Abstract
Asthma is a ubiquitous disease with a broad range of clinical phenotypes. To better understand the complex genetic and environmental interactions underlying asthma, we compared the gene–gene interactions of four genetically distinct mouse strains that demonstrate biologically distinct responses to allergen. Using DNA microarrays and knock-out mouse studies, we showed that CCR5 plays a definitive role in the development of ovalbumin-induced allergic airway inflammatory disease. In addition, gene expression profiling data have revealed other potential novel targets for therapeutics-based research and has enhanced the understanding of the molecular mechanisms underlying the etiology of “asthma.”
Keywords: airway hyperresponsiveness, asthma, CCR5, microarray, multistrain
Nearly 20 million Americans are afflicted by asthma and, alarmingly, the incidence and severity of this disease is on the rise in industrialized nations. Asthma is a complex genetic disease characterized by reversible airflow obstruction as well as airway inflammation, hyperresponsiveness, and remodeling. These symptoms are the result of many biologically unique gene–gene and gene–environment interactions. Given the complexity of these interactions in humans, identification of asthma susceptibility genes has been extremely difficult.
To simplify the complex genetic and environmental interactions that underlie the pathogenesis of asthma, researchers have used animal models of allergic inflammatory airway disease, where both the genetics and environment can be controlled, and DNA microarray analysis to provide a global picture of gene expression in the lung (1, 2). However, microarray-based gene expression analysis usually reveals many more candidate genes than can be reasonably pursued by functional analysis.
To address this problem, we compared multiple strains of mice that exhibit specific asthma phenotypes to identify a concise list of candidate genes that regulate airway hyperresponsiveness and/or lung eosinophilia. One such candidate is CCR5, a member of the chemokine receptor family. To validate the biologic and physiologic significance of CCR5, we ovalbumin (OVA)-treated CCR5+/+ and CCR5−/− mice and confirmed the genetic inference that CCR5 regulates the development of airway hyperresponsiveness, but not eosinophilic inflammation, in a mouse model of allergic airway inflammatory disease. In addition, the gene expression profiling data have provided several other potential novel targets for therapeutics-based research, and have enhanced the understanding of the molecular mechanisms underlying the etiology of “asthma.”
MATERIALS AND METHODS
General Protocol
To develop a refined list of asthma candidate genes, we used the novel approach of comparing the genetic array profile of four strains of mice that demonstrate unique phenotypic responses to OVA sensitization and challenge (Table 1). We previously showed that 129/SvIm mice display “double responsiveness” to OVA; that is, they develop both increased airway hyperresponsiveness and increased lung eosinophilia, two of the primary symptoms of allergic asthma in humans (3). In contrast, the CAST/Ei mice were nonresponders developing neither OVA-induced airway hyperresponsiveness nor eosinophilia. By setting the data mining criteria to exclude any genes that were similarly regulated (up or down) in these two strains of mice, we were able to favorably reduce the list of candidate genes. Similarly, by using partial responder strains (C57BL/6J and BALB/cJ) to differentially identify genes involved in airway responsiveness or eosinophilic lung inflammation, the candidate list was further refined. In addition, responses were assessed at three time points (24, 48, and 72 h) liberating trends in gene expression rather than “one point” at which to assess the transcriptional regulation of a candidate gene. To validate the importance of these candidates, we selected one, CCR5, that was consistently associated with OVA-induced airway hyperresponsiveness, and determined whether CCR5 affected airway hyperresponsiveness in genetically deficient mice (CCR5−/−; B6129P2-CCR5tm1Kuz/J [4]) versus wild-type mice (CCR5+/+; B6129PF2/J [5]). It is important to note that the CCR5+/+ and CCR5−/− mice are on a mixed 129/J and C57BL/6J background. Although OVA treatment does not induce significant transcription of CCR5, or produce airway hyperresponsiveness, in C57BL/6J mice, the 129/J mice demonstrate a robust response. Thus, the contribution of CCR5 to airway responsiveness can be validated using this mixed-background model. All experiments were conducted in accordance with the National Institutes of Health guidelines for the care and use of animals and with an approved protocol from the Duke University Animal Care and Use Committee.
TABLE 1.
SUMMARY OF GENES SIGNIFICANTLY EXPRESSED DUE TO OVALBUMIN TREATMENT
| Phenotype
|
Significant Genes
|
|||
|---|---|---|---|---|
| Strain | AHR | EOS | Up | Down |
| 129/SvIm | + | + | 209 | 274 |
| C57BL/6J | − | + | 132 | 117 |
| BALB/cJ | + | − | 427 | 135 |
| CAST/Ei | − | − | 40 | 217 |
Definition of abbreviations: AHR, airway hyperresponsiveness; EOS, eosinophil.
Genes with statistically significant changes were determined using SAM, an algorithm whereby temporal changes in gene expression for each strain were calculated using a two-class paired analysis. Normalized Log2ratios (n = 3–4) from samples precontrol and post–ovalbumin (OVA) challenge for individual time points of 24, 48, 72 h were obtained. Results include gene expression changes due to OVA challenge in treated (OVA) and control (alum) animals, and lists reflect unique genes only corrected for gene duplicate found in our microarray clone sets.
Sensitization and Challenge to OVA
Mice were sensitized with a 100 μl intraperitoneal injection of 10 μg OVA and 2 mg alum in saline on Days 0 and 14. Sham-sensitized mice were given 2 mg alum in saline on Days 0 and 14. On Day 21, all mice were exposed to 1% OVA (in saline) aerosol using a 6-jet nebulizer (TSI Instruments; 35 psi, 16 liter/min dilution with air; 3-jets, aerosol particle mean diameter of 0.3 μm) for 1 h. On day 22, physiologic and biologic measurements were made.
Airway Hyperresponsiveness
To test airway responsiveness, increasing doses of methacholine (MCh; 25–100 μg/kg) were sequentially administered intravenously to pentobarbital-anesthetized and doxacurium chloride–paralyzed mice. Peak tracheal pressure was measured under control conditions as a baseline. MCh injection caused increased peak tracheal pressure, and the difference in peak values between control and MCh challenge at each breath was integrated over a 30 s period and designated as airway pressure time index (APTI) (6). Increased APTI is indicative of an increased airway response. Airway hyperresponsiveness, a hallmark feature of asthma in humans, is an umbrella term that describes airway hypersensitivity (a left-ward shift in a bronchoconstrictor dose–response curve) and airway hyperreactivity (an increased slope in a bronchoconstrictor dose–response curve). Although we did not administer a sufficient number of MCh doses to generate a true bronchoconstrictor dose–response curve, we chose two low doses of MCh (25 and 50 μg/kg) to test airway sensitivity and one moderate dose of MCh (100 μg/kg) to assess airway reactivity (7, 8).
Whole Lung Lavage
Mice were killed by sodium pentobarbital overdose (60 mg/kg body weight). Lungs were lavaged via a tracheal cannula with 6 × 1 ml of sterile saline (3). The lung lavage fluid was centrifuged (10 min, room temperature, at 500 × g) and the cell pellet was resuspended in Hank's buffered salt solution and cells counted in a Neubauer hemocytometer (VWR Scientific Products, Darmstadt, Germany). Differential cell counts were performed on cytospin preparations (Cytospin 2; Shandon, Pittsburgh, PA) stained with Hema3 protocol stain (Biochemical Sciences Inc., Swedesboro, NJ) by classification of 200 cells on standard morphologic criteria.
Microarray Fabrication, RNA Labeling, and Hybridization
Microarrays were constructed using the NIA 15k and BMAP mouse cDNA clone sets that, together, contain 27,010 clones representing ∼ 22,000 unique transcript probes. PCR amplicons were prepared for printing as described previously (9). After amplification and purification, amplicons were resuspended at 100–200 nM in 50% DMSO and printed onto UltraGAPs aminosaline-coated slides (Corning, Inc., Corning, NY) using an Intelligent Automation System arrayer (Cambridge, MA). After printing, DNA was cross-linked to the slides by UV irradiation with a Stratalinker UV Crosslinker (Stratagene, La Jolla, CA) and stored in a vacuum chamber until used.
Total RNA was prepared from whole lung tissue using Trizol (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Detailed cDNA target preparation and hybridization protocols are available online at http://pga.tigr.org/protocols.shtml. Briefly, cDNA was generated through incorporation of amino-allyl–labeled deoxyuridine triphosphate (dUTP) via random-primed Superscript II reverse transcription (Invitrogen) using 10 μg of total RNA, followed by coupling to the ester of either the Cy3 or Cy5 fluorescent dye (Amersham-Pharmacia, Piscataway, NJ) (9). Fluorescently labeled cDNAs were purified with Qiagen PCR purification kit (Qiagen, Valencia, CA).
Slides were prehybridized in 1% BSA in 5× SSC (0.75 M NaCl, 75 mM sodium citrate pH 7.0) 0.1% SDS for 45 min at 42°C, after which the slides were washed and dried. Cy3- and Cy5-labeled cDNA was resuspended in 30 ml of 50% formamide, 5× SSC, 0.1% SDS containing 0.5 μg mouse COT1-DNA, 1 μg poly-dA, and hybridized to the microarray at 42°C for 16 h under glass coverslips. After hybridization, slides were washed for 4 min at 42°C in solution containing 1× SSC and 0.2% SDS, followed by a 4-min wash of 0.1× SSC, 0.1% SDS, at ambient temperature, then by two 2.5-min washes of 0.1× SSC, at the ambient temperature. Slides were dried by centrifugation and scanned without delay at 10-μm resolution using an Axon 4000B scanner (Axon; Molecular Devices, Sunnyvale, CA). In this study, each target was cohybridized with a common RNA reference from mouse strains 129/SvIm, CAST/Ei, BALB/cJ, and C57BL/6J to facilitate interstrain comparisons, and a dye reversal approach was used to minimize dye-specific artifacts.
Data Analysis
A repeated-measures analysis of variance was used to determine differences in airway sensitivity at low-dose MCh (25 and 50 μg/kg). To determine if the airway response to 100 μg/kg MCh was significantly affected by genotype, a one-way analysis of variance, in combination with a Tukey honestly significant difference post hoc test, was used. A Student t test was used to determine differences when only two experimental groups existed (bronchoalveolar lavage eosinophils). For all tests, P < 0.05 was considered significant.
Image analysis, data processing, and data handling steps were performed using algorithms found in our TM4 software suite (10) (http://www.tigr.org/software/tm4). Spot intensities from TIFF images for Cy3 and Cy5 flours were processed using TIGR Spotfinder (Dana-Farber Cancer Institute, Boston, MA). The resulting expression data were filtered to remove low-intensity spots and normalized using local lowess (smoothing parameter 0.33) implemented in MIDAS (11). Normalized data were loaded into MeV for statistical and cluster analysis of log2 (treated/control) ratios. Significance analysis of microarrays (12) was used to identify genes significantly induced or repressed due to OVA treatment within pre-control samples (n = 4) and each time point (n = 4) using 75% cut-off and a false discovery rate < 0.05. After evaluating all time points, a list of genes was generated and time courses constructed for subsequent cluster analysis. A subset of 90 unique genes was selected from significant genes that overlapped in strains C57BL/6J and BALB/cJ with 129/SvIm strain for comparisons of temporal expression profiles across all four strains using average-linkage hierarchical clustering with a Euclidean distance metric (13, 14).
Inferring Biological Themes and Pathways
For a functional analysis of expression profiles, statistically significant genes were annotated using gene ontology assignments (15) provided through microarray analysis tool RESOURCERER (http://biocomp.dfci.harvard.edu/cgi-bin/magic/r1.pl) (16), and categorized into summary GO-Slim categories. The Expression Analysis Systematic Explorer (17, 18) bioinformatics software package was used to identify statistically significant functional categories represented in the selected gene set.
Quantitative Real-Time RT-PCR
To confirm microarray gene expression results, quantitative (real-time) RT-PCR was used, as described previously (19). Briefly, primers were designed using Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) using the mouse gene index tentative consensus sequences (http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=mouse) for selected genes (see Table E2 in the online supplement). A total of 2 μg total RNA was assayed using TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA) and QuantiTech SYBR Green PCR kit (Qiagen) methods.
RESULTS
Gene Regulation Response to OVA
Although the development of allergic asthma in mice does not exactly mimic the progression of the human disease, like humans, genetic susceptibility is evident in these mice. For example, in response to OVA treatment, the 129/SvIm mouse strain develops substantial airway hyperresponsiveness and lung eosinophilia, whereas the CAST/Ei mouse strain displays no measurable physiologic or biologic response (Table 1). Yet all strains exposed to OVA show significant changes in the regulation of numerous genes that can be further characterized by fundamental categories (Figure 1). Thus, the challenge is to determine which of the several hundred genes that are transcriptionally regulated by OVA treatment are responsible for the pathophysiologic response.
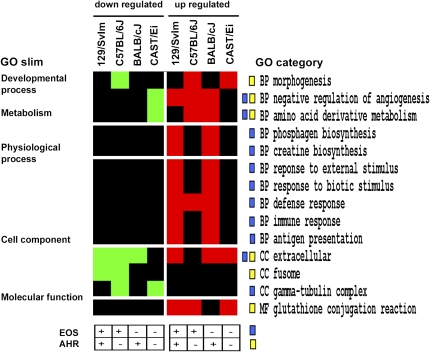
Figure 1.
Functional analysis of significantly expressed genes. Significant genes determined by significance analysis of microarrays (SAM) analysis were annotated with gene ontology (GO) terms for cellular component, biological process, and molecular function, and overrepresented GO categories were determined using Expression Analysis Systematic Explorer software. Here, overrepresented categories among upregulated genes are shown in red and for downregulated genes in green for each of the four mouse strains analyzed in this study; black indicates that a particular category was not found to be significant. GO terms are shown to the right and corresponding GO Slim categories are presented on the left. Phenotypic classifications previously reported for each strain are outlined in Table 1 for eosinophilic airway inflammation (EOS) and airway hyperresponsiveness (AHR). Proposed associations of differential expressions to phenotypes are indicated for each GO category with blue and yellow boxes for eosinophilic infiltration and airway hyperreactivity, respectively.
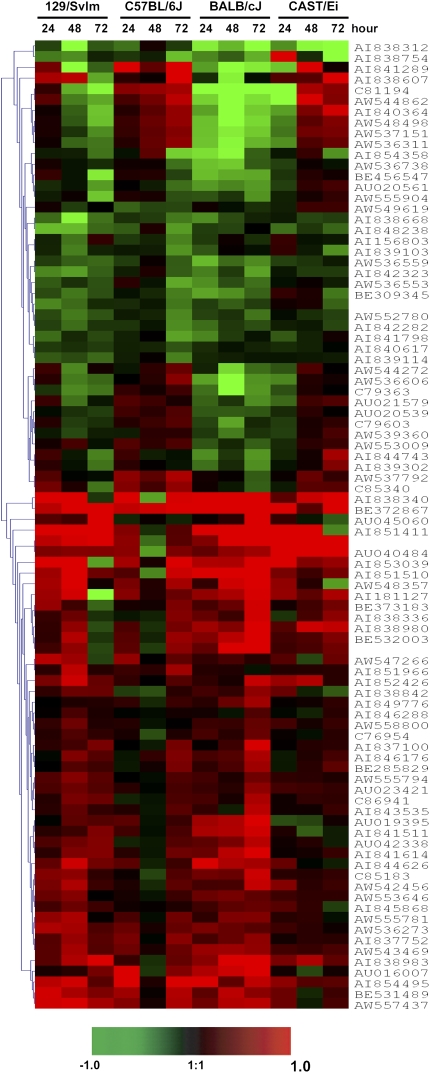
To identify candidate genes that may give rise to OVA-induced airway hyperresponsiveness and/or eosinophilic inflammation, a subset of 90 unique genes that showed expression patterns consistent with phenotypic responses in responder mouse strains 129/SvIm, C57BL/6J, and BALB/cJ were inspected for differences in temporal patterns of gene expression (Table E3). This subset of 90 genes includes 62 encoding previously annotated proteins, 28 genes encoding unknown proteins, and 9 coding genes that map within previously assigned quantitative trait loci (QTL) markers for antigen-induced bronchial hyperresponsiveness on mouse chromosome 2 and 17 (20). Time course expression values for all 4 mouse strains corresponding to these 90 genes were organized using average-linkage hierarchical clustering with an Euclidean distance metric (Figure 2) (13, 14), allowing correlations between patterns of gene expression and strain to be visualized.
Figure 2.
Identification of differential expression patterns. The intersection of genes obtained from our SAM analysis between 129/SvIm, C57BL/6J, and BALB/cJ strains yielded a subset of 90 potential candidate genes involved in airway hyperresponsiveness sorted by average-linkage hierarchical clustering. Log2(ratio) values comparing expression in treated and the corresponding control animals were calculated based on a total of three to four microarray replicates.
Identification of Candidate Genes Based on Strain Responses to OVA
Genes identified in the aforementioned hierarchical clustering analysis were visually inspected for genes showing differential expression patterns among strains and a significant change in expression greater than 1.5-fold (or log2[ratio] of 0.58). Based on these criteria, a subset of 21 candidate genes was selected for further directed analysis (Table 2). Figure 3 illustrates differential expression of individual candidate genes in the mouse lung after OVA challenge, along with proposed associations with OVA model phenotypes including airway hyperresponsiveness and eosinophilia. Functional annotation of these candidate genes with GO terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways is in agreement with pathways inferred from the overrepresentation analysis (Table 2).
TABLE 2.
CANDIDATE GENES SHOWING STRAIN-SPECIFIC DIFFERENTIAL EXPRESSIONS
| Accession No. | Gene Name | Gene Ontology | Metabolic Pathway* |
|---|---|---|---|
| Airway eosinophilia | |||
| AW547266 | Fibronectin precursor | Cell-substrate junction assembly | |
| C85340 | Unknown | ||
| AI851966 | STAT2 | Cytokine- and chemokine-mediated signaling | |
| Protection against airway hyperresponsiveness | |||
| AI841289 | dnaK-type molecular chaperone hsp70 | Nonchaperonin ATPase activity | |
| C81194 | hsp105 | hsp | |
| AW536606 | DNAJ-like 1, hsp40 | Positive regulation of cell proliferation | |
| AI840364 | NEDD4-like ubiquitin ligase 3 | Ubiquitin cycle | |
| AW548498 | Unknown | ||
| AW537151 | Unknown | ||
| AW536311 | Heat shock cognate 71-kDa protein | ||
| AW537792 | 78-kDa glucose regulated protein precursor | Protein folding | |
| Airway hyperresponsiveness | |||
| AW555904 | Complement component receptor precursor C1qRp | Phagocytosis | |
| AU045060 | AU044919 protein | B-cell activation | |
| AI851510 | C-C chemokine receptor 5 | JAK-STAT pathway | Inositol phosphate metabolism |
| AI838983 | Glutathione transferase omega 1 | Glutathione transferase activity | Glutathione metabolism |
| AI838842 | RNA binding motif protein 3 | RNA processing | |
| AU042338 | MHC II H2-M α chain | Immune response | |
| AW555794 | Peroxiredoxin 4 | Peroxidase activity | Glutathione metabolism |
| AU01939 | Solute carrier organic anion transporter | Transporter activity | |
| AI841511 | Carbonic anhydrase II | Gluconeogenesis | Nitrogen metabolism |
| AI841614 | Putative integral membrane protein |
Definition of abbreviations: hsp, heat shock protein; MHC, major histocompatability complex.
Based on KEGG annotation.
Figure 3.
Candidate genes based on the physiologic and biologic response to ovalbumin (OVA). This figure illustrates differential expression of candidate genes in the mouse lung after OVA challenge along with proposed associations with phenotypes in the OVA asthma model. Differentially expressed genes were selected based on a significant change of ± 1.5-fold due to OVA treatment in the time course and, where identified, as responses leading to the promotion or protection of airway hyperresponsiveness (AHR) (129/SvIm and BALB/cJ) or eosinophil (EOS) (129/SvIm and C57BL/6J) phenotypes. Data shown as the Log2 (OVA/alum) from two to four measurements.
Description of Candidate Genes
Genes that were upregulated in eosinophil responder strains 129/SvIm and C57BL/6J include extracellular matrix glycoprotein fibronectin, unknown C85340, and transcription factor STAT2. The role of fibronectin in the development of asthma has been previously described as an important mediator in the migration of small epithelial cells in the airway (21). STAT2 transcription factor is a member of the JAK-STAT signaling cascade elicited by cytokines and chemokines resulting in cell proliferation and inflammatory responses (22).
Eight genes displayed patterns that were associated with a protective airway hyperresponsiveness phenotype. Genes for heat shock protein (hsp) 70, hsp106, hsp40, neuronal precursor cell-expressed developmentally downregulated (Nedd) 41, hsp71, glucose regulated protein (GRP)78, and genes encoding for proteins AW548498 and AW53715 of unknown function, show an increased expression in nonresponder strain C57BL/6J compared with a downregulation in responder strains 129/SvIm and BALB/cJ. Genes for this subphenotype were mainly hsps perhaps required for transport or proper folding of newly synthesized proteins. Aside from hsp activity, studies have shown an association of hsp70 in asthma through an inhibition of NF-κB transcription factor activation (23). The Nedd4 family of proteins has been described as ubiquitin protein ligases, which regulate the activity of epithelial Na+ channel via protein ubiquitination (18). It is not clear how the degradation of this channel may contribute to asthma phenotypes in the mouse.
Transcription of genes leading to the promotion of airway hyperresponsiveness include C1qRp, AU044919 protein, major histocompatability complex II, glutathione S-transferase-1, peroxiredoxin 4, Rbm3, anion transporter, carbonic anhydrase 2, a novel membrane protein, and CCR5. The gene for C1qRp, a subcomponent of C1qR receptor, is downregulated at 72 h and, although the exact role is still unclear, it is involved in the classical complement pathway, an important pathway in the asthmatic response (24, 25). Other immune response genes, including AU044919 protein, MHCII, peroxiredoxin 4, and CCR5, were all upregulated in responder strains with a delayed response for AU044919 protein at 72 h. Genes AU044919 and MHCII encode for immunoglobulin heavy chain and MHCII α chain precursor, which are crucial elements in the antigen presentation by proliferating lymphocytes (26, 27). Upregulation of glutathione S-transferase-1 and peroxiredoxin 4 genes constitute the induction of glutathione metabolism, an important pathway in the response to oxidative stress that subsequently effects expression of inflammatory genes driven by NF-κB transcription factor (28, 29). Although Rbm3 is a cold-response protein, expression of this gene may lead to proper translation of coregulated genes by acting as an RNA chaperone (30). Additional important metabolic pathways that were also inferred include inositol phosphate and nitrogen metabolism represented by CCR5 and carbonic anhydrase II genes, respectively.
Biologic and Physiologic Importance of CCR5 in Mice Treated with OVA
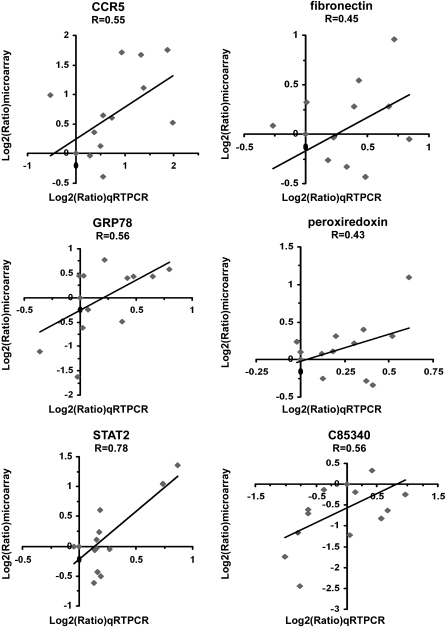
The CCR5 gene was identified as significantly upregulated in mouse strains that demonstrated airway hyperresponsiveness (129/SvIm and BALB/cJ), and not significantly increased in C57BL/6J and CAST/Ei mouse strains, which lacked a measurable increase in airway responsiveness. The microarray gene transcription results for six candidate genes, including CCR5, were verified by quantitative RT-PCR (Figure 4). Values obtained from the quantitative RT-PCR analysis are consistent with differential expression patterns observed in our microarray data, which clearly suggest that the expression of CCR5, a chemokine receptor, is positively related to OVA-induced airway hyperresponsiveness. Furthermore, the gene expression data suggest that CCR5 is unrelated to lung eosinophilic infiltration, because the CCR5 gene is oppositely regulated in two mouse strains that display similar lung eosinophilic responses (C57BL/6J and 129/SvIm).
Figure 4.
Expression of candidate genes, including CCR5, observed by microarray analysis was validated by qRT-PCR. Quantitative RT-PCR analysis was performed (in duplicate) for six significantly regulated genes: CCR5, fibronectin, GRP78, peroxiredoxin, STAT2, and a gene encoding protein of unknown function, and the data compared as log2(treated/control) to their corresponding microarray expression profiles. The temporal expression profiles for all strains show a positive correlation to values measured by qRT-PCR.
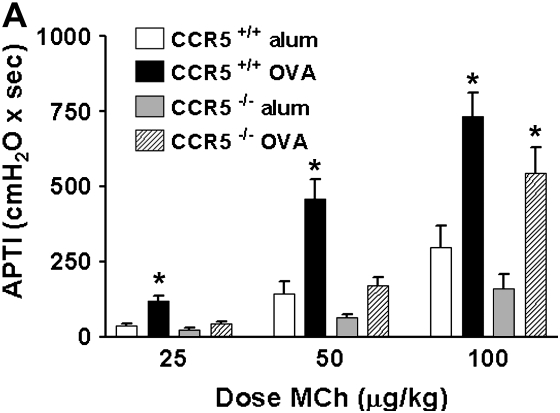
To determine the importance of CCR5 in OVA-induced airway disease, we measured airway responsiveness to the bronchoconstrictor MCh and the concentration of eosinophils in the lower respiratory tract from OVA-treated CCR5+/+ and CCR5−/− mice. Figure 5A shows that OVA treatment resulted in no significant increase in APTI in CCR5−/− mice at 25 and 50 μg/kg MCh compared with their alum-treated controls. This was in stark contrast to the significant increase in APTI observed in OVA-treated CCR5+/+ mice relative to their alum-treated controls. Thus, OVA-treatment had a significantly diminished effect on MCh airway sensitivity in CCR5−/− mice relative to CCR5+/+ mice. However, the APTI response to 100 μg/kg MCh, a moderate dose, was comparably increased by OVA treatment in both CCR5−/− and CCR5+/+ mice. Genotype had no significant effect on the APTI response to any dose of MCh in alum-treated mice.
Figure 5.
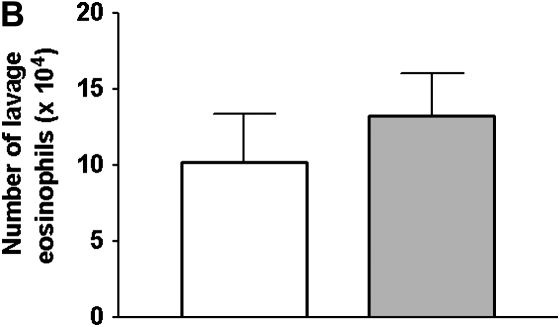
Effect of CCR5 expression on the response to OVA treatment. (A)The airway responsiveness to methacholine (MCh), defined by the time-integrated change in peak airway pressure, or airway pressure time index (APTI), was measured. OVA treatment caused a significant increase in APTI (relative to alum-treated controls, respectively) for CCR5+/+ mice (open bars), but not CCR5−/− mice (stipled bars) at low-dose MCh (25 and 50 ug/kg). OVA treatment enhanced the response to 100 μg/kg MCh similarly in CCR5+/+ (open bars) and CCR5−/− mice (stipled bars) when compared with their respective alum-treated controls. Data are mean ± SEM; CCR5+/+ n = 9; CCR5−/− n = 9 mice per group. *P < 0.05. (B) The number of eosinophils in the lavage fluid in CCR5+/+ and CCR5−/− mice. Data are mean ± SEM; CCR5+/+ n = 9; CCR5−/− n = 9 mice per group. A P value of < 0.05 was considered statistically significant. *Represents statistically significant effect of OVA treatment relative to respective alum-treated genotype control.
We calculated the change in the APTI response between 50 and 100 ug/kg MCh as a measure of airway reactivity. This slope is not different between OVA-treated CCR5−/− and CCR5+/+ mice (mean change was 314 ± 46 and 272 ± 40, respectively). Thus, the airway hyperresponsiveness demonstrated by OVA-treated CCR5+/+ mice is likely composed of airway hypersensitivity and airway hyperreactivity to MCh. In contrast, OVA treatment did not increase airway MCh sensitivity in mice lacking CCR5, despite the fact that increased airway reactivity to MCh was observed.
Consistent with the predictions of the microarray, there was no significant difference (P = 0.566) in the concentration of lung lavage eosinophils between CCR5+/+ and CCR5−/− mice (Figure 5C). Thus, airway responsiveness is independent of eosinophilic inflammation in these mixed-background mice. This is consistent with our previous findings, where neither OVA-treated C57BL/6J, nor 129/J, mice showed a correlation between eosinophil number and airway responsiveness (3).
DISCUSSION
The immune response to an environmental antigen is a complex and highly coordinated process involving myriad gene transcription events. In genetically susceptible individuals, the immune response to an otherwise innocuous antigen can become excessive, and allergic asthma may ensue. One key to understanding asthma is to identify what combination of gene transcription events transforms a normal immune response into a pathologic one.
As demonstrated by others, this study shows that an enormous number of genes are up- or down-regulated in response to antigen exposure. Unique to this study, however, is the distillation of numerous regulated genes into a concise list of those likely to underlie the pathogenesis of asthma. This was achieved through the simultaneous consideration of gene regulation events at multiple time points among several strains of mice displaying distinct “allergic asthma” phenotypes, and a careful analysis of the classes of response represented by these genes and the pathways to which they map.
Our analysis identified 18 genes associated with airway hyperresponsiveness (10 positively and 8 inversely correlated) and 3 genes associated with lung eosinophilic inflammation. One gene, CCR5, the expression of which was positively correlated with airway hyperresponsiveness, was selected, based on these criteria, for further study to elucidate its role in the hyperresponsive phenotype.
The gene for CCR5 encodes a seven-membrane-spanning G protein–coupled chemokine receptor. CCR5 responds to T helper type 1 (TH1)-type chemokines produced in response to viral pathogens, and is best known for its role as an HIV coreceptor (4). Additionally, in allergic asthma, CCR5 is expressed on eosinophils, monocytes, and activated T cells, and mediates the migration of these cells to the lung (31). Despite this, there is only a paucity of information regarding the role for CCR5 in the development of allergic asthma.
Although two earlier studies demonstrated the importance of various CCR5 ligands to the development of both allergic inflammation and airway responsiveness (32, 33), no conclusion as to the role for CCR5 in controlling the allergic asthmatic response could be drawn, as these ligands act promiscuously and activate other chemokine receptors. Others demonstrated that CCR5 positively regulates airway responsiveness and lung eosinophilia in a time-dependent manner in a model of chronic fungal asthma (5). However, the present study is the first to show that CCR5 is involved in the development of allergen-induced airway hyperresponsiveness. Moreover, the present study further defines that this regulation occurs independent of the magnitude of eosinophilic inflammation.
Although CCR5 is a chemokine receptor that can direct cell migration, our data show that CCR5 is not critical for the trafficking of eosinophils to the lung in a mouse model of allergic airway inflammatory disease. This apparent paradox may be partially explained by the fact that eotaxin and regulated upon activation, normal T-cell expressed and secreted (RANTES), as well as other eosinophil chemokines, can act through chemokine receptors other than CCR5 to promote eosinophil migration to the lung (31). However, CCR5 does appear to be important for lung inflammation induced by the fungus, Aspergillus fumigatus (5).
The mechanism by which CCR5 permits airway hypersensitivity in allergic airway inflammatory disease is unknown and is beyond the scope of this article. However, eosinophilic lung inflammation and airway remodeling are not likely factors in the development of airway hyperresponsiveness in this study.
Although this report has focused on CCR5, there are a number of other genes identified in our analysis (Table 2) that may also play a role in modulating the phenotypic response to allergen sensitization and challenge, and consequently may serve as possible targets for therapeutic intervention in asthma.
Upregulation of the fibronectin precursor gene was associated with lung eosinophilia. Fibronectin, an extracellular matrix protein, is one of numerous adhesion molecules that support extravasation of leukocytes, and other cells, into tissue (34). Thus, from the many proteins involved in eosinophil migration to the lung, our genetic analysis has identified one that may potentially be involved in the pathophysiology of asthma.
Upregulation of several genes was associated with protection from developing airway hyperresponsiveness. In particular, the Nedd4-like family of E3 ubiquitin ligases is an interesting candidate. Several of these E3 ubiquitin ligases negatively regulate T-cell growth factor production and proliferation (35). Other Nedd4-like proteins mediate ubiquitination and lysosomal degradation of CXCR4 (36), a chemokine receptor that is integral to the development of symptoms in a mouse model of allergic asthma (37). Thus, there are a number of potential mechanisms by which Nedd4-like proteins may protect against the development of airway hyperresponsiveness in allergic asthma.
Although the antioxidative function of peroxiredoxin 4 would suggest that upregulation of this protein might be associated with protection from airway hyperresponsiveness, our gene profiling data show the opposite trend. Evidence suggests that, dissociated from their role as peroxidases, peroxiredoxins also regulate cell proliferation and differentiation—two processes critically important to the development of allergic inflammatory airway disease that may specifically underlie the development of airway hyperresponsiveness (29).
The present results demonstrate the value of combining detailed phenotypic analysis with gene expression profiling to dissect specific responses and identify candidate genes. In this study the results of the genetic microarray profiling steered the physiologic and biologic experiments. The microarray results provided the rationale to carefully dissect the effect of CCR5 on airway responsiveness in allergic inflammatory airway disease, and revealed CCR5 as a significant and complex mediator of airway hypersensitivity. If applicable to humans, blockade of CCR5 may help return the bronchoconstrictor sensitivity of patients with asthma to that of normal subjects, and thereby reduce debilitating asthma symptoms associated with airway hyperresponsiveness, such as wheezing and coughing.
Supplementary Material
Acknowledgments
The authors thank Don Burdick from the Duke University Statistical Consulting Center for advice on statistical analysis. J.K.L.W. is supported by the Veterans Administration Medical Center.
This work was supported by National Institutes of Health grant U01 HL66580-01.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2005-0314OC on February 10, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Kohl J, Wahl L, Kuperman D, Germer S, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol 2000;1:221–226. [DOI] [PubMed] [Google Scholar]
- 2.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest 2003;111:1863–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead GS, Walker JK, Berman KG, Foster WM, Schwartz DA. Allergen-induced airway disease is mouse strain dependent. Am J Physiol Lung Cell Mol Physiol 2003;285:L32–L42. [DOI] [PubMed] [Google Scholar]
- 4.Nansen A, Christensen JP, Andreasen SO, Bartholdy C, Christensen JE, Thomsen AR. The role of CC chemokine receptor 5 in antiviral immunity. Blood 2002;99:1237–1245. [DOI] [PubMed] [Google Scholar]
- 5.Schuh JM, Blease K, Hogaboam CM. The role of CC chemokine receptor 5 (CCR5) and RANTES/CCL5 during chronic fungal asthma in mice. Faseb J 2002;16:228–230. [DOI] [PubMed] [Google Scholar]
- 6.Walker JK, Peppel K, Lefkowitz RJ, Caron MG, Fisher JT. Altered airway and cardiac responses in mice lacking G protein–coupled receptor kinase 3. Am J Physiol 1999;276:R1214–R1221. [DOI] [PubMed] [Google Scholar]
- 7.Woolcock AJ, Salome CM, Yan K. The shape of the dose–response curve to histamine in asthmatic and normal subjects. Am Rev Respir Dis 1984;130:71–75. [DOI] [PubMed] [Google Scholar]
- 8.Lotvall J, Inman M, O'Byrne P. Measurement of airway hyperresponsiveness: new considerations. Thorax 1998;53:419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques 2000;29:548–550, 552–554, 556 passim. [DOI] [PubMed] [Google Scholar]
- 10.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003;34:374–378. [DOI] [PubMed] [Google Scholar]
- 11.Quackenbush J. Microarray data normalization and transformation. Nat Genet 2002;32:496–501. [DOI] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001;98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi LM, Myers TG, Fan Y, O'Connor PM, Paull KD, Friend SH, Weinstein JN. Mining the National Cancer Institute Anticancer Drug Discovery Database: cluster analysis of ellipticine analogs with p53-inverse and central nervous system–selective patterns of activity. Mol Pharmacol 1998;53:241–251. [DOI] [PubMed] [Google Scholar]
- 14.Chang B, Somogyi R, Fuhrman S. Evidence for shared genetic programs from cluster analysis of hippo-campal gene expression dynamics in development and response to injury. Restor Neurol Neurosci 2001;18:115–125. [PubMed] [Google Scholar]
- 15.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 2004;32:D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai J, Sultana R, Lee Y, Pertea G, Karamycheva K, Antonescu V, Cho J, Parvizi B, Cheung F, Quackenbush J. Genome Biol 2001;2:software0002.1–0002.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol 2003;4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey KF, Kumar S. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol 1999;9:166–169. [DOI] [PubMed] [Google Scholar]
- 19.Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, Quackenbush J. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol 2002;3:research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewart SL, Kuperman D, Schadt E, Tankersley C, Grupe A, Shubitowski DM, Peltz G, Wills-Karp M. Quantitative trait loci controlling allergen-induced airway hyperresponsiveness in inbred mice. Am J Respir Cell Mol Biol 2000;23:537–545. [DOI] [PubMed] [Google Scholar]
- 21.Hocking DC, Chang CH. Fibronectin matrix polymerization regulates small airway epithelial cell migration. Am J Physiol Lung Cell Mol Physiol 2003;285:L169–L179. [DOI] [PubMed] [Google Scholar]
- 22.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol 2003;197:157–168. [DOI] [PubMed] [Google Scholar]
- 23.Yoo CG, Lee S, Lee CT, Kim YW, Han SK, Shim YS. Anti-inflammatory effect of heat shock protein induction is related to stabilization of I kappa B alpha through preventing I kappa B kinase activation in respiratory epithelial cells. J Immunol 2000;164:5416–5423. [DOI] [PubMed] [Google Scholar]
- 24.Henson P. Complementing asthma. Nat Immunol 2000;1:190–192. [DOI] [PubMed] [Google Scholar]
- 25.McGreal E, Gasque P. Structure–function studies of the receptors for complement C1q. Biochem Soc Trans 2002;30:1010–1014. [DOI] [PubMed] [Google Scholar]
- 26.Zakut R, Cohen J, Givol D. Cloning and sequence of the cDNA corresponding to the variable region of immunoglobulin heavy chain MPC11. Nucleic Acids Res 1980;8:3591–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho SG, Attaya M, Monaco JJ. New class II–like genes in the murine MHC. Nature 1991;353:573–576. [DOI] [PubMed] [Google Scholar]
- 28.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol 2004;17:17. [DOI] [PubMed] [Google Scholar]
- 29.Fujii J, Ikeda Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep 2002;7:123–130. [DOI] [PubMed] [Google Scholar]
- 30.Chappell SA, Owens GC, Mauro VP. A 5′ leader of Rbm3, a cold stress-induced mRNA, mediates internal initiation of translation with increased efficiency under conditions of mild hypothermia. J Biol Chem 2001;276:36917–36922. [DOI] [PubMed] [Google Scholar]
- 31.Lloyd C. Chemokines in allergic lung inflammation. Immunology 2002;105:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalo JA, Lloyd CM, Wen D, Albar JP, Wells TN, Proudfoot A, Martinez AC, Dorf M, Bjerke T, Coyle AJ, et al. The coordinated action of CC chemokines in the lung orchestrates allergic inflammation and airway hyperresponsiveness. J Exp Med 1998;188:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukacs NW, Strieter RM, Warmington K, Lincoln P, Chensue SW, Kunkel SL. Differential recruitment of leukocyte populations and alteration of airway hyperreactivity by C–C family chemokines in allergic airway inflammation. J Immunol 1997;158:4398–4404. [PubMed] [Google Scholar]
- 34.Pagani F, Zagato L, Vergani C, Casari G, Sidoli A, Baralle FE. Tissue-specific splicing pattern of fibronectin messenger RNA precursor during development and aging in rat. J Cell Biol 1991;113:1223–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nat Immunol 2004;5:883–890. [DOI] [PubMed] [Google Scholar]
- 36.Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Mol Cell Biol 2004;24:3860–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalo JA, Lloyd CM, Peled A, Delaney T, Coyle AJ, Gutierrez-Ramos JC. Critical involvement of the chemotactic axis CXCR4/stromal cell–derived factor-1 alpha in the inflammatory component of allergic airway disease. J Immunol 2000;165:499–508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.