR product revealed the presence of a variety of different sequences.
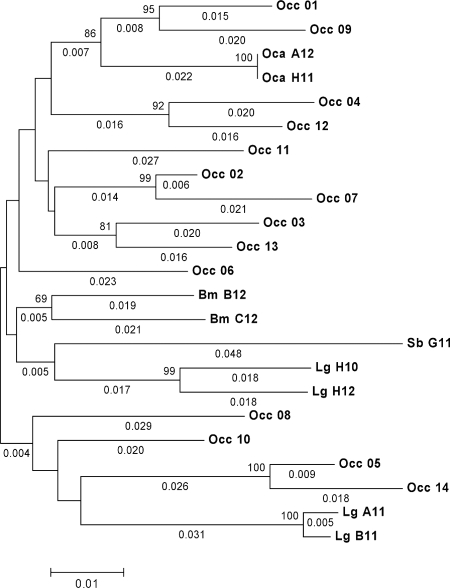
For each species, the cloned sequences showed more than 90% similarity with GagC. These sequences were grouped per species (OCA, BM, SB, and LG; see above) and compared to the 14 WGS assemblies of the archive of Oryctolagus c. cuniculus (Occ01 to Occ14, forming the species group OCC). These assemblies were selected for their complete coverage of the RELIK gag region under study. The distance tree in Fig. 1 visualizes the common history of RELIK-like sequences among the Oryctolagus, Bunolagus, Lepus, and Sylvilagus species. We note that, independently of the tree-building method used (including maximum likelihood) and analysis of synonymous and nonsynonymous distance trees (data not shown), there were no indications of separate clustering according to species origin (i.e., although the two BM sequences form one cluster, they are embedded within the OCC clade). This confirms that the initial genome insertion of the RELIK precursor must have occurred in a common ancestor of these species. Because ratios of nonsynonymous to synonymous evolutionary changes were consistently and significantly smaller than 1 (0.55 to 0.36) (6), we have estimated both synonymous and nonsynonymous divergence times. The pairwise distances among the rabbit WGS sequences were found to differ by almost an order of magnitude, implying that different pairs do not have the same history. In Table 1, we therefore show the range of distance values between the sequence pairs rather than their averages.
FIG. 1.
Evolutionary-distance tree of homologous gag gene fragments of various leporid species. The neighbor-joining tree was obtained using the p distance method provided by the MEGA4 program (13). p distances are obtained by dividing the number of nucleotide differences by the total number of nucleotides compared. All positions containing gaps and missing data were eliminated from the data set (by using the complete deletion option [592 bp]). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown above the branches for percentages above 60. The tree is drawn to scale, with branch lengths shown in the same units as those of the evolutionary distances used to infer the distance tree. Values larger than 0.003 are shown below the branches. Sequences were aligned on the PCR target GagC, ignoring primer-imposed parts and frame-disrupting inserts (658 bp). The sequences were named according to species origin, as follows: Occ, Oryctolagus cuniculus cuniculus; Oca, Oryctolagus c. algirus; Bm, Bunolagus monticularis; Lg, Lepus granatensis; and Sb, Sylvilagus brasiliensis. Occ_01 to Occ_14 are derived from assembled WGS contigs. Their GenBank accession numbers are AAGW01007437, AAGW01043000, AAGW01086182, AAGW01127774, AAGW01133803, AAGW01183526, AAGW01222583, AAGW01228233, AAGW01228525, AAGW01252278, AAGW01327267, AAGW01433545, AAGW01516016, and AAGW01717649, respectively. The remaining sequences are those of cloned PCR products. Their GenBank accession numbers are shown in the text.
TABLE 1.
Ranges of synonymous nucleotide distances between pairs of RELIK elements within and between species groupsa
| Species group (no. of sequences) | Range of nucleotide distances for species group
|
|||
|---|---|---|---|---|
| OCC (WGS) | OCA | BM | LG | |
| OCC (14) | 0.023 ± 0.011-0.130 ± 0.024 | |||
| OCA (2) | 0.022 ± 0.010-0.088 ± 0.020 | NA | ||
| BM (2) | 0.033 ± 0.013-0.113 ± 0.023 | 0.055 ± 0.016-0.091 ± 0.020 | NA | |
| LG (4) | 0.066 ± 0.018-0.141 ± 0.024 | 0.090 ± 0.018-0.116 ± 0.021 | 0.066 ± 0.018-0.099 ± 0.021 | 0.006 ± 0.005-0.131 ± 0.025 |
| SB (1) | 0.086 ± 0.020-0.141 ± 0.024 | 0.109 ± 0.022 | 0.084 ± 0.020-0.097 ± 0.020 | 0.096 ± 0.022-0.125 ± 0.024 |
The synonymous distance between two sequences is obtained by dividing the number of synonymous differences by the total number of synonymous sites. The maximum and minimum synonymous-distance values are shown for pairwise distances within groups and between groups. Standard errors (preceded by “±”) were obtained by a bootstrap procedure (1,000 replicates). Analyses were conducted using the modified Nei-Gojobori method in MEGA4 (13, 15). Positions containing gaps and missing data were eliminated only in pairwise sequence comparisons (by using the pairwise deletion option). There were a total of 211 positions in the final data set. Groups and sequences are identified in the text and in the legend to Fig. 1. NA, insufficient data.
This study was designed to test the primary prediction of the hypothesis of a 10-My or longer endogenous history of RELIK by answering the question of whether or not RELIK was present in lagomorph species other than Oryctolagus c. cuniculus. There are, however, more predictions to be satisfied, some of which appear to be supported by our limited data set. Indeed, pairs of sequences having duplicated in a common ancestor should have accumulated similar quantities of mutations within each of the descending lineages. We note that maximum values for pairwise distances within species tend indeed to be similar among species (i.e., 0.130 ± 0.024 within OCC and 0.131 ± 0.025 within LG) (Table 1). Assuming a neutral substitution rate of 4 × 10−9 for rabbits (6), these values suggest minimum dates of duplication of ∼13 My ago [i.e., (0.130 − 0.024)/2/0.004 = 13.25 My]. This is in tune with the estimated divergence times of Oryctolagus-Bunolagus versus Lepus-Sylvilagus lineages (12 My) (8). At the same time, we expect minimum interspecies distances to be proportional to the ages of the species splits. According to Matthee et al. (8), the split between the Oryctolagus and Bunolagus lineages was about 5 My posterior to that between Oryctolagus and Lepus (8). The minimum distances for OCC versus BM sequence pairs (∼0.033) (Table 1) are indeed significantly smaller than those observed for the OCC-versus-LG and the OCC-versus-SB pairwise comparisons (∼0.066 and ∼0.086, respectively). However, in view of the important within-species variation, as exemplified by the rabbit WGS data, our limited data set does not allow strong conclusions to be drawn.
The failure of amplification of RELIK-gag from Ochotona is in accordance with divergence times estimated by Katzourakis et al. (8), which imply that the RELIK insertion into the leporid ancestor must be largely posterior to the Ochotona-Leporidae split (35 My or 40 to 50 My ago, according to molecular [8] or fossil [9] data, respectively). The presumed absence of RELIK-related sequences in pikas was furthermore supported by the fact that intensive screening of the WGS trace archives representing a twofold coverage of the genome of Ochotona princeps (project 19235) did not reveal a single sequence remotely similar to RELIK. Equally negative results were obtained by BLAST searching the WGS archives for horses, cats, or pikas with the entire RELIK sequence (8.5 kb) rather than with GagC (0.7 kb).
In conclusion, the present results provide factual evidence that, as predicted by the phylogenetic inference methods of Katzourakis et al., RELIK was already present in a common ancestor of the Lepus, Sylvilagus, and Oryctolagus and Bunolagus lineages. It opens the door to more in-depth phylogenetic studies of the ancient history of this important viral group.
Nucleotide sequence accession numbers.
The nucleotide sequences of the clones obtained in this study have been submitted to GenBank and were assigned the following accession numbers: for Oca_H11, FJ531485; for Oca_A12, FJ531486; for Bm_B12, FJ531487; for Bm_C12, FJ531488; for Lg_B11, FJ531489; for Lg_A11, FJ531490; for Lg_H10, FJ531491; for Lg_H12, FJ531492; and for Sb_G11, FJ531493.
Acknowledgments
This work was supported by the Foundation for Science and Technology Portugal (project POCTI/BIA-BDE/61553/2004) and grants SFRH/BPD/27021/2006 and SFRH/BD/31048/2006 to P.J.E. and J.A., respectively.
We thank Conrad A. Matthee, Paulo C. Alves, and Dennis Lanning for providing the samples of Bunolagus monticularis, Lepus granatensis, and Ochotona princeps, respectively.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Brown, E. W., N. Yuhki, C. Packer, and S. J. O'Brien. 1994. A lion lentivirus related to feline immunodeficiency virus: epidemiologic and phylogenetic aspects. J. Virol. 685953-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann, R. S., and A. T. Smith. 2005. Order Lagomorpha, p. 185-211. In D. E. Wilson and D. M. Reeder (ed.), Mammal species of the world: a taxonomic and geographic reference. John Hopkins University Press, Baltimore, MD.
- 3.Holmes, E. C. 2003. Molecular clocks and the puzzle of RNA virus origins. J. Virol. 773893-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes, E. C. 2007. Ancient lentiviruses leave their mark. Proc. Natl. Acad. Sci. USA 1046095-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes, J. F., and J. M. Coffin. 2005. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics 1711183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katzourakis, A., M. Tristem, O. G. Pybus, and R. J. Gifford. 2007. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. USA 1046261-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Martinez, N. 1989. Revision sistematica y biostratigrafica de los lagomorphos (Mammalia) del terciario y cuaternario de España. Mem. Mus. Paleontol. Univ. Zaragoza 31-343. [Google Scholar]
- 8.Matthee, C. A., B. J. van Vuuren, D. Bell, and T. J. Robinson. 2004. A molecular supermatrix of the rabbits and hares (Leporidae) allows for the identification of five intercontinental exchanges during the Miocene. Syst. Biol. 53433-447. [DOI] [PubMed] [Google Scholar]
- 9.Rose, K. D., V. Burke DeLeon, P. Missiaen, R. S. Rana, A. Sahni, L. Singh, and T. Smith. 2008. Early Eocene lagomorph (Mammalia) from Western India and the early diversification of Lagomorpha. Proc. Biol. Sci. 2751203-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozas, J., J. C. Sánchez-DelBarrio, J. C. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalecent and other methods. Bioinformatics 192496-2497. [DOI] [PubMed] [Google Scholar]
- 11.Sharp, P. M., E. Bailes, D. L. Robertson, F. Gao, and B. H. Hahn. 1999. Origins and evolution of AIDS viruses. Biol. Bull. 196338-342. [DOI] [PubMed] [Google Scholar]
- 12.Sharp, P. M., E. Bailes, F. Gao, B. E. Beer, V. M. Hirsch, and B. H. Hahn. 2000. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem. Soc. Trans. 28275-282. [DOI] [PubMed] [Google Scholar]
- 13.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 14.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, Baltimore, MD.
- 15.Zhang, J., H. F. Rosenberg, and M. Nei. 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 953708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]