Abstract
Differential splice site pairing establishes alternative splicing patterns resulting in the generation of multiple mRNA isoforms. This process is carried out by the spliceosome, which is activated by a series of sequential structural rearrangements of its five core snRNPs. To determine when splice sites become functionally paired, we carried out a series of kinetic trap experiments using pre-mRNAs that undergo alternative 5′ splice site selection or alternative exon inclusion. We show that commitment to splice site pairing in both cases occurs in the A complex, which is characterized by the ATP-dependent association of the U2 snRNP with the branch point. Interestingly, the timing of splice site pairing is independent of the intron or exon definition modes of splice site recognition. Using the ATP analog ATPγS, we showed that ATP hydrolysis is required for splice site pairing independent from U2 snRNP binding to the pre-mRNA. These results identify the A complex as the spliceosomal assembly step dedicated to splice site pairing and suggest that ATP hydrolysis locks splice sites into a splicing pattern after stable U2 snRNP association to the branch point.
The splicing of nuclear pre-mRNAs is a fundamental process required for the expression of most metazoan genes. It is carried out by the spliceosome, which recognizes splicing signals and catalyzes the removal of noncoding intronic sequences to assemble protein-coding sequences into mature mRNAs prior to export to the cytoplasm and translation (4). Of the approximately 25,000 genes carried by the human genome (16), more than 70% are believed to produce transcripts that are alternatively spliced, enriching the proteomic diversity of higher eukaryotic organisms (18). Because regulation of this process can determine when and where a particular protein isoform is expressed, changes in alternative splicing patterns modulate many cellular activities. Consequently, the process of splicing must occur with a high degree of specificity and fidelity to ensure the appropriate expression of functional mRNAs. Commitment to splice site pairing constitutes the most crucial point in the generation of mRNA isoforms, because it sets in stone unique splicing patterns of pre-mRNAs.
For several years it has been possible to evaluate the formation of the mammalian spliceosome in vitro (39). At least four distinct complexes can be resolved by nondenaturing gel electrophoresis, in the order E→A→B→C (6, 22). These complexes differ in composition and order of appearance, suggesting a sequential model of spliceosome assembly. This progression requires the activity of more than 150 distinct protein factors and the U1, U2, U4, U5, and U6 snRNAs (20). The first intermediate of this assembly reaction is the E complex, which is characterized by stable interactions of the U1 snRNP with the 5′ splice site (2, 17, 27) and of the U2 auxiliary factor with the polypyrimidine tract (2, 34, 43). ATP hydrolysis then leads to the formation of the A complex, which is characterized by the stable association of the U2 snRNP with the branch point/3′ splice site sequence. The B complex and the catalytically active C complex then form after the incorporation and rearrangement of the U4-U6/U5 tri-snRNP. Even though it is well established that the formation of the first ATP-independent spliceosomal complex commits the pre-mRNA to the general splicing pathway (32), it is unclear if this step also coincides with a commitment to alternative splice site choice.
Several structural studies have shown an association between the 5′ splice site and the 3′ splice site at the E complex (8, 21, 26). Biochemical purification demonstrated that the U2 snRNP is loosely associated with the E complex (7) and that its presence is required for E complex formation (9). The U2 snRNP was found to be in close proximity to the U1 snRNP and to the 5′ and 3′ splice sites (8), suggesting that the U1 and U2 snRNPs already bridge the splice sites in the E complex. We have previously determined that alternative 3′ splice site pairing coincides with A complex formation (25). These results demonstrated that even though a pair of 5′ and 3′ splice sites may be in close proximity in the E complex (8, 21), their association remains dynamic until ATP-dependent formation of the A complex.
In this work, we use a functional splice site pairing assay to determine at which stage during spliceosomal assembly alternative 5′ splice site or alternative exon inclusion patterns are established. We show that splice sites are paired during A complex formation, regardless of the type of alternative splicing assayed. Interestingly, the timing of splice site pairing is independent from the intron or exon definition modes of splice site recognition. Using the slowly hydrolysable ATP analog ATPγS during preincubations, we were able to demonstrate that splice site pairing has not occurred, even though higher-order A complexes are able to form. These results are consistent with the proposal that a dedicated ATP hydrolysis event is utilized for splice site pairing. Together, these data show that the assembly pathways of the spliceosome for different types of alternative splicing converge to a common pathway after A complex formation.
MATERIALS AND METHODS
RNAs.
The substrate for alternative 5′ splice site selection, fruF, was generated as previously described (24). The substrate for testing alternative exon inclusion, βGskip, is a fusion of the human β-globin precursor containing exon 1, intervening sequence 1, and exon 2 (33). The fusion generates a three-exon substrate, which was further modified to increase the 5′ splice site strength of exon 1 and to increase the length of intron 2. Detailed information regarding the generation of βGskip is available in the supplemental material. fruF and βGskip were linearized with XhoI and ClaI, respectively, prior to transcription.
In vitro splicing and kinetic trap assay.
In vitro splicing reactions were carried out in 30% HeLa cell nuclear extracts as previously described (13). For fruF, the presence of saturating amounts of recombinant His-tagged Tra/Tra2 proteins (400 nM) was used to activate female-specific splicing. Recombinant Tra/Tra2 were purified as described previously (35). βGskip reactions were carried out in 60 mM KCl and in the presence or absence of 3% polyvinyl alcohol (PVA) (ICN Biomedicals; molecular weight, 15,000). PVA is a molecular crowding agent that increases the effective concentration of larger molecules, including splicing components and regulators. By including PVA, the effective concentration of a crucial splicing factor (unknown for βGskip) is increased over a threshold to induce preferential exon inclusion (13). To deplete extracts of ATP for E complex enrichment, HeLa cell nuclear extracts were incubated at 37°C for 30 min prior to use (25).
The kinetic trap assay to test commitment to splice site pairing followed published protocols (25). For fruF, all reaction mixtures were assembled with radiolabeled fruF in the presence or absence of Tra/Tra2 and were preincubated in ATP-depleted nuclear extracts at 30°C for 30 min in the absence of ATP and creatine phosphate to enrich the mixture in E complex. The reactions were then chased with ATP, creatine phosphate, and Tra/Tra2 (if needed), and mixtures were incubated at 30°C for 90 min. Competitor pre-mRNA was added when necessary. For reactions marked 5, 10, 15, and 20 min, after E complex enrichment, ATP and creatine phosphate were added and left for the corresponding amount time for a secondary preincubation at 30°C. Following these secondary preincubations, the reactions were chased at 30°C for 90 min with ATP, creatine phosphate, Tra/Tra2, and competitor pre-mRNA when necessary. Reaction lariats were visualized and quantitated by PhosphorImager analysis to monitor changes in splicing. Each kinetic trap assay was repeated at least four times. Kinetic analyses for commitment to splice site pairing were based on the switch from female to cryptic splicing after normalization to the observed ratio at the E complex. Rates were determined as previously described (13).
To test commitment for βGskip, 20-μl reaction mixtures were assembled with radiolabeled βGskip in either the absence or presence of 3% PVA and preincubated in ATP-depleted nuclear extracts at 30°C for 30 min to enrich the mixture in E complex. The chase was initiated by adding 20 μl of a chase mix containing 1 mM ATP, 20 mM creatine phosphate, 3.2 mM MgCl2, 12 mM HEPES (pH 7.9), 60 mM KCl, 30% HeLa nuclear extract, and 6% PVA. The final reaction volume of 40 μl was then incubated for 90 min at 30°C. For reactions marked 5, 10, 15, and 20 min, following E complex enrichment, ATP and creatine phosphate were added for a secondary preincubation at 30°C. Following the secondary preincubation, the chase reaction was initiated by changing the PVA concentration to 3% and continuing incubation at 30°C for 90 min. Reaction lariats were visualized and quantitated by PhosphorImager analysis, and each kinetic trap assay was repeated at least four times. Kinetic analyses for commitment to splice site pairing were based on the switch from internal exon inclusion to exclusion after normalization to the observed ratio at the E complex. Rates were determined as previously described (13).
Kinetic trap assay for intron definition.
To test commitment to splice site pairing in intron definition, a reverse βGskip chase was carried out. Instead of changing conditions from 0% PVA to 3% PVA, the reaction conditions were changed from 4% PVA to 1% PVA. This was achieved by setting up 10-μl preincubation reaction mixtures (at 4% PVA) that were eventually combined with a 30-μl chase reaction mixture (at 0% PVA). All other conditions were identical to those described above.
Kinetic trap assay for ATP hydrolysis.
Reaction mixtures were assembled with radiolabeled fruF pre-mRNA in HeLa nuclear extracts depleted of ATP in the presence of 1 mM ATPγS [adenosine-5′-O-(3-thiotriphosphate); Roche Diagnostics] and were preincubated at 30°C for 30 min; 400 nM Tra/Tra2 was added to a control reaction mixture where specified. Following the preincubation, 400 nM recombinant Tra/Tra2 was added to the reaction mixture where necessary, and ATP, creatine phosphate, and competitor pre-mRNA were added to all reaction mixtures for a 90-min chase incubation at 30°C. Reactions were analyzed as described above.
Spliceosome complex formation.
To analyze E complex formation, radiolabeled fruF or βGskip reaction mixtures were assembled under splicing conditions and resolved by native agarose gel electrophoresis as previously described (6). For analysis of ATP-dependent complexes, radiolabeled fruF and βGskip reaction mixtures were assembled in ATP-depleted extracts to enrich the mixture in E complex. ATP was then added to reaction mixtures and incubated at 30°C for various times. Reactions were then resolved by 3% nondenaturing polyacrylamide gel electrophoresis (PAGE) analysis for 10 h as previously described (22). Complex formation of nuclear extract depleted of U4 snRNA was determined as previously described (14). To analyze complex formation in the presence of ATPγS, reaction mixtures were assembled as described above and incubated at 30°C for 10, 30, and 45 min prior to loading for 3.5% nondenaturing PAGE.
RESULTS
Commitment to alternative 5′ splice site pairing occurs in the A complex.
We have recently demonstrated that alternative 3′ splice site selection coincides with A complex formation (25). To test the generality of this initial observation, we analyzed when commitment to splice site pairing is established for additional alternative splicing events, alternative 5′ splice site selection, and alternative exon inclusion. As a model substrate for alternative 5′ splice site selection, we used a previously characterized minigene of the Drosophila melanogaster fruitless (fruF) gene product (24). In the presence of the SR-like proteins Transformer and Transformer 2 (Tra/Tra2), the female-specific proximal 5′ splice site is activated (Fig. 1A). When these splicing activators are absent, a cryptic distal 5′ splice site is preferentially selected. Importantly, addition of recombinant Tra/Tra2 protein resulted in a significant switch from cryptic to female spliced product (Fig. 2A, lanes 1 and 3).
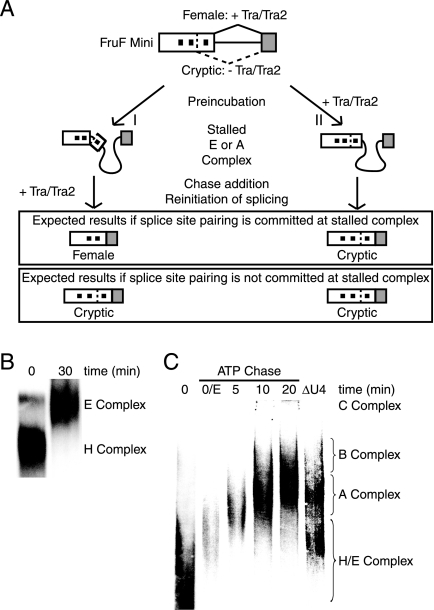
FIG. 1.
Kinetic trap assay to test commitment to alternative 5′ splice site pairing. (A) Schematic of kinetic trap assay to test commitment to alternative 5′ splice site pairing. The common 3′ splice site of fruF competes for the female-specific 5′ splice site (proximal) or the cryptic 5′ splice site (distal). Reaction preferences and expected outcomes in the presence or absence of Tra/Tra2 are indicated. The black boxes within the exon denote Tra/Tra2 binding sites. (B) Native agarose gel electrophoresis of fruF incubated for 0 or 30 min in nuclear extract depleted of ATP. (C) Spliceosomal complex progression of fruF after addition of ATP and creatine phosphate to fruF preincubated in nuclear extract depleted of ATP. The last lane shows complex formation of fruF incubated in nuclear extract depleted of U4 snRNA for 30 min.
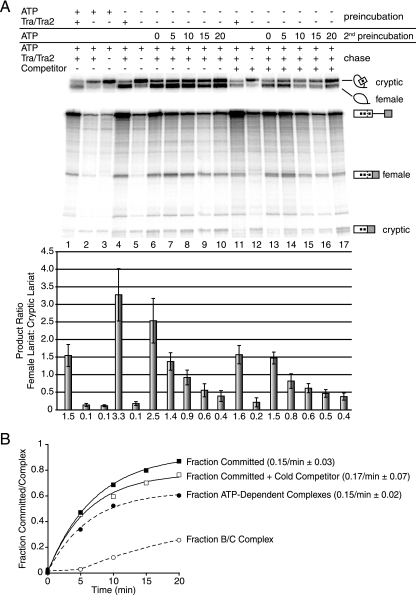
FIG. 2.
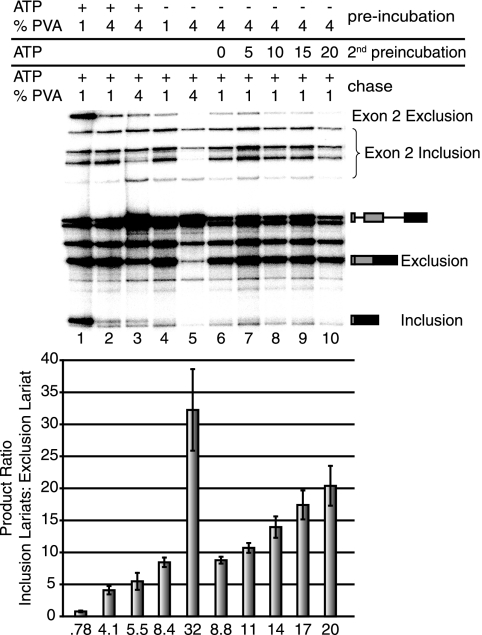
Commitment to alternative 5′ splice site pairing occurs in the A complex. (A) Chase reactions for fruF enriched in E or A complexes. After preincubation in extract depleted of ATP, reactions were chased by the addition of ATP and creatine phosphate. The presence or absence of ATP, Tra/Tra2, and competitor pre-mRNA and the length of ATP incubation prior to chase initiation are indicated above each lane. “Preincubation” refers to the initial reaction conditions, “2nd preincubation” refers to the addition of ATP for the corresponding amounts of time in minutes so ATP-dependent complexes could form, and “chase” refers to changes in reaction conditions following either the primary or secondary preincubation. Error bars represent the standard deviations from at least four independent experiments. The identity of splicing intermediates and products is indicated to the right. Quantitation of female to cryptic splicing is based on lariat formation and is graphed below each lane. The number below each bar is the mean value from four independent experiments. (B) Kinetic analysis of commitment to splice site pairing based on the data shown in panel A. Observed rates are compared to the observed rates for A complex formation from Fig. 1C. The ratio of inclusion to exclusion was normalized to the ratio at time zero and subtracted by one to get the fraction committed.
Prior to evaluating commitment to splice site pairing, we tested whether the efficiency of splicing depended on the addition of Tra/Tra2. Time course experiments demonstrated that the addition of recombinant Tra/Tra2 did not alter the kinetics of spliced product appearance; only splice site choice was affected (see Fig. S1A in the supplemental material). To test commitment to splice site pairing for fruF, we used an in vitro kinetic trap assay that we have recently developed (25). Briefly, by changing reaction conditions (chase) after enrichment of spliceosomal intermediate complexes (preincubation), this assay identifies the spliceosomal assembly step that coincides with commitment to a particular splicing pattern (Fig. 1A). Enrichment of the E complex can be achieved by incubating radiolabeled pre-mRNAs in extracts depleted of ATP. Thus, one of the reactions was assembled in ATP-depleted extract without recombinant Tra/Tra2 (Fig. 1A, pathway I), setting up preferential cryptic splice site selection. After complex enrichment, the reaction was chased by the addition of recombinant Tra/Tra2, which promotes the use of the female 5′ splice site. If commitment to splice site pairing occurred during or prior to formation of the stalled complex in the preincubation, then adding Tra/Tra2 would not change the outcome of the reaction, and the cryptic 5′ splice site would still be preferred. However, if splice site pairing was not established during the preincubation, then the addition of Tra/Tra2 would change the splicing pattern from cryptic to female splicing. As controls, recombinant Tra/Tra2 were added throughout the reaction to demonstrate female 5′ splice site dependence (Fig. 1A, pathway II, and Fig. 2A, lane 1), or the enhancers Tra/Tra2 were not added to show cryptic splicing (Fig. 2A, lane 3).
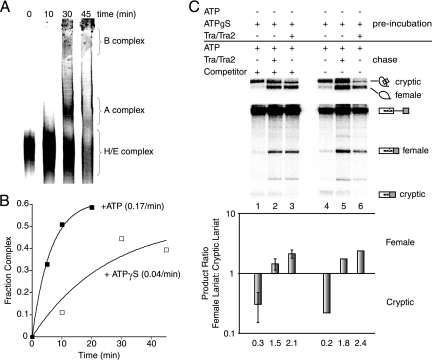
To obtain meaningful data from the kinetic trap assay, it is essential that spliceosomal intermediate complexes can be enriched during the preincubation period. The earliest detectable spliceosomal intermediate complex, the E complex, is easily accumulated by incubating splicing reaction mixtures in nuclear extract depleted of ATP. As demonstrated by native agarose gel electrophoresis, incubation of fruF in nuclear extract depleted of ATP for 30 min resulted in significant enrichment of the E complex (Fig. 1B). To enrich fruF in the A complex, a similar but modified approach was used. We have previously demonstrated A complex enrichment after short incubations of stalled E complex with ATP (19, 25). Due to the large size of fruF, we used 3% native PAGE to follow time-dependent higher-order complex formation after E complex enrichment (Fig. 1C) (22). Incubation of fruF in extracts depleted of U4 snRNP served as a marker for A complex formation. Five minutes after the addition of ATP, only a small fraction of pre-mRNAs progressed from E to A complex. At the later time points the majority of fruF pre-mRNAs converted into A, B, and C complexes. Importantly, B complex formation was observed only after a 5-min lag (Fig. 1C and Fig. 2B). This delay is expected because the spliceosome forms in a stepwise fashion; for example, B complex formation requires prior A complex assembly. Quantitation of these data demonstrated that the A complex formed at a rate of 0.15/minute ± 0.02 (Fig. 1C and Fig. 2B).
To test the hypothesis that commitment to alternative 5′ splice site selection occurs during A complex formation, the kinetic trap assay was carried out with fruF. Reaction mixtures were preincubated to form the E complex, and then either chase was carried out to test if commitment to splice site pairing occurred in the E complex or ATP was added and left for 5, 10, 15, or 20 min to allow higher-order complexes to assemble prior to the initiation of the chase. As expected, in the presence of Tra/Tra2, female-specific splicing was preferred at the E complex (Fig. 2A, lane 4), and in the absence of Tra/Tra2, cryptic 5′ splice site activation was favored (Fig. 2A, lane 5). However, when the E complex was enriched in the absence of activators and then chased with Tra/Tra2, a clear preference for female splicing was observed (Fig. 2A, lane 6). These results were similar to those obtained when Tra/Tra2 were present from the beginning of the reaction, demonstrating that commitment to alternative 5′ splice site pairing had not occurred in the E complex. Once ATP was added to the reaction mixture and left for just 5 min after E complex enrichment, the preference for cryptic splicing increased. This trend continued with the 10-, 15-, and 20-min secondary ATP incubations (Fig. 2A, lanes 7 to 10). From these data points we derived the rate of switching between female-specific to cryptic 5′ splice sites to be 0.15/minute ± 0.03 (Fig. 2B). This rate essentially represents the rate of commitment to splice site pairing. When evaluating the kinetics for commitment to splice site pairing, it is apparent that the rate profile does not contain an obvious lag as has been observed for B complex formation. In addition, the calculated rate for splice site commitment (0.15/minute ± 0.03) is identical to the observed rate for A complex formation (Fig. 2B). We conclude from these observations that commitment to alternative 5′ splice site pairing coincides with A complex formation. To control for the possibility that not all of the fruF pre-mRNA had formed E complex during the preincubation or that the spliceosome reformed on some pre-mRNAs during the chase, we performed the same experiments in the presence of 100 nM unlabeled β-globin pre-mRNA competitor (25, 27). Adding this concentration of pre-mRNA at the beginning of the reaction inhibited all formation of spliced product, as the excess unlabeled substrate squelched the splicing of the radiolabeled substrate (see Fig. S2 in the supplemental material). When unlabeled pre-mRNA was added to the chase incubation of the reaction mixtures, the rate of commitment to alternative 5′ splice site pairing was not altered (Fig. 2A, lanes 11 to 17, and B). These experiments confirm that commitment to a splicing pattern occurred in the A complex for alternative 5′ splice site pairing.
Commitment to alternative exon inclusion occurs during A complex formation.
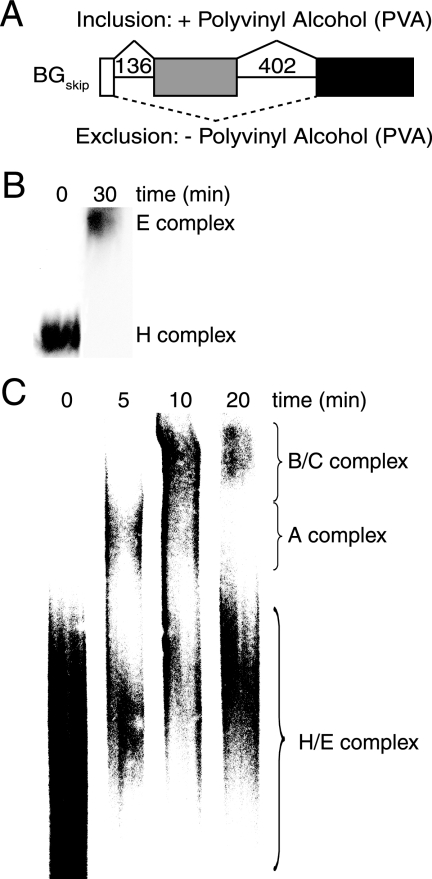
Since alternative 3′ and 5′ splice sites are selected for a particular splicing pattern during A complex formation, we set out to determine if the mechanisms of alternative exon inclusion work in a similar fashion. For this set of experiments, we made use of a pre-mRNA substrate (βGskip) that switches between exon inclusion and exon exclusion upon the addition of the molecular crowding agent PVA (Fig. 3A). PVA increases the effective concentration of larger molecules, including splicing components and regulators. By including PVA, the effective concentrations of splicing factors are increased to induce preferential exon inclusion (13). When PVA is absent from the reaction mixture, both inclusion and exclusion of the internal exon are observed, with exclusion being the favored product (Fig. 4A, lane 3). However, inclusion was favored when PVA was added to the splicing reaction mixture (Fig. 4A, lane 1). As was observed with fruF, the kinetics of splicing product appearance were unaltered in the presence or absence of PVA (see Fig. S1B in the supplemental material). Native agarose gel electrophoresis of βGskip pre-mRNA in the absence of PVA demonstrated essentially complete E complex formation after a 30-min incubation in extract depleted of ATP (Fig. 3B). In addition, the kinetics of A complex formation were analyzed by enriching in the E complex, followed by a secondary incubation with ATP for either 5, 10, or 20 min prior to loading for native 3% PAGE as described for fruF. The A complex was the main complex observed after 5 min, followed by a shift to B/C complexes after 10 min. Rate analysis of the gel shift data showed that the rate of A complex formation was 0.17/minute (Fig. 4B). B/C complex was detected after a short lag (Fig. 4B). Similar rates of formation of higher-order complexes were observed in the presence of PVA (data not shown).
FIG. 3.
E complex enrichment and kinetics of higher-order complex formation for βGskip. (A) Schematic of βGskip pre-mRNA highlighting splicing preferences in the presence or absence of PVA. (B) Enrichment of the E complex. βGskip pre-mRNA was preincubated in extract depleted of ATP prior to native agarose gel electrophoresis. (C) Complex formation of βGskip pre-mRNA after E complex enrichment. Reactions were initiated by the addition of ATP and resolved by native polyacrylamide gel electrophoresis.
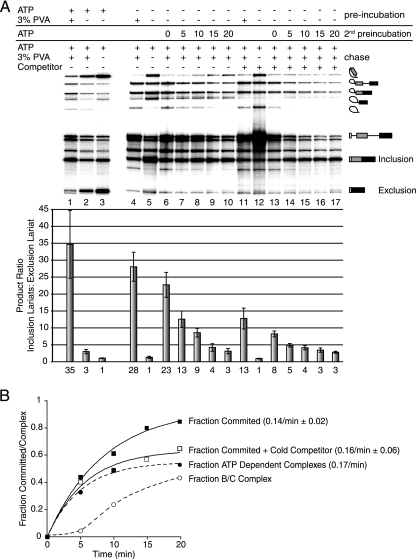
FIG. 4.
Commitment to alternative exon inclusion coincides with A complex formation. (A) Chase reactions for βGskip enriched in E or A complexes. After preincubation in extract depleted of ATP, reactions were chased by the addition of ATP and creatine phosphate. The presence or absence of ATP/creatine phosphate, PVA, and competitor pre-mRNA and the length of ATP incubation prior to chase initiation are indicated above each lane. “Preincubation” refers to the initial reaction conditions, “2nd preincubation” refers to the addition of ATP for the corresponding amounts of time in minutes so ATP-dependent complexes could form, and “chase” refers to changes in reaction conditions following either the primary or secondary preincubation. Error bars represent the standard deviations from at least four independent experiments. The identity of splicing intermediates and products is indicated to the right. Quantitation of the exon inclusion/exclusion ratio is based on lariat formation and is graphed below each lane. The number below each bar is the mean value from four independent experiments. (B) Kinetic analysis of commitment to splice site pairing based on the data shown in panel A. Observed rates of commitment were compared to the observed rates for A complex formation.
To test the hypothesis that commitment to exon inclusion or exclusion coincides with A complex formation, reaction mixtures were preincubated in the absence of PVA and enriched in the E complex. Under these conditions, exon exclusion would be favored (Fig. 4A, lane 5). When PVA is added during the chase, then two different outcomes could be expected depending on whether commitment had occurred or not. If inclusion is the preferred product, then the addition of PVA to the reaction mixture was able to switch splice site choice and commitment to pairing had not yet happened. However, if PVA addition does not alter the exon exclusion preference, then commitment occurred during the initial complex enrichment.
E complex enrichment of βGskip in the presence of PVA during the preincubation resulted in a preference for internal exon inclusion, while barely any excluded product was visible (Fig. 4A, lane 4). In the absence of PVA, there is a preference for the excluded product (Fig. 4A, lane 5). Indeed, enrichment of E complex preincubated in the absence of PVA, followed by a chase with PVA and ATP, resulted in a preference for exon inclusion (Fig. 4A, lane 6), demonstrating that commitment to alternative exon inclusion did not occur during E complex formation. Since commitment did not coincide with E complex formation, short secondary incubations with ATP following E complex enrichment were carried out to analyze the kinetics of commitment during later complex formation. Over the time course of ATP addition prior to chase initiation, we observed a reduction in the preference of inclusion toward more efficient exon exclusion (Fig. 4A, lanes 7 to 10). As was observed for alternative 5′ splice site selection, the observed rate of commitment to splice site pairing (0.14/minute ± 0.02) was similar in magnitude to that of A complex formation (0.17/minute), and it occurred without a lag (Fig. 4B). Furthermore, similar results were observed when 100 nM unlabeled βGskip was added to the reaction mixture (Fig. 4A, lanes 13 to 17, and B). We conclude from these results that commitment to splice site pairing of this three-exon substrate occurred during A complex formation.
The timing of splice site pairing is independent of intron or exon definition.
Previous studies demonstrated that the exon/intron architecture influences the efficiency of splicing (10, 37). When the intron length is less than 250 nucleotides, splice sites are recognized across the intron through intron definition (10). When introns are longer than 250 nucleotides, splice sites are recognized across the exon. This exon definition mechanism may necessitate an extra step to pair splice sites (12). For example, the test substrate fruF contains a long intron, and therefore this construct's splice sites are recognized through exon definition. To test the hypothesis that commitment to splice site pairing occurs prior to A complex formation if splice sites are recognized by intron definition, we investigated alternative exon inclusion for βGskip, which undergoes inclusion when PVA is present and exclusion when the molecular crowding agent is absent. The first intron of βGskip is 136 nucleotides long, while the second intron is 402 nucleotides long (Fig. 3A). In our commitment assays described in Fig. 4, preincubation conditions in the absence of PVA favored exon exclusion. Thus, splice sites in the absence of PVA were recognized through exon definition. To test whether intron definition permits splice site pairing to occur prior to A complex formation, we carried out a reverse chase, with preincubation conditions favoring exon inclusion in the presence of PVA. Under these conditions, the splice sites flanking the first intron are recognized through intron definition. After E complex enrichment in the presence of PVA, a chase reaction was initiated by reducing the PVA concentration from 4% to 1%, a change in concentration that favors internal exon exclusion (Fig. 5, compare lanes 1 and 3). Importantly, when the E complex was enriched in the presence of 4% PVA and then diluted to 1% PVA, exclusion was the preferred product, similar to the case for reactions assembled in 1% PVA (Fig. 5, compare lanes 4 and 6). This result demonstrated that commitment to intron-defined splice site pairing did not occur in the E complex. When ATP was added to reaction mixtures prior to the chase initiation and left for 5, 10, 15, and 20 min, the preference for exclusion switched to inclusion over time, showing that commitment occurs in higher-order complexes, most likely the A complex (Fig. 5). We conclude that commitment to splice site pairing does not occur prior to A complex formation when splice sites are recognized through intron definition.
FIG. 5.
Commitment to splice site pairing is independent of intron or exon definition. Results of chase reactions for βGskip enriched in E or A complexes in the presence of 1% or 4% PVA are shown. After “preincubation” in ATP-depleted extract, ATP and creatine phosphate was added to the indicated reactions for the corresponding amounts of time, referred to as “2nd preincubation.” Reactions were “chased” by the addition of ATP and creatine phosphate and diluting the PVA concentration from 4% to 1%. Error bars represent the standard deviations from at least four independent experiments. The identity of splicing intermediates and products is indicated to the right. Quantitation of the exon inclusion/exclusion ratio is based on lariat formation and is graphed below each lane. The number below each bar is the mean value from four independent experiments.
The ATP requirement for splice site pairing.
The transition from E to A complex requires ATP (27). This transition also coincides with splice site pairing. To evaluate the ATP requirement during splice site pairing, we performed commitment assays using the pre-mRNA substrate fruF in the presence of the ATP analog ATPγS. This analog is slowly hydrolysable and unlikely to support intron excision (28). In agreement with this interpretation are observations that nuclear extracts supplemented with ATPγS are splicing incompetent (38) (see Fig. S2 in the supplemental material). In the first set of experiments, spliceosomal complex formation was analyzed in the presence of ATPγS. Splicing reactions using ATPγS in place of ATP were resolved on native nondenaturing polyacrylamide gels. In agreement with published work, ATP-dependent spliceosomal complexes were able to form in the presence of ATPγS (38) (Fig. 6A). However, rate analyses indicated that ATP-dependent spliceosomal complexes accumulated approximately fourfold faster than when ATPγS was the sole energy source. Interestingly, after a 30-min incubation, more than 50% of fruF pre-mRNAs were converted into higher-order complexes (Fig. 6B). These observations are consistent with the hypothesis that slow hydrolysis of ATPγS reduced the overall efficiency of spliceosomal assembly. An alternative interpretation is that A complex formation can occur without ATP hydrolysis, albeit at reduced rates. We conclude that in the presence of the ATP analog ATPγS, ATP-dependent spliceosome complex formation is supported, although inefficiently.
FIG. 6.
ATP hydrolysis requirement for establishing splice site pairing. (A) Native gel analysis of fruF pre-mRNA incubated with ATPγS in HeLa cell nuclear extract depleted of ATP. (B) Quantitation of the data in panel A. (C) fruF splicing reaction mixtures were assembled with ATPγS and preincubated for 30 min. Following this preincubation in extract depleted of ATP, reactions were chased by the addition of ATP and creatine phosphate. The presence or absence of ATP/creatine phosphate, Tra/Tra2, or 100 nM cold competitor β-globin pre-mRNA is indicated above each lane. “Preincubation” refers to the initial reaction conditions, and “chase” refers to changes in reaction conditions following the preincubation. Error bars represent the standard deviations from at least three independent experiments. The identity of splicing intermediates and products is indicated to the right. Quantitation of the exon inclusion/exclusion ratio is based on lariat formation and is graphed below each lane. The number below each bar is the mean value from three independent experiments. Results of ATPγS kinetic trap reactions are displayed as the ratio of female to cryptic lariat formation.
To examine the role of ATP hydrolysis in splice site pairing, we carried out chase experiments in the presence of ATPγS. If A complex formation alone is sufficient to commit splice sites, we would expect to see commitment occur with the formation of any higher-order complexes in the presence of ATPγS. If a dedicated ATP hydrolysis is required to establish splice site pairing, we would expect to lose commitment in the presence of ATPγS. To test the ATP requirement for splice site pairing, we used the same kinetic trap approach as described for Fig. 2 except that ATPγS was added instead of ATP. A preincubation was initiated in ATP-depleted nuclear extract with the addition of ATPγS. After 30 min, splicing was initiated by supplementing the reaction mixtures with ATP, cold β-globin competitor pre-mRNA to prevent the formation of new splicing complexes, and Tra/Tra2 if appropriate. In the absence of the splicing activators Tra/Tra2, cryptic 5′ splice site selection predominates, similar to results when ATP was added (Fig. 6C, lanes 1 and 4). In the presence of Tra/Tra2, the female 5′ splice site is preferentially selected, as expected (Fig. 6C, lanes 3 and 6). Importantly, when Tra/Tra2 were added after the preincubation, the female-specific 5′ splice site was preferentially used, indicating that at the time of the chase, commitment to splice site pairing was not established (Fig. 6C, lanes 2 and 5). The small fraction that appears to be committed during this chase reaction (Fig. 6C, compare lane 2 with lane 3 or lane 5 with lane 6) is consistent with the observation that at the time of chase initiation, complex formation for some pre-mRNAs has progressed to the B complex (Fig. 6A). At first glance, these results suggest that proper ATP hydrolysis is required to establish splice site pairing even though higher-order complexes had already formed. An alternative explanation for the results is that none of the higher-order complexes observed in the presence of ATPγS are functional complexes and the splicing ratios obtained originated from pre-mRNAs that were assembled in the E complex at the time of chase initiation.
To evaluate if higher-order complexes assembled in ATPγS are functional and to determine to what degree spliceosomal complexes stalled in the E complex contribute to the chase results, we carried out additional pulse-chase experiments. The kinetics of splicing were compared between reactions where a chase with ATP and cold competitor was initiated after E complex enrichment in the absence of any ATP, after preincubation with ATP, and after preincubation with ATPγS (see Fig. S4 in the supplemental material). The results show that only a small fraction of pre-mRNAs enriched in the E complex are chased to spliced products, whereas ATP or ATPγS preincubations result in much more significant levels of splicing. Importantly, the kinetics of intron removal are comparable under all three conditions (see Fig. S4D in the supplemental material). Thus, the chase reaction does not alter the speed of intron removal, but it limits the fraction of pre-mRNAs that have appropriately assembled at the time of chase initiation. We conclude from this comparison that the higher-order complexes that are observed upon ATPγS incubation are not dead-end complexes. However, these experiments do not indicate whether these higher-order complexes remodel prior to going on to execute intron removal. In summary, our experiments using the ATP analog ATPγS are consistent with the interpretation that, independent of higher-order complex formation, splice site pairing requires a dedicated ATP hydrolysis event.
DISCUSSION
In this study, we demonstrate that commitment to splice site pairing coincides with A complex formation. The timing of splice site pairing was investigated in the context of alternative 5′ splice site selection and alternative exon inclusion. In both cases we determined when a splicing pattern is established by using a pulse-chase approach after the enrichment of spliceosomal intermediate complexes. By incubating pre-mRNAs in nuclear extracts depleted of ATP, it is relatively easy to enrich the earliest spliceosomal complex, the E complex. However, functional forms of the subsequent A, B, and C complexes are more difficult to accumulate because of the dynamic and ATP-dependent transitions between assembly steps. In our approach to differentiate whether splice site pairing is established in the A complex or subsequent complexes, we took advantage of the stepwise transition from E to C complex. For example, B complex formation is observed only after a short lag because the preceding A complex needs to be formed prior to the stable association of the tri-snRNP, which is characteristic of the B complex (Fig. 1C, 2B, 3C, and 4B). When comparing the kinetics of pairing commitment with the kinetics of higher-order complex formation, we were able to show that A complex formation is the spliceosomal assembly step that establishes functional splice site pairing. This conclusion is based on the argument that splice site pairing was observed to be efficient and, importantly, without a lag. Moreover, the observed rates of A complex formation, as determined from native gel analysis, and the rate of commitment to splice site pairing were virtually identical (Fig. 2B and 4B). In combination with our previous work (25), these results show that commitment to splice site pairing occurs during A complex formation, regardless of the type of alternative splicing analyzed. These data indicate that during alternative splicing, the assembly pathways of the spliceosome converge to a common pathway, most likely defined after the stable association of the U2 snRNP with the branch point, which is characteristic for A complex formation. Once splice site pairing is firmly established, the remaining components of the splicing machinery associate and rearrange to allow the creation of a functional catalytic center around the paired splice sites.
It has been demonstrated that the efficiency and mechanism of splice site recognition depend on intron length (3, 10). Splice sites flanking introns less than 250 nucleotides in length are typically recognized across the intron (intron definition), with spliceosomal components assembling around the intron that will be excised. Splice sites flanking long introns (>250 nucleotides) are recognized across the exon (exon definition). It is currently unknown how spliceosomal components assembled across exons are combined to define the intron that will be excised. However, one mechanistic difference between the two modes of splice site selection may be the requirement of an additional exon juxtaposition step during exon definition. To test if this presumed difference elicits alternative mechanisms of splice site pairing, we determined if the pairing of intron-defined splice sites occurred earlier than the A complex. The results clearly demonstrated that during intron definition, splicing patterns are not established in the E complex but are established during the formation of ATP-dependent complexes, most likely the A complex. Thus, functional splice site pairing is established after the stable recruitment of the U2 snRNP, regardless of intron or exon definition.
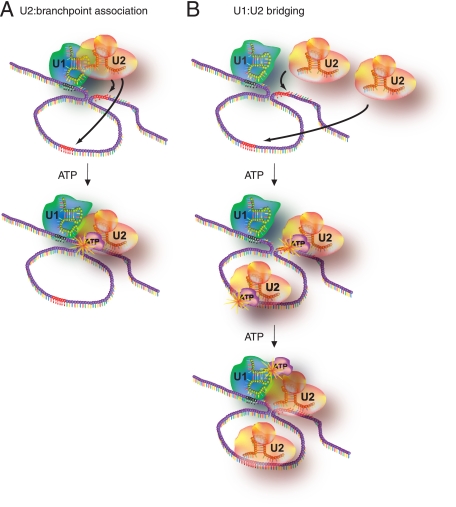
Because the A complex is characterized by stable U1 and U2 snRNP binding to the pre-mRNA, two models were considered for splice site pairing. In the first model, the loose association of the U2 snRNP in the E complex is transitioned into a stable association of the U2 snRNP with the branch point during A complex formation (Fig. 7A). This step requires ATP hydrolysis to induce U2 snRNA conformational changes that permit stable interactions with the branch point (29-31). It is possible that this snRNP rearrangement event alone is sufficient to establish splice site pairing by promoting direct or indirect interactions between components of the U1 and U2 snRNPs. Thus, splice site pairing could be a direct consequence of stable branch point/U2 snRNA interactions. An alternative model proposes a dedicated splice site pairing step after stable U2 snRNP binding to the branch point (Fig. 7B). Considering that splice site pairing increases the order of the prespliceosomal complex, this step would be expected to require energy consumption. To differentiate between these models, we carried out pulse-chase experiments in the presence of ATPγS, a slowly hydrolysable ATP analog. Although ATPγS does not support intron removal, we and others (38) demonstrated that higher-order complexes form, albeit at reduced rates. Surprisingly, our kinetic trap experiments showed that splice site pairing was not established in these spliceosomal intermediate complexes. These results are consistent with the interpretation that a dedicated ATP hydrolysis step is required to functionally pair splice sites. Furthermore, these experiments suggest that the stable association of the U2 snRNP to the branch point and splice site pairing are separate steps during spliceosomal assembly. In this scenario U2 snRNP binding is expected to precede splice site pairing, because ATPγS supports A complex formation without committing to splice site pairing (Fig. 6). While these arguments do not unequivocally prove the existence of a dedicated ATP hydrolysis step for splice site pairing, the results are more consistent with the second model of commitment to splice site pairing (Fig. 7B).
FIG. 7.
Models for splice site pairing. (A) Commitment to a splicing pattern may occur as a consequence of the U2 snRNP's stable association with the branch point sequence during the transition from the E complex to the A complex. (B) Following U2 snRNP's stable association with the branch point sequence (red bases), the functional pairing of splice sites may be established in a distinct step in a separate ATP hydrolysis event. For both pathways, ATP hydrolysis is required to lock U1 snRNP and U2 snRNP into close proximity. The 5′ splice site is highlighted in black, and the 3′ splice sites are in red. The arrows indicate potential interactions between the U2 snRNP and the pre-mRNA.
Recent experiments demonstrated that in some cases regulation of alternative splicing can occur after E complex formation (5, 15, 36). In two cases, it was shown that the presence of the splicing repressors prevented inclusion of an internal exon by stalling at the A complex (15, 36). Thus, stable association of the U2 snRNP was observed; however, splice site pairing was not yet established. Most recently, RBM5 was identified as a regulator of fas exon 6 alternative splicing (5). Similar to the case in other studies, RBM5 interfered with the transition from cross-exon to cross-intron interactions, even though U1 and U2 snRNPs were firmly associated with the regulated exon. It is plausible that the formation of these splicing regulatory complexes stabilizes interactions between U1 and U2 snRNPs across the exon, thereby interfering with the productive pairing of its splice sites across the neighboring introns (23). In light of our proposal that a dedicated ATP hydrolysis step is required to establish splice site pairing, it is conceivable that this inhibition could be achieved through blocking the proposed ATP hydrolysis step for splice site pairing or through preventing snRNP rearrangements that may be necessary prior to splice site pairing across the intron. Regardless of the mechanism, our commitment assay results in the presence of ATPγS support the notion that the spliceosome is equipped with multiple avenues to regulate alternative splicing.
Considering that only the U1 snRNP and U2 snRNP are stably associated with the pre-mRNA during A complex formation, components that make up these snRNPs are reasonable candidates to be involved in mediating commitment to splice site pairing. In addition, the ATP hydrolysis requirement suggests that ATPases are involved in this crucial step. Mass spectroscopy of purified A complex identified four proteins with ATPase activity, and all are part of the U2 snRNP (1, 11, 40). The DEAD box ATPase hPrp5 emerges from this group as an attractive candidate that may be involved in mediating commitment to a splicing pattern. In addition to its role in regulating U2 snRNA secondary structure in yeast (29-31), hPrp5 has been shown to immunoprecipitate both U1 and U2 (41). In yeast, ATP hydrolysis by Prp5 has recently been shown to proofread U2 snRNP/branch point interactions, permitting only highly stable associations (42). This function, however, appears to be less important in metazoans, where limited base pair interactions between U2 snRNA and the branch point are the norm (42). Thus, it is possible that the role of Prp5 has evolved to facilitate cross-intron interactions between the U2 snRNP and components of the U1 snRNP, a proposal consistent with the low conservation of Prp5 from yeast to humans (40). Besides its set of SM core proteins, the U1 snRNP contains only three factors, U1-70k, U1-A, and U1-C. While none of these components have been shown to hydrolyze ATP, U1-70k has been associated with the regulation of alternative splicing and with bridging the upstream 3′ splice site through an interaction with the U2 auxiliary factor in the E complex. Thus, U1-70k may be the best U1 snRNP candidate to complete the cross-intron bridge with the U2 snRNP that establishes splice site pairing. Based on our conclusion that splice sites are functionally paired at the A complex, future experimental efforts will focus on determining the molecular basis for splice site pairing.
Supplementary Material
Acknowledgments
We are grateful to Bianca Lam for generating βGskip and to members of the Hertel laboratory for helpful comments on the manuscript. HeLa cells were obtained from the National Cell Culture Center (Minneapolis, MN).
This work was supported by NIH grant GM 62287 (to K.J.H.).
Footnotes
Published ahead of print on 8 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Behzadnia, N., M. M. Golas, K. Hartmuth, B. Sander, B. Kastner, J. Deckert, P. Dube, C. L. Will, H. Urlaub, H. Stark, and R. Luhrmann. 2007. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 261737-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, M., S. Michaud, J. Kingston, and R. Reed. 1992. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 61986-2000. [DOI] [PubMed] [Google Scholar]
- 3.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 2702411-2414. [DOI] [PubMed] [Google Scholar]
- 4.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. [DOI] [PubMed] [Google Scholar]
- 5.Bonnal, S., C. Martinez, P. Forch, A. Bachi, M. Wilm, and J. Valcarcel. 2008. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol. Cell 3281-95. [DOI] [PubMed] [Google Scholar]
- 6.Das, R., and R. Reed. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 51504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, R., Z. Zhou, and R. Reed. 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell 5779-787. [DOI] [PubMed] [Google Scholar]
- 8.Donmez, G., K. Hartmuth, B. Kastner, C. L. Will, and R. Luhrmann. 2007. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol. Cell 25399-411. [DOI] [PubMed] [Google Scholar]
- 9.Donmez, G., K. Hartmuth, and R. Luhrmann. 2004. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 101925-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox-Walsh, K. L., Y. Dou, B. J. Lam, S. P. Hung, P. F. Baldi, and K. J. Hertel. 2005. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. USA 10216176-16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Luhrmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 9916719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hertel, K. J. 2008. Combinatorial control of exon recognition. J. Biol. Chem. 2831211-1215. [DOI] [PubMed] [Google Scholar]
- 13.Hicks, M. J., B. J. Lam, and K. J. Hertel. 2005. Analyzing mechanisms of alternative pre-mRNA splicing using in vitro splicing assays. Methods 37306-313. [DOI] [PubMed] [Google Scholar]
- 14.Hicks, M. J., C. R. Yang, M. V. Kotlajich, and K. J. Hertel. 2006. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 4e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.House, A. E., and K. W. Lynch. 2006. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat. Struct. Mol. Biol. 13937-944. [DOI] [PubMed] [Google Scholar]
- 16.International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431931-945. [DOI] [PubMed] [Google Scholar]
- 17.Jamison, S. F., A. Crow, and M. A. Garcia-Blanco. 1992. The spliceosome assembly pathway in mammalian extracts. Mol. Cell. Biol. 124279-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch, C. D. Armour, R. Santos, E. E. Schadt, R. Stoughton, and D. D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 3022141-2144. [DOI] [PubMed] [Google Scholar]
- 19.Jurica, M. S., and M. J. Moore. 2002. Capturing splicing complexes to study structure and mechanism. Methods 28336-345. [DOI] [PubMed] [Google Scholar]
- 20.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 125-14. [DOI] [PubMed] [Google Scholar]
- 21.Kent, O. A., and A. M. MacMillan. 2002. Early organization of pre-mRNA during spliceosome assembly. Nat. Struct. Biol. 9576-581. [DOI] [PubMed] [Google Scholar]
- 22.Konarska, M. M., and P. A. Sharp. 1986. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46845-855. [DOI] [PubMed] [Google Scholar]
- 23.Kotlajich, M. V., and K. J Hertel. 2008. Death by splicing: tumor suppressor RBM5 freezes splice-site pairing. Mol. Cell 32162-164. [DOI] [PubMed] [Google Scholar]
- 24.Lam, B. J., A. Bakshi, F. Y. Ekinci, J. Webb, B. R. Graveley, and K. J. Hertel. 2003. Enhancer-dependent 5′-splice site control of fruitless pre-mRNA splicing. J. Biol. Chem. 27822740-22747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim, S. R., and K. J. Hertel. 2004. Commitment to splice site pairing coincides with A complex formation. Mol. Cell 15477-483. [DOI] [PubMed] [Google Scholar]
- 26.Michaud, S., and R. Reed. 1993. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 71008-1020. [DOI] [PubMed] [Google Scholar]
- 27.Michaud, S., and R. Reed. 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 52534-2546. [DOI] [PubMed] [Google Scholar]
- 28.Peck, M. L., and D. Herschlag. 2003. Adenosine 5′-O-(3-thio)triphosphate (ATPgammaS) is a substrate for the nucleotide hydrolysis and RNA unwinding activities of eukaryotic translation initiation factor eIF4A. RNA 91180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perriman, R., and M. Ares, Jr. 2000. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 1497-107. [PMC free article] [PubMed] [Google Scholar]
- 30.Perriman, R., I. Barta, G. K. Voeltz, J. Abelson, and M. Ares, Jr. 2003. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. USA 10013857-13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perriman, R. J., and M. Ares, Jr. 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 21811-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed, R. 1996. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Gen. Dev. 6215-220. [DOI] [PubMed] [Google Scholar]
- 33.Ruskin, B., A. R. Krainer, T. Maniatis, and M. R. Green. 1984. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38317-331. [DOI] [PubMed] [Google Scholar]
- 34.Ruskin, B., P. D. Zamore, and M. R. Green. 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52207-219. [DOI] [PubMed] [Google Scholar]
- 35.Sciabica, K. S., and K. J. Hertel. 2006. The splicing regulators Tra and Tra2 are unusually potent activators of pre-mRNA splicing. Nucleic Acids Res. 346612-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, S., L. A. Kohlstaedt, A. Damianov, D. C. Rio, and D. L. Black. 2008. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 15183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sterner, D. A., T. Carlo, and S. M. Berget. 1996. Architectural limits on split genes. Proc. Natl. Acad. Sci. USA 9315081-15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tazi, J., M. C. Daugeron, G. Cathala, C. Brunel, and P. Jeanteur. 1992. Adenosine phosphorothioates (ATP alpha S and ATP tau S) differentially affect the two steps of mammalian pre-mRNA splicing. J. Biol. Chem. 2674322-4326. [PubMed] [Google Scholar]
- 39.Will, C. L., and R. Luhrmann. 1997. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 9320-328. [DOI] [PubMed] [Google Scholar]
- 40.Will, C. L., H. Urlaub, T. Achsel, M. Gentzel, M. Wilm, and R. Luhrmann. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 214978-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, Y. Z., C. M. Newnham, S. Kameoka, T. Huang, M. M. Konarska, and C. C. Query. 2004. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 23376-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, Y. Z., and C. C. Query. 2007. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell 28838-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zamore, P. D., J. G. Patton, and M. R. Green. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355609-614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.