Abstract
Outer membrane proteins (OMPs) of Gram-negative bacteria are synthesized in the cytosol and must cross the periplasm before insertion into the outer membrane. The 17-kDa protein (Skp) is a periplasmic chaperone that assists the folding and insertion of many OMPs, including OmpA, a model OMP with a membrane embedded β-barrel domain and a periplasmic αβ domain. Structurally, Skp belongs to a family of cavity-containing chaperones that bind their substrates in the cavity, protecting them from aggregation. However, some substrates, such as OmpA, exceed the capacity of the chaperone cavity, posing a mechanistic challenge. Here, we provide direct NMR evidence that, while bound to Skp, the β-barrel domain of OmpA is maintained in an unfolded state, whereas the periplasmic domain is folded in its native conformation. Complementary cross-linking and NMR relaxation experiments show that the OmpA β-barrel is bound deep within the Skp cavity, whereas the folded periplasmic domain protrudes outside of the cavity where it tumbles independently from the rest of the complex. This domain-based chaperoning mechanism allows the transport of β-barrels across the periplasm in an unfolded state, which may be important for efficient insertion into the outer membrane.
Keywords: cavity-based, outer membrane
Many molecular chaperones have evolved cavity-like structures to bind their non-native substrates. Classic examples, such as the chaperonins, bind the substrate in the cavity formed by their open rings and protect them from aggregation. Cycles of ATP binding and hydrolysis, coupled with substrate binding and release, then help the substrate achieve its native conformation (1). Because the cavities of these chaperones have limited capacities, binding substrates larger than the cavity requires a portion of the substrate to be bound inside the cavity while the rest of the substrate remains outside. In these cases, however, the entire substrate remains unfolded while bound to the open ring of the chaperone (2–4).
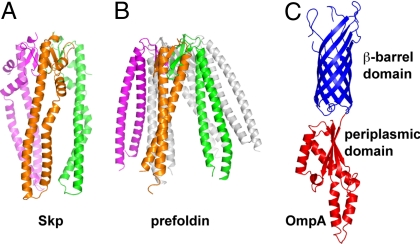
The 17-kDa protein (Skp) is a periplasmic chaperone present in many Gram-negative bacteria involved in the folding and insertion of proteins in the outer membrane (5–7). The crystal structure of Skp revealed a trimer with a “jellyfish” architecture where a central cavity is formed by long tentacle-like helical protrusions emanating from a body domain (Fig. 1A) (8, 9). The Skp structure was unexpectedly similar to that of prefoldin, a cytosolic molecular chaperone present in archea and eukarya (8–10). Although Skp is a trimer (8, 9, 11) and prefoldin is a hexamer (12, 13), both proteins share jellyfish architectures with a central cavity (Fig. 1 A and B). In prefoldin, this cavity has been shown to bind substrate proteins (10, 13). Prefoldin thus belongs to a family of chaperones sometimes referred to as “holdases.” These chaperones are ATP independent and, in contrast to chaperonins, do not directly facilitate folding of their substrates. Instead they protect the substrates from aggregation by holding them in their cavities, and substrate release appears to be mediated by interaction with downstream folding systems such as the CCT/TriC complex in the case of prefoldin (13). The structural similarity between Skp and prefoldin suggests that Skp also belongs to the holdase chaperone family.
Fig. 1.
Structures of Skp, prefoldin, and OmpA. (A) Model of the Skp trimer with subunits colored green, magenta, and orange (8). (B) Structure of the prefoldin hexamer with 3 subunits colored in transparent gray and 3 colored as in A. (C) Structure of the β-barrel domain of OmpA (51) (in blue) and the periplasmic domain of the OmpA-like protein RmpM (23) (in bright red). All models are on the same scale.
Outer membrane proteins (OMPs) are the main substrates of Skp. These integral membrane proteins share a characteristic membrane-embedded β-barrel structure and some have additional nonmembrane domains. One such example is OmpA, a Skp substrate that contains an N-terminal β-barrel in addition to the C-terminal periplasmic domain (Fig. 1C) and has been extensively used as a model to study OMP folding and membrane insertion (14–17). OMPs have to cross the inner membrane and the aqueous periplasmic space before they are inserted in the outer membrane (18). Skp is thought to bind OMPs as they emerge from the translocation machinery in the inner membrane, protect them from aggregation in the periplasm, and deliver them to the outer membrane in a state competent for insertion (8, 18, 19). However, the molecular mechanism remains unclear.
Here, we directly assess the folding state and topology of OmpA while bound to Skp. OmpA represents a class of challenging 2-domain substrates for Skp consisting of a membrane domain (the β-barrel) and a “soluble” domain (the periplasmic domain). In addition, the size of full-length OmpA (≈35 kDa) appears to exceed the capacity of the Skp cavity (Fig. 1), further complicating how the chaperone protects its substrate from aggregation. Using a combination of NMR and biochemical experiments we provide direct evidence that Skp binds the β-barrel domain of OmpA in its cavity and maintains it in an unfolded state while simultaneously allowing folding of the soluble periplasmic domain of OmpA outside of the cavity. This domain-based accommodation of large substrates in the Skp cavity may represent a general paradigm used by other cavity-based chaperones.
Results and Discussion
Skp Prevents the Aggregation of OmpA by Forming a Stable Complex.
Previous studies have shown that Skp can prevent the aggregation of model proteins like lysozyme (8). We next wanted to test Skp function with a true OMP substrate. OmpA is an outer membrane β-barrel protein of Gram-negative bacteria, extensively used as a model to study OMP folding and membrane insertion (14–17). In vivo, Skp is required for proper membrane insertion of OmpA and is therefore an excellent choice to study Skp function (5, 19, 20). When urea-denatured OmpA is diluted into buffer, the protein forms aggregates that elute in the void volume of a size exclusion chromatography (SEC) column (Fig. S1). However, OmpA aggregation is completely prevented when denatured OmpA is diluted in a buffer containing Skp trimers in stoichiometric amounts. In this case, a complex between Skp and OmpA is formed, which can be isolated by SEC (Fig. S1). The complex had an absolute mass [obtained with a multiangle light scattering (MALS) detector] of 82 kDa, indicating that a single OmpA molecule is bound to the Skp trimer (predicted molecular mass, 82.4 kDa). Despite OmpA's size and 2-domain architecture, the Skp trimer forms a 1:1 complex with OmpA even in an excess of Skp (data not shown). This finding is consistent with recent data showing that Skp forms 1:1 complexes with many OMPs (21). There was no indication of the complex falling apart during the chromatographic run, and the complex was stable for days as assayed by SEC (data not shown), which indicates that the complex formed between Skp and OmpA is stable and does not dissociate over time.
The Folding Status of OmpA Bound to Skp.
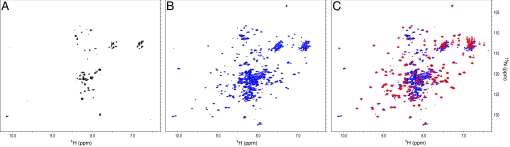
In vivo, Skp has been shown to be important for the folding and insertion of several OMPs (5, 7, 19, 22). These OMPs share the signature β-barrel structure as their integral membrane domain. One model is that periplasmic chaperones like Skp maintain the β-barrel domain of OMPs in an unfolded state to facilitate their insertion in the membrane. However, some studies have suggested that Skp recognizes a conformational motif in its substrates rather than extended, unfolded polypeptides (7, 19), implying that substrate proteins must be at least partially folded when bound by Skp. To directly address this issue, heteronuclear NMR experiments were used to monitor the folding state of the β-barrel domain of OmpA (amino acids 1–177; OmpA177) in complex with Skp. 2D 1H, 15N heteronuclear sequential quantum correlation (HSQC) spectra provide a “fingerprint” of the amide backbone in a protein and represent a valuable tool to determine whether a protein is properly folded. By 15N labeling OmpA177 but not Skp we are able to selectively observe the folding state of the substrate complexed with Skp. The amide cross-peaks in a folded protein show a wide chemical shift dispersion and generally have uniform signal intensity, whereas an unfolded protein will have poor dispersion and a wide range of cross-peak intensities.
Denatured 15N-labeled OmpA177 was diluted into a buffer containing unlabeled Skp to induce binding, and the complex was purified by SEC before NMR data collection. The 2D 1H, 15N transverse-relaxation optimized spectroscopy (TROSY)-HSQC spectrum of this complex shows a wide range of peak intensities and poor chemical shift dispersion (Fig. 2A). Thus, the β-barrel domain of OmpA is maintained in an unfolded state when bound to Skp.
Fig. 2.
2D 1H, 15N TROSY-HSQC of 15N-labeledOmpA and its complexes with Skp. (A) OmpA177 (β-barrel domain) in complex with Skp. (B) Full-length OmpA in complex with Skp. (C) Overlay of the spectrum of the isolated periplasmic domain OmpA177–325 (red) with the spectrum of the OmpAfull-length-Skp complex (blue).
As mentioned above, OmpA has a periplasmic domain in addition to the membrane-embedded β-barrel structure. Using the same NMR approach described above, we probed the folding status of full-length 15N-labeled OmpA in complex with Skp. Surprisingly, the OmpA–Skp complex has well-resolved peaks in the 1H, 15N TROSY-HSQC spectrum (Fig. 2B), indicating that full-length OmpA is at least partially folded in complex with Skp. One explanation for this result is that the C-terminal periplasmic domain of OmpA is well folded in the OmpA–Skp complex, whereas the β-barrel domain is maintained in an unfolded state.
To test this hypothesis a 2D 1H, 15N HSQC-TROSY spectrum was acquired on the isolated 15N-labeled C-terminal periplasmic domain of OmpA (amino acids 177–325; OmpA177–325) (red spectrum in Fig. 2C). This spectrum shows well-resolved cross-peaks indicating a well-folded protein and is consistent with the αβ fold of this domain (23) (Fig. 1C). Furthermore, a superposition of the spectra of isolated OmpA177–325 and the OmpA–Skp complex reveals that most of the well-resolved cross-peaks in the spectrum of the OmpA–Skp complex overlap with cross-peaks in the isolated OmpA177–325 spectrum (Fig. 2C). Therefore, when OmpA is in complex with Skp, the periplasmic domain of OmpA appears to adopt its native structure, whereas the β-barrel domain remains unfolded. In contrast to full-length OmpA, denaturation of OmpA177–325 followed by dilution in a buffer containing Skp did not result in complex formation (data not shown). Thus, the isolated periplasmic domain of OmpA (OmpA177–325) does not interact with Skp and instead this 16-kDa domain appears to refold rapidly into its native conformation (Fig. S2).
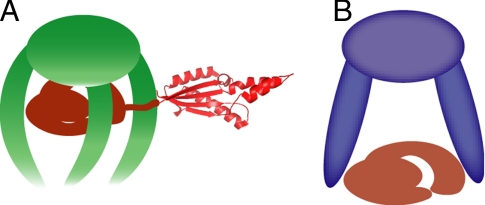
The Skp central cavity appears too small to accommodate both the β-barrel and periplasmic domains of full-length OmpA. The NMR data collected for the complex of Skp and full-length OmpA indicated that the β-barrel remains unfolded, whereas the periplasmic domain is folded to its native conformation (Fig. 2). Therefore, to accommodate larger, 2-domain substrates such as OmpA Skp may be able to bind the β-barrel inside its cavity, keeping it in an unfolded state, whereas the periplasmic domain protrudes outside the cavity where it folds into its native conformation as shown schematically in Fig. 4A. In such a model, the periplasmic domain of OmpA outside of the cavity would orient and tumble in solution relatively independent of the rest of the Skp–OmpA complex.
Fig. 4.
Models of substrate binding by Skp and prefoldin. (A) Model of a Skp–OmpA complex with the periplasmic domain of OmpA folded and protruding out of the Skp cavity. (B) Model of prefoldin with bound substrate based on published data (13).
To test this hypothesis, 15N T1 and T1ρ relaxation experiments were performed on both the isolated 15N-labeled periplasmic domain of OmpA and the full-length 15N-labeled-OmpA–unlabeled Skp complex. TROSY versions of standard gradient selected 2D 1H, 15N T1, and T1ρ experiments were acquired at 25 °C (data not shown) and used to estimate the apparent rotational correlation time, τc, for the periplasmic domain of OmpA in both its free form and the full-length OmpA–Skp complex. The 15N T1 and T1ρ values were determined for well-resolved cross-peaks that correspond to cross-peaks observed in the TROSY-HSQC spectrum of the isolated OmpA177–325 domain. Only cross-peaks that showed good fits to a single exponential decay for the T1 and T1ρ experiments were used in the analysis. The 15N T2 was determined by correcting the T1ρ data for resonance offset effects (see Methods). The T1/T2 ratio was then used to calculate τc. Because both chemical exchange and internal motion can affect the measured 15N T2, any cross-peak that had T1/T2 values >1 SD above or below the mean were not used in the analysis (24). A set of T1/T2 ratios from 37 cross-peaks was used to determine the average apparent τc for the periplasmic domain of OmpA in both the free form and the full-length OmpA–Skp complex. An isotropic tumbling model was assumed, which then yields an apparent τc of 16.4 (+/−1.4) ns for the periplasmic domain of OmpA in the full-length OmpA–Skp complex and 11.7 (+/−1.1) ns for the isolated periplasmic domain.
For a spherical isotropically tumbling molecule the τc can also be calculated from the Stokes–Einstein relation (25), which yields a calculated τc of 30 ns for the 82-kDa full-length OmpA–Skp complex. Therefore, the average τc of 16.4 ns obtained from the 15N relaxation data indicates that, when in complex with Skp, the periplasmic domain of OmpA tumbles much faster than an 82-kDa complex but slightly slower than a free 16-kDa domain. These data are consistent with a model in which the periplasmic domain of OmpA protrudes outside of the Skp cavity and orients relatively independently from the rest of the OmpA bound in the Skp complex. This domain remains tethered to the complex and therefore tumbles slower than the free periplasmic domain.
It is possible, although unlikely, that the unfolded N-terminal domain and the folded C-terminal domain of OmpA are both bound in the Skp cavity and the 2 domains tumble as a unit inside the cavity independently of Skp. This scenario would also yield a τc faster than that calculated for the 82-kDa Skp–OmpA complex but slower than a free C-terminal domain, and thus is compatible with the observed data. We have ruled out this possibility as described below.
Skp Binds β-Barrel Domains in Its Cavity.
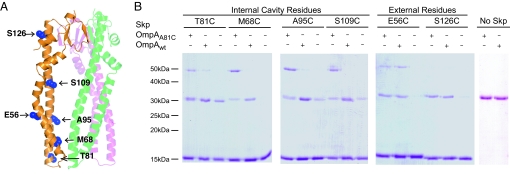
Cross-linking of chaperones to their substrates has been a useful approach in the identification of chaperone substrate binding sites (26–28). Disulfide cross-linking was used to study the topology of OmpA binding to Skp. Because Skp is naturally devoid of Cys residues, we used a structure-guided approach to introduce single Cys residues and probe the topology of OmpA binding to Skp by disulfide cross-linking. The mutations were designed to probe the inside surface of the cavity and positions on the outer surface of Skp. The specific residues mutated to Cys and their location in the structure are indicated in Fig. 3A. All of the mutants showed wild-type levels of activity in a standard lysozyme aggregation assay (8) (data not shown).
Fig. 3.
OmpA binding to Skp. (A) Location of single Cys mutations in the Skp monomer are shaded in blue and labeled. Labels on the right point to residues lining the inside of the cavity. Labels on the left point to residues on the outer surface of Skp. The other 2 Skp subunits are shown in faded green and magenta for reference but the Cys residues are not shown for clarity. (B) Nonreducing SDS/PAGE analysis of the cross-linking products of single-cysteine Skp mutants to wild-type OmpA (OmpAwt) containing 2 cysteines in the periplasmic domain or OmpA A81C (OmpAA81C) containing an additional Cys in the β-barrel domain. Plus and minus signs indicate the presence or absence of the 2 forms of OmpA in each reaction. Bands of ≈15 kDa correspond to Skp monomers and bands of ≈50 kDa correspond to a Skp1–OmpA1 cross-link. Note that a disulfide-linked Skp dimer (≈31 kDa) comigrates with free OmpA in these gels (compare lanes without OmpA and lanes labeled No Skp).
The periplasmic, C-terminal domain of OmpA contains 2 naturally-occurring Cys residues (C290 and C302), whereas the N-terminal β-barrel domain has no Cys residues. A Cys residue was introduced in the OmpA β-barrel domain by an A81C mutation. OmpA wild-type (OmpAwt) and A81C (OmpAA81C) were then used to form complexes with single Cys mutants of Skp as described above, and formation of disulfide bridges was monitored by nonreducing SDS/PAGE.
Because Skp is a homotrimer, the introduced Cys residues were present at 3 positions (Fig. 3A). Adjacent subunits of Skp can form 31-kDa disulfide-cross-linked dimers. These dimers were observed in control experiments in the absence of OmpA for the T81C, A95C, and S109C mutants but were not prevalent in the M68C mutant (Fig. 3B). No Skp dimers were observed in the absence of OmpA for the E56C and S126C mutants on the outside of the cavity (Fig. 3B), which shows that there are no intermolecular cross-links between Skp trimers.
When a Cys residue was present on the inside surface of the Skp cavity, OmpAA81C, but not OmpAwt, was efficiently cross-linked to Skp, yielding a band of ≈50 kDa consistent with a Skp1-OmpA1 product (Fig. 3B, Internal Cavity Residues). This finding indicates that the N-terminal, membrane domain of OmpA is bound inside the Skp cavity in a way that allows disulfide bonds to form. In contrast, the Cys residues in the C-terminal, periplasmic domain of OmpA were not available for cross-linking with Skp cavity residues, suggesting that this OmpA domain is outside of the Skp cavity. Among the Cys residues located outside of the Skp cavity, S126C produced no cross-links with either OmpAwt or OmpAA81C, indicating that this position is far from the substrate binding site (Fig. 3B, External Residues). However, the Skp E56C mutant cross-linked OmpAwt and OmpAA81C with similar efficiency, indicating that the Cys residues in the periplasmic domain of OmpA are available to react with the outward-facing Cys in the E56C Skp mutant. Therefore, the cross-linking data support the model shown in Fig. 4A, in which the N-terminal β-barrel domain of OmpA is bound in the Skp cavity, whereas the periplasmic domain is outside of the cavity where its cysteines can react with Skp E56C mutant.
The strong cross-links between OmpAA81C and Skp mutants M68C, A95C, and S109C indicate that Skp binds its substrates deep within its cavity interacting with the full length of the tentacles. This mode of binding contrasts with that of prefoldin, where substrates interact with the tips of the tentacle domains and not deep in the cavity (Fig. 4B), as shown in both EM reconstructions (10) and biochemical experiments (13). Furthermore, the distribution of hydrophobic patches thought to interact with the substrates is also consistent with these different binding models. Whereas prefoldin has a concentration of hydrophobic residues at the tips of the tentacles (12), there is a more even distribution of hydrophobic residues along the inner face of the tentacles in Skp (8).
In vitro, folding of β-barrels appears to be a slow process coupled to membrane insertion (29, 30), which may explain why Skp recognizes the β-barrel domain of OmpA and maintains it in an unfolded state in the cavity; whereas the periplasmic domain folds into its native conformation outside of the cavity. Skp is capable of delivering its cargo directly to membranes in vitro (29, 30). In vivo, however, the current hypothesis is that periplasmic chaperones such as Skp deliver their substrate OMPs to the YaeT complex in the outer membrane, which works as the insertion machine (18, 31). YaeT (also known as Omp85) is an essential OMP with a large periplasmic domain containing 5 polypeptide-transport- associated (POTRA) domains (32). Recent data suggest that these POTRA domains recognize unfolded OMPs and may serve to nucleate the formation of β-strands in OMPs before their membrane insertion (33–35). Importantly, there is no ATP in the periplasm that could be used to power the unfolding of proteins before their insertion in the membrane. Therefore, we hypothesize that the ability of chaperones such as Skp to maintain β-barrels in an unfolded state is a requirement for efficient transfer to YaeT and competent insertion in the membrane.
Many Gram-negative bacteria, such as Esherichia coli, have a second periplasmic chaperone, SurA, involved in OMP transport and insertion (36). E. coli can survive the deletion of either Skp or SurA, but a double deletion produces a synthetic lethal phenotype (20), which suggests that these chaperones have overlapping functions in E. coli. Thus, the chaperoning mechanism described here for Skp may be also be important for SurA. However, whereas SurA binding to small peptides has been described (37–40), the mechanism for protecting β-barrels from aggregation remains unclear.
Insertion of β-barrels in the outer membrane is observed not only in bacteria, but also in their phylogenetically-related eukaryotic organelles such as mitochondria (41). In these organelles, the Tim9–Tim10 complex of the intermembrane space functions as a chaperone that delivers β-barrel proteins to the Sam complex in the outer membrane for insertion (42). Although Sam is homologous to YaeT (41), Tim9 and Tim10 do not share significant sequence similarity with Skp. However, the structure of the Tim9–Tim10 complex is analogous to Skp with long α-helical tentacles emerging from a body domain and may use a similar mechanism to chaperone β-barrel proteins (43).
In summary, we provide direct evidence that the β-barrel domain of OMPs is maintained in an unfolded state inside the Skp cavity where it is protected from aggregation. We propose that for large substrates Skp partitions individual domains inside and outside of its cavity where nonmembrane domains protrude outside and fold into their native conformation (Fig. 4A). All cavity-based chaperones, including Skp, prefoldin, and the bacterial and eukaryotic chaperonins face a challenge when handling substrates larger than the capacity of their cavities (1, 44, 45). The accommodation of individual domains inside the cavity described here for Skp may represent a general paradigm in the folding of large substrates by other cavity-based chaperones.
Methods
Protein Expression, Purification, and Mutagenesis.
His-tagged Skp was purified and the tag removed as described (8) with the following modifications. Ni-NTA column washes contained 1 M NaCl. After elution from the Ni-NTA column the protein was diluted 5-fold with dialysis buffer 1 [25 mM Hepes (pH 7.5), 500 mM NaCl, 10 mM DTT] and then dialyzed overnight. Tobacco etch virus (TEV) protease was added (1:10 wt/wt) to the protein solution before dialysis to remove the His tag. After 48 h of dialysis and TEV reaction, the buffer was changed to 25 mM Hepes (pH 7.5), 10 mM DTT. After the second dialysis the protein mixture was loaded onto a 1-mL monoS cation exchange column equilibrated with 25 mM Hepes (pH 7.5), 5 mM 2-mercaptoethanol. The column was developed with a gradient from 0 to 0.4 M NaCl over 25 column volumes. Fractions were analyzed with SDS/PAGE and those containing pure protein were pooled and concentrated if necessary. PCR site-directed mutagenesis was used to introduce cysteine mutations into the Skp gene, and the resulting single Cys mutants were purified as the wild-type protein.
OmpA was cloned into a modified pET28 vector after PCR amplification from E. coli genomic DNA. The PCR product was ligated into the vector between NcoI and SacI sites. The resulting plasmid (pMS223) contained the sequence for mature OmpA with no signal sequence and no affinity tag. OmpA was expressed in the cytosol as an inclusion body and purified as follows: The cell pellet was resuspended in 20 mM Tris (pH 8.5), 0.1% Triton X-100, 5 mM EDTA, 5 mM 2-mercaptoethanol, and 0.25 mg/mL lysozyme. The cell suspension was then incubated on ice for 30 min and lysed by sonication. The inclusion body pellet was harvested by centrifugation at 37,000 × g for 30 min, and the resulting pellet was resuspended and washed 3 times in 20 mM Tris (pH 8.5), 0.2% Triton X-100, 5 mM EDTA, and 5 mM 2-mercaptoethanol. After the third Triton X-100 wash the pellet was washed in buffer containing 1% deoxycholate instead of Triton X-100. This final pellet was then resuspended in anion exchange buffer A [20 mM Tris (pH 7.5), 5 mM 2-mercaptoethanol, and 5 M urea]. Insoluble material was separated with an additional 37,000 × g spin, and the supernatant was loaded onto a 12-mL resourceQ column. The column was developed with a gradient of 0 to 0.5 M NaCl over 20 column volumes. Fractions were analyzed with SDS/PAGE and those containing pure protein were pooled and stored at −80 °C. For OmpAA81C, Ala-81 was mutated to cysteine by PCR mutagenesis and the mutant protein was purified as described above.
The N-terminal (β-barrel) fragment of OmpA was made by PCR mutagenesis of Pro-177 to a stop codon in the full-length construct. Purification followed the same protocol as the full-length protein. The periplasmic domain of OmpA was PCR-subcloned from the plasmid containing the full-length gene, resulting in a construct with an N-terminal His6 tag followed by a TEV protease cleavage site. It was purified as described for Skp (8) with the following changes: Ni-NTA column washes contained 300 mM NaCl. After the Ni-NTA purification the protein was dialyzed against 20 mM Hepes (pH 7.5), 150 mM NaCl, and 10 mM DTT. The TEV reaction to remove the His tag was allowed to run overnight with the dialysis, and TEV was used at 1:100 (wt/wt) OmpA177–325. The TEV protease was removed by the addition of Ni-NTA beads (we used His-tagged TEV) followed by centrifugation to pellet the beads. OmpA177–325 without a His tag was then run on a Superdex-200 size exclusion column where it eluted primarily as a monomer species. Fractions containing pure protein as judged by SDS/PAGE were pooled, concentrated, and stored at −80 °C. 15N-labeled OmpA constructs were all expressed in minimal media as described (46) and purified as above.
SEC–MALS Sample Preparation and Data Collection.
OmpA denatured in 8 M urea and 5 mM DTT was diluted 100-fold into a solution containing Skp in 25 mM Hepes (pH 7.5) and 250 mM NaCl. After incubation at room temperature for 10 min they were filtered through a 0.1-μm filter. OmpA and Skp (trimer) were typically 20 μM. Injections (100-μL) were analyzed by using a Shodex KW803 size exclusion column running in SEC buffer (20 mM phosphate buffer at pH 6.5, which was brought to 150 mM [Na+] with Na2SO4) at 1 mL/min. Data were collected with Wyatt Technologies MALS (DAWN EOS) and refractive index (Optilab DSP) detectors. Data were processed with Astra 4.9 software.
Cysteine Cross-Linking.
The cross-linking experiments were carried out as described (47) with minor modifications. Briefly, complexes of Skp cysteine mutants and OmpAwt or OmpA81C, were formed by quick dilution as described above. Initial protein stocks were adjusted to 100 μM with 1 mM DTT and diluted to final concentrations of 0.5 μM OmpA and Skp (trimer) in 25 mM Hepes (pH 7.5), 250 mM NaCl, 5 mM EDTA, and 10% glycerol. Reactions of 100-μL were allowed to incubate at room temperature for 30 min without the addition of an oxidation catalyst, before being quenched in 6× nonreducing Laemmli sample buffer + 40 mM N-ethylmaleimide at 95 °C for 2 min. Samples were then analyzed by nonreducing SDS/PAGE and stained with Coomasie blue.
NMR Sample Preparation and Data Collection.
Complexes for NMR were prepared as described for the SEC–MALS assay, except the complexes were formed in SEC buffer, purified over a Superdex 200 column, and concentrated to ≈150 μM for NMR data collection.
2D 1H, 15N TROSY-HSQC spectra (48) for the OmpA–Skp complexes and HSQC spectra for OmpA177–325 were acquired on a Varian Inova 800-MHz NMR. In all cases Skp was unlabeled and OmpA or its fragments were labeled with 15N. For the OmpA177–Skp complex, 2,048 × 150 complex points were acquired with sweep widths of 10,810 × 2,950 Hz in 1H and 15N, respectively. Full-length OmpA-Skp data were collected by using 9,615 × 2,760-Hz sweep widths and 1,024 × 128 complex points in 1H and 15N. Both TROSY spectra were acquired with a preacquisition delay of 1.6 s and 128 transients per free induction decay (FID). The OmpA177–325 HSQC was collected with 9,950 × 2,760-Hz sweep widths and 840 × 256 complex points in 1H and 15N, a 1.4-s preacquisition delay, and 64 transients per FID. All spectra were processed and figures were made by using NMRPipe software (49).
TROSY versions of T1 and T1ρ 15N relaxation experiments (24, 50) were collected on a Varian VNMRS 800-MHz NMR spectrometer for the purpose of estimating τc values for the free OmpA periplasmic domain and full-length OmpA–Skp complex. Relaxation experiments were run with a 3-s preacquisition delay, 32 transients per FID, and sweep widths of 9,124 × 2,600 Hz and 2,048 × 64 complex points in 1H and 15N, respectively. T1 data were collected with relaxation times of 0, 0.108, 0.216, 0.432, 0.864, 1.621, and 2.809 s. T1ρ data were acquired by using a 15N spinlock field of 2,747 Hz and relaxation times of 0, 5, 10, 20, 30, 40, 60, and 80 ms. Data were processed in NMRPipe by using Lorentzian-to-Gaussian apodization, then peak volumes were measured by using tools in NMRPipe and the relaxation rates were extracted by using the fitting algorithm, modelXY in NMRPipe.
The rotational correlation time for a molecule can be estimated from R1 and R2 relaxation rates (equal to 1/T1 and 1/T2, respectively) by using the following equation:
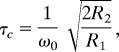 |
where ω0 is the 15N carrier frequency multiplied by 2π. R2 can be estimated from R1 and R1ρ, ignoring chemical exchange, as follows (50):
γB1 is the 15N spinlock field in the T1ρ experiment and Δω is the offset of the 15N resonance from the carrier frequency, both in Hz.
The T1 and T1ρ data were used to estimate the τc of the C-terminal periplasmic domain of OmpA in the full-length OmpA–Skp complex. After extracting rates, the data were pruned in 3 stages as follows. First, R1 and R1ρ values with >10% error were discarded. Second, any residues with R2/R1 ratios >1 SD from the mean were discarded (24). Finally, only R1 and R1ρ values extracted from peaks common to both OmpA–Skp and OmpA177–325 were used to calculate the correlation times The remaining set of 37 cross-peaks was used to calculate the average τc values for the periplasmic domain of OmpA in both the free form and the full-length OmpA–Skp complex.
Supplementary Material
Acknowledgments.
We thank Lisa Warner for help with NMR data analysis. This work was supported in part by National Science Foundation Grant 0719225 (to M.C.S.) and National Institutes of Health Grant AI033098 (to A.P.). Support for T.A.W was provided by National Institutes of Health Training Grant GM65103. The 800-MHz NMR used in this study was purchased with partial support from National Institutes of Health Grant RR16649, National Science Foundation Grant 0230996, and the W. M. Keck Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809275106/DCSupplemental.
References
- 1.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: Physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 2.Horst R, et al. Direct NMR observation of a substrate protein bound to the chaperonin GroEL. Proc Natl Acad Sci USA. 2005;102:12748–12753. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin Z, Rye HS. Expansion and compression of a protein folding intermediate by GroEL. Mol Cell. 2004;16:23–34. doi: 10.1016/j.molcel.2004.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park ES, Fenton WA, Horwich AL. No evidence for a forced-unfolding mechanism during ATP/GroES binding to substrate-bound GroEL: No observable protection of metastable Rubisco intermediate or GroEL-bound Rubisco from tritium exchange. FEBS Lett. 2005;579:1183–1186. doi: 10.1016/j.febslet.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Chen R, Henning U. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol. 1996;19:1287–1294. doi: 10.1111/j.1365-2958.1996.tb02473.x. [DOI] [PubMed] [Google Scholar]
- 6.Missiakas D, Betton JM, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA, and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 7.Schafer U, Beck K, Muller M. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- 8.Walton TA, Sousa MC. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell. 2004;15:367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 9.Korndorfer IP, Dommel MK, Skerra A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat Struct Mol Biol. 2004;11:1015–1020. doi: 10.1038/nsmb828. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Benito J, et al. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 2002;21:6377–6386. doi: 10.1093/emboj/cdf640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlapschy M, et al. The periplasmic E. coli chaperone Skp is a trimer in solution: Biophysical and preliminary crystallographic characterization. Biol Chem. 2004;385:137–143. doi: 10.1515/BC.2004.032. [DOI] [PubMed] [Google Scholar]
- 12.Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. Structure of the molecular chaperone prefoldin: Unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell. 2000;103:621–632. doi: 10.1016/s0092-8674(00)00165-3. [DOI] [PubMed] [Google Scholar]
- 13.Lundin VF, et al. Molecular clamp mechanism of substrate binding by hydrophobic coiled-coil residues of the archaeal chaperone prefoldin. Proc Natl Acad Sci USA. 2004;101:4367–4372. doi: 10.1073/pnas.0306276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surrey T, Jahnig F. Kinetics of folding and membrane insertion of a β-barrel membrane protein. J Biol Chem. 1995;270:28199–28203. doi: 10.1074/jbc.270.47.28199. [DOI] [PubMed] [Google Scholar]
- 15.Danese PN, Silhavy TJ. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genetics. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 16.Kleinschmidt JH, Wiener MC, Tamm LK. Outer membrane protein A of E. coli folds into detergent micelles, but not in the presence of monomeric detergent. Protein Sci. 1999;8:2065–2071. doi: 10.1110/ps.8.10.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem. 2003;278:9092–9099. doi: 10.1074/jbc.M211177200. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 19.Harms N, et al. The early interaction of the outer membrane protein phoe with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J Biol Chem. 2001;276:18804–18811. doi: 10.1074/jbc.M011194200. [DOI] [PubMed] [Google Scholar]
- 20.Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu J, Mayer C, Behrens S, Holst O, Kleinschmidt JH. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J Mol Biol. 2007;374:91–105. doi: 10.1016/j.jmb.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 22.De Cock H, et al. Affinity of the periplasmic chaperone Skp of Escherichia coli for phospholipids, lipopolysaccharides, and non-native outer membrane proteins. Role of Skp in the biogenesis of outer membrane protein. Eur J Biochem. 1999;259:96–103. doi: 10.1046/j.1432-1327.1999.00010.x. [DOI] [PubMed] [Google Scholar]
- 23.Grizot S, Buchanan SK. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol Microbiol. 2004;51:1027–1037. doi: 10.1111/j.1365-2958.2003.03903.x. [DOI] [PubMed] [Google Scholar]
- 24.Farrow NA, et al. Backbone dynamics of a free and a phosphopeptide-complexed Src homology-2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 25.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. 2nd Ed. Amsterdam: Academic; 2007. [Google Scholar]
- 26.Elad N, et al. Topologies of a substrate protein bound to the chaperonin GroEL. Mol Cell. 2007;26:415–426. doi: 10.1016/j.molcel.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Schlieker C, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- 29.Kleinschmidt JH, den Blaauwen T, Driessen AJ, Tamm LK. Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry. 1999;38:5006–5016. doi: 10.1021/bi982465w. [DOI] [PubMed] [Google Scholar]
- 30.Kleinschmidt JH, Tamm LK. Secondary and tertiary structure formation of the β-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J Mol Biol. 2002;324:319–330. doi: 10.1016/s0022-2836(02)01071-9. [DOI] [PubMed] [Google Scholar]
- 31.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 32.Bos MP, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane. Curr Opin Microbiol. 2004;7:610–616. doi: 10.1016/j.mib.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: Conformational flexibility and substrate recognition. Structure (London) 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 35.Knowles TJ, et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Webb HM, Ruddock LW, Marchant RJ, Jonas K, Klappa P. Interaction of the periplasmic peptidylprolyl cis-trans isomerase SurA with model peptides. The N-terminal region of SurA id essential and sufficient for peptide binding. J Biol Chem. 2001;276:45622–45627. doi: 10.1074/jbc.M107508200. [DOI] [PubMed] [Google Scholar]
- 38.Bitto E, McKay DB. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J Biol Chem. 2003;278:49316–49322. doi: 10.1074/jbc.M308853200. [DOI] [PubMed] [Google Scholar]
- 39.Hennecke G, Nolte J, Volkmer-Engert R, Schneider-Mergener J, Behrens S. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J Biol Chem. 2005;280:23540–23548. doi: 10.1074/jbc.M413742200. [DOI] [PubMed] [Google Scholar]
- 40.Xu X, Wang S, Hu YX, McKay DB. The periplasmic bacterial molecular chaperone SurA adapts its structure to bind peptides in different conformations to assert a sequence preference for aromatic residues. J Mol Biol. 2007;373:367–381. doi: 10.1016/j.jmb.2007.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiedemann N, et al. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: Intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem. 2004;279:18188–18194. doi: 10.1074/jbc.M400050200. [DOI] [PubMed] [Google Scholar]
- 43.Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed α-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 44.Chapman E, et al. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci USA. 2006;103:15800–15805. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudhuri TK, Farr GW, Fenton WA, Rospert S, Horwich AL. GroEL/GroES-mediated folding of a protein too large to be encapsulated. Cell. 2001;107:235–246. doi: 10.1016/s0092-8674(01)00523-2. [DOI] [PubMed] [Google Scholar]
- 46.Anderson EM, Halsey WA, Wuttke DS. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: Applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 2007;423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delaglio F, et al. Nmrpipe: A multidimensional spectral processing system based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 50.Kempf JG, Loria JP. Protein dynamics from solution NMR. Cell Biochem Biophys. 2003;37:187–211. doi: 10.1385/cbb:37:3:187. [DOI] [PubMed] [Google Scholar]
- 51.Pautsch A, Schulz GE. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.