Abstract
Background
Current public health efforts often use molecular technologies to identify and contain communicable disease networks, but not for HIV. Here, we investigate how molecular epidemiology can be used to identify highly-related HIV networks within a population and how voluntary contact tracing of sexual partners can be used to selectively target these networks.
Methods
We evaluated the use of HIV-1 pol sequences obtained from participants of a community-recruited cohort (n=268) and a primary infection research cohort (n=369) to define highly related transmission clusters and the use of contact tracing to link other individuals (n=36) within these clusters. The presence of transmitted drug resistance was interpreted from the pol sequences (Calibrated Population Resistance v3.0).
Results
Phylogenetic clustering was conservatively defined when the genetic distance between any two pol sequences was <1%, which identified 34 distinct transmission clusters within the combined community-recruited and primary infection research cohorts containing 160 individuals. Although sequences from the epidemiologically-linked partners represented approximately 5% of the total sequences, they clustered with 60% of the sequences that clustered from the combined cohorts (O.R. 21.7; p=<0.01). Major resistance to at least one class of antiretroviral medication was found in 19% of clustering sequences.
Conclusions
Phylogenetic methods can be used to identify individuals who are within highly related transmission groups, and contact tracing of epidemiologically-linked partners of recently infected individuals can be used to link into previously-defined transmission groups. These methods could be used to implement selectively targeted prevention interventions.
Keywords: molecular epidemiology, HIV, surveillance, contact tracing, drug resistance
Introduction
Molecular techniques can determine the transmission of communicable diseases, such as tuberculosis[1], syphilis[2] and gonorrhea[3], and public health agencies often use contact tracing of individuals with communicable diseases to contain or prevent outbreaks by identifying the source or potential recipients of the index cases' infection[4]. Currently, molecular techniques to monitor HIV transmission dynamics within populations are not commonly used for prevention efforts[4].
The genetic composition of HIV within a person is determined by the high mutation and recombination rates[5] and the genetic background of the infecting virus[6], the immunologic pressures[7, 8] and target cell availability of the host[9], and potentially the pressures of antiretroviral therapy[10]. As a consequence of these factors, the genetic composition of HIV is relatively unique to each infected person. By exploiting the genetic relatedness of HIV between individuals who have transmitted HIV, viral sequence analysis can be used to investigate HIV transmissions in healthcare settings[11], households[12], adult entertainment settings[13], and judiciary settings[11, 14, 15]. HIV sequence analysis has also been used on a population level to define highly related transmission networks[16], associated with stage of HIV infection[17-19], illicit substance use[20], drug resistance[20, 21], and sexually transmitted infections[18]. Efficiently identifying these highly related clusters could be used to discover sexual networks where HIV is being transmitted, and being able to access these networks could be very important for public health efforts to target prevention strategies.
Since the rate of new HIV transmissions has increased or remained constant for over a decade in the United States despite aggressive HIV awareness and behavioral modification campaigns[22], new prevention methods are needed. Therefore, we evaluated how molecular epidemiology coupled with partner contact tracing can be used to identify individuals within a population that belong to highly related HIV transmission groups with the ultimate goal of being able to target prevention efforts to interrupt HIV transmission. Specifically, we evaluated the use of pol sequences generated for the detection of drug resistance among two San Diego cohorts, one recently-HIV infected research cohort and one mainly chronically HIV-infected clinical cohort, for the molecular surveillance of transmission clusters. We further evaluated the effectiveness of contact tracing of epidemiologically-linked source partners of participants in the recently-infected cohort to identify additional HIV-infected individuals who also cluster within the previously defined transmission networks. Using these methods, we outline how pol sequences and contact tracing can be used responsibly to identify groups of individuals with highly related virus, most likely representing social networks engaged in high-risk behaviors and forward HIV transmission that could be targeted for prevention interventions.
Methods
Study Participants
A certificate of confidentiality was obtained for all investigations to help protect the identity of the study participants and ensure that investigators will not be forced to divulge confidential research information without their written permission, even in the face of a court order. Data from three San Diego cohorts were analyzed. (1) The County of San Diego Cohort included all HIV-infected individuals who received genotypic drug resistance testing through a program for antiretroviral-naïve individuals funded by the County of San Diego from January 1, 2006 through May 1, 2007[23]. Access to this program was decided by each patient's clinical provider and patient clinical and demographic data were not collected. Duration of HIV infection was not known for subjects in this cohort, but presumed to be long-standing infection for the majority of participants based upon the sources of referral. (2) The First Choice Program (FCP) cohort included all participants of the San Diego site of the Acute Infection and Early Disease Research Program (AIEDRP) from July 1, 1996 through May 2007 who received genotypic resistance testing for research and clinical purposes at the time of HIV diagnosis. Duration of infection for the FCP cohort was based upon a standardized protocol (www.aiedrp.org)[24]. At the time of their enrollment, participants of the FCP cohort were asked to identify their recent sex partners for evaluation. Contact tracing was voluntary and involved the FCP participant either providing contact information for their recent sex partners to trained outreach workers or contacting them for study recruitment themselves. (3) These epidemiologically-linked partners constituted the third cohort analyzed in this study and underwent the same clinical screening as the FCP participants. All epidemiologically-linked partners who were HIV seropositive by standard enzyme-linked immunoassay testing for HIV antibody also received less-sensitive enzyme-linked immunoassay testing (detuned)[24], unless prior HIV test results suggested chronic infection.
Epidemiologic Linkage
Epidemiologic linkage was defined when a FCP participant supplied contact information on a sexual partner who they believed to be the likely source of their HIV infection, as described above. Contact tracing was performed by personnel trained in partner notification, and all information used for contact tracing was obtained voluntarily from the index FCP participant by the trained personnel. Participants identified as having primary HIV infection and eligible for the FCP were not denied participation in the FCP study if they chose not to supply contact information on their sexual partners. Those FCP participants who consented were offered three options for contact tracing of their partners: 1) Self-disclosure: they disclosed to their partners their recent HIV infection themselves and then referred them to FCP for evaluation, 2) Self-disclosure with assistance: they brought their partner to the FCP and disclosed their recent HIV infection diagnosis to their partner with FCP staff present, and 3) Anonymous third party notification: they supplied contact and identifying information to be used by FCP personnel for partner notification.
Phylogenetic Linkage and Transmission Clusters
Even though contact tracing identified sets of HIV-infected partners, who were epidemiologically-linked, it did not establish whether the HIV transmission took place between the two partners. To investigate if HIV transmission likely occurred, we performed phylogenetic analysis on the HIV-1 pol sequences obtained from each partner (Monogram Biosciences, South San Francisco). Sequences were initially compiled and edited in BioEdit, and aligned using the MUSCLE alignment tool[25, 26]. The alignment was then manually edited to preserve frame insertions and deletions if present. Phylogenetic trees and distance matrices were generated from pol sequences that were obtained from a matrix of synonymous nucleotide distances using the Hasegawa-Kishino-Yang (HKY85) model of evolution with a 2:1 ratio of transversion to transitions in the HyPhy package[27]. In confirmatory phylogenetic analyses, amino acid residue sites associated with drug resistance[28] were stripped from all sequences to evaluate linkage independent of the presence of resistance associated mutations. If HIV transmission occurred between partners, then the genetic distance between the HIV isolates, should be so close as to not be likely to have occurred from a third party[29]. The sequence length necessary to detect and establish clustering was dependent on the average divergence between sequences from unrelated infections and the genetic distance between viral isolates obtained from closely related epidemiologically-linked partners (see results below)[29]. Phylogenetic data were not disclosed to either partner and was used for participant recruitment in any cohort.
Drug Resistance
The presence of drug resistance was interpreted from the pol sequence for each individual using the Monogram Biosciences genotypic interpretation algorithm (GeneSeq™). These results were made available to each participant's primary care provider for their clinical management. To investigate the prevalence of drug resistance among the cohorts, we interpreted the prevalence of drug resistance using the Calibrated Population Resistance (CPR) algorithm and the surveillance drug resistance mutation list (SDRM) that was specifically developed for the epidemiologic surveillance of drug resistant HIV[28] among the sequences from each of the study cohorts.
Statistics
We examined bivariate relationships between cohorts, clustering and drug resistance in contingency table analysis. Analyses were performed using STATA version 9.1. Confidence intervals were obtained using the adjusted Wald method.
Results
Participants
Between January 2006 and May 2007, 268 individuals were enrolled in the San Diego County Resistance Testing Program for newly-diagnosed, antiretroviral-naïve, HIV-infected patients in public health funded clinics in San Diego. Between July 1996 and May 2007, 369 individuals were enrolled in the FCP cohort. Of those FCP participants who consented to partner tracing: 1) most (>90%, n=32) disclosed to their partners themselves and referred them directly to the FCP for evaluation; 2) a small minority (<10%, n=3) brought their partners to the FCP and disclosed their recent HIV infection to their partners with the assistance of FCP personnel; 3) only one FCP participant opted for third party notification, where FCP personnel were supplied contact information by the FCP participant and anonymous third-party notification was performed by trained FCP personnel. In total, voluntary contact tracing of partners of FCP participants identified 36 epidemiologically-linked partner pairs, which is roughly 10% of all FCP participants.
The demographics of the FCP and the epidemiologically-linked partner cohorts were similar, >90% of the individuals were male and sexual exposures with men was the most reported HIV risk factor. The demographics of these two cohorts were also similar to the epidemiology of HIV in the County of San Diego as a whole (Table 1)[30]. The estimated mean duration of HIV infection among the FCP participants was 76 days at the time of enrollment, and 38% of the epidemiologically-linked partners were also found within the first year of their infection, while the rest (62%) were chronically infected when identified. Only two of the epidemiologically-linked partners did not know that they were HIV positive; one was found during early HIV infection (LS EIA OD <0.3, consistent with infection <70 days[31]) and the other was chronically infected. The mean duration of infection among individuals in the San Diego County cohort was most likely multiple years[23].
Table 1.
Demographics of Study Cohorts.
| Cohorts | |||
|---|---|---|---|
| Demographic | Demographics of San Diego County* | First Choice Program** | Epidemiologically-linked Partner** |
| Male sex (%) | 89 | 96 | 92 |
| Age (mean years) | 35 | 34 | 34 |
| Ethnicity (%) | |||
| Caucasian | 62 | 79 | 62 |
| African American | 13 | 6 | 8 |
| Latino/a | 22 | 21 | 21 |
| Other | 3 | 7 | 9 |
| HIV Risk Factor (%) | |||
| MSM | 80 | 92 | 91 |
| MSM+IDU | 7 | 2 | 3 |
| IDU | 4 | 1 | 0 |
| Heterosexual | 4 | 4 | 3 |
| Other | 5 | 1 | 3 |
These demographic data are not the study demographics of the County of San Diego cohort in this study, as those data were not collected in this study. These demographic data are the available reported data from all individuals who tested HIV positive in San Diego County in 2006[30].
First Choice Program and Epidemiologically linked partner demographics are based on time of study enrollment.
Definition of Transmission Clusters
Since the high level of genetic variation of HIV isolated from different individuals can provide information on the transmission network of HIV[16-19], we used sequence data of the protease (codons 1-99) and reverse transcriptase (codons 1-305) coding regions of HIV pol from 268 San Diego County cohort participants, 369 FCP cohort participants and 36 epidemiologically-linked partners. The alignment was without insertions or deletions, and on average, pol sequences were 5.1% genetically different but with a wide range of divergence (0%-11.2%). To define a distance cutoff of sequences as being in a cluster, we first calculated the distances between sequences for the epidemiologically-linked transmitting partners. Twelve epidemiologically-linked partner pairs demonstrated >5% genetic divergence between pol sequences, which is similar to the reported divergence between participants who were epidemiologically-unlinked[13, 18]; therefore, these epidemiologically-linked partner pairs were defined as phylogenetically-unlinked for the purposes of this study. Five of the thirty-six epidemiologically-linked but phylogenetically-unlinked partners clustered with someone else within the cohort. The other 24 epidemiologically-linked partners had a genetic distance of <1% between pol sequences; therefore, these partner pairs were considered epidemiologically and phylogenetically linked. Given these data, transmission clusters were then conservatively defined when pol sequences from any two people were >99% genetically similar (i.e. <1% genetic distance between sequences). To further support the use of this very conservative cutoff, we found that within our combined cohort, the size of the largest cluster and the number of individuals within a cluster did not change appreciably until the genetic divergence between sequences was >1% (data not shown). Based on the <1% cutoff, 160 (30%) individuals were in clusters, which was, of course, less than the proportion of individuals reported to be in clusters as described previously (34-49%), which used an arbitrarily higher cutoff (1.5%)[18, 19]. Using our conservative cut-off, we were less-likely to falsely include participants within transmission networks to which they did not belong, but also more likely to exclude participants from clusters from which they did belong. Since our goal was to most accurately identify transmission networks and to assess how voluntary contact tracing could identify other individuals who fit within these networks, we chose to continue our analyses using the <1% genetic distance cut-off, which was supported by our epidemiologically-linked partner data, who all demonstrated <1% HIV genetic distance between partners.
Clustering Characteristics
The San Diego County cohort consisted of 268 individuals and represented 139 person/years of follow-up. Within this cohort, 17 sequences (6%) were found to group within 3 clusters. The FCP cohort consisted of 369 individuals and represented 1657 person/years of follow-up. Within the FCP cohort, 112 sequences (30%) were found to group within 34 clusters. When sequences from the San Diego County and FCP cohorts were combined (Combined Cohort, Table 2), 160 sequences (25%) were found to group within clusters.
Table 2.
Likelihood that sequences will cluster within the combined San Diego County, FCP and Epidemiologically-linked Partner cohorts.
| Clustering of Combined Cohort* | ||||
|---|---|---|---|---|
| Cohorts | Clustered (n) | Not Clustered (n) | Odds Ratio | 95% C.I.* |
| San Diego County | 43 | 225 | 1 (ref.) | |
| FCP | 117 | 252 | 2.43 | (1.61, 3.67) |
| Epi-linked Partners | 29 | 7 | 21.68 | (8.37, 58.42) |
| Total | 189 | 484 | ||
Transmission clusters were defined after combining the San Diego County and First Choice Program (FCP) cohorts (combined cohorts) but not with the sequences from the Epidemiologically-linked (Epi-linked) partners; however, the epi-linked partners demonstrated the greatest odds of belonging within a transmission cluster that was previously defined. C.I.: confidence interval.
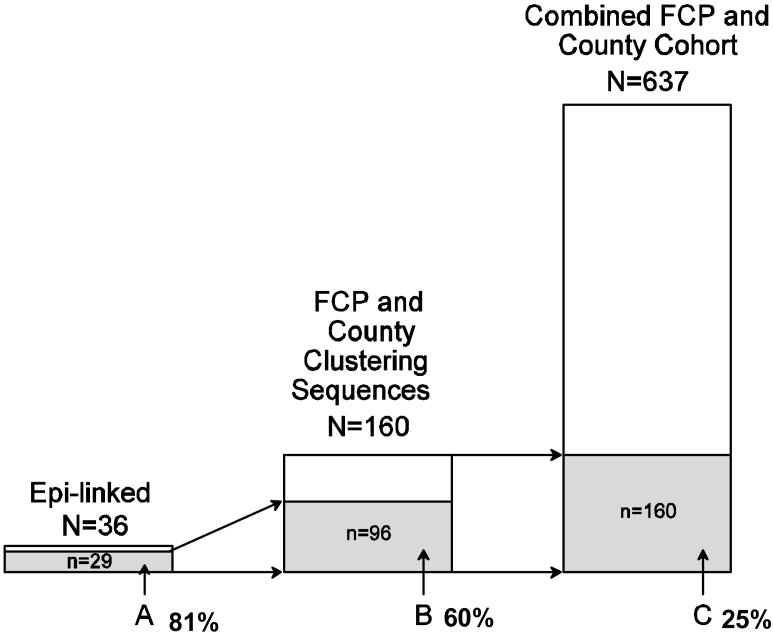
Through voluntary contact tracing of partners of participants of the FCP cohort, 36 epidemiologically-linked partners were enrolled. The success of contact tracing of partners varied considerably by year of FCP enrollment (mean 11% of FCP participants recruited a partner each year, 95% C.I.: 6%-16%). Only about 67% of the epidemiologically-linked partners were found to be phylogenetically-linked (<1% genetic distance) to their FCP partner (n=24); however, the likelihood was extremely high (60%) that epidemiologically-linked partners (whether or not they were phylogenetically-linked) grouped within previously defined clusters of the combined San Diego County and FCP cohorts (Figure 1). Although sequences from the epidemiologically-linked partners represented only about 5% of the total number of sequences, they clustered with 60% of the clustering sequences from the combined San Diego County and FCP cohort sequences (Table 2; O.R.21.7; p=<0.01). When all sequences were combined from the San Diego County and FCP cohorts (Combined Cohort) and analyzed in conjunction with the sequences from the epidemiologically-linked partners, there was a large increase in the number of clustering sequences for the San Diego County cohort (6% to 16%; p < 0.001 Exact), but not for the FCP cohort (30% to 30%; p>0.5), even though the epidemiologically-linked partners were referred from the FCP participants. In total, clustering sequences represented 25% of the combined cohort with the largest cluster containing twelve individuals. Sequences from epidemiologically-linked partners were found within fifteen of these clusters, which included a total of 96 study participants.
Figure 1.
A: Sequences from epidemiologically-linked partners that fell within clusters previously defined from the combined FCP and San Diego County cohorts (n=29)
B: Sequences from the combined FCP and San Diego County cohorts that clustered together and with sequences from epidemiologically-linked partners (n=96)
C: Sequences that clustered within the combined FCP and San Diego County cohorts (n=160)
Prevalence of Drug Resistance
The prevalence of `any' resistance associated mutation, interpreted by the Geneseq™ algorithm, within each of the study cohorts was San Diego County 30%, FCP 19%, epidemiologically-linked partners 22%, and the clustering sequences of the combined cohorts 19%. Overall the San Diego County and epidemiologically-linked partner cohorts tended to have a greater prevalence of drug resistance as compared to the other study cohorts (Table 3). Since these cohorts were largely comprised of chronically infected individuals, the higher prevalence of drug resistance most likely represents the presence of individuals who had received antiretroviral therapy. Even though individuals were supposed to be antiretroviral naïve to be eligible for the San Diego County Cohort, specific treatment data were not collected and eligibility was determined by individual clinical providers; therefore, we were unable to assure that all of these individuals were antiretroviral naïve. Theoretically, if some of the individuals in the San Diego County cohort had received antiretroviral therapy, then that could explain the observed decrease in clustering as compared to the FCP cohort. Although, forward HIV transmission from these individuals would have still been possible as the viral burden in these individuals was at least high enough for viral sequencing[32].
Table 3.
The prevalence of drug resistance in the cohorts.
| Cohort (n) | Drug Resistance % (95% C.I.) | ||||
|---|---|---|---|---|---|
| `Any' | NRTI only | NNRTI only | PI only | 3 or more classes | |
| San Diego County (n=268) | .30 (.25-.36) | .19 (.15-.24) | .18 (.14-.23) | .07 (.05-.11) | .02 (.01-.04) |
| First Choice Program (n=367) | .19 (.15-.23) | .11 (.08-.15) | .10 (.07-.14) | .04 (.02-.07) | .02 (.01-.04) |
| Clustering Sequences (n=160) | .19 (.13-.26) | .09 (.05-.14) | .10 (.06-.16) | .03 (.01-.07) | .01 (0-.05) |
| Epi-linked Partners (n=36) | .22 (.12-.38) | .16 (.08-.32) | .15 (.06-.29) | .07 (.02-.23) | .04 (0-.15) |
C.I.: 95% confidence interval, NRTI: nucleoside reverse transcriptase inhibitors, NNRTI: non-nucleoside reverse transcriptase inhibitors, and PI: protease inhibitors, Epi-linked: Epidemiologically-linked, Clustering Sequences: those sequences that clustered together from the combined San Diego County and First Choice Program cohorts.
Conclusions
These investigations confirm previous reports that pol sequences can be used to define highly related HIV transmission groups or clusters within a population, including the identification of clusters of drug resistance[13, 18-21]. This study extends those observations with the demonstration of methods on how to increase the identification of individuals belonging to HIV transmission clusters. First, pol sequences that are obtained for the clinical evaluation of the presence of transmitted drug resistance can be used to identify transmission clusters among individuals with HIV infection of unknown duration in a population. Second, using sequences from individuals with known recent HIV infection greatly increases the size and number of transmission clusters identified. Third, epidemiologically-linked partners of individuals with recent HIV infection have a very high likelihood of belonging to previously defined transmission clusters. Taken together, this information outlines how molecular surveillance and contact tracing can be used to define transmission clusters within a population and identify individuals who have a high likelihood to belong to these clusters. Future studies will need to be performed to determine how these individuals can then be targeted for prevention measures.
This study has several limitations including investigating only the San Diego population, which is primarily comprised of MSM; therefore, further investigation will be required to assess these methods in other populations. In fact, our study population is similar to previous studies evaluating molecular HIV epidemiology in that they too were predominantly MSM populations[16-18, 20]. This similarity may simply represent a selection bias in the populations studied or may suggest that these methods may offer an opportunity to assist prevention efforts in specific risk populations, such as MSM. Additionally, this study performed contact tracing only for those participants with known recent HIV infection based on the assumption that they would be more likely to be able to access their source partner for contact tracing, although this may not be true. Contact tracing and epidemiologically linking partners may be just as or more successful for people with unknown duration of infection[33], and other methods of finding transmission clusters, like respondent driven or snowball sampling, may be more effective than voluntary contact tracing[34]. It is also likely that voluntary contact tracing will not be popular among all public health settings or all risk groups, and its effectiveness will need to be independently evaluated by region and targeted group. Enrollment of epidemiologically-linked partners was also inconsistent over the eleven years of the FCP cohort (range n=0-11 yearly); however, contact tracing and partner participation were completely voluntary. Despite the overall low success rate of finding and enrolling epidemiologically-linked partners (<10%), the partners who were enrolled clustered within the majority of the highly related transmission groups (>60%), which demonstrates a potentially very effective way of identifying individuals who belong to transmission groups. Although not assessed in this study, the individuals identified as belonging to transmission clusters or perhaps even only identified epidemiologically-linked partners could be targeted for prevention interventions, such as antiretroviral-based and behavioral modification strategies.
The approaches proposed in this study to identify transmission clusters must be used responsibly. To protect the health of their local populations, public health agencies have broad authority to examine medical records without patient consent, perform molecular epidemiology and compulsory contact tracing, and mandate testing and treatment for communicable diseases[4]. There is significant concern that similar public health procedures could place source partners of HIV transmission at legal jeopardy, since in nearly every jurisdiction in the United States it is illegal to transmit or expose someone to HIV[15]. While public health agencies have a long track record of protecting patient privacy, these legal issues must be considered in the context of HIV prevention efforts using molecular epidemiology and partner contact tracing[4]. Even though HIV represents a significant public health threat, it has historically been treated differently than other communicable diseases by public health agencies because of issues of stigma and discrimination; hence, HIV testing is often anonymous and partner notification is rarely performed. Allowing for HIV to be treated differently, public health agencies themselves may be complicit in the propagation of HIV stigma. Therefore, we propose the following public health model of designing prevention trials: 1) increase the identification of individuals with primary HIV infection with nucleic acid testing and less-sensitive antibody testing[35]; 2) perform routine voluntary (versus mandatory) contact tracing of partners for individuals identified with primary HIV infection; 3) require the entities that perform the resistance testing for patients provide the nucleotide sequences in a timely fashion; and 4) perform molecular surveillance in real time and focus outreach and prevention programs (behavioral- and antiretroviral-based) on individuals found within transmission clusters. These efforts would likely be more efficient if centralized. To further the usefulness and safety of these methods, we also propose decriminalization of unintended HIV transmission during consensual exposure and legal recognition that phylogenetic linkage using pol sequences does not prove beyond a reasonable doubt that transmission between partners occurred. Future research needs to explore the effectiveness of these proposed methods in public health settings with the ultimate outcome of reducing HIV transmission.
Acknowledgements
We are grateful to the County of San Diego Department of Health and Human Services and all the participants of the San Diego First Choice Program and their partners whose unwavering generosity allows us to do these investigations. We would also like to thank Heidi Aiem, Tari Gilbert, Paula Potter, and Joanne Santangelo for their clinical assistance. Davey M. Smith has served as a consultant for Symmunity LLC and has received research support from Pfizer. Sergei Kosakovsky-Pond and Simon D.W. Frost have served as a consultant for Monogram Biosciences. Rick L. Pesano and Yolanda O. Lie are employees of Monogram Biosciences. Douglas D. Richman: has consulted for Pfizer, Merck, Bristol Myers Squibb, Gilead, Idenix, Roche, Monogram Biosciences. Susan J. Little: has received research support from Pfizer and Merck, and served as a consultant for Monogram Biosciences and Koronis.
Financial support: This work was supported by National Institutes of Health grants MH083552, AI077304, 5K23AI055276, AI69432, AI38858, AI43638, AI43752, AI29164, AI47745, MH62512, AI047745 and AI57167, the UCSD Center for AIDS Research (AI36214), CDC Contract 200-2002-00656, and the Research Center for AIDS and HIV Infection of the San Diego Veterans Affairs Healthcare System (10-92-035).
Footnotes
Written informed consent was obtained from all patients and the human experimentation guidelines of the U.S. Department of Health and Human Services and the individual institutions were followed in conducting this research. The U.S. Department of Health and Human Services has also issued a Confidentiality Certificate to all of the UCSD studies involving acute and early HIV infection and recruitment of sexual partners.
References
- 1.van Deutekom H, Gerritsen JJ, van Soolingen D, van Ameijden EJ, van Embden JD, Coutinho RA. A molecular epidemiological approach to studying the transmission of tuberculosis in Amsterdam. Clin Infect Dis. 1997;25:1071–1077. doi: 10.1086/516072. [DOI] [PubMed] [Google Scholar]
- 2.Sutton MY, Liu H, Steiner B, Pillay A, Mickey T, Finelli L, et al. Molecular subtyping of Treponema pallidum in an Arizona County with increasing syphilis morbidity: use of specimens from ulcers and blood. J Infect Dis. 2001;183:1601–1606. doi: 10.1086/320698. [DOI] [PubMed] [Google Scholar]
- 3.Thompson DK, Deal CD, Ison CA, Zenilman JM, Bash MC. A typing system for neisseria gonorrhoeae based on biotinylated oligonucleotide probes to PIB gene variable regions. J Infect Dis. 2000;181:1652–1660. doi: 10.1086/315464. [DOI] [PubMed] [Google Scholar]
- 4.Hecht FM, Wolf LE, Lo B. Lessons from an HIV transmission pair. J Infect Dis. 2007;195:1239–1241. doi: 10.1086/512247. [DOI] [PubMed] [Google Scholar]
- 5.Robertson DL, Sharp PM, McCutchan FE, Hahn BH. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 6.Burger H, Weiser B, Flaherty K, Gulla J, Nguyen PN, Gibbs RA. Evolution of human immunodeficiency virus type 1 nucleotide sequence diversity among close contacts. Proc Natl Acad Sci U S A. 1991;88:11236–11240. doi: 10.1073/pnas.88.24.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulder PJ, Brander C, Tang Y, Tremblay C, Colbert RA, Addo MM, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 9.Overbaugh J, Bangham CR. Selection forces and constraints on retroviral sequence variation. Science. 2001;292:1106–1109. doi: 10.1126/science.1059128. [DOI] [PubMed] [Google Scholar]
- 10.Frost SD, Gunthard HF, Wong JK, Havlir D, Richman DD, Leigh Brown AJ. Evidence for positive selection driving the evolution of HIV-1 env under potent antiviral therapy. Virology. 2001;284:250–258. doi: 10.1006/viro.2000.0887. [DOI] [PubMed] [Google Scholar]
- 11.Ou CY, Ciesielski CA, Myers G, Bandea CI, Luo CC, Korber BTM, et al. Molecular Epidemiology of Hiv Transmission in A Dental Practice. Science. 1992;256:1165–1171. doi: 10.1126/science.256.5060.1165. [DOI] [PubMed] [Google Scholar]
- 12.Human immunodeficiency virus transmission in household settings--United States. MMWR Morb Mortal Wkly Rep. 1994;43:347, 353–346. [PubMed] [Google Scholar]
- 13.Brooks JT, Robbins KE, Youngpairoj AS, Rotblatt H, Kerndt PR, Taylor MM, et al. Molecular analysis of HIV strains from a cluster of worker infections in the adult film industry, Los Angeles 2004. Aids. 2006;20:923–928. doi: 10.1097/01.aids.0000218558.82402.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Oliveira T, Pybus OG, Rambaut A, Salemi M, Cassol S, Ciccozzi M, et al. Molecular epidemiology: HIV-1 and HCV sequences from Libyan outbreak. Nature. 2006;444:836–837. doi: 10.1038/444836a. [DOI] [PubMed] [Google Scholar]
- 15.Lazzarini ZFR. Sex, Crimes and HIV. Focus A Guide to AIDS Research and Counseling 2007. 2007 May;:1–4. [PubMed] [Google Scholar]
- 16.Hue S, Pillay D, Clewley JP, Pybus OG. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc Natl Acad Sci U S A. 2005;102:4425–4429. doi: 10.1073/pnas.0407534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pao D, Fisher M, Hue S, Dean G, Murphy G, Cane PA, et al. Transmission of HIV-1 during primary infection: relationship to sexual risk and sexually transmitted infections. AIDS. 2005;19:85–90. doi: 10.1097/00002030-200501030-00010. [DOI] [PubMed] [Google Scholar]
- 19.Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C, et al. High Rates of Forward Transmission Events after Acute/Early HIV-1 Infection. J Infect Dis. 2007;195:951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 20.Cachay ERMN, Kosakovsky Pond SL, Pesano R, Lie YS, Aiem H, Butler DM, Letendre S, Mathews WC, Smith DM. Active methamphetamine use is associated with transmitted drug resistance to non-nucleoside reverse transcriptase inhibitors in individuals with HIV infection of unknown duration. The Open AIDS Journal. 2007;1:5–10. doi: 10.2174/1874613600701010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truong HH, Grant RM, McFarland W, Kellogg T, Kent C, Louie B, et al. Routine surveillance for the detection of acute and recent HIV infections and transmission of antiretroviral resistance. AIDS. 2006;20:2193–2197. doi: 10.1097/01.aids.0000252059.85236.af. [DOI] [PubMed] [Google Scholar]
- 22.Hall HI, Song R, Rhodes P, Prejean J, An Q, Lee LM, et al. Estimation of HIV incidence in the United States. JAMA. 2008;300:520–529. doi: 10.1001/jama.300.5.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith D, Moini N, Pesano R, Cachay E, Aiem H, Lie Y, et al. Clinical utility of HIV standard genotyping among antiretroviral-naive individuals with unknown duration of infection. Clin Infect Dis. 2007;44:456–458. doi: 10.1086/510748. [DOI] [PubMed] [Google Scholar]
- 24.Hecht FM, Wang L, Collier A, Little S, Markowitz M, Margolick J, et al. A multicenter observational study of the potential benefits of initiating combination antiretroviral therapy during acute HIV infection. J Infect Dis. 2006;194:725–733. doi: 10.1086/506616. [DOI] [PubMed] [Google Scholar]
- 25.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis suite. North Carolina State University; Raleigh, North Carolina: 1999. [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2004:bti079. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 28.Shafer RW, Rhee SY, Pillay D, Miller V, Sandstrom P, Schapiro JM, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. Aids. 2007;21:215–223. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchinson SJ, Gore SM, Goldberg DJ, Yirrell DL, McGregor J, Bird AG, Leigh-Brown AJ. Method used to identify previously undiagnosed infections in the HIV outbreak at Glenochil prison. Epidemiol Infect. 1999;123:271–275. doi: 10.1017/s0950268899002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.HIV/AIDS Epidemiology Report 2006. Health and Human Services Agency, Public Health Services; County of San Diego: 2006. [Google Scholar]
- 31.Respess RA, Rayfield MA, Dondero TJ. Laboratory testing and rapid HIV assays: applications for HIV surveillance in hard-to-reach populations. AIDS. 2001;15(Suppl 3):S49–59. doi: 10.1097/00002030-200104003-00007. [DOI] [PubMed] [Google Scholar]
- 32.Butler DM, Smith DM, Cachay ER, Hightower GK, Nugent CT, Richman DD, Little SJ. Herpes simplex virus 2 serostatus and viral loads of HIV-1 in blood and semen as risk factors for HIV transmission among men who have sex with men. AIDS. 2008;22:1667–1671. doi: 10.1097/QAD.0b013e32830bfed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahrens K, Kent CK, Kohn RP, Nieri G, Reynolds A, Philip S, Klausner JD. HIV partner notification outcomes for HIV-infected patients by duration of infection, San Francisco, 2004 to 2006. J Acquir Immune Defic Syndr. 2007;46:479–484. doi: 10.1097/qai.0b013e3181594c61. [DOI] [PubMed] [Google Scholar]
- 34.Magnani R, Sabin K, Saidel T, Heckathorn D. Review of sampling hard-to-reach and hidden populations for HIV surveillance. Aids. 2005;19(Suppl 2):S67–72. doi: 10.1097/01.aids.0000172879.20628.e1. [DOI] [PubMed] [Google Scholar]
- 35.Pilcher CD, McPherson JT, Leone PA, Smurzynski M, Owen-O'Dowd J, Peace-Brewer AL, et al. Real-time, universal screening for acute HIV infection in a routine HIV counseling and testing population. JAMA. 2002;288:216–221. doi: 10.1001/jama.288.2.216. [DOI] [PubMed] [Google Scholar]