Abstract
Nuclear factor erythroid 2–related factor 2 (Nrf2) is a transcription factor critical for protection against electrophilic and oxidative stress. In a recently engineered mouse with knockdown of kelch-like ECH associated protein 1 (Keap1-kd mice), the cytosolic repressor of Nrf2, there is a 55% decrease in Keap1 mRNA and a 200% increase in Nrf2 protein in liver. Experiments with Nrf2-null mice have demonstrated the effects of a lack of Nrf2. However, little is known about the biological effects of more Nrf2 activation. Accordingly, the hepatic phenotype of Keap1-kd mice, as well as the hepatic mRNA expression of cytoprotective genes were compared among wild-type, Nrf2-null, and Keap1-kd mice. Three distinct patterns of hepatic gene expression were identified among wild-type, Nrf2-null, and Keap1-kd mice. The first pattern encompassed genes that were lower in Nrf2-null mice and considerably higher in Keap1-kd mice than wild-type mice, which included genes mainly responsible for the detoxification and elimination of electrophiles, such as NAD(P)H:quinone oxidoreductase 1 and glutathione-S-transferases (Gst), and multidrug resistance–associated proteins. The second pattern encompassed genes that were lower in Nrf2-null mice but not increased in Keap1-kd mice, and included genes, such as epoxide hydrolase-1, UDP-glucuronosyltransferases, aldehyde dehydrogenases, as well as genes important in the detoxification of reactive oxygen species, such as superoxide dismutase 1 and 2, catalase, and peroxiredoxin 1. The third pattern encompassed genes that were not different among wild-type, Nrf2-null, and Keap1-kd mice and included genes such as glutathione peroxidase, microsomal Gsts, and uptake transporters. In conclusion, the present study suggests that increased activation of hepatic Nrf2 is more important for the detoxification and elimination of electrophiles than reactive oxygen species.
Keywords: Nrf2, electrophilic stress, oxidative stress, cytoprotection
Oxidative and electrophilic stresses are implicated in the development of numerous diseases. Oxidative stress is caused by increased production of reactive oxygen and nitrogen species, such as superoxide, hydroxyl radical, and peroxynitrite, or by depletion of protective antioxidants, such as reduced glutathione (GSH), ascorbate, and α-tocopherol. Oxidative stress leads to oxidative damage to critical macromolecules. Superoxide is enzymatically detoxified by first being converted to hydrogen peroxide and then to water by catalase (Cat), GSH peroxidase (Gpx), or peroxiredoxin (Prx) (Fig. 1, left side). Detoxification of the hydroxyl radical is not usually possible because of its short half-life (10−9 s), and thus prevention of its formation by detoxification of its precursor hydrogen peroxide by Cat, Gpx, or Prx is necessary. Detoxification of peroxynitrite to nitrite is carried out by Gpx and Prx; GSH may also be capable of reducing perxoxynitrite to nitrite (Harwood et al., 2006; Trujillo et al., 2008).
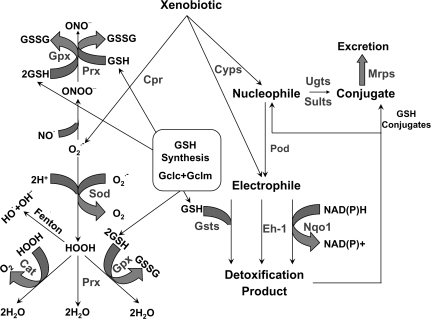
FIG. 1.
Illustration of oxidative and electrophilic stress. Oxidative stress is represented on the left side and electrophilic stress on the right side. Xenobiotics can undergo redox cycling, such as in the case of paraquat, which generates superoxide in a reaction catalyzed by cytochrome p-450 reductase (Cpr). Detoxificaton of superoxide (O2−.) to hydrogen peroxide (HOOH) is mediated by Sod. HOOH is detoxified to H2O by Cat, Prx, and Gpx. GSH is oxidized to glutathione disulfide (GSSG) in the Gpx-catalyzed reaction. If HOOH is not detoxified, it can undergo a Fenton reaction to generate hydroxyl radical (OH.). O2−. can also react with nitric oxide (NO.) to form peroxynitrite (ONOO−), which can be detoxified by Gpx, Prx, or GSH. Biotransformaton of xenobiotics by Cyps and Pod can lead to the formation of nucleophiles or electrophiles. Detoxification of electrophilic intermediates by Nqo1 or Eh-1 creates nucleophiles, which can be conjugated with glucuronic acid via Ugts or conjugated with sulfate via Sults and excreted by Mrps. Electrophiles can also be conjugated with GSH in a reaction catalyzed by Gsts and excreted via Mrps. GSH and its synthesis play important roles in both superoxide and electrophile detoxification. The rate-limiting enzymes in GSH synthesis are glutamate-cysteine ligase catalytic and modifier subunits (Gclc and Gclm, respectively).
Electrophilic stress is caused by effects of reactive metabolites, such as the cytochrome p-450 (Cyp)–mediated formation of N-acetyl-p-benzo-quinoneimine (NAPQI) in acetaminophen hepatotoxicity, or the epoxide formed in benzo[a]pyrene-mediated carcinogenesis. Electrophiles cause injury by covalently binding to and damaging critical macromolecules. Detoxification of reactive intermediates or electrophiles typically occurs through enzymatic processes, such as conjugation with GSH by glutathione-S-transferases (Gsts), reduction of quinones by NAD(P)H:quinone oxidoreductase 1 (Nqo1), or hydrolysis of epoxides by epoxide hydrolase-1 (Eh-1) (Fig. 1, right side). Nucleophilic conjugation mediated by UDP-glucuronosyltransferases (Ugts) or sulfotransferases (Sults) can also prevent the formation of electrophiles by limiting xenobiotic availability for toxification by Cyps or peroxidases (Pods).
Nuclear factor erythroid 2–related factor 2 (Nrf2) is an important transcriptional regulator of antioxidative cellular protection. Upon cellular oxidative/electrophilic insult, Nrf2 initiates a response by upregulating a battery of cytoprotective genes, such as Nqo1, glutamate-cysteine ligase, catalytic and modifier subunits (Gclc and Gclm), heme oxygenase-1 (Ho-1), and drug processing genes, such Gsts, Ugts, and multidrug resistance–associated proteins (Mrps) (Kobayashi and Yamamoto, 2006; Maher et al., 2007; Venugopal and Jaiswal, 1996). Moreover, inductive control over a broad array of genes that facilitate increased detoxification and clearance of reactive metabolites and xenobiotics has earned Nrf2 a crucial role in toxicology and xenobiotic metabolism.
Under physiological conditions, Nrf2 is bound in the cytosol by its repressor Kelch-like ECH associating protein 1 (Keap1). Keap1 functions as an adapter for Cullin 3–based E3 ligase, a scaffold protein that aids in the ubiquitination and subsequent degradation of Nrf2 (Cullinan et al., 2004; Kobayashi et al., 2004). In fact, Nrf2 has a very rapid turnover, with a half-life of approximately 20 min, and thus Nrf2 protein is difficult to detect in unstressed conditions (Itoh et al., 2003; McMahon et al., 2003). When an oxidative or electrophilic insult occurs, reactive cysteines of Keap1 are modified, and Keap1 is unable to target Nrf2 for proteasomal degradation (Dinkova-Kostova et al., 2002). Nrf2 then translocates into the nucleus, heterodimerizes with a small musculo-aponeurotic fibrosarcoma protein, and binds to antioxidant response elements in the upstream promoter region of a variety of cytoprotective genes, promoting their transcription (Itoh et al., 1997).
Nrf2-null mice have lower constitutive expression and an inability to induce cytoprotective genes, such as Nqo1 and Gclc, upon oxidative/electrophlic insult. Nrf2-null mice are extremely susceptible to chemical models of oxidative and electrophilic stress (Aleksunes and Manautou, 2007), contributing to increased hepatotoxicity when administered acetaminophen (Enomoto et al., 2001), ethanol (Lamle et al., 2008), pentachlorophenol (Umemura et al., 2006), or a high-fat diet (Tanaka et al., 2008b). In contrast to Nrf2-null mice, Keap1-null mice were engineered to investigate the effects of increased activation of Nrf2. Unfortunately, even though Keap1-null mice have enhanced activation of Nrf2 and higher constitutive expression of Nrf2-target genes, the mice died at weaning due to malnutrition from hyperkeratosis of the esophagus and forestomach (Wakabayashi et al., 2003). In an attempt to circumvent the postnatal lethality in Keap1-null mice, a hepatocyte-specific Keap1-null mouse was engineered, utilizing an Albumin-Cre loxP system. This mouse is viable, has enhanced activation of Nrf2 and Nrf2-target genes in liver, and decreased susceptibility to acetaminophen hepatotoxicity (Okawa et al., 2006). However, later it was discovered that mice homozygous for Keap1 loxP sites (no Albumin-Cre transgene) have decreased or a “knockdown” of Keap1 (Keap1-kd), leading to increased activation of Nrf2 in multiple organs (Okada et al., 2008).
A considerable amount of knowledge has accumulated on the effect of a lack of Nrf2 by doing experiments in Nrf2-null mice. However, very little is known about the biological effects of increased Nrf2 expression. Therefore, the purpose of this study was to determine the hepatic phenotype of Keap1-kd mice, as well as the mRNAs of genes whose protein products participate in xenobiotic metabolism and/or are thought to protect against oxidative and electrophilic stress, and compare them with Nrf2-null and wild-type mice. The model used in the present study for increased activation of Nrf2 is the Keap1-knockdown (Keap1-kd) mouse. Both mouse models, Nrf2-null and Keap1-kd, have been backcrossed into C57BL/6 mice to > 99% congenicity, allowing for comparisons among wild-type, Nrf2-null, and Keap1-kd mice without the added variable of strain differences.
METHODS
Reagents.
Nrf2 antibody was purchased from Santa Cruz Biotechnology (sc-30915, Santa Cruz, CA). β-Actin antibody was purchased from Abcam (Ab8227, Cambrdige, MA). PolyADP-ribose Polymerase (PARP) antibody, originally purchased from BD Pharmingen (cat. # 556362, San Jose, CA), was a gift from Dr John Robertson (University of Kansas Medical Center, Kansas City, KS). All other chemicals, unless otherwise specified, were purchased from Sigma-Aldrich (St Louis, MO).
Animals and husbandry.
Eight-week-old male mice were used for this study. Mice were euthanized by cervical dislocation, unless otherwise noted. C57BL/6 mice were purchased from Charles River Laboratories, Inc. (Wilmington, MA). Nrf2-null mice were obtained from Dr Jefferson Chan (University of California, Irvine, Irvine, CA) (Chan et al., 1996). Keap1-kd mice were supplied by Dr Masayuki Yamamoto (Tohoku University, Aoba-ku, Sendai, Japan) (Okada et al., 2008). In an attempt to make a hepatocyte-specific Keap1-null mouse, utilizing a loxP, Alb-Cre system (Okawa et al., 2006), a Keap1-kd mouse, in which Keap1 was decreased throughout the body, was engineered (Okada et al., 2008).
Nrf2-null and Keap1-kd mice were backcrossed into the C57BL/6 background, and > 99% congenicity was confirmed by the speed congenics group at Jackson Laboratories (Bar Harbor, ME). Animals were housed in a temperature-, light-, and humidity-controlled environment and had access to Teklad Rodent Diet #8604 (Harlan Laboratories, Madison, WI) and water ad libitum. The housing facility is an American Animal Associations Laboratory Animal Care-accredited facility at the University of Kansas Medical Center, and all procedures were preapproved in accordance with Institutional Animal Care and Use Committee guidelines.
Bile collection.
Wild-type, Nrf2-null, and Keap1-kd mice (n = 5) were anesthetized by injection of ketamine/midazolam (100 and 5 mg/kg, respectively, i.p.). Body temperature was maintained at 37°C by rectal-probe controlled heating pads. Subsequently, the common bile duct was cannulated with the shaft of a 30-gauge needle attached to PE-10 tubing through a high abdominal incision. Bile was collected for 15 min into preweighed 0.6-ml microcentrifuge tubes and immersed in ice. The volumes of bile samples were determined gravimetrically, using 1.0 as specific gravity for bile.
Serum and bile enzyme analyses.
Glucose, alanine transaminase (ALT), triglycerides, cholesterol, phospholipids, and total bilirubin concentrations were quantified spectrophotometrically using commercially available kits (Pointe Scientific, Canton, MI).
Hepatic enzyme analyses.
Livers were collected from wild-type, Nrf2-null, and Keap1-kd mice (n = 5) in the morning (prior to 11:00 A.M.). Livers were homogenized in ice-cold 10mM sucrose–250mM Tris buffer (pH 7.5), and microsomal suspensions were prepared by differential centrifugation. Microsomal protein concentrations were determined by bicinchoninic acid (BCA) Assay (Pierce Biotechnology, Rockford, IL). Microsomal cytochrome p450 (Cyp) activities were determined by XenoTech, LLC (Lenexa, KS) using established methods. Briefly, Cyp1a1 and 2b1 activities were quantified by fluorimetric analysis of 7-ethoxyresorufin O-deethylation and 7-pentoxyresorufin O-deethylation, respectively. Cyp2e1 activity was determined by spectrophotometric analysis of 4-nitrophenol hydroxylation. Lastly, Cyp3a11 and 4a14 activities were measured by liquid chromatography-mass spectrometry/mass spectrometry analysis of testosterone 6β-hydroxylation and lauric acid 12-hydroxylation, respectively.
Cytosolic extracts were prepared with the NE-PER nuclear extraction kit according to the manufacturer's directions (Pierce Biotechnology). Protein concentrations were determined by the BCA Assay (Pierce Biotechnology). Hepatic GPx and Cat activity in cytosolic fractions were quantified spectrophotometrically, as adapted from previously described methods (Flohe and Gunzler, 1984; Luck, 1963; Tappel, 1978). Superoxide dismutase (Sod) activity was quantified spectrophotometrically utilizing a commercially available kit (Cayman Chemical, Ann Arbor, MI).
GSH quantification.
Reduced GSH in livers and bile were quantified utilizing the GSH-Glo Glutathione Assay from Promega (Madison, WI) according to their protocol.
Nrf2 protein expression in hepatic nuclear extracts.
Nuclear extracts were prepared with the NE-PER nuclear extraction kit according to their protocol (Pierce Biotechnology). Protein concentrations were determined with the BCA Assay Kit from Pierce Biotechnology. Nuclear proteins (40 μg protein per lane) were electrophoretically resolved using polyacrylamide gels (4% stacking and 10% resolving). Gels were transblotted overnight at 4°C onto polyvinylidene fluoride membranes. Membranes were then washed with PBS-buffered saline containing 0.05% Tween-20 (PBS-T). Membranes were blocked for 1 h at room temperature with 5% nonfat milk in PBS-T. Blots were then incubated with primary antibody (1:1000 dilution in 2% nonfat milk in PBS-T) for 3 h at room temperature. Blots were washed in PBS-T and incubated with species-appropriate secondary antibody conjugated with horseradish peroxidase (1:2000 dilution) in 2% nonfat milk in PBS-T buffer for 1 h at room temperature. Blots were then washed with PBS-T. Protein-antibody complexes were detected using an ECL chemiluminescent kit (Pierce Biotechnology) and exposed to X-ray film (Denville Scientific, Metuchen, NJ). Equal protein loading was confirmed by β-actin and PARP. Nrf2, β-actin, and PARP proteins migrated the same distance as proteins of approximately 110, 45, and 116 kDa, respectively. Intensity of protein bands was quantified by Discovery Series Quantity One 1-D Analysis Software (Bio-Rad Laboratories, Hercules, CA). Individual blot densities were normalized to that of wild-type mice.
Quantification of hepatic nonesterified fatty acids, free cholesterol, and triglycerides.
Lipids were extracted from livers as described previously (Tanaka et al., 2008b). In brief, 100 mg of liver was homogenized in 1 ml of buffer containing 18mM Tris, pH 7.5, 300mM mannitol, 50mM ethylene glycol tetraacetic acid, and 0.1mM phenylmethylsulfonyl fluoride. Five hundred microliters of homogenate was mixed with 4 ml of chloroform/methanol (2:1) and incubated overnight at room temperature with occasional shaking. The next day, 1 ml of H2O was added, vortexed, and centrifuged for 5 min at 3000 × g. The lower lipid phase was collected and concentrated by vacuum. The lipid pellets were dissolved in PBS with 0.01% Triton X-100. Liver lipid samples were analyzed spectrophotometrically for free cholesterol (500 nm), nonesterified fatty acids (550 nm), and triglycerides (500 nm) using commercially available kits (Pointe Scientific, Canton, MI and Wako Diagnostics, Richmond, VA).
Messenger RNA quantification.
Mouse liver mRNA expression was determined from liver tissue homogenates prepared as described by the manufacturer's protocol utilizing Quantigene Plex 2.0 Technology (Panomics, Fremont, CA). Individual bead-based oligonucleotide probe sets, specific for each gene examined, were developed by Panomics, Inc. Genes and accession numbers are freely available at http://www.panomics.com. Samples were analyzed using a Bio-Plex 200 System Array reader with Luminex 100 X-MAP technology, and data were acquired using Bio-Plex Data Manager Software Version 5.0 (Bio-Rad). Assays were performed according to the manufacturer's protocol. The mRNA of Cyp2b10 and Mrp4 was quantified using liver tissue homogenates with Quantigene 1.0 Technology (Panomics), according to previously described methods (Maher et al., 2005; Petrick and Klaassen, 2007). All data were standardized to the internal control glyceraldehyde 3-phosphate dehydrogenase.
Statistical analysis.
All data were analyzed using one-way analysis of variance (ANOVA) followed by Duncan's multiple range test (p ≤ 0.05).
RESULTS
Nrf2 Protein in Hepatic Nuclear Fractions and Hepatic mRNA Expression of Nrf2 and Keap1
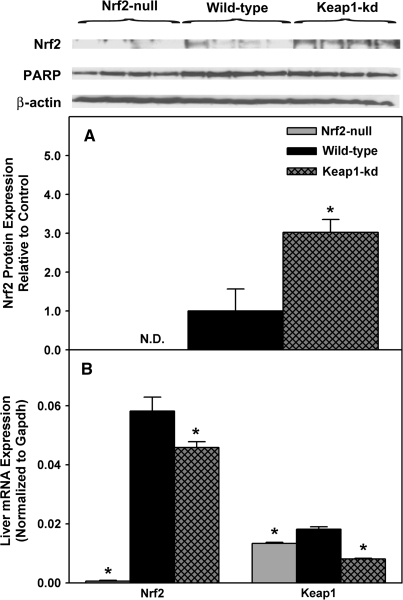
Nrf2 protein in hepatic nuclear fractions of wild-type, Nrf2-null, and Keap1-kd mice was quantified using immunoblotting (Fig. 2A). As expected, Nrf2 protein was not detected in Nrf2-null mice. However, Nrf2 protein in hepatic nuclear fractions from Keap1-kd mice was 200% higher than wild-type mice. Both β-actin and PARP were used to confirm equal loading.
FIG. 2.
Nrf2 protein expression in hepatic nuclear fractions in wild-type, Nrf2-null, and Keap1-kd mice (n = 5) (A). Intensity of protein bands was quantified, and individual blot densities were normalized to wild-type and expressed as mean ± SEM. Messenger RNA expression of Nrf2 and Keap1 in wild-type, Nrf2-null, and Keap1-kd mice (n = 5) (B). Values are expressed as mean ± SEM Asterisks (*) indicate a statistically significant difference from wild-type mice (p ≤ 0.05).
Nrf2 mRNA expression was not detected in Nrf2-null mice and was 22% lower in Keap1-kd mice than in wild-type mice (Fig. 2B). Keap1 mRNA expression was lower in Nrf2-null (-24%) and Keap1-kd mice (-55%) than in wild-type mice.
Serum, Liver, and Bile Parameters
Various serum, liver, and bile parameters of hepatic function in the three genotypes of mice were quantified (Table 1). Serum ALT, glucose, triglyceride, and cholesterol concentrations were not different among genotypes, neither were hepatic histology (data not shown), nonesterified fatty acids, free cholesterol, nor triglycerides. Bile flow tended to be lower in Nrf2-null mice and was higher in Keap1-kd mice. The biliary excretion of GSH was 42% lower in Nrf2-null mice and 43% higher in Keap1-kd mice. The biliary excretion of total bilirubin was similar in the three genotypes. There were no differences in the biliary excretion of cholesterol and phospholipids between Nrf2-null and wild-type mice. In contrast, the biliary excretion of cholesterol and phospholipids was higher in Keap1-kd mice than wild-type mice.
TABLE 1.
Serum, Liver, and Bile Parameters of Hepatic Function in Wild-Type, Nrf2-null, and Keap1-kd Mice
| Analyte (units) | Nrf2-null | Wild-type | Keap1-kd |
| Serum | |||
| ALTs (IU/l) | 20.9 ± 1.3 | 19.5 ± 1.6 | 22.7 ± 3.9 |
| Glucose (mg/dl) | 176 ± 6 | 185 ± 7 | 206 ± 10 |
| Triglycerides (mg/dl) | 66.0 ± 8.9 | 56.7 ± 4.9 | 64.1 ± 4.0 |
| Cholesterol (mg/dl) | 57.0 ± 1.5 | 65.8 ± 2.0 | 61.4 ± 5.4 |
| Liver | |||
| Nonesterified fatty acids (mEq/g liver) | 0.018 ± 0.002 | 0.025 ± 0.003 | 0.020 ± 0.003 |
| Free cholesterol (mg/g liver) | 19.0 ± 4.2 | 23.0 ± 4.2 | 26.3 ± 3.9 |
| Triglycerides (mg/g liver) | 34.2 ± 1.3 | 40.2 ± 7.2 | 36.9 ± 10.7 |
| Bile | |||
| Bile flow (ml/min/kg) | 106 ± 5 | 124 ± 1 | 167 ± 12* |
| GSH (nmol/min/kg) | 371 ± 24* | 644 ± 51 | 922 ± 54* |
| Bilirubin (mg/min/kg) | 7.57 ± 0.42 | 7.05 ± 0.52 | 6.65 ± 1.05 |
| Cholesterol (mg/min/kg) | 35.5 ± 0.6 | 40.1 ± 1.7 | 54.3 ± 3.9* |
| Phospholipids (mg/min/kg) | 37.0 ± 3.2 | 35.0 ± 2.6 | 54.34 ± 2.2* |
Note. Values are expressed as mean ± SEM. Asterisks (*) indicate a statistically significant difference from wild-type mice (p ≤ 0.05).
Hepatic GSH Concentrations
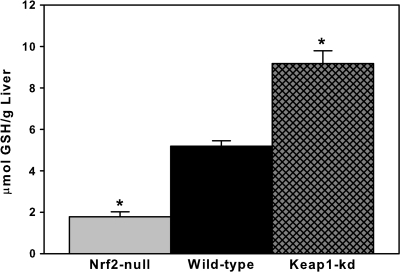
Gclc and Gclm are Nrf2-dependent genes important in the de novo synthesis of GSH (Moinova and Mulcahy, 1999). Therefore, reduced GSH content was quantified among the three genotypes (Fig. 3). Nrf2-null mice had 65% lower and Keap1-kd mice had 77% higher GSH concentrations in liver than wild-type mice.
FIG. 3.
Hepatic reduced liver GSH content in wild-type, Nrf2-null, and Keap1-kd mice (n = 5). Values are expressed as mean ± SEM. Asterisks (*) indicate a statistically significant difference from wild-type mice (p ≤ 0.05).
Hepatic mRNA Expression of Classical Oxidative and Electrophilic-Stress Genes
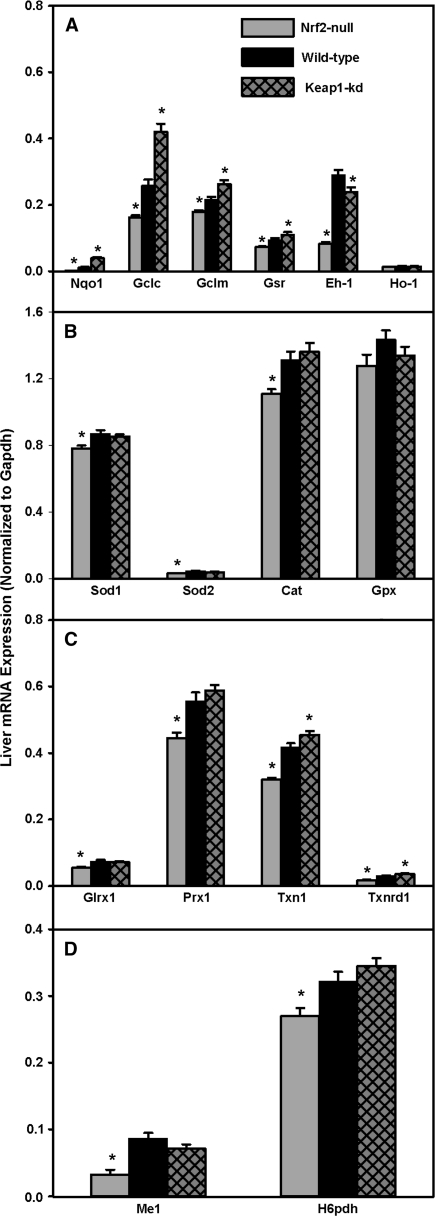
Hepatic mRNA expression of prototypical Nrf2-target genes is depicted in Figure 4A. The expression of mRNAs for Nqo1, Gclc, Gclm, and GSH reductase (Gsr) was lower in Nrf2-null mice (-88, -38, -16, and -23%, respectively) and higher in Keap1-kd mice (222, 63, 22, and 17%, respectively). Eh-1 was 72% lower in Nrf2-null mice and 18% lower in Keap1-kd mice. Ho-1 mRNA expression was not different among the three genotypes.
FIG. 4.
Messenger RNA expression of prototypical Nrf2 targets (A), superoxide and hydrogen peroxide reducing enzymes (B), redoxins (C), and NADPH generating enzymes (D) in wild-type, Nrf2-null, and Keap1-kd mice (n = 5). Values are expressed as mean ± SEM. Asterisks (*) indicate a statistically significant difference from wild-type mice (p ≤ 0.05).
Hepatic mRNA expression of superoxide and hydrogen peroxide metabolizing enzymes is shown in Figure 4B. There was less than a 15% decrease in mRNA for Sod1, Sod2, and Cat in Nrf2-null mice, whereas there was no difference between Keap1-kd and wild-type mice. Gpx mRNA was not different among the three genotypes. In addition, there were no differences in Sod, Cat, or Gpx enzyme activities among the three genotypes (data not shown).
Hepatic mRNA expression of redoxins is shown in Figure 4C. Glutaredoxin 1 (Glrx1) and Prx1 mRNA were lower in Nrf2-null mice (-27 and -20%, respectively), whereas there were no differences between wild-type mice and Keap1-kd mice. Thioredoxin 1 (Txn1) and thioredoxin reductase 1 (Txnrd1) mRNA were lower in Nrf2-null mice (24 and 41%, respectively) and higher in Keap1-kd mice (9 and 23%, respectively).
Hepatic mRNA of malic enzyme 1 (Me1) and hexose-6-phosphate dehydrogenase (H6pdh), enzymes important in generating nicotinamide adenine dinucleotide phosphate (NADPH), were quantified (Fig. 4D). Messenger RNAs for both Me1 and H6pdh were lower in Nrf2-null mice (62 and 16%, respectively), but were unchanged between Keap1-kd and wild-type mice.
Cytochrome P-450 and Major Transcription Factor mRNA Expression and Cyp Activity of Liver Microsomes
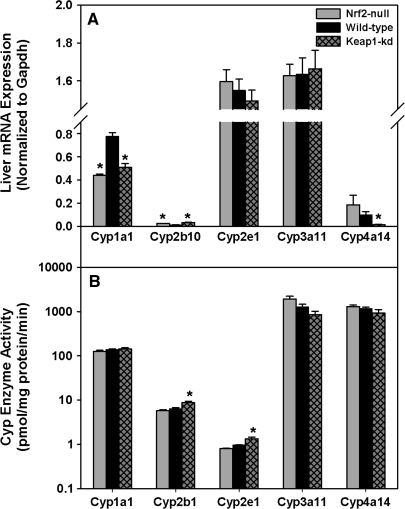
Hepatic mRNA expression and enzyme activity of major Cyps were quantified (Fig. 5A). Cyp1a1 mRNA was lower in both Nrf2-null (43%) and Keap1-kd (34%) mice than wild-type mice. Cyp2b10 mRNA was higher in both Nrf2-null (92%) and Keap1-kd (155%) mice than wild-type mice. The amounts of Cyp2e1 and 3a11 mRNAs were not different among the three genotypes. Cyp4a14 mRNA tended to be higher in Nrf2-null and was much lower in Keap1-kd (85%) mice than wild-type mice. Enzyme activities of Cyp1a1, 2b1, 2e1, 3a11, and 4a14 were quantified (Fig. 5B). There were no differences in Cyp1a1, 3a11, and 4a14 enzyme activity among genotypes. Cyp2b1 and 2e1 enzyme activities were unchanged between Nrf2-null and wild-type mice, but increased 36 and 38%, respectively, in Keap1-kd mice. Also, there were no differences in mRNA expression of the transcription factors aryl hydrocarbon receptor (AhR), pregnane X receptor (PXR), and peroxisome proliferator–activated receptor-alpha among genotypes (data not shown). However, constitutive androstane receptor (CAR) mRNA was 59% lower in Nrf2-null mice, whereas there were no differences between Keap1-kd and wild-type mice (data not shown).
FIG. 5.
Messenger RNA expression (A) and enzyme activity (B) of cytochrome p450s in wild-type, Nrf2-null, and Keap1-kd mice (n = 5). Values are expressed as mean ± SEM. Asterisks (*) indicate a statistically significant difference from wild-type mice (p ≤ 0.05).
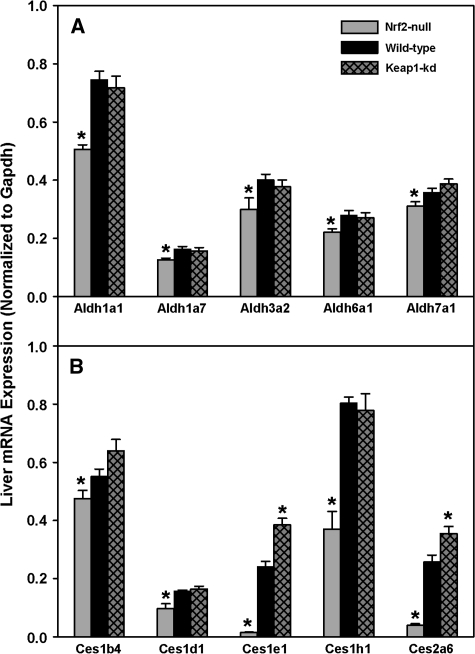
Hepatic mRNA Expression of the Phase-I Genes: Aldehyde Dehydrogenases and Carboxylesterases
Hepatic mRNAs of aldehyde dehydrogenase 1a1 (Aldh1a1), 1a7, 3a2, 6a1, and 7a1 were decreased in Nrf2-null mice (14-33%), whereas there were no differences between Keap1-kd and wild-type mice (Fig. 6A). Aldh1b1, 2, 4a1, 8a1, and 9a1 mRNA were not different among the three genotypes (data not shown).
FIG. 6.
Messenger RNA expression of Aldhs (A) and carboxylesterases (B) in wild-type, Nrf2-null, and Keap1-kd mice (n = 5). Values are expressed as mean ± SEM. Asterisks (*) indicate a statistically significant difference from wild-type mice (p ≤ 0.05).
Hepatic mRNAs of carboxylesterase 1b4 (Ces1b4), 1d1, and 1h1 were lower in Nrf2-null mice (14-46%), whereas there were no differences between Keap1-kd and wild-type mice (Fig. 6B). Ces1e1 and 2a6 mRNAs were markedly lower in Nrf2-null mice (94 and 85%, respectively) and higher in Keap1-kd mice (38 and 59%, respectively) than wild-type mice. The expression of paraoxonase 1 (Pon1) and Pon3 mRNAs were not different among the three genotypes (data not shown).
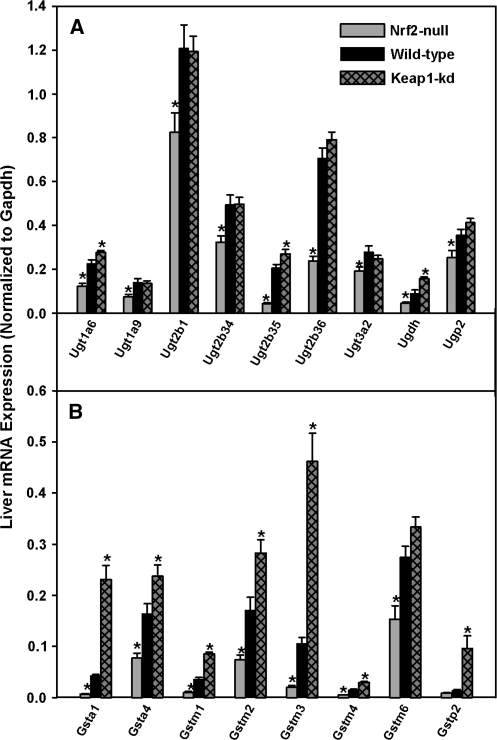
Hepatic mRNA Expression of the Phase-II Genes: Ugts, Gsts, and Sults
Messenger RNAs for Ugt1a9, 2b1, 2b34, 2b36, 3a2, and the uridine 5′-diphospho-glucuronic acid (UDP-GA) synthesizing enzyme UDP-glucose pyrophosphorylase (Ugp2) were lower in Nrf2-null mice (28-66%), whereas there were no differences between wild-type and Keap1-kd mice (Fig. 7A). Messenger RNAs of Ugt1a6, 2b35 and the UDP-GA synthesizing enzyme UDP-glucose dehydrogenase (Ugdh) were lower in Nrf2-null mice (45-78%) and higher in Keap1-kd mice (23-75%). There were no differences in mRNA expression of Ugt1a1, 1a5, or 2a3 among the three genotypes (data not shown).
FIG. 7.
Messenger RNA expression of Ugts (A) and Gsts (B) in wild-type, Nrf2-null, and Keap1-kd mice (n = 5). Values are expressed as mean ± SEM. Asterisks (*) indicate a statistically significant difference from wild-type mice (p ≤ 0.05).
Hepatic mRNAs of Gsta1, a4, m1, m2, m3, m4, m6, and p2 were lower in Nrf2-null mice (40-84%) and higher in Keap1-kd mice (45-585%) than in wild-type mice (Fig. 7B). There were no differences in mRNA expression for microsomal Gst1 (mGst1), mGst2, mGst3, Gstt1, or Gstt2 among the three genotypes (data not shown).
Messenger RNAs of hepatic Sults were quantified (Alnouti and Klaassen, 2006). There were no differences among the three genotypes for mRNAs of Sult1a1, 1d1, and the Sult cosubstrate 3′-phosphoadenoside 5′-phosphosulfate (PAPS) synthesizing enzyme, PAPS synthetase 1 (Papps1) (data not shown). Sult5a1 mRNA expression was 10% higher in Keap1-kd mice, whereas its expression in Nrf2-null mice did not differ from wild-type mice (data not shown). In addition, Papps2 mRNA was 25% higher in Nrf2-null mice than wild-type mice, whereas its expression in Keap1-kd mice was not different from wild-type mice (data not shown).
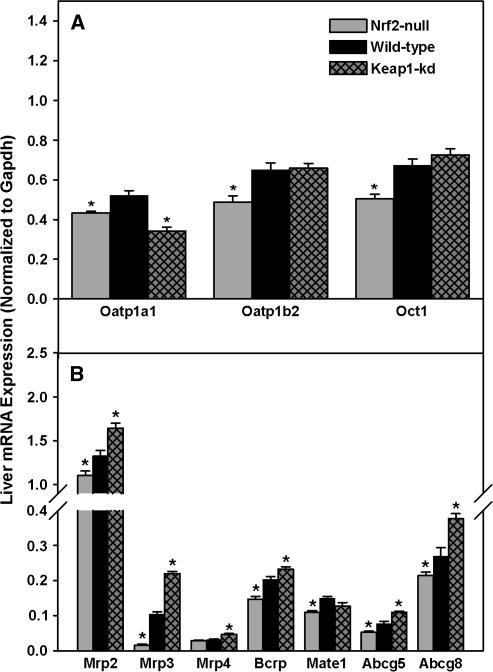
Hepatic mRNA Expression of Uptake and Efflux Transporters
Hepatic mRNA expression of uptake transporters in livers of the three genotypes of mice was quantified (Fig. 8A). Hepatic mRNAs of organic anion transporting polypeptide 1a1 (Oatp1a1), 1b2, and organic cation transporter 1 (Oct1) were lower (17-25%) in Nrf2-null mice. Surprisingly, in Keap1-kd mice, Oatp1a1 mRNA was 35% lower, whereas there were no differences in Oatp1b2 and Oct1 mRNA expression between Keap1-kd and wild-type mice. In addition, there were no differences in the mRNA expression among the three genotypes for organic anion transporter 2 (Oat2), Oatp1a4, 2b1, equilibrative nucleoside transporter 1 (Ent1), or sodium taurocholate cotransporting polypeptide (Ntcp) (data not shown).
FIG. 8.
Messenger RNA expression of uptake (A) and efflux (B) transporters in wild-type, Nrf2-null, and Keap1-kd mice (n = 5). Values are expressed as mean ± SEM. Asterisks (*) indicate a statistically significant difference from wild-type mice (p ≤ 0.05).
Hepatic mRNAs of various efflux transporters were also quantified (Fig. 8B). The mRNA of Mrp2, Mrp3, breast cancer resistance protein (Bcrp), and the two cholesterol and plant sterol efflux ATP-binding cassette transporters, Abcg5 and Abcg8, were lower in Nrf2-null (17, 85, 27, 29, and 20%, respectively) and higher in Keap1-kd mice (24, 111, 16, 45, and 40%, respectively). Mrp4 mRNA was not differently expressed between wild-type and Nrf2-null mice, but increased 55% over wild-type mice in Keap1-kd mice. Messenger RNA of multidrug and toxin extrusion 1 (Mate1) was 26% lower in Nrf2-null mice, whereas there was no difference between wild-type mice and Keap1-kd mice. There were no differences in mRNA among genotypes for multidrug resistance protein 2 (Mdr2), Mrp6, ATP-binding cassette transporter a1 (Abca1), or bile salt export pump (Bsep) (data not shown).
DISCUSSION
Nrf2 is a transcription factor that upon activation by oxidative/electrophilic insult induces a battery of cytoprotective genes. Nrf2-null mice are highly susceptible to a variety of oxidative/electrophilic stress-induced pathologies because of reduced basal and inducible expression of detoxification enzymes (Aleksunes and Manautou, 2007). In contrast, chemicals that activate Nrf2 protect rodents from pathologies linked to or caused by oxidative/electrophilic stress, leading to the hypothesis that Nrf2 is a target for chemoprevention (Lee and Surh, 2005). However, because chemical compounds typically have many off-target effects, it would be more informative to study activation of Nrf2 in a mouse model of increased Nrf2 activation. For example, the Nrf2 activator oltipraz, which has been used in the identification of possible Nrf2-target genes, also activates the CAR, inducing its target gene Cyp2b10 (Merrell et al., 2008). Therefore, the present study utilized genetic models of Nrf2 under- and overactivation to determine which genes have decreased mRNA expression with lack of Nrf2, and which genes have more mRNA expression when there is increased activation of Nrf2.
Upon activation, Nrf2 translocates into the nucleus, as reflected by the amount of Nrf2 protein in the nucleus in wild-type, Nrf2-null, and Keap1-kd mice. As shown in Figure 2, wild-type mice have very little Nrf2 protein in the nucleus, similar to previously published results (Kwak et al., 2001; Maher et al., 2007). Low Nrf2 protein in the nucleus in wild-type mice is most likely due to the rapid turnover of Nrf2 under basal or unchallenged conditions (Kobayashi et al., 2004). As expected, no hepatic Nrf2 mRNA or protein was detected in Nrf2-null mice, similar to previous results (Tanaka et al., 2008a). Nrf2-null mice also have lower Keap1 mRNA expression, most likely due to loss of Nrf2 binding to a functional ARE in the promoter region of Keap1 in a proposed negative feedback mechanism (Lee et al., 2007). In contrast, Keap1-kd mice have 55% lower Keap1 mRNA expression, which is likely the mechanism for tripling the amount of Nrf2 protein in the nucleus of Keap1-kd mice. Keap1-kd mice also have a modest decrease in Nrf2 mRNA expression, which suggests a possible negative feedback pathway for Nrf2 mRNA expression when Nrf2 is overactivated.
Most basal hepatic physiological parameters in serum, liver, and bile were not different among wild-type, Nrf2-null, and Keap1-kd mice (Table 1). However, bile flow tended to be lower in Nrf2-null mice and was significantly higher in Keap1-kd mice. Increased bile flow in Keap1-kd mice is due to increased biliary excretion of GSH, as GSH excretion regulates bile-acid–independent bile flow (Ballatori and Truong, 1989). Increased expression of GSH synthetic enzymes (Gclc and Gclm), as discussed in detail below, likely contributes to increased GSH concentrations in liver and biliary excretion of GSH in Keap1-kd mice. There were no differences in total biliary bile acids among genotypes (data not shown). Biliary excretion, possibly as a result of increase bile flow, but not biliary concentrations of phospholipids and cholesterol were increased in Keap1-kd mice.
Hepatic concentrations of GSH in the three mouse genotypes were proportional to the amount of activated Nrf2. GSH is a prominent cellular antioxidant that protects against oxidative/electrophilic stress by directly scavenging reactive oxygen species or acting as a cosubstrate for Gst-mediated detoxification of reactive electrophiles (Meister and Anderson, 1983). The rate-limiting enzymatic process in GSH synthesis is catalyzed by glutamate-cysteine ligase, which is made up of the catalytic and modifier subunits, Gclc and Gclm, respectively. Nrf2-null mice have lower, whereas Keap1-kd mice have higher hepatic mRNA expression of Gclc and Gclm, genes positively regulated by Nrf2 (Lee et al., 2007; Moinova and Mulcahy, 1999; Wild et al., 1999) (Fig. 4A). Furthermore, mRNA expression of Gsr, the enzyme responsible for the reduction of oxidized glutathione (GSSG) to reduced GSH, is lower in Nrf2-null mice and higher in Keap1-kd mice. Because Gclc, Gclm, and Gsr mRNA expression is lower in Nrf2-null mice and higher in Keap1-kd mice, the amount of GSH in the liver was determined. Nrf2-null mice have less than half, whereas Keap1-kd mice have almost double the hepatic reduced GSH as wild-type mice (Fig. 3). Higher GSH concentrations in the Keap1-kd mouse should increase resistance to a variety of oxidative/electrophilic stress-induced pathologies, whereas lower reduced GSH concentrations render the Nrf2-null mice more susceptible to such pathologies.
With few exceptions, typical Nrf2-target genes are lower in Nrf2-null mice and higher in Keap1-kd mice. Nqo1 is a Nrf2-dependent flavoprotein that catalyzes the two-electron reduction and detoxification of electrophilic quinones and its derivatives (Jaiswal, 2000). Nqo1 is a prototypical Nrf2 target gene and is expressed lower in Nrf2-null mice and higher in Keap1-kd mice (Fig. 4A). Eh-1 is induced by sulforophane and 3H-1,2-dithiole-3-thione (D3T) in wild-type but not Nrf2-null mice (Kwak et al., 2001; Thimmulappa et al., 2002). However, Eh-1 mRNA is not increased in Keap1-kd mice, suggesting that other mechanisms aside from Nrf2 nuclear translocation are important for Eh-1 mRNA induction in the liver. Ho-1 catalyzes the breakdown of heme into iron, carbon monoxide, and biliverdin. Biliverdin is then reduced to bilirubin, an antioxidant, via biliverdin reductase. Ho-1 is induced, in mice, by many Nrf2 activators or stressors, such as CDDO-Im (Liby et al., 2005), butylated hyroxyanisole (Keum et al., 2006), hyperoxia (Cho et al., 2002), and arsenite (Gong et al., 2002). However, no difference in Ho-1 expression was observed among wild-type, Nrf2-null, and Keap1-kd mice, a result that is consistent with previous reports (Okawa et al., 2006). The lack of induction of Ho-1 in Keap1-kd mice is attributed to the presence of Bach1, a protein that suppresses the activity of Maf proteins, which are important heterodimers for Nrf2-ARE binding and subsequent Ho-1 gene transcription (Keum et al., 2006).
The superoxide and hydrogen peroxide detoxifying enzymes Sod1, Sod2, Cat, and Gpx are inducible in response to Nrf2 activators, such as D3T (Zhu et al., 2005), phenolic acids (Yeh and Yen, 2006), and shear stress (Jones et al., 2007). In the present study, a minor decrease in hepatic mRNA expression of Sod1, Sod2, and Cat was observed in Nrf2-null mice (10-30%), whereas there was no increase in Keap1-kd mice. Gpx mRNA expression was not different among the three genotypes (Fig. 4B). There were also no differences among the three genotypes in Sod, Cat, or Gpx enzyme activities. The lack of induction of Sods, Cat, and Gpx in Keap1-kd mice suggests that nuclear translocation of Nrf2 does not induce enzymes capable of detoxifying superoxide and related reactive oxygen species in liver.
Redoxins are important for protein repair, as they are responsible for the reduction of protein disulfide bonds that could disrupt proper protein folding and function. Peroxiredoxins reduce hydrogen peroxide to water, protecting against hydroxyl radical formation. Glrx1, Prx1, Txn1, and Txnrd1 have lower hepatic mRNA expression in Nrf2-null mice, whereas only Txn1 and Txnrd1 are induced in Keap1-kd mice (Fig. 4C). NADPH is a cofactor for many oxidoreductase reactions that are important in detoxifying reactive oxygen species. Me1 and H6pdh are enzymes that regenerate NADPH after it has been oxidized. Both Me1 and H6pdh are lower in Nrf2-null mice but not increased in Keap1-kd mice (Fig. 4D).
Chemicals, such as acetaminophen, benzo[a]pyrene, carbon tetrachloride, and benzene, are activated to reactive intermediates by Cyps, which can result in toxicity. Knowledge of Cyp activity in toxicity studies utilizing Nrf2-null and Keap1-kd mice is critical, because changes in Cyp activity could alter the amount of toxic metabolite generated, leading to an incorrect interpretation of results, that is, less injury caused by decreased toxic metabolite formation and not increased detoxification of the metabolite. Even though there were differences among genotypes in mRNA expression of Cyp1a1, 2b10, and 4a14, minor differences, if any, were detected in the enzyme activity of major Cyps. Thus, Cyps most likely will not play a role in the interpretation of results from toxicity studies involving Nrf2-null and Keap1-kd mice.
Aldhs catalyze the oxidation of a wide variety of electrophilic aliphatic and aromatic aldehydes to carboxylic acids (Parkinson and Ogilvie, 2008). Nrf2-null mice have lower expression of Aldh1a1, 1a7, 3a2, 6a1, and 7a1, whereas Keap1-kd mice were not different from wild-type mice (Fig. 6A). The lack of induction of Aldhs in Keap1-kd mice suggests that Aldhs are not induced by Nrf2 in this mouse model.
Carboxylesterases (Cess) catalyze the hydrolysis of ester- and amide-containing chemicals and are important in the activation of prodrugs (Parkinson and Ogilvie, 2008). Produrg strategies allow for improvement of oral bioavailability of poorly absorbed drugs. Expression of Ces1b4, 1d1, 1e1, 1h1, and 2a6 were lower in Nrf2-null mice, whereas only Ces1e1 and 2a6 were higher in Keap1-kd mice (Fig. 6B). An increase in expression of Cess in Keap1-kd mice suggests that activation of Nrf2 increases expression of Cess and metabolism of xenobiotics by Cess. An increase in Ces1e1 also might increase the amount of prodrug (i.e., the chemotherapeutics floxuridine and gemcitabine) converted to the active form, providing a more efficacious result (Landowski et al., 2006; Marsh et al., 2004; Taketani et al., 2007).
Hepatic mRNA expression of phase-II enzymes is also altered in Nrf2-null and Keap1-kd mice. Ugts catalyze the conjugation of a glucuronosyl group from UDP-GA to a variety of substrate molecules, making them more water soluble and readily excreted. Increased excretion of xenobiotics decreases the amount of compound available for biotransformation to a toxic electrophilic metabolite. In addition, Ugt-catalyzed reactions are responsible for approximately 35% of all drugs metabolized by phase-II enzymes (Evans and Relling, 1999). In general, Ugt mRNA expression is lower in Nrf2-null mice, whereas Keap1-kd mice are not different from wild-type mice, with the exception of a minor increase in mRNA expression of Ugt1a6 and Ugt2b35 (Fig. 7A). The availability of the cosubstrate UDP-GA is required for the enzyme activity of Ugts. Both Ugdh and Ugp2 mRNA expression were lower in Nrf2-null mice, whereas Ugdh was significantly higher and Ugp2 tended to be higher in Keap1-kd mice.
Sults catalyze approximately 20-25% of phase-II reactions by transferring a sulfonic acid group from the cosubstrate PAPS (Evans and Relling, 1999). There are few differences in the amount of the various Sult mRNAs among wild-type, Nrf2-null, and Keap1-kd mice, suggesting that Nrf2 does not play a major transcriptional role in the expression of Sults.
Gsts catalyze the conjugation of nucleophilic GSH with reactive and potentially damaging electrophiles (Parkinson and Ogilvie, 2008). Substrates for Gsts include hydroperoxides of fatty acids, phospholipids, cholesterol, and quinone-containing compounds (Hayes et al., 2005). The expression of many Gsts is dependent on Nrf2, with most Gsts having 41-85% lower expression in Nrf2-null mice and 45-585% higher expression in Keap1-kd mice (Fig. 7B). A decrease in hepatic GSH concentration and Gst mRNA expression contributes to Nrf2-null mice being highly susceptible to electrophilic stress. In contrast, Keap1-kd mice have increased hepatic GSH concentrations and Gst mRNA expression and most likely have an increased resistance against damaging electrophiles that can be neutralized via GSH conjugation. The ability to produce and use more GSH appears to be one of the most important benefits of increased hepatic activated Nrf2, as observed in Keap1-kd mice.
Uptake transporters are important for the hepatic uptake and clearance of xenobiotics, an important process in the first-pass effect (Klaassen and Lu, 2008). In general, mRNA expression of hepatic uptake transporters was similar among genotypes, with the exception of Oatp1a1, 1b2, and Oct1, which exhibit relatively minor differences (< 25%) in expression (Fig. 8A). Of note, Oatp1a1 expression is lower in Keap1-kd mice, which is consistent with what has been observed upon administration of Nrf2 activators (Cheng et al., 2005). Furthermore, Oatp and Oct1 mRNA are generally not altered by Nrf2 activators or other microsomal enzyme inducers (Cheng et al., 2005). Thus, this data suggests that uptake transporters in the liver are not activated by Nrf2.
Hepatic efflux transporters are important for the elimination and overall clearance of xenobiotics from the liver. Mrps are a group of ATP-dependent transporters that are important in cytoprotection, because Mrps can remove potentially toxic xenobiotics, metabolites, and endogenous substrates from cells (Maher et al., 2007). Mrp2 and Mrp3 mRNA expression are lower in Nrf2-null mice, but higher in Keap1-kd mice. Mrp4 mRNA was lowly expressed and not different between Nrf2-null and wild-type mice, but was increased 55% over wild-type in Keap1-kd mice (Fig. 8B). Bcrp is an efflux transporter for substrates, such as mitoxantrone, anthracyclines, camptothecins, topotecan, and SN-38, the active metabolite of irinotecan (Brangi et al., 1999; Doyle et al., 1998; Litman et al., 2000; Miyake et al., 1999; Ross et al., 1999). Bcrp mRNA expression is 28% lower in Nrf2-null mice and 15% higher in Keap1-kd mice. Mate1 effluxes organic cations, such as metformin and tetraethylammonium, from hepatocytes into bile (Hiasa et al., 2006; Terada et al., 2006). Mate1 mRNA is expressed at a 27% lower level in Nrf2-null mice. There was no difference in Mate1 mRNA expression between Keap1-kd and wild-type mice, similar to a previous report in which Nrf2 activators did not induce Mate1 mRNA expression (Lickteig et al., 2008). Abcg5 and Abcg8, transporters involved in the efflux of cholesterol and potentially toxic plant sterols from the liver into bile, were lower in Nrf2-null mice and higher in Keap1-kd mice. The increase of mRNA expression of efflux transporters may provide Keap1-kd mice the ability to increase the clearance of potentially toxic xenobiotics, thereby decreasing time of exposure and toxicity.
Table 2 categorizes mRNA expression of detoxifying and transporter genes among wild-type, Nrf2-null, and Keap1-kd mice into three different patterns. The first pattern encompasses genes that have decreased mRNA expression in Nrf2-null mice and increased expression in Keap1-kd mice compared with wild-type mice. Pattern 1 genes include Nqo1, Gsts, and Mrps, which detoxify and eliminate electrophiles. The second pattern consists of genes that have decreased mRNA expression in Nrf2-null mice but no difference between Keap1-kd and wild-type mice. Prominent genes in pattern 2 are the superoxide detoxifying enzymes Sod1, Sod2, Cat, and Prx1, as well as some Ugts and Aldhs. The third pattern includes genes that were not different among wild-type, Nrf2-null, and Keap1-kd mice and includes genes such as some Ugts, Sults, some Aldhs, Gpx, Ho-1, and uptake transporters. It should be noted that the genes in pattern 3, with the exception of Ho-1, are not known Nrf2-target genes.
TABLE 2.
Categorization of Antioxidant, Phase-I, Phase-II, and Transporter mRNA Expression
| Pattern 1 |
Pattern 2 |
Pattern 3 |
||||
| Decrease in Nrf2-null mice and increase in Keap1-kd mice |
Decrease in Nrf2-null mice and nonsignificant change in Keap1-kd mice |
No changes among genotypes | ||||
| Gene | Nrf2-null % change | Keap1-kd % change | Gene | Nrf2-null % change | Keap1-kd % change | Gene |
| Gstp2* | −40 | 586 | Ugp2 | −29 | 16 | Ho-1 |
| Gsta1 | −84 | 448 | Ces1b4 | −14 | 16 | Gpx |
| Gstm3 | −81 | 336 | Ugt2b36 | −66 | 12 | Pon1 |
| Nqo1 | −88 | 222 | Txn1 | −23 | 9 | Pon3 |
| Gstm1 | −74 | 140 | Aldh7a1 | −13 | 8 | Aldh1b1 |
| Mrp3 | −85 | 111 | Oct1 | −25 | 8 | Aldh2 |
| Gstm4 | −67 | 102 | H6pdh | −16 | 7 | Aldh4a1 |
| Ugdh | −50 | 75 | Prx1 | −20 | 6 | Aldh8a1 |
| Gstm2 | −57 | 65 | Ces1d1 | −38 | 4 | Aldh9a1 |
| Gclc | −37 | 63 | Cat | −15 | 4 | Ugt1a1 |
| Ces1e1 | −94 | 59 | Oatp1b2 | −25 | 1 | Ugt1a5 |
| Mrp4* | −3 | 55 | Ugt2b34 | −35 | 0 | Ugt2a3 |
| Gsta4 | −53 | 45 | Ugt2b1 | −32 | −1 | |
| Abcg5 | −29 | 45 | Glrx1 | −27 | −1 | Gstt1 |
| Abcg8 | −20 | 40 | Sod1 | −10 | −2 | Gstt2 |
| Ces2a6 | −85 | 38 | Ugt1a9 | −47 | −3 | mGst1 |
| Ugt2b35 | −79 | 32 | Ces1h1 | −54 | −3 | mGst2 |
| Mrp2 | −17 | 24 | Aldh6a1 | −21 | −3 | mGst3 |
| Ugt1a6 | −46 | 23 | Aldh1a1 | −32 | −4 | Sult1a1 |
| Txnrd1 | −41 | 23 | Aldh1a7 | −23 | −5 | Sult1d1 |
| Gclm | −16 | 22 | Aldh3a2 | −25 | −6 | Papps1 |
| Gsr | −23 | 17 | Ugt3a2 | −31 | −11 | Oat2 |
| Bcrp | −27 | 16 | Sod2 | −32 | −14 | Oatp1a4 |
| Mate1 | −26 | −15 | Oatp2b1 | |||
| Me1 | −62 | −18 | Ent1 | |||
| Eh-1 | −72 | −18 | Ntcp | |||
| Oatp1a1 | −17 | −35 | Mdr2 | |||
| Mrp6 | ||||||
| Abca1 | ||||||
| Bsep | ||||||
Note. Percent change from wild-type in Nrf2-null and Keap1-kd mice are presented. The first pattern represents genes that had significant decrease in Nrf2-null mice (p ≤ 0.05) and a significant increase in Keap1-kd mice (p ≤ 0.05). The second pattern represents genes that had significant decrease in Nrf2-null mice (p ≤ 0.05) and no change from wild-type in Keap1-kd mice (p > 0.05). The third pattern of gene expression represents genes that were not different among wild-type, Nrf2-null, and Keap1-kd mice. *No significant difference between Nrf2-null and wild-type mice (p > 0.05).
Keap1 has been shown in vitro to bind and regulate the ubiquitination and subsequent proteasomal degradation of one other protein, phosphoglycerate mutase family member 5 (PGAM5) (Lo and Hannink, 2006). Phosphoglycerate mutases catalyze the conversion of 3-phosphoglycerate to 2-phosphoglycerate, which is an important substrate in glycolysis. Therefore, because Keap1 is capable of binding other proteins, at least in vitro, it is possible that some of the genes that are upregulated in Keap1-kd mice could be independent of Nrf2. However, a follow-up study by Lo et al. demonstrated in vitro that PGAM5 tethers a ternary complex containing both Keap1 and Nrf2 in mitochondria (Lo and Hannink, 2008). Lo and Hannick (2008) also hypothesized that Keap1 may use other proteins, such as PGAM5, to regulate Nrf2 at other subcellular localizations, such as the mitochondria. In addition, PGAM5 has not been shown to transcriptionally regulate any gene. Therefore, it seems unlikely that PGAM5 would be responsible for the upregulation of genes in Keap1-kd mice.
In conclusion, this study has shown that whereas Nrf2-null and Keap1-kd mice have normal livers under standard institutional animal care conditions, baseline defenses against electrophilic stress are lower in Nrf2-null mice and higher in Keap1-kd mice. In addition, classical reactive oxygen species reducing enzymes, such as Cat, Gpx, and Sods, were not induced in livers of Keap1-kd mice, whereas genes, such as Gsts, Nqo1, and Mrps, important in detoxifying and eliminating electrophiles are markedly increased in Keap1-kd mice. The major advantage Keap1-kd mice have against reactive oxygen and nitrogen species in the liver appears to be an increase in hepatic GSH concentrations. Collectively, these results suggest that activation of Nrf2 in liver results in induction of genes, whose protein products, are more important in the direct detoxification of highly reactive electrophilies formed from xenobiotic exposure than for detoxification of reactive oxygen species.
FUNDING
National Institute of Health (ES09716, ES07079, ES013714, ES09649, and RR021940).
Acknowledgments
We thank the members of the Klaassen laboratory for their technical support and manuscript preparation. Nrf2-null mice were graciously provided by Dr Jefferson Chan (University of California-Irvine, Irvine, CA) and Keap1-kd mice by Dr Masayuki Yamamoto (Tohoku University, Aoba-ku, Sendai, Japan). Also, the authors thank XenoTech, LLC (Lenexa, KS) for assistance with Cyp enzyme activity assays.
References
- Aleksunes LM, Manautou JE. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol. Pathol. 2007;35:459–473. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol. Sci. 2006;93:242–255. doi: 10.1093/toxsci/kfl050. [DOI] [PubMed] [Google Scholar]
- Ballatori N, Truong AT. Relation between biliary glutathione excretion and bile acid-independent bile flow. Am. J. Physiol. 1989;256:G22–G30. doi: 10.1152/ajpgi.1989.256.1.G22. [DOI] [PubMed] [Google Scholar]
- Brangi M, Litman T, Ciotti M, Nishiyama K, Kohlhagen G, Takimoto C, Robey R, Pommier Y, Fojo T, Bates SE. Camptothecin resistance: Role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res. 1999;59:5938–5946. [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Maher J, Dieter MZ, Klaassen CD. Regulation of mouse organic anion-transporting polypeptides (Oatps) in liver by prototypical microsomal enzyme inducers that activate distinct transcription factor pathways. Drug Metab. Dispos. 2005;33:1276–1282. doi: 10.1124/dmd.105.003988. [DOI] [PubMed] [Google Scholar]
- Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O'Connor T, Harada T, Yamamoto M. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- Evans WE, Relling MV. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- Flohe L, Gunzler WA. Assays of glutathione peroxidase. Methods Enzymol. 1984;105:114–121. doi: 10.1016/s0076-6879(84)05015-1. [DOI] [PubMed] [Google Scholar]
- Gong P, Stewart D, Hu B, Vinson C, Alam J. Multiple basic-leucine zipper proteins regulate induction of the mouse heme oxygenase-1 gene by arsenite. Arch. Biochem. Biophys. 2002;405:265–274. doi: 10.1016/s0003-9861(02)00404-6. [DOI] [PubMed] [Google Scholar]
- Harwood DT, Kettle AJ, Winterbourn CC. Production of glutathione sulfonamide and dehydroglutathione from GSH by myeloperoxidase-derived oxidants and detection using a novel LC-MS/MS method. Biochem. J. 2006;399:161–168. doi: 10.1042/BJ20060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Hiasa M, Matsumoto T, Komatsu T, Moriyama Y. Wide variety of locations for rodent MATE1, a transporter protein that mediates the final excretion step for toxic organic cations. Am. J. Physiol. Cell. Physiol. 2006;291:C678–C686. doi: 10.1152/ajpcell.00090.2006. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- Jaiswal AK. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic. Biol. Med. 2000;29:254–262. doi: 10.1016/s0891-5849(00)00306-3. [DOI] [PubMed] [Google Scholar]
- Jones CI, 3rd, Zhu H, Martin SF, Han Z, Li Y, Alevriadou BR. Regulation of antioxidants and phase 2 enzymes by shear-induced reactive oxygen species in endothelial cells. Ann. Biomed. Eng. 2007;35:683–693. doi: 10.1007/s10439-007-9279-9. [DOI] [PubMed] [Google Scholar]
- Keum YS, Han YH, Liew C, Kim JH, Xu C, Yuan X, Shakarjian MP, Chong S, Kong AN. Induction of heme oxygenase-1 (HO-1) and NAD[P]H:quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm. Res. 2006;23:2586–2594. doi: 10.1007/s11095-006-9094-2. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Lu H. Xenobiotic transporters: Ascribing function from gene knockout and mutation studies. Toxicol. Sci. 2008;101:186–196. doi: 10.1093/toxsci/kfm214. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol. Med. 2001;7:135–145. [PMC free article] [PubMed] [Google Scholar]
- Lamle J, Marhenke S, Borlak J, von Wasielewski R, Eriksson CJ, Geffers R, Manns MP, Yamamoto M, Vogel A. Nuclear factor-eythroid 2-related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology. 2008;134:1159–1168. doi: 10.1053/j.gastro.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Landowski CP, Lorenzi PL, Song X, Amidon GL. Nucleoside ester prodrug substrate specificity of liver carboxylesterase. J. Pharmacol. Exp. Ther. 2006;316:572–580. doi: 10.1124/jpet.105.092726. [DOI] [PubMed] [Google Scholar]
- Lee OH, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J. Biol. Chem. 2007;282:36412–36420. doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- Lee JS, Surh YJ. Nrf2 as a novel molecular target for chemoprevention. Cancer Lett. 2005;224:171–184. doi: 10.1016/j.canlet.2004.09.042. [DOI] [PubMed] [Google Scholar]
- Liby K, Hock T, Yore MM, Suh N, Place AE, Risingsong R, Williams CR, Royce DB, Honda T, Honda Y, et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- Lickteig AJ, Cheng X, Augustine LM, Klaassen CD, Cherrington NJ. Tissue distribution, ontogeny and induction of the transporters multidrug and toxin extrusion (MATE) 1 and MATE2 mRNA expression levels in mice. Life Sci. 2008;83:59–64. doi: 10.1016/j.lfs.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman T, Brangi M, Hudson E, Fetsch P, Abati A, Ross DD, Miyake K, Resau JH, Bates SE. The multidrug-resistant phenotype associated with overexpression of the new ABC half-transporter, MXR (ABCG2) J. Cell. Sci. 2000;113(Pt 11):2011–2021. doi: 10.1242/jcs.113.11.2011. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J. Biol. Chem. 2006;281:37893–37903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck H. Catalase. In: Bergmeyer HU, editor. Methods of Enzymatic Analysis. New York: Academic Press; 1963. pp. 885–888. [Google Scholar]
- Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, et al. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Cherrington NJ, Cheng X, Klaassen CD. Tissue distribution and hepatic and renal ontogeny of the multidrug resistance-associated protein (Mrp) family in mice. Drug Metab. Dispos. 2005;33:947–955. doi: 10.1124/dmd.105.003780. [DOI] [PubMed] [Google Scholar]
- Marsh S, Xiao M, Yu J, Ahluwalia R, Minton M, Freimuth RR, Kwok PY, McLeod HL. Pharmacogenomic assessment of carboxylesterases 1 and 2. Genomics. 2004;84:661–668. doi: 10.1016/j.ygeno.2004.07.008. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- Meister A, Anderson ME. Glutathione. Annu. Rev. Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- Merrell MD, Jackson JP, Augustine LM, Fisher CD, Slitt AL, Maher JM, Huang W, Moore DD, Zhang Y, Klaassen CD, et al. The Nrf2 activator oltipraz also activates the constitutive androstane receptor. Drug Metab. Dispos. 2008;36:1716–1721. doi: 10.1124/dmd.108.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K, Mickley L, Litman T, Zhan Z, Robey R, Cristensen B, Brangi M, Greenberger L, Dean M, Fojo T, et al. Molecular cloning of cDNAs which are highly overexpressed in mitoxantrone-resistant cells: Demonstration of homology to ABC transport genes. Cancer Res. 1999;59:8–13. [PubMed] [Google Scholar]
- Moinova HR, Mulcahy RT. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem. Biophys. Res. Commun. 1999;261:661–668. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- Okada K, Shoda J, Taguchi K, Maher JM, Ishizaki K, Inoue Y, Ohtsuki M, Goto N, Takeda K, Utsunomiya H, et al. Ursodeoxycholic acid stimulates Nrf2-mediated hepatocellular transport, detoxification, and antioxidative stress systems in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;295:G735–G747. doi: 10.1152/ajpgi.90321.2008. [DOI] [PubMed] [Google Scholar]
- Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- Parkinson A, Ogilvie B. Biotransformation. In: Klaassen CD, editor. Casarett and Doull's Toxicology: The Basic Science of Poisons. New York: McGraw-Hill; 2008. pp. 161–304. [Google Scholar]
- Petrick JS, Klaassen CD. Importance of hepatic induction of constitutive androstane receptor and other transcription factors that regulate xenobiotic metabolism and transport. Drug Metab. Dispos. 2007;35:1806–1815. doi: 10.1124/dmd.107.015974. [DOI] [PubMed] [Google Scholar]
- Ross DD, Yang W, Abruzzo LV, Dalton WS, Schneider E, Lage H, Dietel M, Greenberger L, Cole SP, Doyle LA. Atypical multidrug resistance: Breast cancer resistance protein messenger RNA expression in mitoxantrone-selected cell lines. J. Natl. Cancer Inst. 1999;91:429–433. doi: 10.1093/jnci/91.5.429. [DOI] [PubMed] [Google Scholar]
- Taketani M, Shii M, Ohura K, Ninomiya S, Imai T. Carboxylesterase in the liver and small intestine of experimental animals and human. Life Sci. 2007;81:924–932. doi: 10.1016/j.lfs.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Aleksunes LM, Goedken MJ, Chen C, Reisman SA, Manautou JE, Klaassen CD. Coordinated induction of Nrf2 target genes protects against iron nitrilotriacetate (FeNTA)-induced nephrotoxicity. Toxicol. Appl. Pharmacol. 2008a;231:364–373. doi: 10.1016/j.taap.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Aleksunes LM, Yeager RL, Gyamfi MA, Esterly N, Guo GL, Klaassen CD. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J. Pharmacol. Exp. Ther. 2008b;325:655–664. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- Tappel AL. Glutathione peroxidase and hydroperoxides. Methods Enzymol. 1978;52:506–513. doi: 10.1016/s0076-6879(78)52055-7. [DOI] [PubMed] [Google Scholar]
- Terada T, Masuda S, Asaka J, Tsuda M, Katsura T, Inui K. Molecular cloning, functional characterization and tissue distribution of rat H+/organic cation antiporter MATE1. Pharm. Res. 2006;23:1696–1701. doi: 10.1007/s11095-006-9016-3. [DOI] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- Trujillo M, Ferrer-Sueta G, Radi R. Peroxynitrite detoxification and its biologic implications. Antioxid. Redox Signal. 2008;10:1607–1620. doi: 10.1089/ars.2008.2060. [DOI] [PubMed] [Google Scholar]
- Umemura T, Kuroiwa Y, Kitamura Y, Ishii Y, Kanki K, Kodama Y, Itoh K, Yamamoto M, Nishikawa A, Hirose M. A crucial role of Nrf2 in in vivo defense against oxidative damage by an environmental pollutant, pentachlorophenol. Toxicol. Sci. 2006;90:111–119. doi: 10.1093/toxsci/kfj076. [DOI] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Wild AC, Moinova HR, Mulcahy RT. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- Yeh CT, Yen GC. Induction of hepatic antioxidant enzymes by phenolic acids in rats is accompanied by increased levels of multidrug resistance-associated protein 3 mRNA expression. J. Nutr. 2006;136:11–15. doi: 10.1093/jn/136.1.11. [DOI] [PubMed] [Google Scholar]
- Zhu H, Itoh K, Yamamoto M, Zweier JL, Li Y. Role of Nrf2 signaling in regulation of antioxidants and phase 2 enzymes in cardiac fibroblasts: Protection against reactive oxygen and nitrogen species-induced cell injury. FEBS Lett. 2005;579:3029–3036. doi: 10.1016/j.febslet.2005.04.058. [DOI] [PubMed] [Google Scholar]