Abstract
Genetic variation in immune-regulating components such as cytokines may lead to interindividual differences in immunosuppression response and susceptibility to melanoma. We evaluated the associations between genetic variants in 5 interleukin (IL) and IL receptor genes (IL-4, IL-4R, IL-6, IL-6R, and IL-10) and the risk of melanoma. Twenty-five single nucleotide polymorphisms (SNPs) were selected that are functionally relevant or tagging SNPs of each gene. We conducted a nested case-control study of 219 female cases and 219 matched controls within the Nurses' Health Study. We observed that in the IL-6R gene, 4 SNPs in linkage disequilibrium were associated with an increased risk of melanoma. Three are located in introns (rs6684439, rs4845618, and rs4845622), and one is a nonsynonymous SNP in exon 9 (rs8192284 [Asp358Ala]). An elevated risk of melanoma was observed in the heterozygous groups of these SNPs with odds ratio of 1.74 ((95% confidence interval, 1.07, 2.81) for rs6684439, 1.72 (1.04, 2.84) for rs4845618, 1.69 (1.03, 2.75) for rs4845622, and 1.68 (1.04, 2.73) for rs8192284. But these associations were not observed in the homozygous variant group with odds ratios ranging from 0.93 to 1.03. We did not find significant results for the SNPs in the other 4 genes. These data suggest the involvement of IL-6R in melanoma development. Further studies are needed to confirm these findings.
Keywords: interleukins, interleukin receptors, single nucleotide polymorphism, melanoma
Introduction
Melanoma is the most serious form of skin cancer, and its incidence rate has been increasing in recent decades in the United States. Ultraviolet (UV) exposure is the most important risk factor for melanoma[1].
Studies with immune-suppressed renal transplant patients suggest that immune suppression contributes to melanoma susceptibility[2]. Cytokines, secreted in response to immune stimuli, are important in mediating and regulating immune response. IL-10 is an anti-inflammatory factor produced by multiple cell types such as keratinocytes. IL-10 acts as a promoter of human melanoma by inducing melanoma cell proliferation[3] and down-modulating the antitumor immune response[4, 5]. Conversely, IL-10 exerts antitumor and antimetastatic activity by inhibiting angiogenesis[6]. IL-4, secreted by CD15+ neutrophils and Th2 cells, can induce IL-10 secretion[7]. IL-6, a pro-inflammatory factor secreted by keratinocytes and macrophages, acts as a tumor growth inhibitor for early-stage melanomas but as a growth factor for advanced-stage tumor cells[8]. These cytokines act by binding to specific membrane receptors.
Genetic variation of these cytokines and their receptor genes may lead to different immune responses and susceptibility to melanoma. However, only a few studies have examined single nucleotide polymorphisms (SNPs) in these genes in relation to melanoma susceptibility, and the results are mixed. Howell et al.[9] reported that a low-expression genotype in the IL-10 promoter region is a risk factor for more advanced/poorer prognosis melanoma and may confer susceptibility to melanoma in British Caucasians (165 cases and 158 controls). No significant results were obtained for the IL-4 -590 C/T and IL-6 –174 G/C SNPs in regard to melanoma risk among 169 cases and 261 controls by the same group[10]. Recently, Penka et al.[11] found that the genotype frequency distribution of 3 IL-10 SNPs and one IL-6 SNP in 122 Caucasian melanoma patients were significantly different from those reported in healthy Caucasians.
We investigated the association between 5 cytokine/cytokine receptor genes (IL-4, IL-4R, IL-6, IL-6R, and IL-10) and melanoma susceptibility in a nested case-control study of 219 incident melanoma cases and 219 matched controls within the Nurses' Health Study (NHS) Cohort. We selected 25 SNPs in these genes (2 in the IL-4 gene, 2 in the IL-4R gene, 7 in the IL-6 gene, 10 in the IL-6R gene, and 4 in the IL-10 gene) and evaluated their association with melanoma risk.
Materials and methods
Study population
The NHS was established in 1976, when 121,700 female registered nurses between the ages of 30 and 55, residing in 11 larger U.S. states, completed a self-administered questionnaire on their medical histories and baseline health-related exposures. Updated information has been obtained by questionnaire every 2 years. During 1989 and 1990, blood samples were collected from 32,826 of the cohort members. The distribution of risk factors for skin cancer in the subcohort of those who donated blood samples was similar to that in the overall cohort. Eligible cases in this study consisted of women with incident melanoma from the subcohort who had given a blood specimen, with a diagnosis any time after blood collection up to June 1, 2000, and with no previously diagnosed skin cancer. One control per case was randomly selected from participants who gave a blood sample and were free of diagnosed skin cancer up to and including the questionnaire cycle in which the case was diagnosed. Controls were matched to cases by year of birth (±1 year) and self-reported race (Caucasian/missing). Fewer than 5% of cases and controls lacked race/ethnicity data. The nested case-control study consisted of 219 melanoma cases and 219 controls. The study protocol was approved by the Committee on Use of Human Subjects of the Brigham and Women's Hospital, Boston, MA.
Exposure data
Information regarding skin cancer risk factors was obtained from the prospective biennial questionnaires and the retrospective supplementary questionnaire. Questions on natural hair color and childhood and adolescent tanning tendency were asked in the 1982 prospective questionnaire and for the ethnic group in the 1992 questionnaire. In the skin cancer nested case-control study, natural skin color and other sun exposure-related information were collected by the retrospective supplementary questionnaire in 2002. In addition, the 11 states of residence of cohort members at baseline were grouped into 3 regions: Northeast (Connecticut, Massachusetts, Maryland, New Jersey, New York, and Pennsylvania), Northcentral (Michigan and Ohio), and West and South (California, Texas, and Florida). Estimation of past sunlight exposure for each subject was described previously [12].
SNP selection
Twenty-five common SNPs (minor allele frequency greater than 5%) were selected for the 5 interleukin genes (Table 2). For IL-6, we included 5 tagging SNPs (rs2069827, rs1554606, rs2069849, rs2069861, and rs1818879) from the SeattleSNPs database (http://pga.gs.washington.edu/) and 2 well-studied SNPs associated with other diseases (rs1800797 and rs1800795). The detailed information was described elsewhere[13]. For IL-6R, 10 tagging SNPs were chosen with the HapMap CEU population as the reference panel and an r2 of 0.8 as the threshold using a pairwise linkage disequilibrium (LD) tagging approach by the Tagger program (http://www.broad.mit.edu/mpg/tagger/)[14]. For genes IL-4, IL-4R, and IL-10, potential functional SNPs were chosen based on published papers[15-17]. The positions of these SNPs are annotated according to the SNP500Cancer database (http://snp500cancer.nci.nih.gov).
Table 2. Description of polymorphisms genotyped in the study.
| rs number | Gene region | Position 1 | Allele | MAF 2 (%) |
|---|---|---|---|---|
| IL-4 | ||||
| rs2243248 | Promoter | -1098 | T/G | 6.6 |
| rs2070874 | 5′UTR | 338 | C/T | 12.9 |
| IL-4R | ||||
| rs1805010 | Exon5, nonsynonymous | 75 3 | Ile/Val | 48.4 |
| rs1801275 | Exon11, nonsynonymous | 576 3 | Gln/Arg | 20.5 |
| IL-6 | ||||
| rs2069827 | Promoter | -1426 | G/T | 11.1 |
| rs1800797 | Promoter | -661 | A/G | 41.0 |
| rs1800795 | Promoter | -237 | G/C | 41.2 |
| rs1554606 | Intron 3 | 1825 | C/A | 47.1 |
| rs2069849 | Exon5, synonymous | 201 3 | Phe/Phe | 3.1 |
| rs2069861 | 3′UTR | 4772 | C/T | 11.2 |
| rs1818879 | 3′UTR | 5845 | G/A | 32.9 |
| IL-6R | ||||
| rs4845617 | 5′UTR | -208 | G/A | 37.1 |
| rs12083537 | Intron 1 | 2963 | T/C | 19.6 |
| rs4075015 | Intron 1 | 11090 | T/A | 41.9 |
| rs6684439 | Intron 1 | 17733 | C/T | 38.8 |
| rs4845618 | Intron 1 | 21909 | A/C | 44.0 |
| rs4845622 | Intron 6 | 33313 | T/G | 39.5 |
| rs8192284 | Exon 9, nonsynonymous | 358 3 | Asp/Ala | 39.2 |
| rs4329505 | Intron 9 | 54314 | T/C | 17.3 |
| rs4240872 | Intron 9 | 58089 | A/G | 22.6 |
| rs2229238 | 3′UTR | 59790 | G/A | 19.3 |
| IL-10 | ||||
| rs1800890 | Promoter | -3584 | T/A | 39.0 |
| rs1800896 | Promoter | -1116 | A/G | 45.3 |
| rs1800871 | Promoter | -853 | C/T | 23.8 |
| rs1800872 | Promoter | -626 | C/A | 23.0 |
Relative to the translation start point.
MAF: minor allele frequency in the controls.
Amino acid position
Laboratory assays
We performed genotyping on OpenArray™ SNP Genotyping System (BioTrove, Woburn, MA). Laboratory personnel were blinded to case-control status, and 42 blinded quality control samples were inserted to validate genotyping procedures; concordance for the blinded samples was >99%. Primers, probes, and conditions for genotyping assays are available upon request.
Statistical analyses
We used SAS v9.0 (SAS Institute, Cary, NC) for all statistical analyses. PROC ALLELE was used to test whether the genotype frequency distribution for each SNP was a departure from Hardy-Weinberg Equilibrium (HWE) among the controls.
Because conditional and unconditional logistic regression analyses yielded similar results, to increase statistical power, we used unconditional logistic regression to evaluate the association of genotype and melanoma risk. We first did simple analyses adjusting for matching factors (age and ethnicity), and then multivariate analyses adjusting for matching factors, constitutional susceptibility score, family history of melanoma, the number of lifetime severe sunburns that blistered (0, 1-5, 6-11, or >11), sunlamp use or tanning salon attendance (yes/no), cumulative sun exposure while wearing a bathing suit, and geographic region. For each SNP, the odds ratio (OR) and 95% confidence interval (CI) were calculated for the heterozygous group (genotype with one common allele and one rare allele) and the homozygous variant group (genotype with two rare alleles) separately using the wild type (genotype with two common alleles) as the reference group. The p value was obtained from a 2-degree of freedom (df) test. For some SNPs with the sparse homozygous variant group (the expected number was fewer than 5 for either cases or controls), we combined the heterozygous and homozygous variant groups and calculated the OR and 95% CI for the combined group. The p value was obtained from a 1-df test. Constitutional susceptibility score was a summarized multivariate confounder score using multiple skin cancer risk factors such as natural skin color, natural hair color, childhood or adolescent tendency to burn, and the number of palpably raised moles on arms as the predictors. The detailed information for constructing the constitutional susceptibility score and the cumulative sun exposure while wearing a bathing suit was described previously[18].
In the evaluation of gene-environment interactions between the genetic variations and the non-genetic risk factors (cumulative sun exposure with a bathing suit, number of lifetime severe sunburns, and family history of melanoma) on melanoma susceptibility, we added one interaction term between the risk factors (coded as ordinal) and SNP variables (coded as 0, 1, 2) to the logistic regression models with main effects only. The p value of this interaction term was used to evaluate whether the effect of the SNP on melanoma susceptibility differed significantly by these risk factors.
We used several major bioinformatics tools to predict the potential functions of 4 SNPs in the IL-6R gene. For the SNP rs8192284 (Asp358Ala; a nonsynonymous SNP in exon 9), we used 4 major bioinformatic methods: (i) BLOSUM62[19], (ii) PolyPhen[20], (iii) SIFT[21], and (iv) SNPs3D[22]. For the other 3 SNPs in the intron region (rs6684439, rs4845618, rs4845622), we first used SNPHunter [23] to retrieve the 5′ and 3′ 35-bp or 300-bp (for CpG island analysis) flanking sequences for each of the 3 intronic SNPs. Then, we employed 4 in silico prediction programs, (i) NNSPLICE 0.9 version (http://www.fruitfly.org/seq_tools/splice.html) [24], (ii) Branch-Site Analyzer (http://ast.bioinfo.tau.ac.il/BranchSite.htm), (iii) CpG Islands Searcher[25], and (iv) TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) to predict whether each SNP could be located in donor or acceptor splice site, the branch site or the polypyrimidine tract of the intron, CpG island or transcriptional factor binding site, respectively.
Results
The mean age at diagnosis of melanoma cases was 63.4 years, the same as that of controls. Cases were more likely than controls to live in the West and South area (25.1% vs. 18.7%), to have a family history of melanoma (15.3% vs. 6.0%), and to use a sunlamp or attend a tanning salon (23.1% vs. 13.1%). Cases also had a higher constitutional susceptibility risk score, cumulative sun exposure with a bathing suit, and mean number of lifetime severe sunburns (Table 1).
Table 1. Characteristics of melanoma cases and controls in the nested case-control study.
| Characteristic | Controls (n=219) |
Melanoma cases (n=219) |
P value |
|---|---|---|---|
| Age at diagnosis (mean, years) | 63.4 | 63.4 | matched |
| Geographic region at baseline (%) | 0.10 | ||
| Northeast | 57.5 | 58.0 | |
| Northcentral | 23.7 | 16.9 | |
| West and South | 18.7 | 25.1 | |
| Sunlamp use or tanning salon attendance (%) | 13.1 | 23.1 | 0.01 |
| Family history of melanoma (%) | 6.0 | 15.3 | 0.004 |
| Number of lifetime severe sunburns (mean) | 5.0 | 9.6 | <0.0001 |
| Median above constitutional susceptibility risk score (%) | 49.8 | 67.1 | 0.0002 |
| Median above cumulative sun exposure with a bathing suit (%) | 50.2 | 62.1 | 0.01 |
Except for SNP rs4845617 in the IL-6R gene (p=0.01), no departure from HWE was observed among controls for the other 24 SNPs. We observed a higher risk of melanoma in the heterozygous group of 4 linked SNPs of the IL-6R gene, but not in the homozygous variant group. The OR (95% CI) of the heterozygous group was 1.74 (1.07-2.81) for rs6684439, 1.72 (1.04-2.84) for rs4845618, 1.69 (1.03-2.75) for rs4845622, and 1.68 (1.04-2.73) for rs8192284 (Table 3). The former 3 SNPs are in intron regions, and rs8192284 is a nonsynonymous SNP in exon 9. There was no material difference between simple and multivariate analyses. We illustrate the positions and the LD structure of these SNPs in Figure 1. We did not find significant results for SNPs in the other 4 genes.
Table 3. Polymorphisms in selected candidate genes and melanoma risk.
| SNP | Wild type | Heterozygous/variant carriers | Homozygous variant | p | ||
|---|---|---|---|---|---|---|
| case/control | case/control | OR(95% CI) | case/control | OR(95% CI) | ||
| IL-4 | ||||||
| rs2243248 | 180/185 | 31/27 | 1.27 (0.69, 2.36) | 0.45 | ||
| rs2070874 | 160/165 | 52/43 | 1.16 (0.69, 1.94) | 5/6 | 1.12 (0.28, 4.48) | 0.85 |
| IL-4R | ||||||
| rs1805010 | 67/57 | 104/110 | 0.80 (0.48, 1.33) | 43/50 | 0.71 (0.38, 1.32) | 0.52 |
| rs1801275 | 120/121 | 64/65 | 1.10 (0.68, 1.80) | 11/7 | 1.70 (0.58, 4.96) | 0.61 |
| IL-6 | ||||||
| rs2069827 | 149/156 | 48/38 | 1.40 (0.81, 2.41) | 0.22 | ||
| rs1800797 | 68/71 | 102/100 | 1.08 (0.66, 1.75) | 28/34 | 0.89 (0.45, 1.75) | 0.84 |
| rs1800795 | 69/69 | 106/102 | 1.05 (0.65, 1.70) | 32/33 | 0.93 (0.48, 1.81) | 0.93 |
| rs1554606 | 60/55 | 102/92 | 0.98 (0.58, 1.64) | 40/44 | 0.78 (0.41, 1.46) | 0.69 |
| rs2069849 | 202/200 | 9/13 | 0.76 (0.28, 2.04) | 0.58 | ||
| rs2069861 | 169/162 | 39/43 | 0.89 (0.52, 1.54) | 0.68 | ||
| rs1818879 | 100/91 | 79/85 | 0.92 (0.57, 1.47) | 25/23 | 1.18 (0.58, 2.40) | 0.79 |
| IL-6R | ||||||
| rs4845617 | 75/74 | 110/116 | 0.74 (0.46, 1.19) | 28/20 | 0.98 (0.46, 2.05) | 0.40 |
| rs12083537 | 125/135 | 74/66 | 1.04 (0.66, 1.65) | 16/8 | 2.16 (0.82, 5.69) | 0.30 |
| rs4075015 | 78/70 | 108/104 | 1.22 (0.75, 1.97) | 27/36 | 0.89 (0.45, 1.77) | 0.55 |
| rs6684439 | 64/80 | 125/97 | 1.74 (1.07, 2.81) | 28/33 | 0.93 (0.47, 1.83) | 0.035 |
| rs4845618 | 58/66 | 121/93 | 1.72 (1.04, 2.84) | 31/42 | 1.03 (0.53, 1.98) | 0.06 |
| rs4845622 | 62/78 | 119/97 | 1.69 (1.03, 2.75) | 28/34 | 0.98 (0.50, 1.93) | 0.06 |
| rs8192284 | 63/81 | 118/97 | 1.68 (1.04, 2.73) | 30/35 | 1.01 (0.52, 1.95) | 0.06 |
| rs4329505 | 153/146 | 62/65 | 0.82 (0.51, 1.31) | 0.40 | ||
| rs4240872 | 96/79 | 58/56 | 0.81 (0.47, 1.39) | 0.44 | ||
| rs2229238 | 153/139 | 64/73 | 0.86 (0.55, 1.35) | 0.51 | ||
| IL-10 | ||||||
| rs1800890 | 87/72 | 96/111 | 0.74 (0.47, 1.19) | 29/26 | 0.92 (0.47, 1.82) | 0.45 |
| rs1800896 | 57/64 | 112/103 | 1.25 (0.76, 2.07) | 46/44 | 1.13 (0.61, 2.09) | 0.68 |
| rs1800871 | 115/114 | 84/83 | 0.99 (0.63, 1.56) | 11/7 | 1.43 (0.50, 4.09) | 0.79 |
| rs1800872 | 121/124 | 83/80 | 1.05 (0.67, 1.65) | 9/9 | 0.98 (0.35, 2.77) | 0.97 |
Unconditional logistic regression adjusted for the matching variables (age and race [Caucasian and missing]), constitutional susceptibility score, family history of melanoma, the number of lifetime severe sunburns that blistered, sunlamp use or tanning salon attendance (yes/no), cumulative sun exposure while wearing a bathing suit, and geographic region.
We estimated the odds ratios (OR) of heterozygous and homozygous variant groups separately with the wild type as the reference and the p values were obtained from 2-degree of freedom (df) tests. For some SNPs with expected cell count in homozygous variant group of fewer than 5 for either cases or controls, we combined heterozygote and homozygous variant into one category (variant carriers), and the p values are based on 1- df tests.
The number of participants does not sum to total women because of missing data on genotype.
Figure 1.
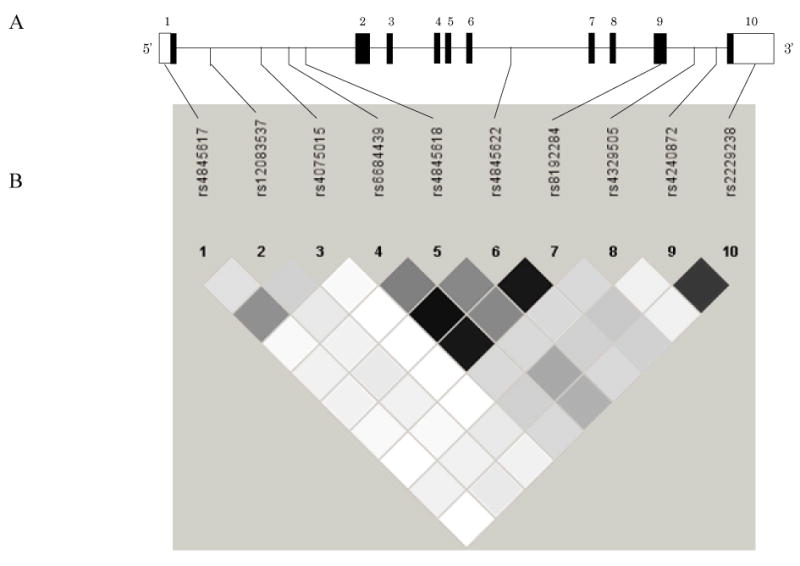
(A) The location of IL-6R gene polymorphisms: black bars represent exons; lines represent introns; white bars represent untranslated regions. (B) Pairwise r2 between selected tagging SNPs. Black squares represent high LD, white squares represent low LD. r2 = 0.49 between SNP4 (rs6684439) and SNP5 (rs4845618); r2 = 0.92 between SNP4 and SNP6 (rs4845622); r2 = 0.89 between SNP4 and SNP7 (rs8192284); r2 = 0.46 between SNP5 and SNP6; r2 = 0.44 between SNP5 and SNP7; r2 = 0.89 between SNP6 and SNP7.
We found no evidence of gene-environment interactions between these 5 genes and melanoma risk factors (cumulative sun exposure with a bathing suit, the number of lifetime severe sunburns, and family history of melanoma) on melanoma susceptibility.
Through the functional prediction of the rs8192284 (Asp358Ala), we found a score of - 2 by the BLOSUM62, indicating that the Asp358Ala is likely to be deleterious to the protein structure. The PolyPhen PSIC score difference (Δ) is 1.667, which is considered “possibly damaging” as well. On the other hand, the score of 0.17 by the SIFT TI (with a MSCS = 3.66) and the svm profile score of 0.04 by the SNPs3D suggest that this SNP is possibly tolerated or not deleterious.
The 3 intronic SNPs, rs6684439, rs4845618, and rs4845622, are located > 1000 bp from either the 5′ or the 3′ boundaries of introns 1 or 6: rs6684439 is located at the 17649 bp position in intron 1 (size=23481 bp), rs4845618 is located at the 21825 bp position in intron 1 (size=23481 bp), and rs4845622 is located at the 2833 bp position in intron 6 (size=12014 bp). Results of NNSPLICE showed that none of the 3 intronic SNPs were located in donor or acceptor splice sites. Results of Branch-Site Analyzer showed that none of them were located in any branch sites or polypyrimidine tracts in their respective introns. Results of CpG Islands Searcher indicated that none of them were located in CpG islands. Results of TFSEARCH indicated that only rs6684439 is located in 2 putative transcriptional binding sites, GATA-1 (TRANSFAC accession number M00075) and deltaEF1 (TRANSFAC accession number M00073). Taken together, these results suggest that rs6684439 could have a functional impact on IL-6R transcriptional regulation by altering 2 putative transcriptional binding sites, but rs4845618 and rs4845622 are likely to be nonfunctional.
Discussion
Genetic variations in cytokine genes may influence melanoma susceptibility. However, only a few epidemiological studies have examined the associations of IL-10[9, 11], IL-4[10], and IL-6[10, 26] genes with melanoma risk. The IL-4R and IL-6R genes have not been previously examined. We evaluated the association of 25 SNPs in these 5 genes with melanoma susceptibility in a nested case-control study of 219 cases and 219 matched controls within NHS.
IL-6 acts as a tumor growth inhibitor for early-stage melanoma and a growth factor for tumor cells of advanced stage in vitro[8]. Among metastatic malignant melanoma patients treated with biochemotherapy, those with a higher pretreatment serum IL-6 level have a shorter survival time[27]. However, in two epidemiological studies, no significant association was observed between IL-6 -174 G/C, a SNP regulating IL-6 production[28], and melanoma susceptibility or prognosis[10, 26]. Our study assessed 5 tagging SNPs in this gene and 2 well-studied SNPs, one of which is IL-6 -174 G/C (rs1800795). We found no significant association between these SNPs and melanoma risk. IL-6 exerts its biological effects by binding to IL-6R, which then signals the cell via second messengers to alter gene expression. We found that 4 linked SNPs in the IL-6R gene were associated with melanoma risk; the heterozygous groups had higher risk than wild types with ORs ranging from 1.68 to 1.74. This significantly increased risk was not observed for their homozygous variant groups with the lower bounds of the CIs 0.45-0.53, and the upper bounds 1.83-1.95. There are several interpretations for this observation: it is possible that the significant associations in the heterozygous groups were false-positive (due to the multiple testing). Another explanation is that the null results in the homozygous variant groups may be due to the wide confidence interval (the OR ranged from about 0.5 to 2.0). Therefore, replication of this finding in other studies is warranted. Among these 4 SNPs, rs8192284 (Asp358Ala) is a well-studied nonsynonymous SNP in exon 9. Prediction by two bioinformatics tools, BLOSUM62 and PolyPhen, suggests that this polymorphism could possibly damage the protein structure of IL-6R. According to an in vitro study of human cells, Asp358Ala occurs in a major position close to the transmembrane region of the protein, and this amino acid substitution could alter the shedding of the membrane-bound IL-6R to the soluble IL-6R[29]. Substitution of the Asp with Gly at position 358 reduced the shedding by 26%[29]. In a study of 115 healthy volunteers, Galicia et al. reported that Ala carriers had higher serum soluble IL-6R levels[30]. In terms of the other 3 SNPs in the intron regions, the SNP rs6684439 in the intron 1 was predicted to be located in 2 putative transcriptional factor (GATA-1 or deltaEF1) binding sites by the TFSEARCH algorithm. We cannot eliminate the possibility of false-positive observations in the heterozygous groups. However, considering these potential functions of the rs8192284 and rs6684439, it is worthwhile to perform replication and functional studies to determine the role of the IL-6R gene and its products in melanoma susceptibility and prognosis.
IL-10 is an important cytokine with multiple functions in terms of tumor progress. As a potent immunosuppressive cytokine, IL-10 has potential cancer-promoting function by inhibiting the T cell immune response against tumor cells[5]. IL-10 can also act as an autocrine growth factor for melanoma cells[3]. Conversely, the antitumor activity of IL-10 has been reported as well in the form of inhibition of angiogenesis[6]. Several case-control studies were performed for this gene with melanoma susceptibility and prognosis, and the results with susceptibility were inconsistent. Howell et al.[9] found that a low-expression genotype in the IL-10 promoter region (-1082 AA) is a risk factor for more advanced/poorer prognosis melanoma and may confer susceptibility to melanoma. According to Alonso et al.[31], the SNP -1082 A/G was not associated with melanoma susceptibility but the researchers observed associations among males of older age at diagnosis, and with the overall survival rate in patients of advanced disease. Vuoristo et al.[32] reported a haplotype carrier (A in - 1082, T in -819, and A in -592) was associated with the susceptibility to melanoma, and with longer survival in advanced melanoma patients. In this study, we did not find a significant association between 4 IL-10 SNPs (including the 3 SNPs well studied previously) and melanoma risk. We did not have stage or other prognostic indicators, which limited our ability to evaluate the relation of the IL-10 gene with melanoma prognosis.
IL-4 can induce IL-10 secretion by peripheral blood lymphocytes[7]. Howell et al.[10] did not find an association between the SNP IL-4 -590 and melanoma susceptibility or prognosis. Likewise, no significant results were observed in our study for the 2 IL-4 SNPs and 2 nonsynonymous IL-4R SNPs in relation to melanoma risk.
It is worth noting that for IL-10, IL-4, and IL-4R genes, we selected SNPs according to published papers rather than a tagging SNP approach. For some cytokines such as IL-10 and IL-6, the functions are complex and may vary in different melanoma stages. However, we do not have enough information to do subgroup analyses.
In conclusion, the IL-6R gene may be involved in melanoma susceptibility; replication in larger samples and further functional study are warranted. Hence, when studying cytokine genes, the cytokine receptor genes may be worth investigating as well.
Acknowledgments
The authors thank Pati Soule and Drs. Hardeep Ranu and David Cox for their assistance in genotyping melanoma samples. We are indebted to the participants in the Nurses' Health Study.
Grant sponsor: NIH; Grant number, CA132175.
Footnotes
No financial conflict of interest
References
- 1.American Cancer Society. Cancer Facts & Figures 2007. Atlanta: American Cancer Society; 2007. [Google Scholar]
- 2.Penn I. Depressed immunity and skin cancer. Immunol Today. 1984;5:291–3. doi: 10.1016/0167-5699(84)90152-X. [DOI] [PubMed] [Google Scholar]
- 3.Yue FY, Dummer R, Geertsen R, Hofbauer G, Laine E, Manolio S, et al. Interleukin-10 is a growth factor for human melanoma cells and down-regulates HLA class-I, HLA class-II and ICAM-1 molecules. Int J Cancer. 1997;71(4):630–7. doi: 10.1002/(sici)1097-0215(19970516)71:4<630::aid-ijc20>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 4.Del Prete G, De Carli M, Almerigogna F, Giudizi MG, Biagiotti R, Romagnani S. Human IL-10 is produced by both type 1 helper (Th1) and type 2 helper (Th2) T cell clones and inhibits their antigen-specific proliferation and cytokine production. J Immunol. 1993;150(2):353–60. [PubMed] [Google Scholar]
- 5.Chen Q, Daniel V, Maher DW, Hersey P. Production of IL-10 by melanoma cells: examination of its role in immunosuppression mediated by melanoma. Int J Cancer. 1994;56(5):755–60. doi: 10.1002/ijc.2910560524. [DOI] [PubMed] [Google Scholar]
- 6.Huang S, Ullrich SE, Bar-Eli M. Regulation of tumor growth and metastasis by interleukin-10: the melanoma experience. J Interferon Cytokine Res. 1999;19(7):697–703. doi: 10.1089/107999099313532. [DOI] [PubMed] [Google Scholar]
- 7.Shreedhar V, Giese T, Sung VW, Ullrich SE. A cytokine cascade including prostaglandin E2, IL-4, and IL-10 is responsible for UV-induced systemic immune suppression. J Immunol. 1998;160(8):3783–9. [PubMed] [Google Scholar]
- 8.Lu C, Vickers MF, Kerbel RS. Interleukin 6: a fibroblast-derived growth inhibitor of human melanoma cells from early but not advanced stages of tumor progression. Proc Natl Acad Sci U S A. 1992;89(19):9215–9. doi: 10.1073/pnas.89.19.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell WM, Turner SJ, Bateman AC, Theaker JM. IL-10 promoter polymorphisms influence tumour development in cutaneous malignant melanoma. Genes Immun. 2001;2(1):25–31. doi: 10.1038/sj.gene.6363726. [DOI] [PubMed] [Google Scholar]
- 10.Howell WM, Turner SJ, Theaker JM, Bateman AC. Cytokine gene single nucleotide polymorphisms and susceptibility to and prognosis in cutaneous malignant melanoma. Eur J Immunogenet. 2003;30(6):409–14. doi: 10.1111/j.1365-2370.2003.00425.x. [DOI] [PubMed] [Google Scholar]
- 11.Nikolova PN, Pawelec GP, Mihailova SM, Ivanova MI, Myhailova AP, Baltadjieva DN, et al. Association of cytokine gene polymorphisms with malignant melanoma in Caucasian population. Cancer Immunol Immunother. 2007 Mar;56(3):371–9. doi: 10.1007/s00262-006-0193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Colditz GA, Hunter DJ. Risk factors for skin cancers: a nested case-control study within the Nurses' Health Study. Int J Epidemiol. 2006 Dec;35(6):1514–21. doi: 10.1093/ije/dyl197. [DOI] [PubMed] [Google Scholar]
- 13.Qi L, van Dam RM, Meigs JB, Manson JE, Hunter D, Hu FB. Genetic variation in IL6 gene and type 2 diabetes: tagging-SNP haplotype analysis in large-scale case-control study and meta-analysis. Hum Mol Genet. 2006 Jun 1;15(11):1914–20. doi: 10.1093/hmg/ddl113. [DOI] [PubMed] [Google Scholar]
- 14.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005 Nov;37(11):1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 15.Landi S, Bottari F, Gemignani F, Gioia-Patricola L, Guino E, Osorio A, et al. Interleukin-4 and interleukin-4 receptor polymorphisms and colorectal cancer risk. Eur J Cancer. 2007 Mar;43(4):762–8. doi: 10.1016/j.ejca.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 16.Brown EE, Fallin MD, Goedert JJ, Hutchinson A, Vitale F, Lauria C, et al. Host immunogenetics and control of human herpesvirus-8 infection. J Infect Dis. 2006 Apr 15;193(8):1054–62. doi: 10.1086/501470. [DOI] [PubMed] [Google Scholar]
- 17.Howell WM, Rose-Zerilli MJ. Cytokine gene polymorphisms, cancer susceptibility, and prognosis. J Nutr. 2007 Jan;137(1 Suppl):194S–9S. doi: 10.1093/jn/137.1.194S. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Colditz GA, Liu JS, Hunter DJ. Genetic variation in XPD, sun exposure, and risk of skin cancer. Cancer Epidemiol Biomarkers Prev. 2005 Jun;14(6):1539–44. doi: 10.1158/1055-9965.EPI-04-0846. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–9. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001 Mar 15;10(6):591–7. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 21.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003 Jul 1;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Liu S, Niu T, Xu X. SNPHunter: a bioinformatic software for single nucleotide polymorphism data acquisition and management. BMC Bioinformatics. 2005;6:60. doi: 10.1186/1471-2105-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997 Fall;4(3):311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 25.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3(3):235–40. [PubMed] [Google Scholar]
- 26.Martinez-Escribano JA, Moya-Quiles MR, Muro M, Montes-Ares O, Hernandez-Caselles T, Frias JF, et al. Interleukin-10, interleukin-6 and interferon-gamma gene polymorphisms in melanoma patients. Melanoma Res. 2002;12(5):465–9. doi: 10.1097/00008390-200209000-00008. [DOI] [PubMed] [Google Scholar]
- 27.Soubrane C, Rixe O, Meric JB, Khayat D, Mouawad R. Pretreatment serum interleukin-6 concentration as a prognostic factor of overall survival in metastatic malignant melanoma patients treated with biochemotherapy: a retrospective study. Melanoma Res. 2005 Jun;15(3):199–204. doi: 10.1097/00008390-200506000-00009. [DOI] [PubMed] [Google Scholar]
- 28.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998 Oct 1;102(7):1369–76. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullberg J, Oberthur W, Lottspeich F, Mehl E, Dittrich E, Graeve L, et al. The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J Immunol. 1994 May 15;152(10):4958–68. [PubMed] [Google Scholar]
- 30.Galicia JC, Tai H, Komatsu Y, Shimada Y, Akazawa K, Yoshie H. Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes Immun. 2004 Sep;5(6):513–6. doi: 10.1038/sj.gene.6364120. [DOI] [PubMed] [Google Scholar]
- 31.Alonso R, Suarez A, Castro P, Lacave AJ, Gutierrez C. Influence of interleukin-10 genetic polymorphism on survival rates in melanoma patients with advanced disease. Melanoma Res. 2005 Feb;15(1):53–60. doi: 10.1097/00008390-200502000-00009. [DOI] [PubMed] [Google Scholar]
- 32.Vuoristo MS. The polymorphisms of interleukin-10 gene influence the prognosis of patients with advanced melanoma. Cancer Genet Cytogenet. 2007 Jul 1;176(1):54–7. doi: 10.1016/j.cancergencyto.2007.03.002. [DOI] [PubMed] [Google Scholar]


