Abstract
The herb feverfew is a folk remedy for various symptoms including inflammation. Inflammation has recently been implicated in the genesis of many diseases including cancers, atherosclerosis and rheumatoid arthritis. The mechanisms of action of feverfew in the human body are largely unknown. To determine the cellular targets of feverfew extracts, we have utilized oligo microarrays to study the gene expression profiles elicited by feverfew extracts in human monocytic THP-1 cells. We have identified 400 genes that are consistently regulated by feverfew extracts. Most of the genes are involved in cellular metabolism. However, the genes undergoing the highest degree of change by feverfew treatment are involved in other pathways including chemokine function, water homeostasis and heme-mediated signaling. Our results also suggest that feverfew extracts effectively reduce Lipopolysaccharides (LPS)-mediated TNF-α and CCL2 (MCP-1) releases by THP-1 cells. We hypothesize that feverfew components mediate metabolism, cell migration and cytokine production in human monocytes/macrophages.
Keywords: feverfew, herbal medicine, immune response, microarray, monocyte
Introduction
Feverfew (Tanacetum parthenium), a member of the family Compositae, is an aromatic perennial herb with yellowish-green chrysanthemum-like leaves and small daisy-like flowers. Feverfew has traditionally been used, through consumption of leaves or the aerial parts, for numerous symptoms including inflammation, fever, rheumatism, asthma and stomachache (1). During the last few decades, feverfew has also been used for migraine prophylaxis although results from early clinical trials were mixed due to inadequate trial design and low power (2). A recent study with randomized, double-blind, placebo-controlled, multi-center, parallel-group design, using feverfew extracts in capsules, has suggested that feverfew may indeed reduce the frequency of migraine attack (3).
Although several mechanisms have been proposed to explain the therapeutic effect of feverfew in migraine (4), the mechanism of action of feverfew remains largely unknown. Due to the complex chemical composition of feverfew extracts and its historically multi-purpose usage, it is reasonable to expect that the mechanisms and pathways by which it functions in human cells are complex. Sesquiterpene lactone parthenolide long believed to be one of the main active components of feverfew extracts, has multiple physiologic effects, including anti-tumor activity, inhibition of DNA synthesis, inhibition of cell proliferation in different cancer cell lines (5,6), mediation of inflammation via cytokines (Tumor Necrosis Factor -alpha (TNF-α), Interleukin-1β (IL-β) and Interleukin-6 (IL-6)) (7), chemokines (IL-β) (8), lipids (prostaglandins) (9,10), COX-2 (7,11) and leukotrienes (11).
In spite of these reports, the mechanisms by which parthenolide acts on cells remain unclear. Although an early study in rats suggested that TNF-α production in alveolar macrophages was substantially inhibited by parthenolide (7), a recent study in mice, reported that expression of IL-6 was moderately suppressed by parthenolide but that expression of TNF-α, IL-1β and COX-2 was not affected (12). Further, while clinical trials using whole extracts have produced promising results, a clinical trial with primary focus on parthenolide enrichment for prevention of migraine failed to demonstrate a significant beneficial effect (13). It is therefore, unclear whether parthenolide is indeed the primary active ingredient of feverfew extracts.
Biological systems function in general by interactions at multiple levels and among many cellular components. Microarray technology allows simultaneous measurement of the gene expression of the entire genome and can provide a global view of gene networking and biological processes. Most importantly, microarray analysis does not require a priori assumptions or knowledge of the biological system and can lead to new hypotheses based on the patterns of gene expression. Microarray analysis has been successfully applied to research on various diseases and systems including prostate cancer (14,15), breast cancer (16), leukemia and lymphoma (17), lung diseases (18), cardiovascular diseases (19,20), infectious diseases (21), toxicology (22), nutrition (23) and the effect of herbs (24). DNA microarrays are extremely useful in elucidation of molecular mechanism of action of an herb (25) because herbal medicine by its complex nature contains a broad spectrum of chemicals that may simultaneously affect multiple cellular targets. In this study, we utilized oligonucleotide microarray technology to study pathways in the response of the human monocytic cell line THP-1 to feverfew extracts. Since monocytes/macrophages are key mediators of inflammation and widely distributed in the body (26–28), monocytic cell lines present an appropriate model system to study immune responses to feverfew extracts.
Methods
Cell Culture and Feverfew Extracts
The human monocytic cell line THP-1 (ATCC 202-TIB) (29) was obtained from ATCC (American Type Culture Collection, Manassas, VA, USA) and cultured according to their recommendations. The cells express various receptors that are found in normal monocytes and have been a model system for macrophage biology and leukemia since 1980. The feverfew extracts were gifts from Phyto-Technologies (Woodbine, IA, USA). They were prepared by sonicating dry feverfew powder in 90% ethanol (30 min at room temperature) followed by carbon dioxide supercritical fluid extraction (SFE, 400 bar, 60°C). The final stock concentration was 20% dry weight (w/v) in dichloromethane (as solvent) and typically contained 0.981 mg/ml of parthenolide (the presumptive active ingredient), estimated by HPLC fingerprinting. Of note, literature search did not reveal any evidence that exposure to dichloromethane changes structural components of feverfew extracts.
Endotoxin And Cytotoxicity Assays
Endotoxins are strong immunological stimuli and are part of the outer cell wall of Gram-negative bacteria. Since, herbs under natural growing conditions are associated with bacteria, we assayed the levels of endotoxin in the herb extracts. The levels of endotoxin of three different extracts of feverfew were less than the lowest detectable level, (0.005 EU/ml) using the kinetic chromogenic LAL method (Cambrex, Walersville, MD, USA), the most sensitive (down to 0.005 EU/ml) method available. Cytotoxicity studies indicated that when THP-1 cells were treated for 3 h at doses equivalent to 0.01%, 0.02%, or 0.04% dry weight feverfew, cell viability did not vary from the control (data not shown). The viability of THP-1 cells exposed to feverfew extracts decreased starting at doses equivalent to 0.04% dry weight after 24 h incubation. Viability further decreased to ∼20% of controls at 0.1% dry weight after 24 h incubations (data not shown). On the basis of these analyses, exposure to the equivalent of 0.01% dry weight feverfew for 3 h was used for experiments unless otherwise noted.
Human Gene Expression Microarrays
THP-1 cells were treated with dichloromethane (solvent control) or feverfew extracts (SFE) equivalent to 0.01% (w/v, dry weight) for 3 h. Four independent experiments were performed for each treatment. To maximize the contrast between samples, we implemented a loop experimental design (30) with dye swap. Total RNA was isolated using the Agilent Total RNA Isolation Mini Kit (Agilent Technologies, Wilmington, DE, USA). RNA samples were labeled with Cy3 or Cy5 fluorescent dye and hybridized for 17 h with Agilent Whole Human Genome Oligo Microarray containing over forty thousand human genes and transcripts (31). Microarray slides were washed and scanned by Agilent G2505B microarray scanner. Image processing and fluorescence intensity were interpreted and analyzed by Agilent Feature Extraction (version 8.5).
Data Processing and Statistical Analysis
Data normalization was performed by a non-linear LOWESS method that utilizes gene intensity and spatial information (32–34). Gene expression fold changes were calculated as ratio of treatment (i.e. feverfew) divided by control (solvent only). For statistical analysis, the fold change ratios were transformed into logarithmic scale. To select the genes that were differentially expressed, we utilized the concept of false discovery rate (FDR) (35–37). For each treatment, a modified t-test was performed for four independent experiments using the software program, Significance Analysis of Microarray (SAM) (38). Experimental data were also analyzed using the statistical package SYSTAT11 (SYSTAT Inc., Richmond, CA, USA). After statistical analysis, the results were converted back to fold change for easy comparison.
Gene Annotation by Gene Ontology (GO)
The Gene Ontology database (39,40) is a large collaborative public database of controlled biological vocabularies (i.e. ontologies) that describe gene products based on their functionalities in the cell. In general, over- or under-representation of GO terms were evaluated using the Fisher Exact Probability Test. We utilized the on-line program FatiGO (41) to produce (1) percentage and number of genes appearing in a GO category; (2) P-values from the Fisher exact test for each GO term associated with the gene.
RT–PCR Primers
Both the oligos and the online tool for design of primer sequences were from Integrated DNA Technologies (Coralville, IA, USA). Our results suggested that mRNAs for the GAPDH gene were not affected by feverfew treatments (data not shown). The GAPDH gene was therefore used as the reference gene. Genes of interest and reference genes (i.e. GAPDH) were PCR-amplified for construction of standard curves.
RNA Isolation for RT–PCR
Total RNA for both treatment and control groups was isolated using the Absolutely RNA RT–PCR Miniprep Kit (Stratagene, La Jolla, CA). Four independent samples (different from that of the microarrays) were obtained. Brilliant SYBR Green QRT-PCR Master Mix Kit (1-Step) from Stratagene was used to prepare the reaction mix. Reactions were carried out on an Mx3000 RT–PCR machine from Stratagene and programmed using the following parameters: reverse transcription at 50°C for 30 min, initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 30 s, 50–60°C for 1 min and 72°C for 30 s. Each PCR experiment per sample was performed in triplicate on the real-time PCR instrument. Changes in expression of genes were calculated by the standard curve method and normalized to the reference gene GAPDH.
ELISA Assays for Cytokines
Commercial enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, Minneapolis, MN, USA) were used to quantify TNF-α and CCL2. The absorbance at 450 nm was read by a microplate reader (model 680; Bio-Rad Laboratories, Mississauga, ON, Canada) with the wavelength correction set at 550 nm. To calculate the concentration of TNF-α and CCL2, a standard curve was constructed using serial dilutions of cytokine standards provided with the kit.
Results
Gene Expression Profiling of THP-1 Treated By Feverfew Extracts
We have conducted microarray analyses to identify the pathways involved in the response of human macrophages to feverfew extract (Supplementary Fig. 1). Statistical analysis by the SAM software (38) suggested that there were about 400 genes significantly altered in cells treated with feverfew extracts (Supplementary Fig. 1). There were 245 up-regulated genes (increases ranging from 17 to 1.21-fold over control) and 155 down-regulated genes (decreases ranging from 0.8 to 0.3-fold compared with controls).
Biological Pathways Affected By Feverfew Extracts
To gain insights into the mechanisms of human monocytic responses to feverfew, we utilized Gene Ontology (GO) annotation (Methods) to search for over-representation of specific biological processes in the list of differentially expressed genes. The percentage for each category was calculated by the number of genes found in that GO category divided by the number of total genes available for annotation. Pathways prominent in this analysis are ‘nucleobase, nucleoside, nucleotide and nucleic acid metabolism’ (41%), ‘regulation of cellular metabolism’ (35%), ‘alcohol metabolism’ (7.6%), ‘lipid metabolism’ (7%) and ‘response to unfolded protein (4%) (Supplementary Table 1).
Genes Highly Impacted By Feverfew Extracts
It would be expected that the THP-1 genes that are highly responsive to feverfew represent the primary targets of feverfew action. We have constructed a list containing the 50 genes most highly affected by feverfew (25 induced and 25 suppressed) (Table 1). Among the most up-regulated genes are the genes coding for heme oxygenase HMOX-1 (induced >17-fold, compared with control); the heat shock protein, HSPA1A (10-fold increase) and the chemokine IL-8 (9-fold increase). The most highly suppressed genes include the genes coding for SEMA4C (0.3-fold of control, function unknown); RGS16 (regulator of G-protein signaling); the orphan chemokine receptor CMKOR1 (also called RDC10) and the inflammatory chemokine CCL2. Interestingly, the expression of the gene AQP1 (water channel-forming protein) was also significantly inhibited.
Table 1.
Fifty genes in the human monocytic cell line THP-1 whose expression is highly impacted by feverfew extracts
| Gene name | Description | Fold change | Gene name | Description | Fold change |
|---|---|---|---|---|---|
| HMOX1 | Heme oxygenase (decycling) 1 | 17.7 | PSCD4 | Pleckstrin homology, Sec7 and coiled-coil domains 4 | 0.5 |
| HSPA1A | Heat shock 70 kDa protein 1A | 9.8 | DKFZP434H132 | DKFZP434H132 protein, mRNA | 0.5 |
| IL8 | Interleukin 8 | 9.0 | AQP1 | Aquaporin 1 (channel-forming integral protein, 28 kDa) | 0.5 |
| DNAJB4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | 6.5 | ID3 | Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 0.5 |
| DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 5.2 | GADD45B | Growth arrest and DNA-damage- inducible, beta | 0.5 |
| SRXN1 | Sulfiredoxin 1 homolog (S. cerevisiae) (SRXN1) | 4.6 | CXXC5 | CXXC finger 5 | 0.5 |
| LOC344887 | mRNA; cDNA DKFZp686B 14224 (from clone DKFZp686B14224) | 4.5 | TFRC | Transferrin receptor (p90, CD71) | 0.5 |
| EGR2 | Early growth response 2 (Krox-20 homolog, Drosophila) | 4.2 | THC2320257 | Unknown | 0.5 |
| ADM | Adrenomedullin | 4.0 | ENC1 | Ectodermal-neural cortex (with BTB-like domain) | 0.5 |
| BCL6 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | 3.9 | LOC389119 | Similar to RIKEN cDNA 6530418L21 | 0.5 |
| SLC7A11 | Solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 | 3.8 | ENST00000332281 | Similar to snail homolog 3 (Drosophila) | 0.5 |
| THC2303268 | HUMCATF catalase, partial (39%) | 3.6 | ENST00000354543 | mRNA; cDNA DKFZp586C0721 (from clone DKFZp586C0721) | 0.5 |
| GCLM | Glutamate-cysteine ligase, modifier subunit | 3.5 | GIMAP1 | GTPase, IMAP family member 1 | 0.5 |
| ATF3 | Activating transcription factor 3 | 3.3 | BC080552 | cDNA clone IMAGE: 6254031 | 0.5 |
| THC2310563 | Unknown | 3.2 | CR593492 | Full-length cDNA clone CS0DI054YC18 of Placenta Cot 25-normalized of (human) | 0.5 |
| HSPH1 | Heat shock 105 kDa/110 kDa protein 1 | 3.1 | HOXA9 | Homeo box A9 | 0.5 |
| HSPA6 | Heat shock 70 kDa protein 6 (HSP70B′) | 2.9 | BCOR | BCL6 co-repressor | 0.5 |
| DUSP1 | Dual specificity phosphatase 1 | 2.9 | LFNG | Lunatic fringe homolog (Drosophila) | 0.5 |
| CR598364 | Full-length cDNA clone CS0CAP007YJ17 of Thymus of (human) | 2.9 | PCANAP6 | Prostate cancer associated protein 6 | 0.5 |
| FBXO30 | F-box protein 30 | 2.8 | ID2 | Inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 0.4 |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 2.7 | FLJ45187 | FLJ45187 protein | 0.4 |
| CR618687 | Full-length cDNA clone CS0CAP008YE23 of Thymus of (human) | 2.6 | CCL2 | Chemokine (C-C motif) ligand 2 | 0.4 |
| PMAIP1 | Phorbol-12-myristate-13-acetate- induced protein 1 | 2.6 | CMKOR1 | Chemokine orphan receptor 1 | 0.4 |
| BAG3 | BCL2-associated athanogene 3 | 2.6 | RGS16 | Regulator of G-protein signalling 16 sema domain, immunoglobulin domain (Ig), | 0.4 |
| ARRDC4 | Arrestin domain containing 4 | 2.5 | SEMA4C | Transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C | 0.3 |
Fold changes in gene expression were calculated as values after feverfew treatment divided by control (solvent only) values.
Verification by RT–PCR of Important Genes Responding to Feverfew Extracts
Since each methodology has its own intrinsic limitations and biases, it was important to verify the feverfew-responsive genes identified through microarray analysis using a different technology and independent samples. We therefore, performed RT-PCR, isolating RNAs for 10 representative feverfew-responsive THP-1 genes. Four independent experiments were performed using cells exposed to feverfew under the same conditions as in the microarray experiments. The results from real-time PCR were similar in trend to the microarray experiments, (Table 2). For example, AQP1 expression was reduced 0.5-fold on average by feverfew extracts in microarray experiments and was similarly reduced 0.4-fold in RT-PCR experiments. The consistency of the microarray and RT-PCR results reinforces the validity and power of our microarray analyses.
Table 2.
Changes in expression of 10 representative feverfew-responsive THP-1 genes: Comparison of microarray and RT–PCR data
| Gene Name | Microarray (fold change) | RT-PCR (fold change) |
|---|---|---|
| AQP1 | 0.5 | 0.4 |
| HMOX1 | 17.7 | 25.3 |
| HOXA10 | 0.6 | 0.6 |
| HSD17B7 | 2.1 | 1.9 |
| HSPA1A | 9.8 | 4.4 |
| ID2 | 0.4 | 0.3 |
| LDLR | 2.4 | 2.6 |
| PTAFR | 0.7 | 0.5 |
| RGS16 | 0.4 | 0.2 |
| SPRY2 | 0.6 | 0.5 |
Each fold change is an average of four independent biological replicates for microarray experiments, and of four independent biological replicates for RT–PCR experiments.
Inhibition of Cytokine TNF-α and CCL2 Production by Feverfew Extracts
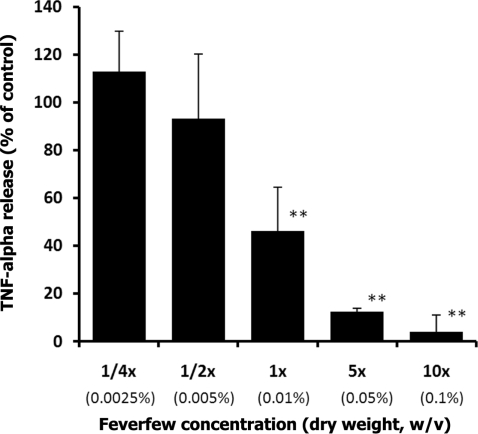
To elucidate the functional roles of feverfew, we tested whether feverfew extracts can interfere with cytokine production in THP-1 cells. Although TNF-α, a key cytokine involved in inflammation, does not appear in the list of feverfew-responsive genes in microarray analysis, treatment with parthenolide has been reported to inhibit LPS mediated TNF-α production in rodent models (7,42). In activated macrophages, a major source of TNF, high levels of TNF-α are produced in response to bacteria or parasitic proteins (as well as to other stimuli) (43). We therefore tested whether feverfew extracts mediate the release of TNF-α stimulated by LPS. Pre-treatment of cells with feverfew extract dramatically reduced LPS-mediated TNF-α release in THP-1 cells in a dose dependent manner (Fig. 1). LPS-mediated TNF-α release in THP-1 cells treated with the 1× concentration (equivalent to 0.01% dry weight) of feverfew was 46% of control cells untreated with feverfew.
Figure 1.
Dose-dependent inhibition of LPS mediated production of TNF-α by feverfew extracts. For each concentration treatment, the level of TNF-α release is represented as a percentage of the control set at 100%. TNF-α release was significantly inhibited (46% of control) by 1X Feverfew (0.01%, w/v, dry weight). **denotes P < 0.01 (Dunnett's Multiple Comparison Test), compared to control. The bar heights represent the values of mean ± S.D. (standard deviation) from three independent ELISA experiments. Control (Ctrl) cells received only solvent.
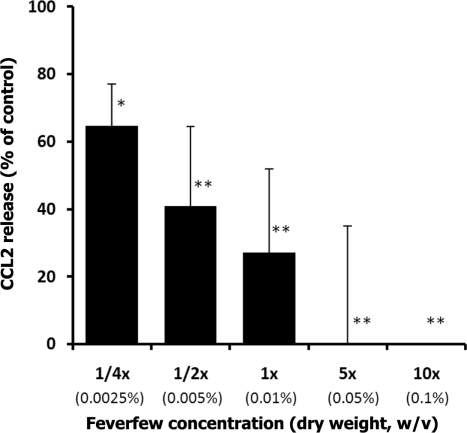
CCL2, (also known as monocyte chemoattractant protein 1, MCP-1), appears in the list of feverfew-responsive genes. We have observed that CCL2 is among the genes whose expression is highly inhibited by the feverfew extract (mRNA expression 40% of controls). To determine whether inhibition of CCL2 expression is reflected at the protein level, we studied LPS-mediated CCL2 release by THP-1 cells. As expected, LPS-mediated CCL2 release by THP-1 was strongly reduced in a dose-dependent manner (Fig. 2). LPS-mediated CCL2 production was 27% of control in cells treated with the 1× concentration (0.01% dry weight) of feverfew extract. At the 5×concentration of feverfew, no CCL2 production was detected.
Figure 2.
Dose-dependent inhibition of LPS mediated production of CCL2 by feverfew extracts. For each concentration treatment, the level of CCL2 release is represented as a percentage of the control set at 100%. CCL2 release was significantly inhibited (27% of control) by 1X Feverfew (0.01%, w/v, dry weight). *denotes P < 0.05 significant level and **P < 0.01 significant level, compared to control (Dunnett's Multiple Comparison Test). The bar heights represent the values of mean ± S.D. (standard deviation) from three independent ELISA experiments. Control (Ctrl) cells received only solvent.
Discussion and Conclusions
Since monocytes/macrophages are key mediators of inflammation and widely distributed in the body (26–28), monocytic cell lines present an appropriate model system to study immune responses to feverfew extracts. We initiated a genomic approach (microarray analysis) to search for molecular mechanisms by which feverfew affects functions in human monocytic cells (THP-1). It should be noted that among the 400 genes displaying a differential response to feverfew in this study, only about half (197 genes) contained annotated functions. In spite of this limitation, it is clear that most of the identified genes are primarily involved in cellular metabolism: regulation of metabolism, nucleotide metabolism, alcohol (i.e. alkyl compounds containing a hydroxyl group) metabolism and lipid metabolism. One possible interpretation is that at the level of gene expression, feverfew in human monocytes may not primarily or directly target known anti-inflammatory or anti-infectious genes, but rather modulate pathways for regulation of cellular metabolism.
However, the pre-ponderance of cellular metabolism genes in the response pattern to feverfew by no means negates a role for feverfew in anti-inflammatory or other functions because cellular metabolism can impact other pathways, including inflammation. This is apparent in atherosclerosis, an inflammatory disease in which lipid metabolism, e.g. oxidative modifications of LDL in serum and lipid-laden macrophages, strongly impacts the progression of atherosclerosis (44–46). Many of the multiple feverfew functions (including some anti-inflammatory effects) may derive from downstream targets of diverse metabolic pathways. Another possibility is that the action of feverfew may be focused on a few key molecules/genes in anti-inflammatory pathways. For example, in our microarray analyses, responsiveness to feverfew was highest for HMOX-1 (Table 1). Previous studies from several laboratories suggest that HMOX-1 mediates signaling pathways related to NF-κB (47), a key component in inflammation.
Relevantly, our studies also suggest that feverfew extracts are potent inhibitors of two pro-inflammatory proteins, TNF-α and CCL2, in human THP-1 monocytes (Figs. 1and 2). While previous studies have suggested that parthenolide (a major constituent of feverfew) decreases TNF-α release in rodent models (7,42), to our knowledge, our study is the first to demonstrate the ability of feverfew extracts to reduce LPS-mediated TNF-α and CCL2 release in human monocytes. It should be noted that the reduction by feverfew of LPS-induced TNF-α release and LPS-induced CCL2 release probably occurs by different mechanisms. Feverfew extracts exert an inhibitory effect on LPS-induced TNF-α release (Fig. 1), but do not appear to regulate the gene expression of TNF-α. In contrast, feverfew inhibits transcription of the CCL2 gene (Table 1) as well as LPS-induced CCL2 release.
Monocytes/macrophages are major producers of diverse cytokines and chemokines (48,49). It is likely that TNF-α and CCL2 represent only a very small fraction of cytokines and chemokines targeted by feverfew and that the different effects of feverfew in the human body may be due to its ability to target cytokines and chemokines of diverse functions (including inflammatory or anti-inflammatory effects).
Our cytokine and chemokine assays were based on the stimulation of monocytes by LPS; a strong agonist for toll-like receptor (TLR, especially TLR4) – mediated signaling. Previous studies on other herbs have suggested a role for TLRs in immune gene responses to herbal medicine (50–52). It would be fruitful to investigate whether TLRs play a role in mediating the effect of feverfew in human cells.
In addition to CCL2, a gene related to chemokine function, genes showing the greatest changes after feverfew exposure include genes related to several complex pathways. Heme regulation (HMOX1), chemokine function (IL-8, and CMKOR1) and several heat shock proteins (HSPA1A, DNAJB4, DNAJB1, HSPH1 and HSPA6) are highly elevated following feverfew treatment. In contrast, expression of CCL2 and AQP1, a gene involved in water homeostasis, is decreased following feverfew treatment.
Of note, a common theme among the functions of the AQP1, CCL2 and HMOX-1 is involvement in cell migration. AQP1 (Aquaporin 1) not only functions as a molecular water channel protein (53) but also localizes to the leading edge of the cell membrane in many cells (54). Cell migration of AQP1-deficient endothelial cells is significantly slower than in WT cells (55).
HMOX-1 (heme oxygenase 1, also called HO-1) not only is an essential enzyme in heme catabolism but also plays a critical role in cell migration. HMOX-1 can mediate cell migration by regulating the expression of adhesion molecules. It has been shown that HMOX-1 induction down-modulates H2O2-mediated induction of P-selectin and subsequently decreases leukocyte binding in vivo (56). HMOX-1 induction also inhibits microvascular leukocyte adhesion through the action of its metabolites, bilirubin and CO (57–59). In addition, increase of HMOX-1 expression inhibits leukocyte infiltration in an in vivo mouse model of inflammatory angiogenesis (60). Similarly, higher expression of HMOX-1 correlates with decreased migration of peritoneal macrophages and bone marrow cells harvested from morphine-receiving mice (61).
The CCL2 protein is a CC chemokine that mediates monocyte recruitment and entry into vessel walls at sites of atherosclerosis (46). CCL2 is produced by multiple cell types, including macrophages, endothelial cells and smooth muscle cells, all of which are important players in atherogenesis (60). In addition, CCL2 may play a role in multiple sclerosis (MS) (61) and in cancer pathobiology (62). Interestingly, the reduction of CCL2 expression might well be a direct result of HMOX-1. In human U937 monocytes, increase of HMOX-1 by hemin is correlated with the suppression of CCL2 mRNA expression (62). Similarly, in other cell types over-expression of HMOX-1 is usually accompanied by reduction of CCL2 (63,64). Together these data suggest that these three genes represent a possible pathway by which feverfew acts on cell migration.
In summary, this study utilized a human monocytic cell line as a model system to identify gene responses to feverfew. We are aware that the THP-1 cell line was established from a single individual and that there is genetic variability of the gene responses to feverfew extracts in human populations. We are therefore preparing to confirm the effects of feverfew on select genes in vivo. It is important to establish feverfew concentrations in blood samples of patients undertaking feverfew treatments and to determine concentrations of feverfew effective for modulating gene responses in vivo.
Supplementary Data
Supplementary data are available at eCAM Online.
Acknowledgement
We are indebted to Dr Zena Indik at the University of Pennsylvania School of Medicine for critically reading of the manuscript. We thank Dr Albert Leung and Shannon Ehlers at Phyto-Technologies for preparation of feverfew extracts.
References
- 1.Knight DW. Feverfew: chemistry and biological activity. Nat Prod Rep. 1995;12:271–6. doi: 10.1039/np9951200271. [DOI] [PubMed] [Google Scholar]
- 2.Pittler MH, Ernst E. Feverfew for preventing migraine. Cochrane Database Syst Rev. 2004;CD002286 doi: 10.1002/14651858.CD002286.pub2. [DOI] [PubMed] [Google Scholar]
- 3.Diener HC, Pfaffenrath V, Schnitker J, Friede M, Henneicke-von Zepelin HH. Efficacy and safety of 625 mg tid feverfew CO2-extract MIG-99 in migraine prevention–a randomized double-blind multicentre placebo-controlled study. Cephalalgia. 2005;25:1031–41. doi: 10.1111/j.1468-2982.2005.00950.x. [DOI] [PubMed] [Google Scholar]
- 4.Klepser TB, Klepser ME. Unsafe and potentially safe herbal therapies. Am J Health Syst Pharm. 1999;56:125–38. doi: 10.1093/ajhp/56.2.125. quiz 139–41. [DOI] [PubMed] [Google Scholar]
- 5.Ross JJ, Arnason JT, Birnboim HC. Low concentrations of the feverfew component parthenolide inhibit in vitro growth of tumor lines in a cytostatic fashion. Planta Med. 1999;65:126–9. doi: 10.1055/s-1999-13972. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Ong CN, Shen HM. Critical roles of intracellular thiols and calcium in parthenolide-induced apoptosis in human colorectal cancer cells. Cancer Lett. 2004;208:143–53. doi: 10.1016/j.canlet.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 7.Hwang D, Fischer NH, Jang BC, Tak H, Kim JK, Lee W. Inhibition of the expression of inducible cyclooxygenase and proinflammatory cytokines by sesquiterpene lactones in macrophages correlates with the inhibition of MAP kinases. Biochem Biophys Res Commun. 1996;226:810–18. doi: 10.1006/bbrc.1996.1433. [DOI] [PubMed] [Google Scholar]
- 8.Mazor RL, Menendez IY, Ryan MA, Fiedler MA, Wong HR. Sesquiterpene lactones are potent inhibitors of interleukin 8 gene expression in cultured human respiratory epithelium. Cytokine. 2000;12:239–45. doi: 10.1006/cyto.1999.0526. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill LA, Barrett ML, Lewis GP. Extracts of feverfew inhibit mitogen-induced human peripheral blood mononuclear cell proliferation and cytokine mediated responses: a cytotoxic effect. Br J Clin Pharmacol. 1987;23:81–3. doi: 10.1111/j.1365-2125.1987.tb03012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pugh WJ, Sambo K. Prostaglandin synthetase inhibitors in feverfew. J Pharm Pharmacol. 1988;40:743–5. doi: 10.1111/j.2042-7158.1988.tb07010.x. [DOI] [PubMed] [Google Scholar]
- 11.Sumner H, Salan U, Knight DW, Hoult JR. Inhibition of 5-lipoxygenase and cyclo-oxygenase in leukocytes by feverfew Involvement of sesquiterpene lactones and other components. Biochem Pharmacol. 1992;43:2313–20. doi: 10.1016/0006-2952(92)90308-6. [DOI] [PubMed] [Google Scholar]
- 12.Smolinski AT, Pestka JJ. Comparative effects of the herbal constituent parthenolide Feverfew on lipopolysaccharide-induced inflammatory gene expression in murine spleen and liver. J Inflamm Lond. 2005;2:6. doi: 10.1186/1476-9255-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Weerdt CJ, Bootsma HPR, Hendriks H. Herbal medicines in migraine prevention randomized double-blind placebo-controlled crossover trial of a feverfew preparation. Phytomedicine. 1996;3:225–30. doi: 10.1016/S0944-7113(96)80057-2. [DOI] [PubMed] [Google Scholar]
- 14.Adhami VM, Ahmad N, Mukhtar H. Molecular targets for green tea in prostate cancer prevention. J Nutr. 2003;133:S2417–24. doi: 10.1093/jn/133.7.2417S. [DOI] [PubMed] [Google Scholar]
- 15.Nelson PS. Predicting prostate cancer behavior using transcript profiles. J Urol. 2004;172:S28–32. doi: 10.1097/01.ju.0000142067.17181.68. discussion S33. [DOI] [PubMed] [Google Scholar]
- 16.Cleator S, Ashworth A. Molecular profiling of breast cancer: clinical implications. Br J Cancer. 2004;90:1120–24. doi: 10.1038/sj.bjc.6601667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greiner TC. mRNA microarray analysis in lymphoma and leukemia. Cancer Treat Res. 2004;121:1–12. doi: 10.1007/1-4020-7920-6_1. [DOI] [PubMed] [Google Scholar]
- 18.Meyerson M, Franklin WA, Kelley MJ. Molecular classification and molecular genetics of human lung cancers. Semin Oncol. 2004;31:4–19. doi: 10.1053/j.seminoncol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Cheek DJ, Cesan A. Genetic predictors of cardiovascular disease: the use of chip technology. J Cardiovasc Nurs. 2003;18:50–6. doi: 10.1097/00005082-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Napoli C, Lerman LO, Sica V, Lerman A, Tajana G, de Nigris F. Microarray analysis: a novel research tool for cardiovascular scientists and physicians. Heart. 2003;89:597–604. doi: 10.1136/heart.89.6.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryant PA, Venter D, Robins-Browne R, Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect Dis. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- 22.Shioda T. Application of DNA microarray to toxicological research. J Environ Pathol Toxicol Oncol. 2004;23:13–31. doi: 10.1615/jenvpathtoxoncol.v23.i1.20. [DOI] [PubMed] [Google Scholar]
- 23.Page GP, Edwards JW, Barnes S, Weindruch R, Allison DB. A design and statistical perspective on microarray gene expression studies in nutrition: the need for playful creativity and scientific hard-mindedness. Nutrition. 2003;19:997–1000. doi: 10.1016/j.nut.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Bigler D, Gulding KM, Dann R, Sheabar FZ, Conaway MR, Theodorescu D. Gene profiling and promoter reporter assays: novel tools for comparing the biological effects of botanical extracts on human prostate cancer cells and understanding their mechanisms of action. Oncogene. 2003;22:1261–72. doi: 10.1038/sj.onc.1206242. [DOI] [PubMed] [Google Scholar]
- 25.Chavan P, Joshi K, Patwardhan B. DNA microarrays in herbal drug research. Evid Based Complement Alternat Med. 2006;3:447–57. doi: 10.1093/ecam/nel075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbas AK, Lichtman AH. Cellular and Molecular Immunology. Philadelphia: WB Saunders; 2003. [Google Scholar]
- 27.Roitt IM, Delves PJ. Roitt's Essential Immunology. Oxford: Blackwell Science Inc.; 2001. [Google Scholar]
- 28.Gilroy DW, Lawrence T, Perretti M, Rossi AG. Inflammatory resolution: new opportunities for drug discovery. Nat Rev Drug Discov. 2004;3:401–6. doi: 10.1038/nrd1383. [DOI] [PubMed] [Google Scholar]
- 29.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line THP-1. Int J Cancer. 1980;26:171–6. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 30.Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32(Suppl):490–5. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- 31.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–7. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 32.Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 33.Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat Rev Genet. 2002;3:579–88. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- 34.Leung YF, Cavalieri D. Fundamentals of cDNA microarray data analysis. Trends Genet. 2003;19:649–59. doi: 10.1016/j.tig.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 36.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–42. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- 37.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–45. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology GO database and informatics resource. Nucleic Acids Res. 2004;32:D258–61. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–80. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 42.Smolinski AT, Pestka JJ. Modulation of lipopolysaccharide-induced proinflammatory cytokine production in vitro and in vivo by the herbal constituents apigenin chamomile ginsenoside Rb1 ginseng and parthenolide feverfew. Food Chem Toxicol. 2003;41:1381–90. doi: 10.1016/s0278-6915(03)00146-7. [DOI] [PubMed] [Google Scholar]
- 43.Russo C, Polosa R. TNF-alpha as a promising therapeutic target in chronic asthma: a lesson from rheumatoid arthritis. Clin Sci Lond. 2005;109:135–42. doi: 10.1042/CS20050038. [DOI] [PubMed] [Google Scholar]
- 44.Glass CK, Witztum JL. Atherosclerosis the road ahead. Cell. 2001;104:503–16. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 45.Daugherty A, Webb NR, Rateri DL, King VL. Thematic review series: The immune system and atherogenesis Cytokine regulation of macrophage functions in atherogenesis. J Lipid Res. 2005;46:1812–22. doi: 10.1194/jlr.R500009-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Hansson GK. Inflammation atherosclerosis and coronary artery disease. N Engl J Med. 2005;352:1685–95. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 47.Maines MD, Gibbs PE. 30 some years of heme oxygenase: from a “molecular wrecking ball” to a “mesmerizing” trigger of cellular events. Biochem Biophys Res Commun. 2005;338:568–77. doi: 10.1016/j.bbrc.2005.08.121. [DOI] [PubMed] [Google Scholar]
- 48.Ross JA, Auger MJ. The biology of the macrophages. In: Burke B, Lewis CE, editors. The Macrophage. Oxford: Oxford University Press; 2002. [Google Scholar]
- 49.Janeway CA, Travers P, Walport M, Shlomchik MJ. Immunobiology. New York: Garland Science Publishing; 2005. [Google Scholar]
- 50.Kasai H, He LM, Kawamura M, Yang PT, Deng XW, Munkanta M, et al. IL-12 Production Induced by Agaricus blazei Fraction H ABH Involves Toll-like Receptor TLR. Evid Based Complement Alternat Med. 2004;1:259–67. doi: 10.1093/ecam/neh043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon YD, Han SB, Kang JS, Lee CW, Park SK, Lee HS, et al. Toll-like receptor 4-dependent activation of macrophages by polysaccharide isolated from the radix of Platycodon grandiflorum. Int Immunopharmacol. 2003;3:1873–82. doi: 10.1016/j.intimp.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 52.Cooper EL. Role of TOLL-like Receptors in Adjuvant-Augmented Immune Therapies by T Seya. Evid Based Complement Alternat Med. 2006;3:133–7. doi: 10.1093/ecam/nek010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.King LS, Kozono D, Agre P. From structure to disease: the evolving tale of aquaporin biology. Nat Rev Mol Cell Biol. 2004;5:687–98. doi: 10.1038/nrm1469. [DOI] [PubMed] [Google Scholar]
- 54.Schwab A. Function and spatial distribution of ion channels and transporters in cell migration. Am J Physiol Renal Physiol. 2001;280:F739–47. doi: 10.1152/ajprenal.2001.280.5.F739. [DOI] [PubMed] [Google Scholar]
- 55.Saadoun S, Papadopoulos MC, Hara-Chikuma M, Verkman AS. Impairment of angiogenesis and cell migration by targeted aquaporin-1 gene disruption. Nature. 2005;434:786–92. doi: 10.1038/nature03460. [DOI] [PubMed] [Google Scholar]
- 56.Hayashi S, Takamiya R, Yamaguchi T, Matsumoto K, Tojo SJ, Tamatani T, et al. Induction of heme oxygenase-1 suppresses venular leukocyte adhesion elicited by oxidative stress: role of bilirubin generated by the enzyme. Circ Res. 1999;85:663–71. doi: 10.1161/01.res.85.8.663. [DOI] [PubMed] [Google Scholar]
- 57.Morisaki H, Katayama T, Kotake Y, Ito M, Handa M, Ikeda Y, et al. Carbon monoxide modulates endotoxin-induced microvascular leukocyte adhesion through platelet-dependent mechanisms. Anesthesiology. 2002;97:701–9. doi: 10.1097/00000542-200209000-00025. [DOI] [PubMed] [Google Scholar]
- 58.Zampetaki A, Minamino T, Mitsialis SA, Kourembanas S. Effect of heme oxygenase-1 overexpression in two models of lung inflammation. Exp Biol Med Maywood. 2003;228:442–46. doi: 10.1177/15353702-0322805-02. [DOI] [PubMed] [Google Scholar]
- 59.Keshavan P, Deem TL, Schwemberger SJ, Babcock GF, Cook-Mills JM, Zucker SD. Unconjugated bilirubin inhibits VCAM-1-mediated transendothelial leukocyte migration. J Immunol. 2005;174:3709–18. doi: 10.4049/jimmunol.174.6.3709. [DOI] [PubMed] [Google Scholar]
- 60.Bussolati B, Ahmed A, Pemberton H, Landis RC, Di Carlo F, Haskard DO, et al. Bifunctional role for VEGF-induced heme oxygenase-1 in vivo: induction of angiogenesis and inhibition of leukocytic infiltration. Blood. 2004;103:761–66. doi: 10.1182/blood-2003-06-1974. [DOI] [PubMed] [Google Scholar]
- 61.Patel K, Bhaskaran M, Dani D, Reddy K, Singhal PC. Role of heme oxygenase-1 in morphine-modulated apoptosis and migration of macrophages. J Infect Dis. 2003;187:47–54. doi: 10.1086/346042. [DOI] [PubMed] [Google Scholar]
- 62.Shokawa T, Yoshizumi M, Yamamoto H, Omura S, Toyofuku M, Shimizu Y, et al. Induction of heme oxygenase-1 inhibits monocyte chemoattractant protein-1 mRNA expression in U937 cells. J Pharmacol Sci. 2006;100:162–66. doi: 10.1254/jphs.sc0040188. [DOI] [PubMed] [Google Scholar]
- 63.Abraham NG, Scapagnini G, Kappas A. Human heme oxygenase: cell cycle-dependent expression and DNA microarray identification of multiple gene responses after transduction of endothelial cells. J Cell Biochem. 2003;90:1098–111. doi: 10.1002/jcb.10736. [DOI] [PubMed] [Google Scholar]
- 64.Sacerdoti D, Colombrita C, Ghattas MH, Ismaeil EF, Scapagnini G, Bolognesi M, et al. Heme oxygenase-1 transduction in endothelial cells causes downregulation of monocyte chemoattractant protein-1 and of genes involved in inflammation and growth. Cell Mol Biol Noisy-le-grand. 2005;51:363–70. [PubMed] [Google Scholar]