Abstract
The leader proteinase (Lpro) of foot-and-mouth disease virus (FMDV) is involved in antagonizing the innate immune response by blocking the expression of interferon (IFN) and by reducing the immediate-early induction of IFN-β mRNA and IFN-stimulated genes. In addition to its role in shutting off cap-dependent host mRNA translation, Lpro is associated with the degradation of the p65/RelA subunit of nuclear factor κB (NF-κB). Bioinformatics analysis suggests that Lpro contains a SAP (for SAF-A/B, Acinus, and PIAS) domain, a protein structure associated in some cases with the nuclear retention of molecules involved in transcriptional control. We have introduced a single or a double mutation in conserved amino acid residues contained within this domain of Lpro. Although three stable mutant viruses were obtained, only the double mutant displayed an attenuated phenotype in cell culture. Indirect immunofluorescence analysis showed that Lpro subcellular distribution is altered in cells infected with the double mutant virus. Interestingly, nuclear p65/RelA staining disappeared from wild-type (WT) FMDV-infected cells but not from double mutant virus-infected cells. Consistent with these results, NF-κB-dependent transcription was not inhibited in cells infected with double mutant virus in contrast to cells infected with WT virus. However, degradation of the translation initiation factor eIF-4G was very similar for both the WT and the double mutant viruses. Since Lpro catalytic activity was demonstrated to be a requirement for p65/RelA degradation, our results indicate that mutation of the SAP domain reveals a novel separation-of-function activity for FMDV Lpro.
Foot-and-mouth disease virus (FMDV) is the etiologic agent of FMD, a highly contagious disease that affects wild and domestic cloven-hoofed animals, including swine and cattle (19). The virus is the prototype member of the aphthovirus genus of the Picornaviridae family and consists of a positive-strand RNA genome of about 8 kb surrounded by an icosahedral capsid containing 60 copies each of four structural proteins. Upon infection, the viral RNA is translated as a single polyprotein that is concurrently processed by three virus-encoded proteinases, leader (Lpro), 2A, and 3Cpro into precursors and mature structural (VP1, VP2, VP3, and VP4) and nonstructural (Lpro, 2A, 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol) proteins (44). Translation of the polyprotein is initiated at two different AUGs which are separated by 84 nucleotides (nt), yielding two alternative forms of Lpro. Initiation at the first AUG results in Lab, an Lpro form of 201 amino acids, and initiation at the second AUG results in Lb, an Lpro form of 173 amino acids that is predominantly produced (6, 40). Lpro is a well-characterized papainlike proteinase (27, 40, 43) that self-cleaves from the nascent polyprotein precursor and also cleaves the host translation initiation factor eIF-4G, an event that results in the shutoff of host cap-dependent mRNA translation, a hallmark of picornaviruses infection (15, 26, 35). Determination of the Lpro crystal structure has provided insights into the mechanism of action for both these scissions (21).
Studies in our laboratory have demonstrated that Lpro plays a critical role in the pathogenesis of FMDV. Viruses lacking the Lpro coding region (leaderless) are attenuated in vitro and in vivo (5, 8, 39). One of the reasons for this attenuation is the inability of the leaderless virus to block host cell translation, in particular, translation of type I alpha/beta interferon (IFN-α/β) (9). In most cell types, expression of IFN is induced in response to viral infection. Subsequently, IFN protein is secreted and binds to specific cell surface receptors acting in an autocrine or paracrine manner. The interaction between IFN and its receptor induces a series of signal transduction events that lead to the expression of IFN-stimulated genes (ISGs) which have antiviral and/or antiproliferative properties (22, 23). Among the ISGs, the IFN-induced double-stranded RNA-dependent protein kinase (PKR) and the IFN-induced RNase L (RNase L) have been shown to inhibit FMDV replication (10, 13). Therefore, the general block in cap-dependent cellular translation induced by Lpro results in the limited synthesis of IFN protein and IFN-triggered antiviral effects.
Recent data have demonstrated that Lpro also inhibits the induction of transcription of various cellular genes including IFN-β, regulated upon activation, normal T-cell expressed, and secreted (RANTES) and tumor necrosis factor alpha (TNF-α) (13). In uninfected cells, transcription of IFN-β is not detectable, but upon viral infection latent transcription factors, including nuclear factor κB (NF-κB), IFN regulatory factors 3 and 7 (IRF3 and IRF7) and the activating transcription factor 2/cellular Jun protein complex (ATF2/c-Jun, also named AP-1) are activated and translocated from the cytoplasm to the nucleus, where they bind to their respective IFN-β enhancer elements, thereby inducing gene expression (23). Several studies have shown that one of the mechanisms used by different viruses to antagonize the innate immune response is the inhibition of the induction of IFN-β transcription (11, 22). Among picornaviruses, it has been reported that poliovirus causes the degradation of several proteins, including the p65/RelA subunit of NF-κB and the RNA helicase MDA-5, resulting in reduced IFN-β transcription (2, 38). Furthermore, for Cardiovirus, the only other genus of the Picornaviridae family that contains an L protein at the beginning of its open reading frame, and also for poliovirus, the induction of nucleocytoplasmic traffic disorder is associated with an inhibitory effect on host translation and IFN responses (4, 12, 30, 49).
Our group has shown that during FMDV infection downregulation of IFN-β transcription is associated with Lpro- dependent degradation of p65/RelA (13, 14). Interestingly, our studies showed that Lpro translocates to the nucleus of infected cells, and there is a correlation between the translocation of Lpro and the decrease in the amount of nuclear p65/RelA. However, it still remains unclear how FMDV Lpro induces p65/RelA degradation since highly conserved Lpro cleavage sites have not been found in the protein primary sequence, nor have defined p65/RelA degradation products been detected during FMDV infection (14).
Bioinformatics analysis of the protein sequence of FMDV Lpro reveals that there is a putative SAP (for SAF-A/B, Acinus, and PIAS) domain between amino acids 47 and 83 (following the numbering from the Lb form of Lpro). SAP domains are usually present in eukaryotic proteins that bind DNA and are involved in multiple steps of DNA metabolism, including replication, transcription, repair, etc. (1). Embedded within the Lpro putative SAP domain, the IQKL amino acid sequence is related to the LXXLL signature motif that is found in most members of the protein inhibitor of activated STAT (PIAS) protein family (46). We mutated conserved Lpro residues within the putative SAP domain and found that a double mutation abolished the retention of Lpro in the nuclei of FMDV-infected cells. Interestingly, this mutant virus was unable to induce degradation of nuclear p65/RelA. In contrast to infection with wild-type (WT) virus, infection with the SAP double mutant virus did not prevent the induction of transcription of NF-κB-dependent mRNAs. Since processing of the translation initiation factor eIF-4G was not significantly affected by the Lpro SAP mutation, our results suggest that, in addition to the proteinase activity, subcellular localization is another important determinant for Lpro inhibition of the early innate immune response.
MATERIALS AND METHODS
Domain annotation.
A consensus sequence of Lpro was obtained from the alignment of all available FMDV amino acid sequences in GenBank. The consensus sequence was used in the annotation of protein domains with the simple modular architecture research tool (SMART) (29).
Cells.
Bovine kidney (LF-BK) (48) and porcine kidney (IBRS-2) cell lines were obtained from the Foreign Animal Disease Diagnostic Laboratory at the Plum Island Animal Disease Center. Secondary porcine kidney (PK) cells and primary bovine embryonic kidney cells (EBK) were provided by the Animal, Plant, and Health Inspection Service, National Veterinary Service Laboratory, Ames, IA. These cells were maintained in minimal essential medium (MEM; Gibco-BRL/Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum and supplemented with 1% antibiotics and nonessential amino acids. BHK-21 cells (baby hamster kidney cells strain 21, clone 13, ATCC CL10) obtained from the American Type Culture Collection (Rockville, MD) were used to propagate virus stocks and to measure virus titers. BHK-21 cells were maintained in MEM containing 10% calf serum and 10% tryptose phosphate broth supplemented with 1% antibiotics and nonessential amino acids. Cell cultures were incubated at 37°C in 5% CO2.
Viruses.
FMDV A12-WT was generated from the full-length serotype A12 infectious clone, pRMC35 (42), and A12-LLV2 (leaderless virus) was derived from the infectious clone lacking the Lb coding region, pRM-LLV2 (39). The A12#47, A12#48, and A12#49 mutant viruses were derivatives of A12-WT constructed by site-directed mutagenesis as described below. Theiler's murine encephalomyelitis virus (TMEV) and a chimeric TMEV containing the Lb coding region of FMDV A12 (TMEV-Lb) were previously described (41). Mutant TMEV-LbC23A was derived from TMEV-Lb and was constructed as described below. Viruses were propagated in BHK-21 cells and were concentrated by polyethylene glycol precipitation, titrated on BHK-21 cells, and stored at −70°C.
Construction of mutant viruses.
Mutant FMDV and TMEV-Lb viruses were constructed by introducing specific nucleotide changes in the cDNA of the respective infectious clones utilizing a QuikChange mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's directions. For FMDV mutants, plasmid pRMC35 and oligonucleotide pairs that annealed to nt 147 to 188, considering the AUG start codon of Lb as nt 1, were used as follows: I55A_FW (5′-CTCACACTAG CAGCCGCCAAACAGCTGGAGGAACTCACAGGG) and I55A_RW (5′-CCCTGTGAGTTCCTCCAGCTGTTTGGCGGCTGCTAGTGTGAG) for A12#47, L58A_FW (5′-CTCACACTAGCAGCCATCAAACAGGCGGAGGAACTCACAGGG) and L58A_RW (5′-CCCTGTGAGTTCCTCCGCCTGTTTGATGGCTGCTAGTGTGAG) for A12#48, and I55A,L58A_FW (5′-CTCACACTAGCAGCCGCCAAACAGGCGGAGGAACTCACAGGG) and I55A,L58A_RW (5′-CCCTGTGAGTTCCTCCGCCTGTTTGGCGGCTGCTAGTGTGAG) for A12#49. For the TMEV-LbC23A mutant, plasmid pDAFSCC1-Lb (41) and the oligonucleotide pair that anneals between nt 47 and 90 of FMDV-Lb, LbC23A_FW (5′- GGCCCAACAACCACGACAACGCTTGGTTGAACACCATCCTCCAG) and LbC23A_RW (5′-CTGGAGGATGGTGTTCAACCAAGCGTTGTCGTGGTTGTTGGGCC), were used.
FMDV cell infections.
Cultured cell monolayers were infected with FMDV or TMEV at the indicated multiplicity of infection (MOI) for 1 h at 37°C. After adsorption, cells were rinsed and incubated with MEM at 37°C. For kinetics of growth or indirect immunofluorescence analyses of FMDV-infected cells, unabsorbed virus was removed by washing the cells with a solution containing 150 mM NaCl in 20 mM morpholineethanesulfonic acid (MES; pH 6.0), before adding MEM and proceeding with the incubation. When indicated, 200 nM leptomycin B (LMB; Sigma Aldrich, St. Louis, MO) was added throughout the infection.
Analysis of mRNA.
A quantitative real-time reverse transcription-PCR (RT-PCR) assay was used to evaluate the mRNA levels of porcine IFN-β, IRF7, Mx1, RANTES, and TNF-α, as previously described (14, 37). Primers and probes sequences are listed in Table 1.
TABLE 1.
Oligonucleotide primer and probe sequences for real-time RT-PCR
| Gene | Primer and probe setsa | Sequence | NCBI GenBank accession no. |
|---|---|---|---|
| GAPDH | Porcine GAPDH-327F | CGTCCCTGAGACACGATGGT | AF017079 |
| Porcine GAPDH-380R | CCCGATGCGGCCAAAT | ||
| Porcine GAPDH-348T | AAGGTCGGAGTGAACG | ||
| IFN-β | Porcine IFN-β-11F | AGTGCATCCTCCAAATCGCT | M86762 |
| Porcine IFN-β-69R | GCTCATGGAAAGAGCTGTGGT | ||
| Porcine IFN-β-32T | TCCTGATGTGTTTCTC | ||
| IRF7 | Porcine IRF7-418F | CTGCGATGGCTGGATGAA | TC224060 |
| Porcine IRF7-511R | TAAAGATGCGCGAGTCGGA | ||
| Porcine IRF7-450T | CCGCGTGCCCTGGAAGCACTT | ||
| Mx1 | Porcine Mx1-803F | GAGGTGGACCCCGAAGGA | M65087 |
| Porcine Mx1-859R | CACCAGATCCGGCTTCGT | ||
| Porcine Mx1-824T | AGGACCATCGGGATC | ||
| RANTES | Porcine RANTES-54F | TGGCAGCAGTCGTCTTTATCA | F14636 |
| Porcine RANTES-125R | CCCGCACCCATTTCTTCTC | ||
| Porcine RANTES-101T | TGGCACACACCTGGCGGTTCTTTC | ||
| TNF-α | Porcine TNF-α-338F | TGGCCCCTTGAGCATCA | NM214022 |
| Porcine TNF-α-405R | CGGGCTTATCTGAGGTTTGAGA | ||
| Porcine TNF-α-356T | CCCTCTGGCCCAAGGACTCAGATCA |
As reported by Moraes et al. (37). Numbering in the primer name refers to the nucleotide positions in the specific gene coding region. Suffix letters: F, forward primer; R, reverse primer; T, TaqMan FAM-MGB probe.
Protein analysis. (i) Western blotting.
Cytoplasmic and nuclear cell fractions were prepared as described previously (14). Proteins were resolved in 4 to 20% NuPAGE Novex Tris-acetate gels (Invitrogen), transferred to polyvinylidene difluoride membranes, and detected by Western blotting using an Immun-Star HRP chemiluminescent kit (Bio-Rad, Hercules, CA) according to the manufacturer's directions. eIF-4G (also named p220) and NF-κB-p65/RelA, were detected with rabbit polyclonal antibodies (Ab) anti-p220 (15) and Ab-1 RB-1638 (NeoMarkers; Lab Vision, Freemont, CA), respectively. FMDV VP1 and tubulin-α were detected with monoclonal Ab (MAb) 6HC4 (3) and Ab-2 MS-581 (clone DM1A; NeoMarkers; Lab Vision), respectively.
(ii) Analysis of IFN protein.
A porcine IFN-α (pIFNα) double-capture enzyme-linked immunosorbent assay (ELISA) previously developed in our laboratory was used to quantitate IFN-α protein in the supernatants of infected cells (36). pIFNα MAb K9 and F17 were purchased from R&D Systems (Minneapolis, MN). MAb K9 (1 μg/ml) was used for antigen capture, and biotinylated MAb F17 (0.35 μg/ml) in conjunction with horseradish-peroxidase-conjugated streptavidin (KPL, Gaithersburg, MD) were used for detection. pIFNα concentrations were determined by extrapolation on a standard curve prepared with recombinant pIFNα (PBL Biomedical Laboratories, Piscataway, NJ).
(iii) Radioimmunoprecipitation of FMDV-infected cell lysates.
[35S]methionine-labeled FMDV proteins from LF-BK-infected cells (250,000 cpm/sample) were immunoprecipitated with bovine convalescent-phase serum. The samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 15% gel and visualized by autoradiography.
Indirect immunofluorescence analyses (IFA).
Subconfluent cell monolayers prepared in 12-mm glass coverslips were infected with FMDV or TMEV at an MOI of 10 or treated with 25 μg of poly[IC] and Lipofectamine 2000 (Invitrogen)/ml for the indicated time. The cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 (Sigma) in phosphate-buffered saline (PBS), blocked with blocking buffer (PBS, 2% bovine serum albumin, 5% normal goat serum, 10 mM glycine), and then incubated overnight at 4°C with the respective primary Abs. FMDV VP1 was detected with mouse MAb 6HC4 (3), Lpro was detected with a rabbit polyclonal Ab elicited against bacterially expressed recombinant protein (39), TMEV VP1 was detected with MAb DAmAb2 (28), and NF-κB-p65/RelA was detected with rabbit polyclonal Ab-1 RB-1638 (NeoMarkers; Lab Vision). Alexa Fluor 488 and Alexa Fluor 594 (Molecular Probes, Invitrogen)-conjugated secondary Abs were used for detection. Nuclei were visualized by DAPI (4′,6′-diamidino-2-phenylindole) staining included in ProLong Gold Antifade mounting medium (Invitrogen). Cells were examined in an Olympus BX40 fluorescence microscope, and the images were taken with a DP-70 digital camera using DP-BSW v2.2 software (Olympus America, Central Valley, PA).
RESULTS
FMDV Lpro coding region contains a putative SAP domain.
Analysis of the Lpro coding sequence utilizing SMART showed that from amino acids 47 to 83 (following the numbering of Lb) there is a conserved sequence motif that resembles a previously defined SAP domain (1) (Fig. 1). Although the homology with any specific sequence of SAP domains is lower than 25% percent, the homology with the consensus SAP domain sequence is greater than 60%. When the Lpro sequence was compared to the sequence profile of the SAP domains, more than 80% the of the Lpro amino acids within this region were found in the profile. In addition, the three-dimensional structure of the Lpro sequence (21) shared the same α-helix-turn-α-helix structure found in SAP domains (1). Despite the presence of a two-amino-acid insertion between the two α-helices in Lpro, the data support the presence of a SAP domain within Lpro.
FIG. 1.
Alignment of FMDV Lpro partial amino acid sequence. Lpro protein sequence was aligned to all available sequences utilizing SMART software. Depicted are sequences in single letters between amino acids 47 and 88, and a schematic diagram displays the approximate location of predicted α-helices (green ovals). Asterisks mark the location of amino acids targeted by mutagenesis. Summary of color coding including consensus sequence: + (positive sign in blue), positive charged amino acids (H, K, and R); b (lowercase letter “b” highlighted in yellow), amino acids with a large or bulky side chain (E, F, H, I, K, L, M, Q, R, W, and Y); c (lowercase letter “c” in pink font), charged amino acids (D, E, H, K, and R); l (lowercase letter “l” highlighted in yellow), aliphatic amino acids (I, L, and V); p (lowercase letter “p” in blue font), polar amino acids (C, D, E, H, K, N, Q, R, S, and T); s (lowercase letter “s” in green font), amino acids with a small side chain (A, C, D, G, N, P, S, T, and V); and gray highlighted, amino acids with ≥60% of the homology in the alignment.
Mutations of Lpro SAP domain partially affect virus growth.
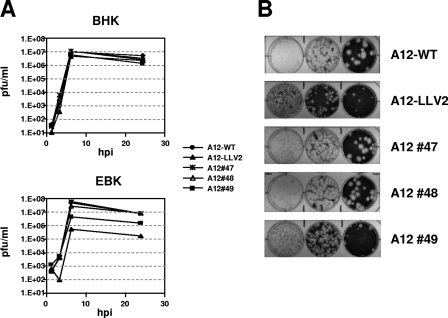
In order to determine whether the putative SAP domain is important for Lpro function, we mutated two residues at positions 55 or 58, individually (A12#47 and A12#48) or in combination (A12#49). We selected these amino acids based on previous studies with PIAS3, a SAP-containing protein, where it was reported that mutation of a similar region altered PIAS3 nuclear localization and retention (16). Viruses derived from transfected cells were passaged four times in BHK-21 cells, and the Lpro coding region of the resulting viruses was sequenced to confirm that the only changes were at the mutated sites. Figure 2 shows the kinetics of growth of the mutant viruses in two different cell types. In BHK-21 cells all viruses grew with similar kinetics, reaching final titers with differences of less than a half log with respect to the WT. In EBK cells the growth differences between WT and some of the mutant viruses were more pronounced. As previously reported, A12-LLV2 grew to a final titer ∼50-fold lower than that of the WT virus (9). Interestingly, A12#49 (the double SAP mutant) grew to a final titer of ∼5-fold lower than WT virus, whereas A12#47 and A12#48 (single SAP mutants) grew to titers similar to that of the WT virus. Regarding plaque size, A12#47 and A12#48 resembled A12-WT virus, and A12#49 was more related to the small-plaque phenotype of A12-LLV2. These results indicated that disruption of the predicted signature motif of the SAP domain requires at least mutations at two sites and affects the growth characteristics of FMDV, resulting in a partially attenuated phenotype.
FIG. 2.
Kinetics of growth and plaque morphology. (A) Growth curves. BHK-21 or EBK cells were infected with the indicated viruses and, after 1 h, unabsorbed virus was removed by washing with 150 mM NaCl-20 mM MES (pH 6.0), followed by the addition of complete medium. Samples were taken at 1, 3, 6, and 24 hpi, and virus titers were determined by plaque assay on BHK-21 cells. (Reported values display one out of three representative experiments with similar results.) (B) BHK-21 cells were infected with similar amounts of viruses and treated as described in panel A, but medium with a gum tragacanth overlay was added and the plaques were stained at 40 hpi.
Mutation of Lpro SAP domain affects nuclear retention.
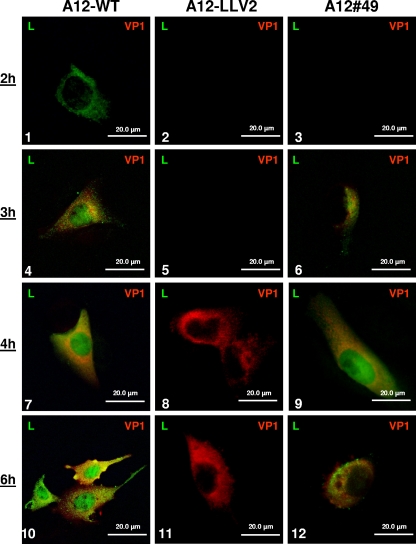
We have previously reported that during FMDV infection Lpro progressively translocates to the nucleus of infected cells (14). We analyzed the subcellular localization of Lpro in the SAP mutants. In LF-BK cells infected with the single mutant viruses (A12#47 and A12#48) the translocation of Lpro into the nucleus was indistinguishable from A12-WT (data not shown). In contrast, the double SAP mutant (A12#49) displayed a distinct phenotype. At the beginning of the infection and up to 4 h postinfection (hpi), Lpro of A12#49 was observed in the cytoplasm and then progressively appeared in the nucleus of the infected cells (Fig. 3, panels 3, 6, and 9) similarly to A12-WT (Fig. 3, panels 1, 4, and 7). We observed that nuclear translocation of Lpro was slightly delayed for A12#49. However, by 6 hpi almost no Lpro nuclear staining was detected in cells infected with A12#49 in contrast to cells infected with A12-WT (Fig. 3, panels 12 and 10, respectively). Most of Lpro was distributed throughout the cytoplasm concentrated in granules, as eventually observed for several other FMDV viral proteins when infection is well established. As a control, infection with A12-LLV2 did not display any Lpro staining (Fig. 3, panels 2, 5, 8, and 11). These observations led us to conclude that mutation of the predicted SAP domain prevented Lpro nuclear accumulation during the course of infection.
FIG. 3.
IFA of Lpro during FMDV infection. LF-BK cells were infected at an MOI of 10 with FMDV A12-WT (panels 1, 4, 7, and 10), A12-LLV2 (panels 2, 5, 8, and 11), or double SAP mutant A12#49 (panels 3, 6, 9, and 12) and were fixed at different times postinfection. Viral protein Lpro was detected using a rabbit polyclonal Ab and an Alexa Fluor 488-conjugated secondary Ab. Viral protein VP1 was detected using mouse MAb 6HC4 and an Alexa Fluor 594-conjugated secondary Ab.
p65/RelA is not degraded after infection with mutant FMDV.
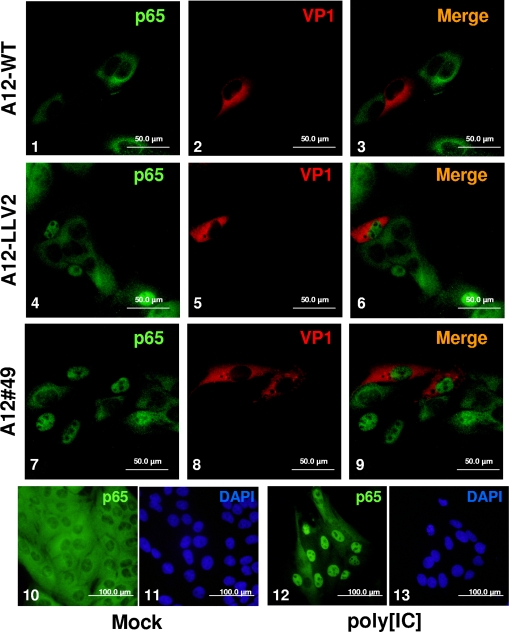
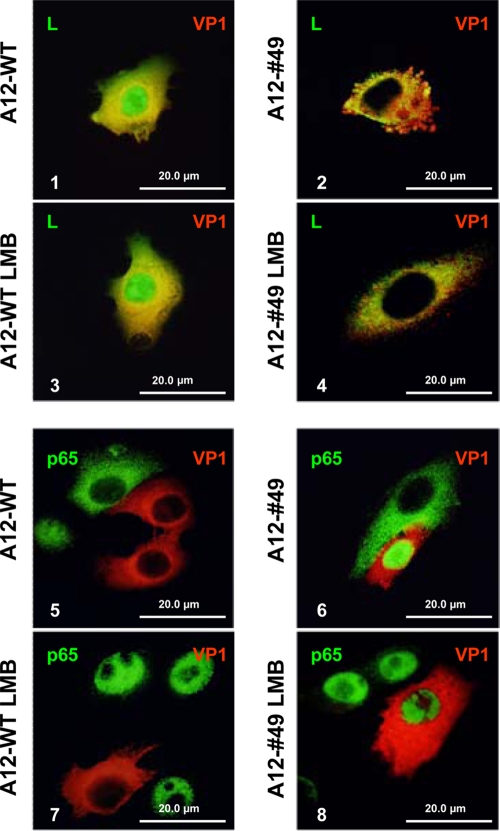
In our previous studies we demonstrated that p65/RelA degradation correlated with nuclear accumulation of Lpro (14). Figure 4 shows the IFA results of p65/RelA in LF-BK cells infected with A12-WT, A12-LLV2, and A12#49. As seen previously in infected PK cells, by 6 hpi the p65/RelA signal was almost absent from the nucleus of WT-infected cells (Fig. 4, panels 1 and 3) and accumulated in the nucleus of A12-LLV2-infected cells (Fig. 4 panels 4 and 6). Interestingly, the pattern for A12#49 (Fig. 4, panels 7 and 9) exactly resembled the pattern for A12-LLV2, with a bright p65/RelA staining concentrated in the nuclei of infected cells. Western blot analysis of cytoplasmic and nuclear extracts of infected LF-BK cells showed similar results (data not shown). Since there is a correlation between p65/RelA degradation and Lpro nuclear accumulation, we decided to add LMB, an inhibitor of the CRM1-dependent pathway of active nuclear export (18). We hypothesized that p65/RelA would be degraded if the exit of A12#49 Lpro from the nucleus was blocked. However, there was no nuclear accumulation of Lpro after the addition of LMB to A12-WT- or A12#49-infected cells (Fig. 5, panels 3 and 4). The addition of LMB did increase the number of cells displaying nuclear p65/RelA staining (Fig. 5, panels 5 to 7 compared to panels 6 to 8) as previously described (33).
FIG. 4.
IFA of p65/RelA during FMDV infection. LF-BK cells were infected at an MOI of 10 with WT (panels 1 to 3), A12-LLV2 (panels 4 to 6), and the double SAP mutant A12#49 (panels 7 to 9). As a control, cells were mock infected (panels 10 and 11) or treated with synthetic dsRNA poly[IC] (25 μg/ml) and Lipofectamine (panels 12 and 13). At 6 hpi, cells were fixed and stained. p65/RelA was detected using a rabbit polyclonal Ab (Abcam RB-1638) and an Alexa Fluor 488-conjugated secondary Ab. Viral protein VP1 was detected using mouse MAb 6HC4 and an Alexa Fluor 594-conjugated secondary Ab. Nuclei were stained with DAPI (panels 11 and 13).
FIG. 5.
Addition of LMB does not block the nuclear export of FMDV Lpro. LF-BK cells were infected at an MOI of 10 with A12-WT (panels 1, 3, 5, and 7) and the double SAP mutant A12#49 (panels 2, 4, 6, and 8). Where indicated, 200 nM LMB was added to the culture medium. At 6 hpi, cells were fixed and stained. For panels 1 to 4, viral protein Lpro was detected using a rabbit polyclonal Ab and an Alexa Fluor 488-conjugated secondary Ab. Viral protein VP1 was detected using mouse MAb 6HC4 and an Alexa Fluor 594-conjugated secondary Ab. For panels 5 to 8, p65/RelA was detected using rabbit polyclonal Ab Abcam RB-1638 and an Alexa Fluor 488-conjugated secondary Ab.
These results suggested that nuclear accumulation of Lpro is required for p65/RelA degradation and that the CRM1-dependent nuclear export pathway is not involved in the exit of Lpro from the nuclei of A12#49-infected cells.
Mutation of Lpro SAP domain prevents Lpro inhibition of IFN expression.
We earlier reported that Lpro antagonizes the innate immune response by blocking the expression of IFN (9, 10, 13, 14). To test whether the SAP double mutant had a similar effect, we analyzed the expression of IFN-β, the proinflammatory cytokine TNF-α, the chemokine RANTES, and the ISGs Mx1 and IRF7 in virus-infected cells. We used secondary PK cells in this assay, rather than secondary EBK or LF-BK cells, because we have previously optimized standard procedures of real-time PCR in PK cells (13, 14) and FMDV SAP mutants behave consistently in these cell types (Fig. 2 to 4 and data not shown). By 4 hpi, there was no significant difference in the induction of any of the analyzed genes after infection with A12-WT, A12-LLV2, or A12#49 (Fig. 6A). For IFN-β we observed at most a twofold difference for A12#49 or A12-LLV2 relative to A12-WT. However, by 8 hpi, there was an increase in IFN-β expression of ∼10-fold for A12#49 and 14-fold for A12-LLV2 compared to A12-WT. A similar pattern was observed for all of the analyzed genes, although the differences were slightly lower, varying from 3- to 10-fold higher for the mutants compared to WT. ELISA quantitation of secreted IFN-α protein in the supernatants of infected PK cells followed similar kinetics (Fig. 6B). By 24 hpi, we detected 6- to 12-fold-higher amounts of IFN-α protein for A12-LLV2 and A12#49, respectively, compared to A12-WT.
FIG. 6.
Analysis of IFN expression. (A) The expression of IFNβ, TNF-α, RANTES, Mx1, and IRF7 mRNAs was measured by real-time RT-PCR in secondary PK cells infected with A12-WT, A12-LLV2, or A12#49 FMDV at an MOI of 2 for the indicated times. Porcine GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control. The results are expressed as the fold increase in gene expression for virus-infected with respect to mock-infected cells. (Reported values display the findings for one out of three representative experiments with similar results.) (B) pIFNα expression in the supernatants of PK-infected cells determined by ELISA. The values are presented as the mean ± the standard deviation of three independent determinations.
These results indicated that mutations of the SAP domain prevented the inhibitory effect of Lpro on NF-κB-dependent transcriptional activity and IFN protein expression.
Cleavage of translation initiation factor eIF-4G is not affected by mutation of the Lpro SAP domain.
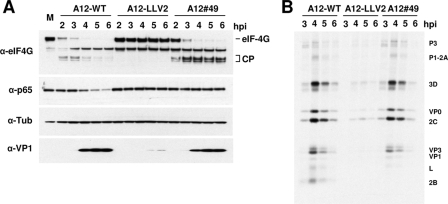
Lpro is responsible for cleaving the translation initiation factor eIF-4G and shutting off host cell translation, a hallmark of picornavirus infection (44). We examined the kinetics of eIF-4G cleavage in LF-BK-infected cells by Western blot analysis. Figure 7A shows that, as expected, eIF-4G (p220) was completely processed in A12-WT-infected cells by 4 hpi. Interestingly, only a minor delay, 1 to 2 h, on eIF-4G processing was detected for double SAP mutant (A12#49). Comparable amounts of viral VP1 suggested that the stage of infection was similar for A12-WT and A12#49. Although complete eIF-4G cleavage was achieved for A12#49 by 5 to 6 hpi, low-molecular-weight cleavage products persisted throughout the course of infection. No eIF-4G processing was observed in mock-infected or A12-LLV2-infected cells. By 4 hpi the p65/RelA signal was significantly reduced in the cytoplasm of cells infected with A12-WT. In contrast, the p65/RelA signal did not decrease in extracts of A12-LLV2- or double SAP mutant A12#49-infected cells.
FIG. 7.
Processing of cellular proteins during infection with WT and mutant FMDV. (A) LF-BK cells were infected with WT (A12-WT), leaderless (A12-LLV2), and SAP mutant (A12#49) at MOIs of 10 for 6 h. At the indicated times, cytoplasmic extracts were prepared and analyzed by Western blotting using rabbit polyclonal Ab anti-eIF-4G (p220), rabbit polyclonal Ab anti-p65/RelA (RB-1638), mouse MAb 6HC4 (VP1), and mouse MAb anti-tubulin-α (Ab-2 MS-581). CP, p220 cleavage products. (B) Radioimmunoprecipitation of FMDV-infected cell lysates at different times postinfection. [35S]methionine-labeled cell lysates from FMDV-infected LF-BK cells were immunoprecipitated with serum from a convalescent bovine. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and developed by autoradiography.
Analysis of viral polyprotein processing using a radioimmunoprecipitation assay revealed no major differences between A12-WT and A12#49 (Fig. 7B). Processing into mature products, L, VP0, VP1,VP3, 2B, 2C, and 3D proceeded almost equivalently by 4 hpi. As previously reported, viral protein synthesis in A12-LLV2-infected cells was significantly delayed (39).
These results indicated that disruption of the Lpro SAP domain selectively prevented p65/RelA processing without affecting the ability of Lpro to cleave eIF-4G.
Lpro catalytic activity is required for p65/RelA processing.
In our previous studies, using a recombinant Theiler's virus that expresses Lpro in the absence of any other FMDV protein (TMEV-Lb), we demonstrated that the presence of Lpro is necessary and sufficient for p65/RelA degradation (14). Using the same recombinant virus, we examined whether the catalytic activity of Lpro was a requirement for p65/RelA disappearance. For this purpose, we mutated the catalytic cysteine residue of Lpro (C23) to alanine, creating TMEV-LbC23A. Mutation of Lpro C23 did not affect the pattern of localization previously observed for WT Lpro (Fig. 8, panels 4 and 6 compared to panels 1 and 3). Interestingly, this mutation abolished the ability of FMDV Lpro to cause p65/RelA degradation (Fig. 8, panels 10, 11, and 12 compared to panels 7, 8, and 9), indicating that the protease activity of Lpro is essential for this function.
FIG. 8.
FMDV Lpro catalytic activity is required for p65/RelA degradation. IBRS-2 cells were infected with TMEV-Lb-containing WT or mutant C23A Lpro at an MOI of 10. At 24 hpi, p65/RelA and viral proteins were visualized by IFA. p65/RelA was detected with rabbit polyclonal Ab (RB-1638) and Alexa Fluor 488-conjugated secondary Ab. Lpro was visualized a rabbit polyclonal Ab and Alexa Fluor 488-conjugated secondary Ab. TMEV VP1 was detected with MAb DAmAb2 (28) and Alexa Fluor 594-conjugated secondary Ab.
DISCUSSION
We have earlier reported that Lpro antagonizes the innate immune response by blocking the expression of IFN. At least two mechanisms are involved in this function: (i) the shutoff of host cell translation resulting in lower levels of IFN protein expression and (ii) the interference in the induction of IFN-β transcription. Lpro cleaves the eukaryotic translation initiation factor eIF-4G, which is required for cap-dependent mRNA translation without affecting the internal ribosome entry site-dependent translation of viral RNA, and thus the virus takes advantage of decreased levels of IFN protein to establish a productive infection (9, 20, 26). In addition, Lpro induces the degradation of the p65/RelA subunit of the transcription factor NF-κB, and this degradation is associated with Lpro nuclear localization (14). A block in the upregulation of IFN-β transcription also results in lower levels of IFN protein (13).
The availability of multiple FMDV protein sequences (7), the high-resolution crystal structure of Lpro (21), and powerful software tools (29) have allowed us to predict that a conserved SAP domain is situated between amino acids 47 and 83 of Lb. In the present study we demonstrated that this domain is important for Lpro function. Double mutation of the SAP domain resulted in an attenuated virus phenotype yielding lower titers and a smaller plaque size. Although the phenotype was not as clear-cut as in the case of the Lpro deletion in leaderless virus, it was indicative of a role of this domain in FMDV virulence. Early translocation of mutant Lpro from the cytoplasm to the nucleus of infected cells was only slightly delayed. However, by 6 hpi, mutant Lpro, in contrast to WT Lpro, was absent from the nuclei of infected cells. Failure in nuclear retention has been reported for another SAP-containing protein, PIAS3L, when this domain was mutated (16). PIAS3L requires an intact SAP box, in conjunction with a RING and a PINIT domain, for proper nuclear localization and retention (16). Perhaps FMDV Lpro depends on an intact SAP domain for docking in the nucleus of infected cells, allowing for interactions with host proteins that might be involved in regulating an antiviral response.
One of the most interesting observations in the present study was the absence of NF-κB degradation upon infection with an FMDV double SAP mutant even though these mutations did not affect the catalytic activity of Lpro. With the exception of leaderless virus, no other viable FMDV Lpro mutant has been previously reported. In vitro studies using mutant recombinant protein or plasmid transient transfection have been very informative, demonstrating that residue C23 is required for the protease enzymatic activity and is utilized by Lpro for self-cleavage from the viral polyprotein and cleavage of the translation initiation factor eIF-4G (15, 34, 40, 43). Recently, Mayer et al. (34), using rabbit reticulocyte lysates, have shown that Lpro residue L115 is also a determinant of self-cleavage and eIF-4G cleavage specificity. Utilizing a recombinant cardiovirus expressing Lpro in the absence of any other FMDV protein, we also demonstrated that the catalytic activity of Lpro is required for NF-κB degradation; mutation of the catalytic residue C23 prevented degradation. Unfortunately, the mechanism used by Lpro to cause NF-κB degradation is still unclear.
As mentioned above, Lpro SAP mutations did not affect Lpro enzymatic activity; self-processing and eIF-4G processing proceeded almost normally. We did observe that degradation of the eIF-4G cleavage products was delayed in cells infected with the double mutant. Mutation of the SAP domain may partially affect the interaction between Lpro and eIF-4G. Quantitative kinetics studies should be performed to verify this hypothesis. It has been reported that FMDV 3C can also cleave eIF-4G at later times after FMDV infection (47); thus, it is possible that the Lpro SAP mutant interferes with the 3C/eIF-4G interaction. Our results, however, suggest that 3C from double mutant SAP virus behaves normally since processing of the viral polyprotein proceeded similarly for the mutant and WT viruses.
The levels of several transcripts, including cytokines, chemokines, and ISGs, were significantly higher after infection with FMDV Lpro SAP mutant #49 compared to WT infection, indicating that disruption of the SAP domain prevented Lpro inhibition of NF-κB-dependent transcription. SAP domains are involved in protein-protein interactions. This motif is required for the repressive activity of PIASy on STAT1-mediated gene activation (31), and it has been proposed that several members of the PIAS protein family negatively regulate NF-κB and STAT signaling, affecting the expression of more than 60 genes (45). Furthermore, the specific role of PIAS proteins in the regulation of NF-κB activity has been examined in vivo utilizing PIAS1-null mice (32). These studies demonstrated that in the absence of PIAS only a subset of NF-κB-regulated genes is affected (ca. 48%), suggesting that there might be alternative mechanisms, independent of PIAS1, for NF-κB regulation.
More interestingly, Jang et al. (24) have provided evidence that the N-terminal region of PIAS3, which contains a SAP domain, is necessary for binding to the p65/RelA subunit of NF-κB, thereby blocking the transcriptional activation. Furthermore, an LXXLL signature motif of PIAS3 is involved in this physical interaction. Although not identical, this motif resembles the IQKL sequence present in FMDV Lpro. Our results suggest that this putative interaction may be involved in docking Lpro in the nucleus of infected cells where Lpro-dependent p65/RelA degradation takes place during FMDV infection. We are currently testing this hypothesis.
SAP domains are also found in several proteins displaying DNA-binding activity, and the contact with defined A/T rich sequences found in matrix attachment regions (MARs) of chromatin is mediated by the predicted α-helices delimited by the SAP box (25). Protein interactions with MARs regions determine the chromatin architecture in zones of interactions with the nuclear matrix. Interestingly, several viral proteins have been shown to localize to these regions, leading to the proposal that viral protein interaction with MARs regions may have a role in blocking host antiviral activities (17). The presence of a SAP domain may allow FMDV Lpro to localize to similar nuclear regions globally affecting the function of transcription factors situated in close proximity during viral infection.
Our results provide new insights into the mechanism used by FMDV to escape the immune response. Structure-function analysis of Lpro has demonstrated that, in addition to the proteinase activity, an intact protein motif, SAP, is required for FMDV virulence. A more detailed understanding of the interactions between FMDV Lpro and/or other viral proteins and the host at the molecular level should help in the development of specific antiviral strategies that could limit virus spread.
Acknowledgments
We thank Z. Lu for technical assistance with DNA sequencing.
This study was supported in part by the Plum Island Animal Disease Research Participation Program administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Department of Agriculture (appointment of F.D.-S.S. and C.C.A.D.) and by CRIS Project no. 1940-32000-052-00D, ARS, USDA (T.D.L.S., J.Z., and M.J.G.).
Footnotes
Published ahead of print on 3 December 2008.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2000. SAP: a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25112-114. [DOI] [PubMed] [Google Scholar]
- 2.Barral, P. M., J. M. Morrison, J. Drahos, P. Gupta, D. Sarkar, P. B. Fisher, and V. R. Racaniello. 2007. MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 813677-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxt, B., D. O. Morgan, B. H. Robertson, and C. A. Timpone. 1984. Epitopes on foot-and-mouth disease virus outer capsid protein VP1 involved in neutralization and cell attachment. J. Virol. 51298-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belov, G. A., Lidsky, P. V., Mikitas, O. V., Egger, D., Lukyanov, K. A., Bienz, K., and V. I. Agol. 2004. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 7810166-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, C. C., M. E. Piccone, P. W. Mason, T. S. McKenna, and M. J. Grubman. 1996. Pathogenesis of wild-type and leaderless foot-and-mouth disease virus in cattle. J. Virol. 705638-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, X., I. E. Bergmann, R. Fullkrug, and E. Beck. 1995. Functional analysis of the two alternative translation initiation sites of foot-and-mouth disease virus. J. Virol. 69560-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrillo, C., E. R. Tulman, G. Delhon, Z. Lu, A. Carreno, A. Vagnozzi, G. F. Kutish, and D. L. Rock. 2005. Comparative genomics of foot-and-mouth disease virus. J. Virol. 796487-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinsangaram, J., P. W. Mason, and M. J. Grubman. 1998. Protection of swine by live and inactivated vaccines prepared from a leader proteinase-deficient serotype A12 foot-and-mouth disease virus. Vaccine 161516-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinsangaram, J., M. E. Piccone, and M. J. Grubman. 1999. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J. Virol. 739891-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinsangaram, J., M. Koster, and M. J. Grubman. 2001. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J. Virol. 755498-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conzelmann, K.-K. 2005. Transcriptional activation of alpha/beta interferon genes: interference by nonsegmented negative-strand RNA viruses. J. Virol. 795241-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delhaye, S., V. van Pesch, and T. Michiels. 2004. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 784357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de los Santos, T., S. de Avila Botton, R. Weiblen, and M. J. Grubman. 2006. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J. Virol. 801906-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de los Santos, T., F. Diaz-San Segundo, and M. J. Grubman. 2007. Degradation of nuclear factor κB during foot-and-mouth disease virus infection. J. Virol. 8112803-12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devaney, M. A., V. N. Vakharia, R. E. Lloyd, E. Ehrenfeld, and M. J. Grubman. 1988. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 624407-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duval, D., G. Duval, C. Kedingerc, O. Pocha, and H. Boeufa. 2003. The PINIT motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 554111-118. [DOI] [PubMed] [Google Scholar]
- 17.Everett, R. D., and M. K. Chelbi-Alix. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89819-830. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390308-311. [DOI] [PubMed] [Google Scholar]
- 19.Grubman, M. J., and B. Baxt. 2004. Foot-and-mouth disease. Clin. Microbiol. Rev. 17465-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grubman, M. J., M. P. Moraes, F. Diaz-San Segundo, L. Pena, and T. de los Santos. 2008. Evading the host immune response: how foot-and-mouth disease virus has become an effective pathogen. FEMS Immunol. Med. Microbiol. 538-17. [DOI] [PubMed] [Google Scholar]
- 21.Guarné, A., J. Tormo, R. Kirchweger, D. Pfistermueller, I. Fita, and T. Skern. 1998. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition EMBO J. 177469-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honda, K., H. Yanai, A. Takaoka, and T. Taniguchi. 2006. Regulation of the type I IFN induction: a current view. Int. Immunol. 171367-1378. [DOI] [PubMed] [Google Scholar]
- 24.Jang, H. D., K. Yoon, Y. J. Shin, J. Kim, and S. Y. Lee. 2004. PIAS3 suppresses NF-κB mediated transcription by interacting with the p65/RelA subunit. J. Biol. Chem. 27924873-24880. [DOI] [PubMed] [Google Scholar]
- 25.Kipp, M., F. Göhring, T. Ostendorp, C. M. van Drunen, R. van Driel, M. Przybylski, and F. O. Fackelmayer. 2000. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 207480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchweger, R., E. Ziegler, B. J. Lamphear, D. Waters, H. D. Liebig, W. Sommergruber, F. Sobrino, C. Hohenadl, D. Blaas, R. E. Rhoads, and T. Skern. 1994. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4 gamma. J. Virol. 685677-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleina, L. G., and M. J. Grubman. 1992. Antiviral effects of a thiol protease inhibitor on foot-and-mouth disease virus. J. Virol. 667168-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong, W. P., G. D. Ghadge, and R. P. Roos. 1994. Involvement of cardiovirus leader in host cell-restricted virus expression. Proc. Natl. Acad. Sci. USA 911796-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lidsky P. V., S. Hato, M. V. Bardina, A. G. Aminev, A. C. Palmenberg, E. V. Sheval, V. Y. Polyakov, F. J. van Kuppeveld, and V. I. Agol. 2006. Nucleocytoplasmic traffic disorder induced by cardioviruses. J. Virol. 802705-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, B., M. Gross, J. ten Hoeve, and K. Shuai. 2001. A transcriptional corepressor of STAT1 with an essential LXXLL signature motif. Proc. Natl. Acad. Sci. USA 983203-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, B., R. Yang, K. A. Wong, C. Getman, N. Stein, M. A. Teitell, G. Cheng, H. Wu, and K. Shuai. 2005. Negative regulation of NF-κB signaling by PIAS1. Mol. Cell. Biol. 251113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loewe, R., W. Holnthoner, M. Gröger, M. Pillinger, F. Gruber, D. Mechtcheriakova, E. Hofer, K. Wolff, and P. Petzelbauer. 2002. Dimethylfumarate inhibits TNF-induced nuclear entry of NF-κB/p65 in human endothelial cells. J. Immunol. 1684781-4787. [DOI] [PubMed] [Google Scholar]
- 34.Mayer, C., D. Neubauer, A. T. Nchinda, R. Cencic, K. Trompf, and T. Skern. 2008. Residue L143 of the foot-and-mouth disease virus leader proteinase is a determinant of cleavage specificity. J. Virol. 824656-4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina, M., E. Domingo, J. K. Brangwyn, and G. J. Belsham. 1993. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology 19455-359. [DOI] [PubMed] [Google Scholar]
- 36.Moraes, M. P., J. Chinsangaram, M. C. S. Brum, and M. J. Grubman. 2003. Immediate protection of swine from foot-and-mouth disease: a combination of adenoviruses expressing interferon alpha and a foot-and-mouth disease virus subunit vaccine. Vaccine 22268-279. [DOI] [PubMed] [Google Scholar]
- 37.Moraes, M. P., T. de los Santos, M. Koster, T. Turecek, H. Wang, V. G. Andreyev, and M. J. Grubman. 2007. Enhanced antiviral activity against foot-and-mouth disease virus by a combination of type I and II porcine interferons. J. Virol. 817124-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neznanov, N., K. M. Chumakov, L. Neznanova, A. Almasan, A. K. Banerjee, and A. V. Gudkov. 2005. Proteolytic cleavage of the p65-RelA subunit of NF-κB during poliovirus infection. J. Biol. Chem. 28024153-24158. [DOI] [PubMed] [Google Scholar]
- 39.Piccone, M. E., E. Rieder, P. W. Mason, and M. J. Grubman. 1995. The foot-and-mouth disease virus leader proteinase gene is not required for viral replication. J. Virol. 695376-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piccone, M. E., M. Zellner, T. F. Kumosinski, P. W. Mason, and M. J. Grubman. 1995. Identification of the active-site residues of the L proteinase of foot-and-mouth disease virus. J. Virol. 694950-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piccone, M. E., H.-H. Chen, R. R. Roos, and M. J. Grubman. 1996. Construction of a chimeric Theiler's murine encephalomyelitis virus containing the leader gene of foot-and-mouth disease virus. Virology 226135-139. [DOI] [PubMed] [Google Scholar]
- 42.Rieder, E., T. Bunch, F. Brown, and P. W. Mason. 1993. Genetically engineered foot-and-mouth disease viruses with poly(C) tracts of two nucleotides are virulent in mice. J. Virol. 675139-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, P. J., and G. J. Belsham. 1995. Identification of critical amino acids within the foot-and-mouth disease virus leader protein, a cysteine protease. Virology 213140-146. [DOI] [PubMed] [Google Scholar]
- 44.Rueckert, R. R. 2007. Picornaviridae: the viruses and their replication, p. 795-948. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 45.Shuai, K., and B. Liu. 2005. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 5593-605. [DOI] [PubMed] [Google Scholar]
- 46.Shuai, K. 2006. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 16196-202. [DOI] [PubMed] [Google Scholar]
- 47.Strong, R., and G. J. Belsham. 2004. Sequential modification of translation initiation factor eIF4GI by two different foot-and-mouth disease virus proteases within infected baby hamster kidney cells: identification of the 3Cpro cleavage site. J. Gen. Virol. 852953-2962. [DOI] [PubMed] [Google Scholar]
- 48.Swaney, L. M. 1988. A continuous bovine kidney cell line for routine assays of foot-and-mouth disease virus. Vet. Microbiol. 181-14. [DOI] [PubMed] [Google Scholar]
- 49.Zoll, J., W. J. Melchers, J. M. Galama, and F. J. van Kuppeveld. 2002. The mengovirus leader protein suppresses alpha/beta interferon production by inhibition of the iron/ferritin-mediated activation of NF-κB. J. Virol. 769664-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]