Abstract
CD1 proteins constitute a distinct lineage of antigen-presenting molecules specialized for the presentation of lipid antigens to T cells. In contrast to the extensive sequence polymorphism characteristic of classical MHC molecules, CD1 proteins exhibit limited sequence diversity. Here, we describe the identification and characterization of CD1d alleles in wild-derived mouse strains. We demonstrate that polymorphisms in CD1d affect the presentation of endogenous and exogenous ligands to CD1d-restricted T cells, including type I (Vα14i) and type II (non-Vα14i) natural killer T (NKT) cells. Using congenic mice, we found CD1d polymorphisms affect the thymic selection of type I NKT cells and induce allogeneic T cell responses. Collectively, results from these studies demonstrate a role for polymorphisms in influencing the development and function of CD1d-restricted T cells.
Keywords: T cell activation, T cell development
In conjunction with MHC class I and class II molecules, CD1 proteins constitute a distinct, third lineage of antigen-presenting molecules (1). CD1 molecules have been described in a variety of mammals and were recently described in chickens, increasing significantly the age of CD1 in vertebrate immunity (2, 3). The CD1 gene family was originally defined in humans and consists of 5 genes designated CD1A–E. The 5 CD1 proteins can be divided into 3 groups (4). Group 1 (CD1a, CD1b, and CD1c) and group 2 (CD1d) CD1 sample intracellular compartments and traffic to the cell surface to present lipid antigens to T cells. In contrast, CD1e, the sole member of group 3 CD1, functions as a chaperone intracellularly and does not traffic to the cell surface (5). Mice have a simple CD1 system that consists of 2 highly homologous genes (CD1D1 and CD1D2) that exhibit sequence similarity to hCD1D.
The majority of CD1d-restricted T cells are type I natural killer T (NKT) cells that express an invariant Vα14-Jα18 T cell antigen receptor (TCR) α chain preferentially paired with a Vβ8.2, Vβ7, or Vβ2 (6). A homologous T cell population that expresses an invariant Vα24-Jα18 TCRα chain is also present in humans (7). Murine type I NKT cells are either CD4+ or CD4−CD8− double-negative (DN), and exhibit an activated/memory phenotype. Stimulation of type I NKT cells results in the rapid production of cytokines, including IFN-γ and IL-4. Although a number of endogenous and pathogen-derived ligands capable of activating type I NKT cells have been described (8), the prototypical ligand remains α-galactosylceramide (α-GalCer) that was originally isolated from the marine sponge Agelas mauritianus (9). α-GalCer-CD1d tetramers can be used to specifically identify type I NKT cells in mice and humans (10).
In addition to type I NKT cells, other CD1d-restricted T cell populations have been described (11). CD1d-restricted NKT cells that do not express the Vα14i TCRα chain are classified as type II NKT cells (12). Type II NKT cells do not bind α-GalCer-CD1d tetramers, and their phenotype can be quite heterogeneous, including CD4+, CD8+, or DN NKT cells. Similar to type I NKT cells, type II NKT cells are capable of rapid production of both IFN-γ and IL-4 and may perform important regulatory functions (13, 14). Recent reports have described roles for type II NKT cells in tumor immunity (15), microbial infection (16), and experimental autoimmune encephalomyelitis (17). Lipid antigens recognized by type II NKT cells may be distinct from those that stimulate type I NKT cells, because type II NKT cell activation does not require CD1d trafficking through endosomes (18, 19).
A hallmark feature of classical MHC class I and II proteins is the extensive polymorphism in residues that constitute the peptide-binding groove. Allelic variation among MHC molecules is the basis for differential peptide binding, thymic repertoire bias, and allograft rejection (20, 21). In contrast, CD1d1 is nonpolymorphic among inbred strains of mice (22). We report here the identification and characterization of CD1d alleles from wild-derived strains of mice, including Mus castaneus and Mus spretus. Using congenic mice, we demonstrate that polymorphisms in CD1d affect the selection of type I NKT cells. Additionally, CD1d polymorphisms affect the presentation of endogenous and exogenous antigens and the subsequent activation of type I and type II NKT cells.
Results
Identification and Characterization of Murine CD1d Alleles.
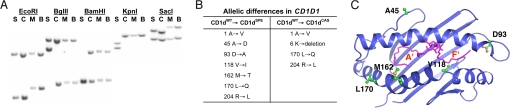
To search for CD1d polymorphisms, we studied the CD1d repertoire in wild-derived mouse strains. Southern blot analysis of genomic DNA isolated from WT C57BL/6 (M. musculus) and wild-derived strains of mice using a CD1-specific probe revealed that restriction fragment length polymorphisms (RFLPs) could be readily detected among B6, Mus castaneus, and M. spretus, whereas no RFLP was observed between B6 and Mus molossinus (Fig. 1A).
Fig. 1.
Identification and characterization of CD1d alleles. (A) Genomic DNA from M. spretus (S), M. castaneus (C), M. molossinus (M), and B6 (B) mice were digested with the indicated restriction enzymes and analyzed in a Southern blot using a CD1-specific probe. (B) The location of polymorphic residues present in CD1dCAS and CD1dSPE. (C) A ribbon diagram of the α1α2 domain of CD1dWT in complex with α-GalCer highlighting polymorphic residues identified in the CD1dSPE. Image was constructed based on the structure of murine CD1d/PBS-25 (35) superimposed with α-GalCer from the structure of human CD1d/α-GalCer (36) by using the PyMOL Molecular Graphics System (www.pymol.org). The polymorphism at position 1 is not included because it is absent from the coordinates used to generate this image.
To examine the wild-derived CD1d repertoire in more detail, we cloned and sequenced CD1D1 genes from both M. castaneus and M. spretus and assigned the allelic forms of CD1d1 to be CD1dCAS and CD1dSPE, respectively, to distinguish them from CD1dWT. We focused our study on CD1d1 because CD1d2 does not play a significant role in the development of type I NKT cells despite low levels of expression in the thymus of WT mice (23). Compared with CD1dWT, the extracellular domains of CD1dCAS contained 3 amino acid substitutions and 1 deletion, whereas 7 amino acid substitutions were found in CD1dSPE (Fig. 1B). Six of the 7 amino acid substitutions between CD1dSPE and CD1dWT are located in the α1 and α2 domains of CD1d (Fig. 1C), which may affect ligand binding and/or CD1d/TCR interactions.
CD1d Polymorphisms Affect the Development of Type I NKT Cells in B6.CD1dSPE Congenic Mice.
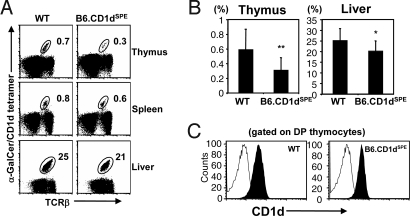
As both of the amino acid changes identified in the α1α2 domains of CD1dCAS were present in CD1dSPE (Fig. 1B), we focused on studying the effects of the polymorphisms in the CD1dSPE allele. To examine the effects of the CD1dSPE polymorphisms on the development and function of CD1d-restricted T cells, we generated CD1d congenic mice on the B6 background: B6-SPE-CD1dSPE (B6.CD1dSPE). We first examined the type I NKT cell compartment in B6.CD1dSPE mice by using α-GalCer-CD1d tetramers. We detected a statistically significant decrease in the proportion of type I NKT cells in the thymus and liver of B6.CD1dSPE mice compared with WT (Fig. 2 A and B). Similar differences were observed when NKT cells were identified by using the traditional mAb combination of anti-TCRβ and anti-NK1.1 (Fig. S1). Although there was a trend toward a lower proportion of type I NKT cells in the spleen of B6.CD1dSPE mice, this difference was not statistically significant within the sample set analyzed (Fig. 2A). These results highlight a defect in the selection of type I NKT cells in the thymus of B6.CD1dSPE mice. Because comparable levels of CD1d were detected on thymocytes isolated from B6 and B6.CD1SPE mice, the reduced proportion of type I NKT cells in B6.CD1dSPE is unlikely caused by differential CD1d expression (Fig. 2C).
Fig. 2.
Polymorphisms in CD1d affect type I NKT cell development. (A) Single-cell suspensions from the indicated organs were stained with anti-TCRβ and CD1d-αGalCer tetramer and analyzed by FACS. (B) Bar graphs depict mean ± SD for the proportion of type I NKT cells in WT (thymus n = 15; liver n = 12) and B6.CD1dSPE (thymus n = 16; liver n = 13) mice in the indicated organs. Statistically significant differences: *, P < 0.05; **, P < 0.01. (C) Representative histograms of CD1d expression on thymocytes of WT and B6.CD1dSPE mice. Thymocytes were stained with either a control mAb (open) or anti-CD1d (filled) and analyzed by flow cytometry.
CD1d Polymorphisms Affect the Activation of Type I NKT Cells.
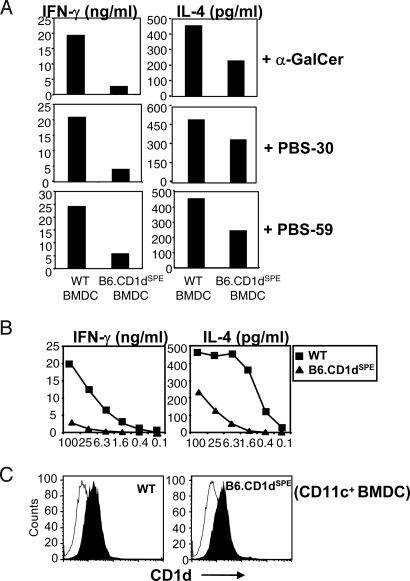
Several of the polymorphisms identified in CD1dSPE are located in the antigen-binding pocket, raising the possibility that these allelic variations may affect antigen presentation by CD1d and its recognition by NKT cells. To address this issue, we examined the ability of bone marrow-derived dendritic cells (BMDC) from WT and B6.CD1dSPE mice to present exogenous antigens to type I NKT cells. Three different exogenous antigens were tested, including α-GalCer, PBS-30 (α-glucuronosylceramide), and PBS-59 (α-galacturonosylceramide), synthetic versions of Sphingomonas cell wall glycosylceramides that activate type I NKT cells in the context of CD1d (24–26). For each of the exogenous antigens tested, the production of IFN-γ and IL-4 by type I NKT cells after stimulation with antigen-pulsed B6.CD1dSPE BMDC was significantly reduced compared with WT BMDC (Fig. 3A). Analysis of type I NKT cell activation by WT and B6.CD1dSPE BMDC pulsed with titrating concentrations of α-GalCer revealed lower levels of cytokine production in response to B6.CD1dSPE BMDC at all concentrations tested (Fig. 3B). The dose–response curve suggests that polymorphisms in CD1dSPE affect interactions with the type I NKT TCR. Indeed, the decreases in cytokine production are unlikely caused by aberrant CD1d expression, because CD1d levels at the cell surface were comparable between WT and B6.CD1dSPE BMDC (Fig. 3C). Furthermore, these results are unlikely to be caused by global antigen presentation defects in B6.CD1dSPE BMDC because the activation of OT-II splenocytes by WT and B6.CD1dSPE BMDC pulsed with OVA323–339 peptide was comparable. Taken together, these results suggest that polymorphisms in CD1d affect the presentation and recognition of exogenous glycolipid antigens and the subsequent activation of type I NKT cells.
Fig. 3.
Impaired type I NKT cell activation by exogenous glycolipid antigens presented by B6.CD1dSPE BMDC. (A) Purified type I NKT cells were stimulated with WT or B6.CD1dSPE BMDC pulsed with the antigen indicated. The amount of IFN-γ and IL-4 was quantitated by ELISA. Bar graphs depict mean values from duplicate wells. The data shown are representative of 2 independent experiments. (B) Line graph depicts IFN-γ and IL-4 production by Vα14-transgenic TCRα knockout splenocytes in response to WT and B6.CD1dSPE BMDC pulsed with titrating concentrations of α-GalCer. Data shown are representative of 3 independent experiments. (C) Histograms depict CD1d expression on WT and B6.CD1dSPE BMDC used in the assay. BMDC were stained with mAb to CD11c and a control mAb (open) or mAb to CD1d (filled) and analyzed by flow cytometry.
CD1d Polymorphisms Affect Type II NKT Cell Function.
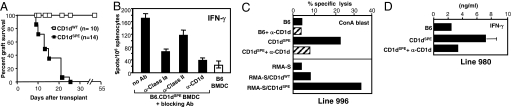
Allogeneic MHC molecules have been demonstrated to be potent immunogens for a variety of MHC-restricted T cells (27). To determine whether the polymorphisms in CD1dSPE induce allogeneic responses, tail skin from either B6.CD1dSPE mice or WT littermates was grafted onto B6 recipient mice. As shown in Fig. 4A, all skin grafts from B6.CD1dSPE mice were rapidly rejected, whereas control WT grafts survived for at least 2 months. In an effort to characterize allogeneic T cells elicited by B6.CD1dSPE skin grafts, splenocytes from B6 recipients were isolated and either analyzed directly ex vivo or maintained in culture by weekly restimulation with irradiated B6.CD1dSPE splenocytes. For the ex vivo analysis, B6 recipient splenocytes were stimulated with B6.CD1dSPE or WT BMDC in the absence or presence of blocking mAb to MHC class I, class II, or CD1d. Splenocyte activation was assessed by IFN-γ production in ELISPOT assays. Splenocytes produced IFN-γ in response to stimulation with B6.CD1dSPE BMDC, but not WT BMDC, indicating this response was allospecific (Fig. 4B). The allogeneic response is likely composed of both CD1d-restricted and conventional T cells, because blocking mAbs specific for CD1d, MHC class I, or MHC class II all significantly decreased the number of IFN-γ-producing cells (Fig. 4B).
Fig. 4.
Preferential recognition of the CD1dSPE allele by type II NKT cells. (A) Tail skins from B6.CD1dSPE mice and control littermates were grafted onto B6 mice. Rejection was defined by complete necrosis of the skin graft. (B) Splenocytes from graft recipients were stimulated with either B6.CD1dSPE or WT BMDC in the absence or presence of blocking mAb to MHC class I, class II, or CD1d. The activation of splenocytes was measured by IFN-γ production in ELISPOT assays. Results shown are representative of 3 independent experiments. (C) The specificity of T cell line 996 was evaluated in a 51Cr release assay. (Upper) ConA blasts from mice indicated were used as targets in the absence or presence of anti-CD1d. (Lower) RMAS cells or RMAS transfectants expressing either CD1dWT or CD1dSPE were used as targets. Bar graphs depict percentage specific lysis at a 10:1 effector-to-target (E/T) cell ratio. Data shown are representative of 3 independent experiments. (D) The specificity of T cell line 980 was evaluated by measuring IFN-γ production after stimulation with irradiated splenocytes from WT and B6.CD1dSPE mice in the absence or presence of anti-CD1d. Data shown are representative of 3 independent experiments.
Two T cell lines, designated 996 and 980, were established from B6 recipients of B6.CD1dSPE skin grafts. Line 996 exhibits preferential cytolytic activity against the CD1dSPE allele compared with CD1dWT (Fig. 4C). The recognition of CD1dSPE-expressing targets by line 996 was CD1d-restricted, because cytolytic activity could be inhibited by mAb to CD1d (Fig. 4C). Additionally, when RMAS-CD1d transfectants were used as targets line 996 exhibited preferential killing activity against CD1dSPE-expressing transfectants compared with CD1dWT (Fig. 4C). These data suggest that line 996 recognized CD1dSPE as an intact molecule and not as a CD1dSPE-derived peptides presented by conventional MHC molecules. Similarly, line 980 produced more IFN-γ in response to stimulation by B6.CD1dSPE splenocytes compared with WT splenocytes (Fig. 4D). Production of IFN-γ by line 980 was also CD1d-restricted, because IFN-γ production decreased in the presence of mAb to CD1d (Fig. 4D). Flow cytometric and RT-PCR analysis revealed that these 2 cell lines do not express NK cell receptors or Vα14-Jα18 message. Taken together, these results suggest that CD1d polymorphisms affect the function of type II NKT cells.
Contribution of Individual CD1dSPE Polymorphisms to the Activation of CD1d-Restricted NKT Cells.
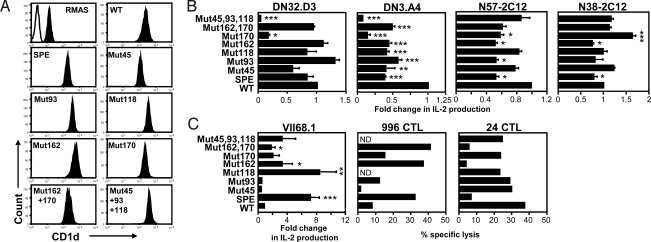
To determine the contribution of individual CD1d polymorphisms to the altered activation of CD1d-restricted T cells, we generated a panel of CD1d mutants incorporating one or a combination of the polymorphisms identified in CD1dSPE. These CD1d mutants were transfected into RMAS cells (Fig. 5A), which were then used to stimulate a panel of CD1d-restricted T cells. Four of 6 type I NKT cell hybridomas tested produced less IL-2 in response to stimulation with the RMAS-CD1dSPE compared with RMAS-CD1dWT (Fig. 5B), whereas the other 2 hybridomas did not show significant differences in the recognition of CD1dWT and CD1dSPE. DN3.A4 (Vβ8.2/Jβ2.1) and N57–2C12 (Vβ14/Jβ1.2) were more sensitive to allelic differences in CD1d compared with DN32.D3 (Vβ8.2/Jβ2.4) and N38–2C12 (Vβ8.2/Jβ2.5), suggesting that CDR2β and CDR3β regions may be able to modulate the TCR affinity of type I NKT cells to CD1d/glycolipid complexes. Interestingly, all type I NKT cells tested were sensitive to changes at residue 170, because mutation resulted in either a significant decrease (DN32.D3, DN3.A4, and N57–2C12) or increase (N38–2C12) in IL-2 production compared with CD1dWT (Fig. 5B). Changes at residue 162 diminished the reactivity of 3 hybridomas, except DN32.D3. The effect of the remaining polymorphic residues varied more, depending on the cell tested.
Fig. 5.
Effect of individual CD1d polymorphisms on T cell activation. (A) Histograms depict CD1d expression by various RMAS transfectants. Cells were stained with a control mAb (open) or mAb to CD1d (filled) and analyzed by flow cytometry. (B) Bar graphs depict mean ± SD of fold change in IL-2 production by type I NKT cell hybridomas after stimulation with a panel of RMAS-CD1d transfectants pulsed with α-GalCer (100 ng/mL). IL-2 production by each hybridoma in response to CD1dSPE and specific mutants was normalized to the CD1dWT allele. Bar graphs depict mean ± SD of fold change in IL-2 production obtained from 3 independent experiments. (C) Bar graphs depict mean ± SD of fold change in IL-2 production or percentage-specific lysis of the indicated RMAS-CD1d transfectants by type II NKT cells. E/T ratios are 1:1 and 60:1 for 996 and 24 CTL, respectively. Statistically significant differences: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We then evaluated the effects of the individual CD1d polymorphisms on the type II NKT cells (Fig. 5C). Interestingly, VII68.1 exhibited preferential recognition of CD1dSPE compared with CD1dWT. We found that mutation of CD1dWT at residue 118 alone was sufficient to impart recognition by VII68.1 to levels comparable with CD1SPE. The CD1dSPE-specific T cell line 996 exhibited enhanced cytolytic activity against several mutants compared with CD1dWT transfectants (Fig. 5C). Finally, similar to type I NKT cells, 24αβ cytotoxic T lymphocyte (CTL) exhibited greater reactivity to CD1dWT compared with CD1dSPE. Type II NKT cells were particularly sensitive to changes at residue 162 as mutation at this position resulted in either enhanced (VII68.1 and 996) or reduced (24αβ) reactivity compared with CD1dWT. Taken together, results from these experiments reveal critical roles for specific residues in CD1d-mediated T cell activation.
Discussion
Limited polymorphism is a hallmark feature of MHC class Ib and class I-like genes (28). In the various inbred strains of mice commonly used in immunological studies, CD1d1 is nonpolymorphic. However, by studying CD1D of wild-derived strains of mice, we identified additional CD1D1 alleles from M. castaneus and M. spretus. DNA sequence analysis revealed that the allelic differences of CD1D genes are much lower than those in MHC class Ia genes, suggesting strong conservation within CD1D genes. The divergence among CD1d alleles is comparable with that seen in MHC class Ib molecules that perform specialized immune functions, including TL, M3, and Qa-1 (29–31).
Polymorphism in MHC class Ia proteins is found selectively concentrated in the peptide-binding regions of the α1 and α2 domains where it affects the spectrum and conformation of MHC-bound peptides (32). Previous reports (33, 34) used visual inspection or computer modeling to identify specific residues in the CD1d antigen-binding groove that are important for the presentation of α-GalCer to type I NKT cells. Interestingly, none of the residues studied previously were polymorphic in CD1dSPE or CD1dCAS. The majority of substitutions between CD1dSPE and CD1dWT are located in the α1α2 domains (Fig. 1C), which may affect ligand binding and antigen presentation. Consistent with this finding, CD1d-restricted T cells exhibited differential reactivity against transfectants expressing either CD1dWT or CD1dSPE. Although incorporation of CD1dSPE polymorphisms appears to decrease the reactivity of type I NKT cells, reactivity by type II NKT cells was either enhanced or reduced. The differential effect of CD1d polymorphisms on type II NKT cells may be caused by the more diverse TCR Vα repertoire of type II NKT cells. Several type I NKT cell hybridomas used in this study express the same Vβ chain (i.e., Vβ8.2). However, they responded differently to some CD1d mutants, suggesting the involvement of CDR3β in the recognition of CD1d/glycolipid complexes. The crystal structure of CD1dWT reveals that the amino acid side chains of loop residues 45 and 93 point away from the antigen-binding groove and unlikely affect antigen binding, but they might affect T cell recognition (35–37). Residue 118, which is located in the β-sheet and points into the F′ pocket, may interact with the acyl chain of lipid antigens. Residue 162 is located in the α2 helix with its side chain pointed up. Substitution of Met to Thr may gain hydrogen-bond interaction with the carbonyl group of residue 158, which may influence the ligand binding/presentation. Interestingly, we found that changes at residue 162 alone were sufficient to impart preferential recognition of type II NKT cells to a specific CD1 allele. Mutation of position 162 also affected the reactivity of several type I NKT cell hybridomas. Residue 170 is located in α2 helix and forms hydrophobic interactions with Phe-171 and Leu-58 and may be involved in maintaining the hydrophobic environment of the A′ pocket. Indeed, we found that the reactivity of both type I and type II NKT cells was significantly affected by mutation of residue 170. A crystal structure of a human NKT TCR bound to α-GalCer/human CD1d identified regions of interaction in the CD1d α1α2 domains (38). Although none of the polymorphisms in the CD1dSPE allele would predict to be involved in direct binding with the NKT TCR, they are in close proximity to the CD1d/NKT TCR contact zone. Our study suggests that subtle changes of these residues might induce allosteric changes by communicating with the networks of residues in the lipid-binding groove to modulate ligand presentation to the TCR (Fig. S2) (39).
The highly-conserved nature of CD1d and the type I NKT TCR suggests there is strong evolutionary pressure to maintain an efficient interaction between CD1d+ APCs and type I NKT cells. Indeed, recognition of CD1d by type I NKT cells is maintained even across species barriers (40). Consistent with this finding, we found that the CD1dSPE allele was able to mediate type I NKT cell development in congenic B6.CD1dSPE mice. However, the proportions of type I NKT cells in B6.CD1dSPE mice were significantly lower than WT. These differences were more pronounced in the thymus than in the periphery. It is possible that lower affinity for the CD1dSPE allele might result in less effective positive selection of developing type I NKT cells. This notion is supported by the lower reactivity of type I NKT cell hybridomas to CD1dSPE compared with CD1dWT-expressing cells. Similarly, type I NKT cells from B6.CD1dSPE mice also exhibited higher reactivity to DC expressing CD1dWT compared with B6.CD1dSPE DC (Fig. S3). Interestingly, B6.CD1dSPE mice showed modest, but statistically significant, increases in the proportion of CD4+ type I NKT cell subsets in all organs tested compared with WT, suggesting interaction with this coreceptor may influence the overall avidity of the type I NKT cell TCR for CD1d. Type I NKT cells have been shown to develop in mice expressing transgenic human CD1d, which exhibits higher affinity for the TCR of murine type I NKT cells than mouse CD1d (41). Type I NKT cells in these mice are almost exclusively Vβ8+, suggesting differences in affinity may influence the TCR repertoire of type I NKT cells. However, we did not detect significant differences in TCR Vβ usage by type I NKT cells between WT and B6.CD1dSPE mice. In addition, we cloned and sequenced the Vα14-Jα18 message from B6.CD1dSPE mice and found that type I NKT cells in these mice express the same Vα-Jα junctions as WT type I NKT cells. Although the function of type I NKT cells in B6.CD1dSPE mice is not impaired, decreased type I NKT cell number in B6.CD1dSPE mice resulted in lower levels of serum IFN-γ production and less pronounced expansion of type I NKT cells in B6.CD1dSPE mice compared with B6 mice, after administration of α-GalCer-pulsed BMDC (Fig. S4). These results suggest that CD1d polymorphisms may affect the therapeutic efficacy of α-GalCer.
All type II NKT cells described to date are autoreactive as they recognize endogenous lipids such as phospholipids and sulfatide in the context of CD1dWT (17, 42). One of the more interesting findings of this work is the allele-specific recognition of CD1dSPE. T cell lines derived from B6 recipients of CD1dSPE skin grafts were CD8+ and demonstrated CD1dSPE-specific cytolytic activity and IFN-γ production. To determine whether CD1dWT could also be recognized as an alloantigen, we immunized B6.CD1dSPE mice with WT DC and were able to generate a T cell line from immunized B6.CD1dSPE mice that exhibited preferential recognition of CD1dWT, suggesting that it, too, can serve as an alloantigen (Fig. S5). These results may have implications for clinical settings in which T cell alloreactivity is of significant interest, including graft-versus-host disease and allogeneic hematopoietic stem cell transplantation (27, 43).
Single-nucleotide polymorphisms have been identified in all 5 human CD1 genes (44). For CD1b and CD1c these mutations are silent, whereas substitutions in CD1a, CD1d, and CD1e result in amino acid replacements (44). Although the functional significance of human group 1 CD1 polymorphisms remains unknown, polymorphisms in CD1e were recently reported to affect lipid antigen presentation (45). Results from our studies should serve as the impetus to continue to study polymorphisms in human CD1 proteins, because these changes have the potential to influence the repertoire and function of CD1-restricted T cells.
Materials and Methods
Mice.
Congenic B6.CD1dSPE was established by backcrossing an interspecific consomic strain, which carries a single introgressed M. spretus chromosome 3 in the B6 background, with B6 mice for an additional 10 generations. During backcrosses, animals were selected for the presence of specific CD1 allele by Southern blot analysis of EcoRI-digested genomic DNA with a probe for the CD1 exon 4 region as described (46). All animal work was approved by the Institutional Animal Care and Use Committee.
Cloning, Mutagenesis of CD1d1 cDNA, and Generation of RMAS-CD1d Transfectants.
CD1d1 cDNA from M. spretus and M. castaneus were obtained by RT-PCR with primers specific to exon 1 and exon 4 of CD1d as described (23). The PCR products were cloned into pCRScript, and DNA samples were sequenced. For mutagenesis studies, the full-length CD1d1 cDNA from B6 was subcloned into the pBluescript KS+ vector, and mutagenesis was conducted by using the QuikChange site-directed mutagenesis kit (Stratagene). The mutant CD1d1 fragments were sequenced and cloned into the pSR-Neo expression vector and transfected into RMAS cells.
Abs, CD1d Tetramers, and Flow Cytometry.
The following Abs were purchased from BD PharMingen: anti-TCRβ, anti-CD4, anti-NK1.1, anti-CD8, and hamster anti-TNP. The CD1d-specific mAbs 3H3 and 5C6 and the generation of α-GalCer-CD1d tetramers have been described (10, 47). Thymocytes, splenocytes, and hepatic lymphocytes were isolated and stained as described (48). After staining, cells were washed and analyzed by flow cytometry using a FACSCanto and analyzed with FlowJo software.
Activation and Analysis of Cytokine Production from Type I NKT Cells and CD1d-Restricted Hybridomas.
Type I NKT cells were enriched from the splenocytes of Vα14-transgenic TCRα−/− mice by incubating cells with M5114-FITC and B220-FITC, followed by depletion of FITC-labeled cells using anti-FITC microbeads and magnetic columns. BMDC were generated, MACS-purified, and pulsed with various glycolipid antigens as described (48). Antigen-pulsed BMDC were incubated with enriched NKT cells for 48 h, and the levels of IL-4 and IFN-γ in supernatants were quantitated by ELISA. To determine the effect of amino acid substitutions on NKT cell activation, CD1d-restricted T cell hybridomas were cocultured with various RMAS-CD1d transfectants. After 24 h, IL-2 production was quantitated by ELISA.
Skin Grafting.
Female B6.CD1dSPE and control littermates (CD1dWT) were used as donors. Full-thickness skin sections (≈1 × 1 cm) were harvested from donor tails and grafted onto the dorsal side of female B6 mice. Bandages were removed on day 7 posttransplant, and grafts were monitored for 55 days for evidence of rejection. Rejection was defined as complete necrosis of the skin graft.
ELISPOT Assays.
Multiscreen-IP plates were coated with anti-IFN-γ, washed, and blocked with RPMI medium10. Splenocytes from B6 recipients of B6.CD1dSPE skin grafts were cultured with BMDC from either B6 or B6.CD1dSPE mice in the absence or presence of mAb for MHC class Ia (B22 and Y3), MHC class II (M5114), or CD1d (3H3). Plates were incubated for 18 h, and IFN-γ-producing cells were quantitated with an ImmunoSpot reader.
Generation of CD1dSPE-Specific T Cell Lines.
WT recipients of B6.CD1dSPE skin grafts were killed, and single cell suspensions were prepared from lymph nodes and spleens. Suspensions were cultured with irradiated B6.CD1dSPE splenocytes in RPMI medium 10 containing blocking mAbs specific for MHC class I and class II. One week later, cultures were restimulated with B6.CD1dSPE splenocytes and maintained in supplemented Mischell Dutton media with IL-2 supplement.
CTL Assays.
Targets (1 × 106) were labeled with 50 μCi [51Cr] sodium chromate for 1 h at 37 °C. Targets were cultured with effectors for 4 h at 37 °C. The percentage of specific lysis was calculated as (experimental release − spontaneous release)/(maximal release − spontaneous release) × 100. For Ab blocking studies, the killing activity of T cell lines against various targets was assayed in the presence of 25% of supernatant containing specific mAbs. Con A blasts were prepared by stimulating splenocytes with ConA (2.5 μg/mL) for 72 h in RPMI medium 10.
Statistical Analysis.
Mean values were compared by using unpaired Student's t tests. All statistical analyses were performed with the PRISM program (GraphPad).
Supplementary Material
Acknowledgments.
We thank Dr. Jean-Louis Guénet (Pasteur Institute, Paris, France) for chromosome 3 consomic mice; Dr. Albert Bendelac (University of Chicago, Chicago, IL) for a breeding pair of Vα14-transgenic mice, DN32.D3 NKT cell hybridoma, and PBS-30 and PBS-59; Dr. Susanna Cardell (Goteborg University, Goteborg, Sweden) for VII68.1 T cell hybridoma and 24αβ TCR Tg mice; Dr. Richard Locksley (University of California, San Francisco, CA) for DN3.A4 NKT cell hybridoma; Drs. Sebastian Joyce and Kyoko Hayakawa (Vanderbilt, Nashville, TN; Fox Chase Cancer Center, Philadelphia, PA) for NKT cell hybridomas N38-2C12 and N57-2C12; Dr. Steve Porcelli (Albert Einstein College of Medicine, Bronx, NY) for pSR-Neo vector; Kirin Brewery (Gunma, Japan) for α-GalCer; Dr. Wei-Jen Tang for advice during construction of the CD1d ribbon diagram; and Tara King, Ashley Rohr, Chunting Yang, Jessica Rojas, and Sharmila Shanmuganad for technical assistance. This work was supported by National Institutes of Health Grant R01 AI43407 (to C.-R.W) and National Research Service Award F32 Fellowship HL083744 (to M.I.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808476106/DCSupplemental.
References
- 1.Brigl M, Brenner MB. CD1: Antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 2.Salomonsen J, et al. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc Natl Acad Sci USA. 2005;102:8668–8673. doi: 10.1073/pnas.0409213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller MM, et al. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc Natl Acad Sci USA. 2005;102:8674–8679. doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barral DC, Brenner MB. CD1 antigen presentation: How it works. Nat Rev Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 5.de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 6.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 7.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8-α/β T cells demonstrates preferential use of several V β genes and an invariant TCRα chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brutkiewicz RR. CD1d ligands: The good, the bad, and the ugly. J Immunol. 2006;177:769–775. doi: 10.4049/jimmunol.177.2.769. [DOI] [PubMed] [Google Scholar]
- 9.Morita M, et al. Structure–activity relationship of α-galactosylceramides against B16-bearing mice. J Med Chem. 1995;38:2176–2187. doi: 10.1021/jm00012a018. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behar SM, Cardell S. Diverse CD1d-restricted T cells: Diverse phenotypes, and diverse functions. Semin Immunol. 2000;12:551–560. doi: 10.1006/smim.2000.0273. [DOI] [PubMed] [Google Scholar]
- 12.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: What's in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 13.Duarte N, et al. Prevention of diabetes in nonobese diabetic mice mediated by CD1d-restricted nonclassical NKT cells. J Immunol. 2004;173:3112–3118. doi: 10.4049/jimmunol.173.5.3112. [DOI] [PubMed] [Google Scholar]
- 14.Skold M, Faizunnessa NN, Wang CR, Cardell S. CD1d-specific NK1.1+ T cells with a transgenic variant TCR. J Immunol. 2000;165:168–174. doi: 10.4049/jimmunol.165.1.168. [DOI] [PubMed] [Google Scholar]
- 15.Terabe M, et al. A nonclassical non-Vα14Jα18 CD1d-restricted (type II) NKT cell is sufficient for down-regulation of tumor immunosurveillance. J Exp Med. 2005;202:1627–1633. doi: 10.1084/jem.20051381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duthie MS, Kahn M, White M, Kapur RP, Kahn SJ. Critical proinflammatory and antiinflammatory functions of different subsets of CD1d-restricted natural killer T cells during Trypanosoma cruzi infection. Infect Immun. 2005;73:181–192. doi: 10.1128/IAI.73.1.181-192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahng A, et al. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu YH, et al. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat Immunol. 2002;3:55–60. doi: 10.1038/ni740. [DOI] [PubMed] [Google Scholar]
- 19.Roberts TJ, et al. Recycling CD1d1 molecules present endogenous antigens processed in an endocytic compartment to NKT cells. J Immunol. 2002;168:5409–5414. doi: 10.4049/jimmunol.168.11.5409. [DOI] [PubMed] [Google Scholar]
- 20.Lawlor DA, Zemmour J, Ennis PD, Parham P. Evolution of class-I MHC genes and proteins: From natural selection to thymic selection. Annu Rev Immunol. 1990;8:23–63. doi: 10.1146/annurev.iy.08.040190.000323. [DOI] [PubMed] [Google Scholar]
- 21.Rothbard JB, Gefter ML. Interactions between immunogenic peptides and MHC proteins. Annu Rev Immunol. 1991;9:527–565. doi: 10.1146/annurev.iy.09.040191.002523. [DOI] [PubMed] [Google Scholar]
- 22.Gozalbo-Lopez B, Perez-Rosado A, Parra-Cuadrado JF, Martinez-Naves E. Identification of a new mouse Cd1d2 allele. Eur J Immunogenet. 2004;31:1–3. doi: 10.1111/j.1365-2370.2004.00431.x. [DOI] [PubMed] [Google Scholar]
- 23.Chen YH, et al. Expression of CD1d2 on thymocytes is not sufficient for the development of NK T cells in CD1d1-deficient mice. J Immunol. 1999;162:4560–4566. [PubMed] [Google Scholar]
- 24.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 25.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 26.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- 27.Welniak LA, Blazar BR, Murphy WJ. Immunobiology of allogeneic hematopoietic stem cell transplantation. Annu Rev Immunol. 2007;25:139–170. doi: 10.1146/annurev.immunol.25.022106.141606. [DOI] [PubMed] [Google Scholar]
- 28.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 29.Davis BK, Cook RG, Rich RR, Rodgers JR. Hyperconservation of the putative antigen recognition site of the MHC class I-b molecule TL in the subfamily Murinae: Evidence that thymus leukemia antigen is an ancient mammalian gene. J Immunol. 2002;169:6890–6899. doi: 10.4049/jimmunol.169.12.6890. [DOI] [PubMed] [Google Scholar]
- 30.Doyle CK, Davis BK, Cook RG, Rich RR, Rodgers JR. Hyperconservation of the N-formyl peptide binding site of M3: Evidence that M3 is an old eutherian molecule with conserved recognition of a pathogen-associated molecular pattern. J Immunol. 2003;171:836–844. doi: 10.4049/jimmunol.171.2.836. [DOI] [PubMed] [Google Scholar]
- 31.Hermel E, et al. Polymorphism and conservation of the genes encoding Qa1 molecules. Immunogenetics. 2004;56:639–649. doi: 10.1007/s00251-004-0722-x. [DOI] [PubMed] [Google Scholar]
- 32.Reche PA, Reinherz EL. Sequence variability analysis of human class I and class II MHC molecules: Functional and structural correlates of amino acid polymorphisms. J Mol Biol. 2003;331:623–641. doi: 10.1016/s0022-2836(03)00750-2. [DOI] [PubMed] [Google Scholar]
- 33.Burdin N, et al. Structural requirements for antigen presentation by mouse CD1. Proc Natl Acad Sci USA. 2000;97:10156–10161. doi: 10.1073/pnas.97.18.10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamada N, et al. Crucial amino acid residues of mouse CD1d for glycolipid ligand presentation to Vα14 NKT cells. Int Immunol. 2001;13:853–861. doi: 10.1093/intimm/13.7.853. [DOI] [PubMed] [Google Scholar]
- 35.Zajonc DM, et al. Structure and function of a potent agonist for the semiinvariant natural killer T cell receptor. Nat Immunol. 2005;6:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch M, et al. The crystal structure of human CD1d with and without α-galactosylceramide. Nat Immunol. 2005;6:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 37.Zeng Z, et al. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 38.Borg NA, et al. CD1d-lipid-antigen recognition by the semiinvariant NKT T cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 39.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 40.Brossay L, et al. CD1d-mediated recognition of an α-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumann J, et al. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Vα14i NKT cells. J Immunol. 2005;175:7303–7310. doi: 10.4049/jimmunol.175.11.7303. [DOI] [PubMed] [Google Scholar]
- 42.Gumperz JE, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 43.Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 44.Han M, Hannick LI, DiBrino M, Robinson MA. Polymorphism of human CD1 genes. Tissue Antigens. 1999;54:122–127. doi: 10.1034/j.1399-0039.1999.540202.x. [DOI] [PubMed] [Google Scholar]
- 45.Tourne S, et al. A naturally occurring mutation in CD1e impairs lipid antigen presentation. J Immunol. 2008;180:3642–3646. doi: 10.4049/jimmunol.180.6.3642. [DOI] [PubMed] [Google Scholar]
- 46.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 47.Mandal M, et al. Tissue distribution, regulation and intracellular localization of murine CD1 molecules. Mol Immunol. 1998;35:525–536. doi: 10.1016/s0161-5890(98)00055-8. [DOI] [PubMed] [Google Scholar]
- 48.Zimmer MI, et al. A cell-type-specific CD1d expression program modulates invariant NKT cell development and function. J Immunol. 2006;176:1421–1430. doi: 10.4049/jimmunol.176.3.1421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.