Abstract
Protein–protein interactions play key roles in virtually all cellular processes, often forming complex regulatory networks. A powerful tool to study interactions in vivo is fluorescence resonance energy transfer (FRET), which is based on the distance-dependent energy transfer from an excited donor to an acceptor fluorophore. Here, we used FRET to systematically map all protein interactions in the chemotaxis signaling pathway in Escherichia coli, one of the most studied models of signal transduction, and to determine stimulation-induced changes in the pathway. Our FRET analysis identified 19 positive FRET pairs out of the 28 possible protein combinations, with 9 pairs being responsive to chemotactic stimulation. Six stimulation-dependent and five stimulation-independent interactions were direct, whereas other interactions were apparently mediated by scaffolding proteins. Characterization of stimulation-induced responses revealed an additional regulation through activity dependence of interactions involving the adaptation enzyme CheB, and showed complex rearrangement of chemosensory receptors. Our study illustrates how FRET can be efficiently employed to study dynamic protein networks in vivo.
Keywords: bacteria, chemotaxis, FRET, protein network, signal transduction
Introduction
The chemotaxis pathway in Escherichia coli senses gradients of attractants and repellents to direct cellular movement toward favorable environments. Being a comparatively simple system with few components, chemotaxis in E. coli is an excellent model for the detailed quantitative study of signaling networks (Sourjik, 2004; Wadhams and Armitage, 2004). The sensory complex is formed by ligand-specific membrane-associated receptors (Tsr, Tar, Trg, Tap and Aer), histidine kinase CheA and coupling protein CheW. Ligand binding to receptor homodimers alters the autophosphorylation activity of the receptor-associated CheA. The kinase subsequently donates the phosphoryl group to response regulator CheY, which diffuses to flagellar motors and modulates their rotation. Dephosphorylation of CheY is catalyzed by the phosphatase CheZ. Adaptation is exerted by methyltransferase CheR and its antagonist, methylesterase CheB. CheB consists of a C-terminal catalytic domain and an N-terminal CheY-like regulatory domain, which is subject to activatory phosphorylation by CheA. Adaptation enzymes tune the ability of receptors to activate CheA by adjusting the level of receptor methylation on four specific glutamate residues in an activity-dependent manner, thereby returning kinase activity to a constant steady-state level under conditions of continuous stimulation. Two of the four glutamates at the methylation sites are originally translated as glutamines, which are functionally equivalent to methylated glutamates, and are converted to glutamates by the deamidase activity of CheB. Immunoelectron (Maddock and Shapiro, 1993), fluorescence (Sourjik and Berg, 2000) and cryo-electron (Zhang et al, 2007) microscopy have shown that thousands of chemoreceptors are organized in polar and lateral clusters, to which all other chemotaxis proteins localize, forming a large sensory machinery (Sourjik, 2004; Wadhams and Armitage, 2004). Allosteric interactions between receptors in clusters appear to play a central role in the amplification and integration of chemotactic signals (Li and Weis, 2000; Gestwicki and Kiessling, 2002; Sourjik and Berg, 2002b, 2004; Lai et al, 2005).
To obtain a comprehensive view of the network dynamics in living cells, we tested all intracellular protein interactions in the pathway and their dependence on chemotactic stimulation by fluorescence resonance energy transfer (FRET). We further quantified concentration dependence and kinetics of stimulation-induced changes in protein interactions. Our results provide a holistic picture of the pathway and of its intracellular dynamics, and demonstrate the efficiency of the FRET-based interaction mapping approach.
Results
FRET mapping of protein interactions
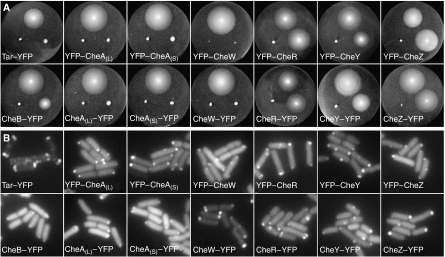
FRET allows the detection of intracellular interactions of fluorescently labeled proteins non-invasively by the energy transfer from an excited donor to an acceptor fluorophore (Wouters and Bastiaens, 2001). The transfer efficiency depends on the distance between the fluorophores as R06/(R6+R06), with the Förster radius R0—at which the energy is transferred with 50% efficiency—being around the size of a typical protein. Such steep dependence on spacing and short characteristic distance make the energy transfer highly specific for proteins that are part of the same complex, either interacting directly or binding to a common scaffolding protein. High distance selectivity, however, can also impair FRET in a protein complex where the spacing between fluorescent labels is above the critical distance, approximately 2 × R0. In our interaction screen, we used cyan and yellow fluorescent proteins (CFP and YFP, respectively) as a donor–acceptor pair with R0∼49 Å, approximately the same size as a fluorescent protein monomer (Tsien and Miyawaki, 1998; Sourjik and Berg, 2002a). To reduce the chances of false negatives due to the large distance or unfavorable orientation of proteins in the complex and to find pairs with strongest FRET efficiency for subsequent investigation of stimulation dependence, we constructed a library of both N- and C-terminal fusions of CFP and YFP to all chemotaxis proteins, the aspartate receptor Tar and the serine receptor Tsr (Supplementary Table SI). E. coli CheA is endogenously expressed from two alternative start codons, yielding a long and a short variant, CheAL and CheAS, respectively (Smith and Parkinson, 1980). To independently analyze both versions of CheA, we made separate fusions to CheA98–655 (CheAS) and to CheAM98I, a mutant of CheAL that does not express CheAS (Sanatinia et al, 1995). Expression of full-length fusions was verified by immunoblot analysis (data not shown). Fusions with the fluorophore at the N terminus of receptors or CheB were omitted from further analysis because of failed membrane insertion or very low expression levels. Fusions to CheY, CheZ, CheR and CheB were fully functional, as tested by their ability to complement respective null mutants for chemotaxis-driven swarming in soft agar (Figure 1A), whereas fusions to the core components of the signaling complex—Tar, CheA and CheW—did not promote swarming. Nevertheless, all fusions localized to the chemosensory clusters (Figure 1B) as expected (Sourjik and Berg, 2000; Shiomi et al, 2002; Banno et al, 2004; Kentner et al, 2006).
Figure 1.
Functionality and localization of YFP fusions. (A) Complementation assay for chemotactic swarming on soft agar plates. Plates were inoculated with wild-type cells (top), the respective mutant containing the fusion (bottom right) and the mutant without the fusion (bottom left). The Tar fusion was tested in the strain UU1250, which lacks all receptor genes, and CheAL and CheAS fusions in the strain VS166, which lacks the entire cheA gene. Respective mutants expressing Tar, CheA and CheW fusions did not swarm at any induction level tested (plate shown contained 50 μM IPTG), other fusions were expressed at 50 μM (CheR–YFP and YFP–CheY), 40 μM (CheY–YFP), 20 μM (CheZ fusions, YFP–CheR) or without (CheB–YFP) IPTG. Although the CheB–YFP construct only partly complemented the ΔcheB mutant even without induction when expressed from the pTrc promoter, complementation was nearly 100% for the same construct expressed under tighter control of the arabinose promoter (not shown). (B) Localization to clusters in the wild-type strain RP437. IPTG inducer levels were 5 μM (CheZ–YFP and CheB–YFP), 10 μM (CheAL–YFP, CheAS–YFP), 20 μM (Tar–YFP, YFP–CheAL and YFP–CheAS) or 50 μM (CheW, CheY and CheR fusions; YFP–CheZ).
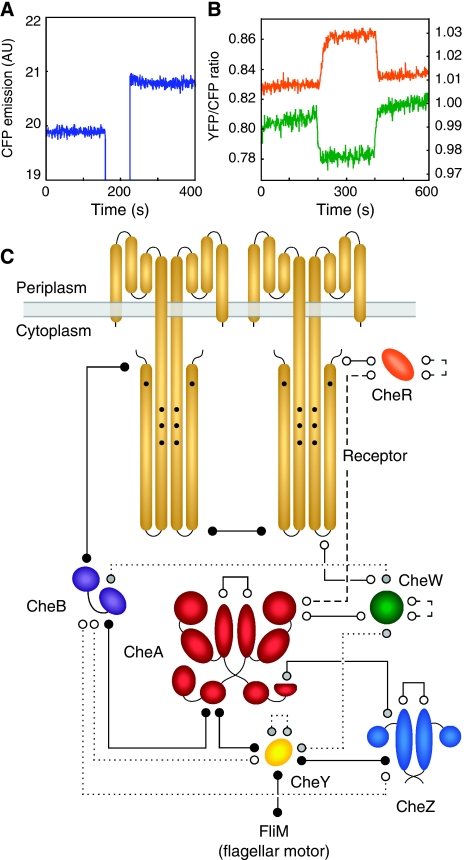
FRET mapping of protein interactions was made by acceptor photobleaching (Figure 2A; Supplementary Figure S1A). For simplicity, all protein pairs were tested in the same wild-type E. coli strain RP437 (Parkinson and Houts, 1982). To avoid false negatives that could arise from competitive binding of native proteins, negative results were confirmed in respective knockout mutants. For pairs including YFP–CheR or CheB–YFP, the respective catalytic mutants, YFP–CheRD154A and CheBS164C–YFP (Barnakov et al, 2002; Shiomi et al, 2002), were also tested, as expression of the enzymatically active fusions influences the receptors' methylation level and activity. After the identification of positive FRET pairs, the dependence of these interactions on chemotactic stimulation was examined using a flow assay (Sourjik et al, 2007), whereby cells were attached to a coverslip in a flow chamber and changes in the YFP/CFP ratio were recorded in response to a stepwise addition and subsequent removal of 1 mM α-methyl-DL-aspartate (MeAsp), a non-metabolizable analog of the Tar-specific attractant aspartate (Figure 2B; Supplementary Figure S1B). This analysis enabled us to picture the entire network of protein interaction in the chemotaxis pathway, with 19 positive FRET pairs being identified out of the 28 possible protein combinations, not counting the identical interactions of CheAL and CheAS separately (Figure 2C; Supplementary Table SII). Most pairs showed FRET even in the absence of all other chemotaxis proteins, arguing for direct interactions (Figure 2C, solid lines), whereas other combinations showed FRET only in the presence of either CheA or receptors (Figure 2C, dotted and dashed lines, respectively), indicating their dependence on a common binding partner. Additionally, several interactions are direct but depend on the kinase activity of CheA (see below).
Figure 2.
FRET analysis of the chemotaxis pathway. (A) FRET measurement by acceptor bleaching. FRET is seen as an increase in CFP emission upon bleaching of YFP for 20 s using a 532-nm laser. Bleaching eliminates energy transfer to the YFP acceptor, causing an unquenching of CFP emission. Example shows CheW–CFP/CheW–YFP pair expressed in Δ[cheA-cheZ] cells. See Materials and methods and Supplementary Figure S1A for details. FRET efficiency for a given pair (Supplementary Tables SII and SIII) was derived from the data as a fractional change in CFP fluorescence. (B) FRET responses to chemostimulation, seen as changes in the YFP/CFP ratio. Examples show VS153 (Δ[cheR-cheZ] Δtsr) cells expressing CFP–CheAS/CheY–YFP (orange line; left Y axis) and CFP–CheAS/CheBS164C–YFP (green line; right Y axis) pairs. Cells were stimulated with 100 μM MeAsp, added at 200 s and removed at 400 s. See Materials and methods and Supplementary Figure S1B for details. (C) FRET interaction map of the chemotaxis pathway. Positive FRET pairs (lines) correspond to direct (solid lines) or presumably indirect interactions that depend either on receptors (dashed lines) or on CheA (dotted lines). Receptors and CheZ are present as homodimers; CheAS and CheAL can form homo- or heterodimers and are depicted as a heterodimer. FRET signal amplitudes are summarized in Supplementary Tables SII and SIII. FRET between Tar and CheW was detected using a truncated Tar1–425 fusion. Receptor–receptor FRET occurs between receptor dimers as it could be measured with both Tar–Tar and Tar–Tsr. Interactions were further classified into stimulation-independent (open circles), direct stimulation-dependent (black circles) and those with indirect or unclear stimulation-dependent (grey circles; see text for details). The stimulus dependence of CheA–CheB and Tar–CheB FRET was measured with a catalytic CheBS164C mutant. The CheY–FliM FRET pair was not included in our initial interaction mapping analysis, but was identified previously (Sourjik and Berg, 2002a).
In most cases where four possible combinations of N- and C-terminal fusions could be tested, interactions were observed with all of these combinations (Supplementary Table SII). A notable exception was CheR, for which only the N-terminal fusion showed interactions with other proteins, although both fusions localized to receptor clusters (Figure 1B). The only previously reported interactions that were not detected in the initial screen were those between Tar and CheW or CheA, which was expected because of the large distance between the C terminus of Tar and the binding site for both CheA and CheW at the signaling domain. A truncated Tar1–425–CFP construct, with CFP being positioned closer to the signaling domain, indeed showed FRET in combination with CheW–YFP, though not with YFP–CheW or with N-terminal fusions to CheAL and CheAS or to a CheA509–655 fragment, which corresponds to the CheW homologous receptor-binding P5 domain of CheA.
The experimentally observed FRET efficiency for individual combinations could be indicative of the relative binding affinity of the respective fusions but also depends on the distance between protein termini in the complex and relative orientation of the fluorophores. It is always lower than the theoretical FRET efficiency for the respective donor–acceptor pair and depends on the expression levels of the donor and acceptor, due to the contribution of the autofluorescent cell background to the overall cyan signal, and also because in most cases the acceptor is not in sufficient excess to saturate all donor molecules.
Stimulation-induced changes in protein interactions
Nine FRET pairs were responsive to chemotactic stimulation, five of which clearly correspond to direct protein interactions (black circles in Figure 2C; Supplementary Table SIII). Among them, CheY–CheZ (Sourjik and Berg, 2002b) and receptor–receptor (Vaknin and Berg, 2006) interactions were previously detected using in vivo FRET, along with an additional direct stimulation-dependent interaction between CheY and FliM (Sourjik and Berg, 2002a). The stimulation dependence of the other interactions involving CheY, CheB and CheZ (grey circles in Figure 2C) could be indirect (see Discussion). Stimulus-induced changes in interactions involving CheY or CheB were phosphorylation dependent, and were not observed for non-phosphorylatable mutants CheYD57A or CheBD56E/S164C, whereas receptor–receptor FRET responses are believed to reflect a change in the spacing between receptor dimers upon ligand binding (Vaknin and Berg, 2006, 2007, 2008). Consistent with that, stimulation-induced fractional changes in FRET were strongest for interactions that involved CheY and weakest for interactions that involved receptors (Supplementary Table SIII).
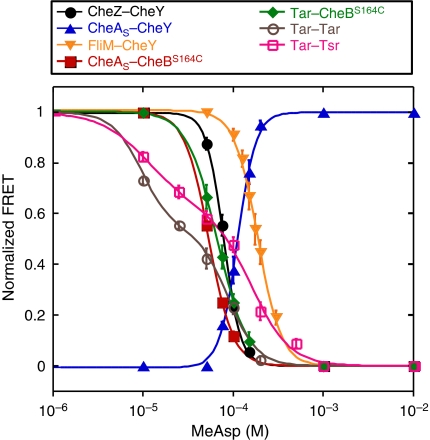
To determine the sensitivity of interactions to stimulation, we recorded response amplitudes across a range of MeAsp concentrations (Figure 3). Although the absolute values of FRET efficiency depend on several parameters, relative changes in FRET allow direct quantification of stimulation effects on the concentration of the FRET complex or on the conformational changes in this complex. Measurements were made with a minimal set of pathway components in the background. Strain VS116, which does not express any chemotaxis genes, was used to measure receptor–receptor responses; strain VS153, which contains only a core sensory complex of Tar, CheA and CheW, along with minor receptors Trg and Aer, was used to investigate the phosphorylation-dependent responses. All pairs except CheA–CheY showed a decrease in FRET with kinase inhibition (addition of attractant) in strains VS116 and VS153, respectively (Figure 3). Notably, the CheA–CheY response also showed a decrease in FRET with kinase inhibition when measured in the wild-type RP437 (CheZ+) cells.
Figure 3.
Dose–response curves for direct stimulation-dependent interactions. Responses to steps of MeAsp for CheZ–CheY (black circles), CheAS–CheY (blue triangles), CheAS–CheBS164C (red squares), Tar–CheBS164C (green diamonds), and FliM–CheY (yellow triangles) fusion pairs in the strain VS153, and Tar–Tar (magenta open circles) and Tar–Tsr (pink open squares) in the strain VS116 (flhC). Cells were equilibrated in the buffer before each stimulation. Response amplitudes were calculated as described in Materials and methods and normalized to the maximal response upon stimulation with 1 mM MeAsp. Smooth curves are fits to the data using a multisite Hill model. Absolute values of stimulation-dependent and total FRET and protein expression levels are summarized in Supplementary Tables SIII and SIV, respectively. Source data is available for this figure at www.nature.com/msb.
FRET between CheBS164C and Tar or CheA responded to stimulation, and we tested whether the responses represent independent changes in the interaction of CheB with each target. The Tar–CheBS164C response was seen in the strain VS177, where CheA lacks the response regulator-binding domain, whereas no FRET was observed in the strain DK1, a derivative of VS153 in which Tar lacks the CheB-binding C-terminal pentapeptide sequence. This confirms that stimulation-dependent Tar–CheBS164C FRET is due to CheBS164C–YFP binding to the C terminus of receptors and not to CheA. On the other hand, the CheAS–CheBS164C FRET response was still present, albeit much weaker, in the strain DK1. FRET was also observed between a CheA98–257 fusion, which cannot bind receptor clusters, and fusions to CheBS164C or to the N-terminal CheB domain, CheB1–134.
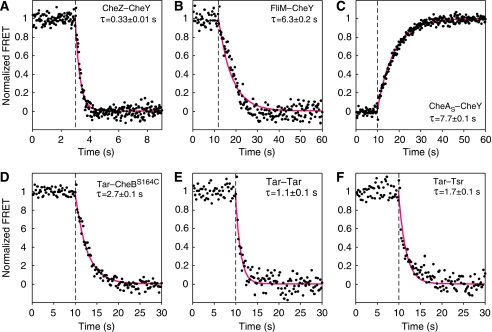
We further determined kinetics of responses to a saturating stimulus of attractant for most direct stimulation-dependent interactions. All kinetics could be well fitted by an exponential decay function, yielding characteristic response times, τ (Figure 4). For all interactions involving CheY, the response kinetics was apparently determined by dephosphorylation of phospho-CheY. CheY–CheZ FRET decayed with τ∼0.33 s (Figure 4A), consistent with the in vivo rate of CheZ-stimulated CheY dephosphorylation estimated before (Sourjik and Berg, 2002a). CheY–CheA and CheY–FliM FRET showed slower increase and decay kinetics, respectively (Figure 4B and C), which corresponded to the autodephosphorylation rate of phospho-CheY in the absence of CheZ. Similarly, decay in the interaction of CheB with Tar (Figure 4D) presumably reflected kinetics of CheB dephosphorylation. The characteristic time, τ=2.7 s, and the derived first-order dephosphorylation rate constant, 1/τ∼0.37 s−1, were close to the previously measured in vitro rate constant for CheB dephosphorylation, 0.35 s−1, at the room temperature (Stewart, 1993). Small response amplitude of the CheB–CheA FRET pair precluded reliable measurement of its kinetics, but it is likely to be similar to that of the CheB–Tar pair. Response kinetics of Tar–Tar and Tar–Tsr pairs had characteristic response times of 1.1 and 1.7 s, respectively (Figure 4E and F). However, monoexponential function did not perfectly fit the data, indicating biphasic response kinetics, which was consistent with the biphasic response observed in dose–response measurements (Figure 3).
Figure 4.
Response kinetics of direct stimulation-dependent interactions. Responses to a saturating step of 10 mM aspartate is shown for CheZ–CheY (A), FliM–CheY (B), CheAS–CheY (C), Tar–CheBS164C (D), Tar–Tar (E) and Tar–Tsr (F). As in Figure 3, receptor pairs were measured in the strain VS116 (flhC), and all other pairs in the strain VS153. FRET signals were normalized to the pre-stimulus value in the buffer. Cells were stimulated at time points indicated by the vertical dashed line. Smooth red curves are fits to the data using an exponential decay model (see Materials and methods). Characteristic response time, τ, is shown for each response kinetics. Source data is available for this figure at www.nature.com/msb.
Discussion
Altogether, our FRET analysis of protein interactions in the chemotaxis pathway of E. coli identified 19 positive FRET pairs out of the 28 possible protein combinations, with 9 pairs being responsive to chemotactic stimulation. The obtained map of protein interactions is in good agreement with previous biochemical data and localization analyses (Sourjik, 2004; Wadhams and Armitage, 2004), and offers a better insight into the pathway regulation.
Core of the sensory complex
The core of the chemosensory complex is formed primarily by the receptor–receptor interactions, and is further stabilized by the binding of CheA and CheW (Maddock and Shapiro, 1993; Kim et al, 1999; Ames et al, 2002; Kentner et al, 2006). This ternary complex is stable on the time scale of signaling and adaptation, with characteristic equilibration time being 12 min for CheA and CheW, and more than 30 min for receptors (Schulmeister et al, 2008). We observed energy transfer among most fusions to the core proteins. Receptor–receptor FRET results primarily from the interactions between homodimers, rather than from an intradimeric energy transfer, as FRET signals were similar for the Tar–Tsr and Tar–Tar pairs (Supplementary Table SII). This is consistent with previous reports (Vaknin and Berg, 2006, 2007, 2008) and suggests that receptors are tightly packed in the cluster, with distances between the C termini of different dimers being shorter that the respective intradimeric distances. We further observed FRET for the CheA–CheA, CheA–CheW and CheW–CheW pairs. CheA–CheA FRET was largely intradimeric, as the strength of the signal was not affected by the lack of all other chemotaxis proteins (Supplementary Table SII). This could mean that CheA dimers are less densely packed in the sensory complexes than receptor dimers, consistent with a high ratio of receptors to CheA in the active complexes (Levit et al, 2002). Energy transfer efficiency was similar for all combinations of CheA fusions, indicating similar distances between all termini in the dimer. FRET between CheA and CheW was also independent of the other proteins and thus reflects the expected direct interaction between the two. FRET between two CheW fusions required receptors, suggesting that CheW fusion proteins come into proximity by binding to receptor oligomers and possibly also to CheA dimers.
Interactions of receptors with CheW and CheA were the only two established protein interactions in the chemotaxis pathway that were initially not detected in our assay. This was not surprising because the CheA- and CheW-binding site is ∼210 Å away from the receptors' C terminus (Kim et al, 1999), meaning that the distance between the fluorophores for these pairs is above the critical range for energy transfer between CFP and YFP (∼100 Å). Using a truncated Tar1–425–CFP construct, FRET was indeed detected in combination with CheW–YFP. However, no signal was obtained with YFP–CheW or with the N-terminal fusions to CheAL, CheAS or the P5 fragment of CheA (CheA509–655). As all of these fusions localize to receptor clusters (Figure 1B), and fusions to CheAS and the P5 fragment have been previously shown to bind to receptors in the absence of CheW (Kentner et al, 2006), it appears that only the C terminus of CheW is positioned in sufficient proximity to the Tar1–425 fluorophore for FRET. Tar–CheA interaction was thus the only false negative in our screen.
Among the interactions in the sensory core, only the Tar–Tar and Tar–Tsr interactions showed stimulation dependence. The decrease in the energy transfer between the C-terminal fusions to receptors upon attractant stimulation is believed to be caused by attractant-induced conformational changes in the receptor complexes rather than by their dissociation (Vaknin and Berg, 2006, 2007, 2008). No stimulation dependence was observed for the other FRET signals, suggesting that there are no major rearrangements in the packing or stoichiometry of the sensory core with stimulation. Furthermore, lack of any detectable response to attractant for the intradimeric CheA FRET indicates that the stimulation-induced conformational changes in this dimer—which are believed to regulate kinase activity—have to be subtle. These conclusions, though, have to be taken with caution as fusions to the sensory core components were not fully functional.
Interactions involving CheZ and CheY
CheZ and CheY were previously shown to localize to receptor clusters through interaction with CheA (Sourjik and Berg, 2000; Cantwell et al, 2003), and we confirmed both of these interactions. CheA–CheY and CheA–CheZ interactions were observed even in the absence of all other proteins, and for fusions to both CheAL and CheAS. Direct interaction between CheZ and CheAL contradicted previous reports that only CheAS is able to bind CheZ (Cantwell et al, 2003), but was confirmed by the localization of CheZ–YFP to receptor clusters in a strain expressing CheAL, but not CheAS or CheY (Supplementary Figure S2). The degree of CheAL-mediated CheZ localization was weaker than in the wild type, which presumably explains why this interaction was missed in the previous fluorescence-microscopy based study (Cantwell et al, 2003). However, FRET signals for the CheZ interactions with C-terminal CFP fusions to both forms of CheA were comparable, possibly indicating differences in CheZ binding to free and cluster-associated CheAL. N-terminal CFP fusion to CheAS indeed showed much stronger FRET than the N-terminal fusion to CheAL, but this might reflect shorter distance between fluorophores in the latter case.
The CheY–CheZ pair also showed FRET, which apparently has two components—one due to the direct interaction between the two proteins and the other mediated by CheA. Phosphorylation-dependent FRET between CheY and CheZ was previously observed (Sourjik and Berg, 2002b), even in mutant strains where CheZ or CheY did not bind to CheA (Vaknin and Berg, 2004; VS, unpublished data). This phosphorylation-dependent interaction between CheY and CheZ could only be observed for the C-terminal CheZ fusions, consistent with the crystal structure of the phospho-CheY complex with CheZ (Zhao et al, 2002). On the other hand, a weak FRET signal between CheY and CheZ was observed for both C- and N-terminal CheZ fusions in the CheA+ cheW strain (Supplementary Table SII). As in this strain CheA is largely inactive and therefore CheY is expected to be unphosphorylated, FRET is likely to be explained by the binding of CheY and CheZ to CheA, which brings them in proximity. No CheY–CheZ FRET was observed in the absence of CheA at the used expression levels.
Consistent with it being a dimer, CheZ also showed FRET with itself even in the absence of all other chemotaxis proteins. Additionally, positive FRET signals were observed for CheW–CheY, CheY–CheB, CheY–CheY and CheZ–CheB pairs. For all of these pairs, energy transfer was not observed in the absence of CheA and presumably results from the proximity of respective fusion proteins when bound to the latter. Indeed, the CheW–CheY interaction was not seen in the strain expressing CheAΔP2, which lacks CheY-binding P2 domain and therefore cannot bind but is still able to phosphorylate CheY. For the CheY–CheY pair, FRET was still observed in the CheAΔP2 strain, but the energy transfer could also take place at the CheZ dimer or at the flagellar motor.
All of the direct interactions that involve response regulator CheY—with the kinase, phosphatase and flagellar motor—exhibited stimulation dependence. CheY phosphorylation decreased its affinity to CheA, and increased the affinity toward CheZ and FliM, in agreement with previous biochemical studies (Li et al, 1995; McEvoy et al, 1999). However, the directionality of the CheA–CheY FRET response was inverted in the presence of CheZ, which can be explained by a strong binding of phospho-CheY to CheZ, which is itself associated with CheAS. Apparently, the increase in FRET caused by association of phospho-CheY with CheA-bound CheZ outbalances the FRET decrease caused by dissociation of phospho-CheY from CheA, consistent with a previous report that described changes in the localization of CheY to chemosensory clusters (Vaknin and Berg, 2004). CheW–CheY and CheY–CheY FRET also showed dependence on CheY phosphorylation, presumably as a result of the change in CheY affinity for CheA. However, we cannot exclude the possibility that the CheY–CheY interaction is direct but requires CheA activity.
No changes in energy transfer upon stimulation could be observed for the CheZ dimer, suggesting that it does not undergo any major structural rearrangements. Although oligomerization of CheZ upon binding phospho-CheY has been previously suggested (Blat and Eisenbach, 1996), later biochemical experiments (Silversmith, 2005) and our in vivo results seem to disprove this idea. Nevertheless, the CheZ dimer does undergo a modest change of conformation and activation (Blat et al, 1998; Silversmith, 2005) upon binding to phospho-CheY. This might result in an increased affinity to CheA and could explain a weak CheY-dependent increase in the energy transfer between CheZ and CheAS with pathway activation. Alternatively, some increase in the CheAS–CheZ FRET may be caused by CheZ binding to the CheA-bound phospho-CheY. In any case, higher local concentration of CheZ would result in an increased association of CheZ with CheAS at the cluster, which has been reported to enhance phosphatase activity (Wang and Matsumura, 1996). Recent Brownian dynamics simulations of the pathway demonstrated that such phospho-CheY-dependent increase in CheZ binding to the receptor cluster would benefit performance of the chemotaxis pathway by sharpening the response, increasing the range of detectable ligand concentrations and improving adaptation and robustness (Lipkow, 2006). The extent of stimulation-dependent changes in CheZ binding to CheA, however, is expected to be modest as no increase in the localization of CheZ to clusters upon stimulation could be previously observed using fluorescence imaging (Vaknin and Berg, 2004).
Interactions involving adaptation enzymes
CheR and CheB showed direct interactions with major receptors, which required the C-terminal pentapeptide sequence and were consistent with physiological and biochemical data (Djordjevic and Stock, 1998b; Okumura et al, 1998; Barnakov et al, 1999). Two other positive FRET pairs that involve CheR—CheA–CheR and CheR–CheR—were not observed in the absence of receptors and are likely to reflect indirect interactions. CheB exhibited positive FRET with a number of other proteins—CheA, CheW, CheY and CheZ. Of these, only its interaction with CheA appears to be direct, and was observed between fusions to the full-length proteins and between CheA98–257 and CheB1–134, confirming that the interaction is mediated by the P2 domain of CheA and the N-terminal domain of CheB (Li et al, 1995). Other interactions involving CheB are presumably mediated by CheA, although it again cannot be excluded that some of them are direct but require kinase activity.
All interactions of CheR were stimulation independent, demonstrating that CheR binding to receptors is not affected by stimulation. In contrast, Tar–CheBS164C FRET increased with kinase activation, indicating that association of CheB with the pentapeptide is phosphorylation dependent. The binding sites targeted by CheB and CheR overlap (Lai et al, 2006), and when the Tar–CheRD154A fusion pair was expressed together with the non-fused CheBS164C, a decrease in FRET was indeed observed with pathway activation, in agreement with a displacement of the CheR fusion by competitive binding of phospho-CheBS164C to the pentapeptide sequence.
Given the homology between CheY and the CheB N-domain, it was expected that phosphorylation of CheB would decrease its affinity for CheA, and such decrease in affinity was suggested based on the indirect evidence (Anand and Stock, 2002). However, we observed that the CheA–CheBS164C FRET increased with kinase activation. As the putative CheA-binding region in the CheB N-domain is buried in the interaction with the C-domain (Djordjevic et al, 1998a), it is conceivable that phosphorylation exposes this region through release of the interdomain linkage and thereby increases the affinity for CheA. The response was still present in a strain lacking the receptor pentapeptide sequence or with a CheA fragment that does not associate with receptors, confirming that the effect does not depend on CheB binding to receptors. However, the response in these strains was much weaker, indicating that the association of CheB with adjacent receptor pentapeptides indeed facilitates the binding to CheA, possibly by raising the local CheB concentration.
Levels of pathway regulation
Recording FRET responses to stimulation also enabled us to distinguish two levels of activity-dependent changes in the pathway: stimulation-induced rearrangement of receptors in the sensory complexes, and downstream changes in the phosphorylation of CheY and CheB. Consistent with a previous report (Vaknin and Berg, 2007), cooperativity—represented by the Hill coefficient—is high for the phosphorylation-dependent FRET pairs, but low for the receptor–receptor responses. Sensitivity, as indicated by the MeAsp concentration eliciting a half-maximal response, is in a similar range for all protein pairs. A somewhat lower sensitivity for the response of FliM–CheY and CheA–CheY could be due to the slow dephosphorylation—and thus higher level—of phospho-CheY in the absence of CheZ. Tar–Tar responses were reported to exhibit lower sensitivity than CheY–CheZ at low levels of receptor modification, but higher sensitivity at high levels of modification. The approximately equal sensitivities in our measurements is presumably due to the intermediate level of receptor modification in the VS153 background, where receptors remain in the original half-modified state with two glutamates and two glutamines that mimic the behavior of methylated glutamates.
Interestingly, the interaction between Tar and the serine receptor Tsr responds to MeAsp with a lower sensitivity than the Tar–Tar interaction. Similar observation was previously made at the level of kinase activity (Sourjik and Berg, 2004) and agrees with the prediction of allosteric models (Duke et al, 2001; Sourjik, 2004; Mello and Tu, 2005; Keymer et al, 2006) that homogeneous sensory complexes should be more sensitive to stimulation. Allosteric interactions are thus involved in signal processing already at the receptor level. Moreover, receptor pairs show a biphasic response in dose–response and kinetics measurements, indicating that either the same receptor complexes undergo two distinct stimulation-induced movements or rearrangements, or that two receptor sub-populations exist in cells, possibly corresponding to weakly and strongly clustered receptors. At present, we can only speculate about the molecular details of these ligand-induced receptor rearrangements at the cluster. Previously observed changes in homo-FRET between receptor fusions have been interpreted in terms of receptor movement in trimers of dimers (Vaknin and Berg, 2006, 2007, 2008), but changes in FRET might as well reflect movement of neighboring trimers or both. Because their kinetics are slower than changes in CheY phosphorylation—reflected by the phosphorylation-dependent interaction of CheY with CheZ—these rearrangements do not seem to directly represent the change in receptor conformation that regulates kinase activity but are rather induced by it.
At the downstream level of regulation, we observed the increased affinity of phosphorylated CheB to both its binding partners at the receptor cluster. Such dependence implies an additional enhancement of a negative feedback from the level of kinase activity to that of receptor activity, which is provided by CheB phosphorylation. This feedback is important to maintain robust output of the chemotaxis system under such perturbations as gene expression noise (Kollmann et al, 2005). Our results suggest that phosphorylation not only largely enhances the methylesterase activity of CheB (Anand and Stock, 2002) but also raises the local concentration of active CheB in the vicinity of methylation sites. CheB activation by binding to the pentapeptide sequence of receptors might enhance the feedback even further (Barnakov et al, 2002). Consistent with its role in the feedback regulation, stimulation-induced changes in the CheB–Tar interaction showed slowest kinetics in the initial pathway response. The dynamic behavior of CheB and its phosphorylation-dependent competition with CheR and CheY in binding receptors and CheA (Li et al, 1995), respectively, may play an important role in determining response kinetics and pathway robustness. Indeed, a recent computational analysis suggested that strong binding of phosphorylated CheB to receptors and weak binding of phosphorylated CheY to CheA are important for robust adaptation in chemotaxis (Matsuzaki et al, 2007).
Conclusions
Despite previous reservations (Phizicky et al, 2003), our study demonstrates that FRET can be successfully used to systematically map and quantify all intracellular protein interactions in a signaling network, including transient interactions, with no false positives and nearly no false negatives. A strong advantage of FRET is that it enables systematic mapping of activity dependence of interactions and measuring response kinetics, and therefore yields a dynamic picture of the network. In addition to direct interactions, protein proximities—mediated by association with a common interaction partner—can be identified, facilitating the characterization of multiprotein complexes and their dynamics in vivo. FRET-based interaction mapping thus stands as a simple and reliable means for the investigation of other protein networks in bacteria or eukaryotes.
Materials and methods
Fluorescent protein fusions
Plasmids used to express fluorescent protein fusions are listed in Supplementary Table SI. With the exception of CheY, CheZ and FliM (Sourjik and Berg, 2002a, 2002b), all fusions were constructed as described before (Kentner et al, 2006): the target gene was amplified by PCR and cloned into respective vectors pDK4, pDK66, pDK2 and pDK85, derivatives of pTrc99a (Pharmacia; pBR ori, pTrc promotor, AmpR) carrying eyfpA206K and ecfpA206K sequences which encode true monomeric versions of YFP and CFP, respectively (Zacharias et al, 2002). CFP fusions were subsequently moved to vector pDK79 (pACYC ori, pBAD promotor, KanR). pDK198 and pDK203 were generated by transferring tar-ecfp from pDK53 to plasmids pDK58 and pDK80, respectively. Mutations in cheRD154A and cheBS164C were generated by PCR, using oligonucleotides encoding the respective amino-acid substitution. Immunoblotting with a monoclonal GFP-specific antibody (JL8; BD Biosciences) was used to confirm that fusions were expressed as full-length proteins with little degradation.
Expression levels of CFP from plasmid pDK2 (26 900±1200 copies per cell) and CheY-YFP from pVS18 (39 400±1400 copies per cell) at full induction were quantified by fluorimetry. Cell cultures were washed and concentrated threefold in tethering buffer, counted in a Neubauer chamber, diluted to the same concentration and lysed by sonication. CFP and YFP signals of the lysates were measured in a fluorimeter, and fluorescent protein numbers were calculated by comparison with a known amount of purified CFP/YFP, which was added to a lysate of control cells not expressing any fluorescent protein. Expression levels of the stimulus-dependent FRET fusion pairs (Supplementary Table SIV) could not be measured the same way, because in some cases the CFP signal was too close to the background. Therefore, CFP and YFP signals of the FRET pairs were quantified from fluorescence microscopy images by measurement of the integrated intensity of about 100–200 single cells, using ImageJ software (W Rasband; http://rsb.info.nih.gov/ij). Cells expressing CFP from pDK2 and CheY-YFP from pVS18, respectively, at full induction were used to calibrate expression levels (CFP/YFP molecules per cell). To convert the absolute molecule numbers per cell to molar concentration, we calculated the cell volume (1.801 × 10−15 l) from the average dimensions of an E. coli cell on fluorescence images.
Strains and their growth
E. coli strains in this study (Supplementary Table SI) are derived from K12 strain RP437. Strains VS166 (ΔcheA), VS167 (ΔcheA Δ[tap-cheZ]) and DK1 (tarΔpp Δ[cheR-cheZ] Δtsr) were made by in-frame deletion of cheA in strains RP437 and RP2893 (Δ[tap-cheZ]), and the tar pentapeptide sequence in the strain VS153 (Δ[cheR-cheZ] Δtsr), respectively. Cells were grown in tryptone broth (TB; 1% tryptone and 0.5% NaCl) with added antibiotics. Ampicillin, chloramphenicol and kanamycin were used at final concentrations of 100, 35 and 50 μg/ml, respectively. Overnight cultures, grown at 30°C, were diluted 1:100 and grown at 34°C for about 4 h, to an OD600 of 0.45–0.5, in the presence of inducers isopropyl β-D-thiogalactoside (IPTG) and arabinose. Standard induction levels for FRET measurements were 0.01% arabinose and 50 μM IPTG, except for pDK58, pDK135, pDK159, pDK203 (20 μM IPTG) and pDK198 (100 μM IPTG). Cells were harvested by centrifugation (4000 r.p.m., 5 min), washed and resuspended in tethering buffer (10 mM potassium phosphate, 0.1 mM EDTA, 1 μM L-methionine, 67 mM sodium chloride, 10 mM sodium lactate, pH 7) prior to FRET measurements.
Chemotaxis tests shown in Figure 1A were performed on TB soft agar plates (1% tryptone, 0.5% NaCl and 0.3% agar) at 30°C overnight.
Fluorescence imaging
Images shown in Figure 1B were obtained by a Zeiss Axio Imager.Z1 microscope, as described before (Kentner et al, 2006).
FRET measurements
Measurements were performed on a custom-modified Zeiss Axiovert 200 microscope. For acceptor photobleaching (Supplementary Figure S1A), cells were concentrated about 10-fold by centrifugation, resuspended in tethering buffer and applied to a thin agarose pad (1% agarose in tethering buffer). Excitation light from a 75 XBO lamp, attenuated by a ND60 (0.2) neutral-density filter, passed through a band-pass (BP) 436/20 filter and a 495DCSP dichroic mirror and was reflected on the specimen by a Z440/532 dual-band beamsplitter (transmission 465–500 and 550–640 nm; reflection 425–445 and 532 nm). Bleaching of YFP was accomplished by a 532 nm diode laser (Rapp OptoElectronic, Hamburg), reflected by the 495DCSP dichroic mirror into the light path. Emission from the field of view, which was narrowed with a diaphragm to the area bleached by the laser, passed through a BP 485/40 filter onto a photon multiplier (Hamamatsu H7421-40; Hamamatsu, Bridgewater, NJ). For each measurement point, photons were counted over 0.5 s using a counter function of the PCI-6034E board, controlled by a custom-written LabView 7.1 program (both from National Instruments, Austin, TX). CFP emission was recorded before and after bleaching of YFP by a 20-s laser pulse, and FRET was calculated as the signal increase divided by the total signal after bleaching, as defined previously (Sourjik et al, 2007). FRET was scored as significant above 0.5% increase in CFP emission.
To measure concentration dependence of FRET responses to chemostimulation (Supplementary Figure S1B), cells were attached to a polylysine-coated coverslip and placed in a flow cell, which was kept under constant flow (500 μl/min) of tethering buffer by a syringe pump (Harvard Apparatus). By rapid exchange of the buffer reservoir, solutions of α-methyl-D,L-aspartate (MeAsp; Sigma) in tethering buffer were added or removed. Excitation was through a BP 436/20 filter and reflected by a 455 dichroic mirror. Emission from a field of 300–500 cells passed through the 455 DC mirror, and was split by a 515 dichroic mirror into two signals, passing through a BP 485/40 cyan filter and a BP 535/30 yellow filter, respectively, on photon multipliers. FRET, defined as the fractional change in cyan fluorescence due to energy transfer, was calculated from changes in the ratios of yellow and cyan fluorescence signals, and the data were fit to a multisite Hill model with either one or two apparent dissociation constants (K0.5), as described before (Sourjik and Berg, 2002a; Sourjik et al, 2007), using KaleidaGraph 3.6 (Synergy Software, Reading, PA).
The same experimental procedure was used for kinetics measurements, except attractant was added at a higher flow rate, 1500 μl/min, and signal integration time was set to 0.03–0.3 s, depending on a FRET pair. The liquid exchange profile in the flow cell at this rate was determined using 100 nM fluorescein solution as a marker, and used to estimate the ramp profile for stimulation with 10 mM aspartate solution (Supplementary Figure S3). Aspartate rather than MeAsp was used, because it binds with >10 × higher affinity to Tar. Under these conditions, saturating attractant concentration for all FRET pairs was reached within 0.1 s, and the observed response times were therefore not limited by the rate of stimulation. FRET values were calculated from the ratios of yellow and cyan fluorescence signals as described above and normalized to the average pre-stimulus FRET value. Characteristic response time, τ, for different FRET pairs (Figure 4) was determined by fitting the data to a model of exponential decay, exp(−(t−t0))/τ), where t−t0 is time after stimulation.
To estimate the stimulus-dependent fraction of the total FRET signal (Supplementary Table SIV), the relative change in the CFP signal in response to a saturating stimulus with 1 mM MeAsp was recorded (stimulus-dependent FRET), followed by subsequent acceptor bleaching (total FRET), using cells in the flow cell and the same microscopic set-up as for the previously described acceptor bleaching measurements.
Supplementary Material
Supplementary figures 1-3 and supplementary tables 1-4
Acknowledgments
We thank Markus Kollmann for helpful discussions. This study was supported by the Deutsche Forschungsgemeinschaft (SO 421/3-1 and SO 421/3-2) and by the EMBO Young Investigator Programme.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alon U, Surette MG, Barkai N, Leibler S (1999) Robustness in bacterial chemotaxis. Nature 397: 168–171 [DOI] [PubMed] [Google Scholar]
- Ames P, Studdert CA, Reiser RH, Parkinson JS (2002) Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci USA 99: 7060–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand GS, Stock AM (2002) Kinetic basis for the stimulatory effect of phosphorylation on the methylesterase activity of CheB. Biochemistry 41: 6752–6760 [DOI] [PubMed] [Google Scholar]
- Banno S, Shiomi D, Homma M, Kawagishi I (2004) Targeting of the chemotaxis methylesterase/deamidase CheB to the polar receptor–kinase cluster in an Escherichia coli cell. Mol Microbiol 53: 1051–1063 [DOI] [PubMed] [Google Scholar]
- Barnakov AN, Barnakova LA, Hazelbauer GL (1999) Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci USA 96: 10667–10672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnakov AN, Barnakova LA, Hazelbauer GL (2002) Allosteric enhancement of adaptational demethylation by a carboxyl-terminal sequence on chemoreceptors. J Biol Chem 277: 42151–42156 [DOI] [PubMed] [Google Scholar]
- Blat Y, Eisenbach M (1996) Oligomerization of the phosphatase CheZ upon interaction with the phosphorylated form of CheY, the signal protein of bacterial chemotaxis. J Biol Chem 271: 1226–1231 [DOI] [PubMed] [Google Scholar]
- Blat Y, Gillespie B, Bren A, Dahlquist FW, Eisenbach M (1998) Regulation of phosphatase activity in bacterial chemotaxis. J Mol Biol 284: 1191–1199 [DOI] [PubMed] [Google Scholar]
- Cantwell BJ, Draheim RR, Weart RB, Nguyen C, Stewart RC, Manson MD (2003) CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J Bacteriol 185: 2354–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic S, Goudreau PN, Xu Q, Stock AM, West AH (1998a) Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc Natl Acad Sci USA 95: 1381–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM (1998b) Chemotaxis receptor recognition by protein methyltransferase CheR. Nat Struct Biol 5: 446–450 [DOI] [PubMed] [Google Scholar]
- Duke TA, Le Novere N, Bray D (2001) Conformational spread in a ring of proteins: a stochastic approach to allostery. J Mol Biol 308: 541–553 [DOI] [PubMed] [Google Scholar]
- Gestwicki JE, Kiessling LL (2002) Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 415: 81–84 [DOI] [PubMed] [Google Scholar]
- Kentner D, Thiem S, Hildenbeutel M, Sourjik V (2006) Determinants of chemoreceptor cluster formation in Escherichia coli. Mol Microbiol 61: 407–417 [DOI] [PubMed] [Google Scholar]
- Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS (2006) Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc Natl Acad Sci USA 103: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH (1999) Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400: 787–792 [DOI] [PubMed] [Google Scholar]
- Kollmann M, Lovdok L, Bartholome K, Timmer J, Sourjik V (2005) Design principles of a bacterial signalling network. Nature 438: 504–507 [DOI] [PubMed] [Google Scholar]
- Lai RZ, Manson JM, Bormans AF, Draheim RR, Nguyen NT, Manson MD (2005) Cooperative signaling among bacterial chemoreceptors. Biochemistry 44: 14298–14307 [DOI] [PubMed] [Google Scholar]
- Lai WC, Barnakova LA, Barnakov AN, Hazelbauer GL (2006) Similarities and differences in interactions of the activity-enhancing chemoreceptor pentapeptide with the two enzymes of adaptational modification. J Bacteriol 188: 5646–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levit MN, Grebe TW, Stock JB (2002) Organization of the receptor–kinase signaling array that regulates Escherichia coli chemotaxis. J Biol Chem 277: 36748–36754 [DOI] [PubMed] [Google Scholar]
- Li G, Weis RM (2000) Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100: 357–365 [DOI] [PubMed] [Google Scholar]
- Li J, Swanson RV, Simon MI, Weis RM (1995) The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 34: 14626–14636 [DOI] [PubMed] [Google Scholar]
- Lipkow K (2006) Changing cellular location of CheZ predicted by molecular simulations. PLoS Comput Biol 2: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L (1993) Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259: 1717–1723 [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Kikuchi S, Tomita M (2007) Robust effects of Tsr–CheBp and CheA–CheYp affinity in bacterial chemotaxis. Artif Intell Med 41: 145–150 [DOI] [PubMed] [Google Scholar]
- McEvoy MM, Bren A, Eisenbach M, Dahlquist FW (1999) Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J Mol Biol 289: 1423–1433 [DOI] [PubMed] [Google Scholar]
- Mello BA, Tu Y (2005) An allosteric model for heterogeneous receptor complexes: understanding bacterial chemotaxis responses to multiple stimuli. Proc Natl Acad Sci USA 102: 17354–17359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura H, Nishiyama S, Sasaki A, Homma M, Kawagishi I (1998) Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J Bacteriol 180: 1862–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE (1982) Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol 151: 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S (2003) Protein analysis on a proteomic scale. Nature 422: 208–215 [DOI] [PubMed] [Google Scholar]
- Sanatinia H, Kofoid EC, Morrison TB, Parkinson JS (1995) The smaller of two overlapping cheA gene products is not essential for chemotaxis in Escherichia coli. J Bacteriol 177: 2713–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V (2008) Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci USA 105: 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Zhulin IB, Homma M, Kawagishi I (2002) Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J Biol Chem 277: 42325–42333 [DOI] [PubMed] [Google Scholar]
- Silversmith RE (2005) High mobility of carboxyl-terminal region of bacterial chemotaxis phosphatase CheZ is diminished upon binding divalent cation or CheY-P substrate. Biochemistry 44: 7768–7776 [DOI] [PubMed] [Google Scholar]
- Smith RA, Parkinson JS (1980) Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci USA 77: 5370–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V (2004) Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol 12: 569–576 [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC (2000) Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol 37: 740–751 [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC (2002a) Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc Natl Acad Sci USA 99: 12669–12674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC (2002b) Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA 99: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC (2004) Functional interactions between receptors in bacterial chemotaxis. Nature 428: 437–441 [DOI] [PubMed] [Google Scholar]
- Sourjik V, Vaknin A, Shimizu TS, Berg HC (2007) In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol 423: 363–391 [DOI] [PubMed] [Google Scholar]
- Stewart RC (1993) Activating and inhibitory mutations in the regulatory domain of CheB, the methylesterase in bacterial chemotaxis. J Biol Chem 268: 1921–1930 [PubMed] [Google Scholar]
- Tsien RY, Miyawaki A (1998) Seeing the machinery of live cells. Science 280: 1954–1955 [DOI] [PubMed] [Google Scholar]
- Vaknin A, Berg HC (2004) Single-cell FRET imaging of phosphatase activity in the Escherichia coli chemotaxis system. Proc Natl Acad Sci USA 101: 17072–17077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin A, Berg HC (2006) Osmotic stress mechanically perturbs chemoreceptors in Escherichia coli. Proc Natl Acad Sci USA 103: 592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin A, Berg HC (2007) Physical responses of bacterial chemoreceptors. J Mol Biol 366: 1416–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin A, Berg HC (2008) Direct evidence for coupling between bacterial chemoreceptors. J Mol Biol 382: 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP (2004) Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5: 1024–1037 [DOI] [PubMed] [Google Scholar]
- Wang H, Matsumura P (1996) Characterization of the CheAS/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol Microbiol 19: 695–703 [DOI] [PubMed] [Google Scholar]
- Wouters FS, Bastiaens PI (2001) Imaging protein–protein interactions by fluorescence resonance energy transfer (FRET) microscopy. Curr Protoc Cell Biol 17: 17.1.1–17.1.15 [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296: 913–916 [DOI] [PubMed] [Google Scholar]
- Zhang P, Khursigara CM, Hartnell LM, Subramaniam S (2007) Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA 104: 3777–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Collins EJ, Bourret RB, Silversmith RE (2002) Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol 9: 570–575 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures 1-3 and supplementary tables 1-4