Abstract
Spinal and bulbar muscular atrophy (SBMA) is a motor neuron disease caused by polyglutamine expansion mutation in the androgen receptor (AR). We investigated whether the mutant protein alters mitochondrial function. We found that constitutive and doxycycline-induced expression of the mutant AR in MN-1 and PC12 cells, respectively, are associated with depolarization of the mitochondrial membrane. This was mitigated by cyclosporine A, which inhibits opening of the mitochondrial permeability transition pore. We also found that the expression of the mutant protein in the presence of ligand results in an elevated level of reactive oxygen species, which is blocked by the treatment with the antioxidants co-enzyme Q10 and idebenone. The mutant protein in MN-1 cells also resulted in increased Bax, caspase 9 and caspase 3. We assessed the effects of mutant AR on the transcription of mitochondrial proteins and found altered expression of the peroxisome proliferator-activated receptor γ coactivator 1 and the mitochondrial specific antioxidant superoxide dismutase-2 in affected tissues of SBMA knock-in mice. In addition, we found that the AR associates with mitochondria in cultured cells. This study thus provides evidence for mitochondrial dysfunction in SBMA cell and animal models, either through indirect effects on the transcription of nuclear-encoded mitochondrial genes or through direct effects of the mutant protein on mitochondria or both. These findings indicate possible benefit from mitochondrial therapy for SBMA.
INTRODUCTION
Spinal and bulbar muscular atrophy (SBMA) results from the expansion of a polyglutamine tract in the androgen receptor (AR) (1). SBMA is one of at least nine diseases with polyglutamine expansion, including Huntington's disease (HD), dentatorubral–pallidoluysian atrophy and six spinocerebellar ataxias (2). SBMA is X-linked and fully affects only males (1). The polyglutamine expansion in the AR results in selective degeneration of motor neurons in the brainstem and spinal cord (3,4). The clinical characteristics of SBMA include muscle cramps, slowly progressive extremity weakness, muscle atrophy, fasciculations, dysphagia and signs of androgen insensitivity, such as gynecomastia and reduced fertility (4,5).
The AR is a member of the nuclear receptor family and a ligand-activated transcription factor (6). In its inactive state, the AR is cytoplasmic and sequestered by heat shock proteins. On ligand binding, it dissociates from the complex of heat shock proteins and translocates to the nucleus where it binds to specific DNA sequences and activates transcription (6). The polyglutamine expansion in the AR likely confers a toxic gain and also a partial loss of AR function (7,8). Aberrant interactions of the mutant AR with other proteins, including transcriptional co-activators, result in transcriptional dysregulation and neuronal toxicity (9,10). There is considerable evidence supporting ligand-dependent toxicity (11–15). For example, reducing androgen levels in transgenic mice rescues the motor impairment and increases survival (12,14). These studies point to altered transcriptional regulation as a disease mechanism, but they do not preclude the possibility of toxicity in the cytoplasmic compartment. The specific mechanism of SBMA remains unclear, and there is currently no effective treatment.
Mitochondrial dysfunction has been implicated in various neurodegenerative diseases, including Huntington's disease (HD), amyotrophic lateral sclerosis (ALS) and Friedreich's ataxia (16–21). It is also known that transcriptional dysregulation in the nucleus can affect mitochondrial turnover and oxidative phosphorylation (22–24). Peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) and its homolog, PGC-1β, are nuclear transcriptional co-activators and important regulators of energy metabolism (23–26). Recent studies in HD show altered PGC-1α to be a likely link between transcriptional dysregulation and mitochondrial defects (27,28). Currently, little is known about mitochondrial involvement in SBMA. Previous studies in cell models suggest alterations in mitochondrial distribution and sequestration of nuclear-encoded mitochondrial proteins in mutant AR aggregates (29,30).
In this study, we investigated whether mutant AR alters mitochondrial function in models of SBMA. We found that the expression of mutant AR in motor neuron-derived (MN-1) cells and with doxycycline induction in pheochromacytoma-derived (PC12) cells is associated with mitochondrial membrane depolarization and increased reactive oxygen species (ROS). Cyclosporine A (CsA) mitigates the membrane depolarization, and the increase in ROS levels is blocked by the antioxidants co-enzyme Q10 (CoQ10) and idebenone. We also found that the mutant AR increases Bax protein levels, and activates caspases 9 and 3. The activation of caspases 9 and 3 is likely mediated by the mitochondrial transition pore. Moreover, we observed decreased levels of PGC-1β and mitochondrial superoxide dismutase (SOD2) in MN-1 cells and in the tissues of a knock-in mouse model of SBMA. We also found in vitro evidence for association of AR with mitochondria. At the ultrastructural level, this association is greater with the AR containing the expanded repeat. Taken together, our results implicate mitochondrial abnormalities and redox imbalance in SBMA. This study provides evidence that mitochondrial modulators and antioxidants may have therapeutic value in SBMA.
RESULTS
Mutant AR activates the mitochondrial caspase pathway
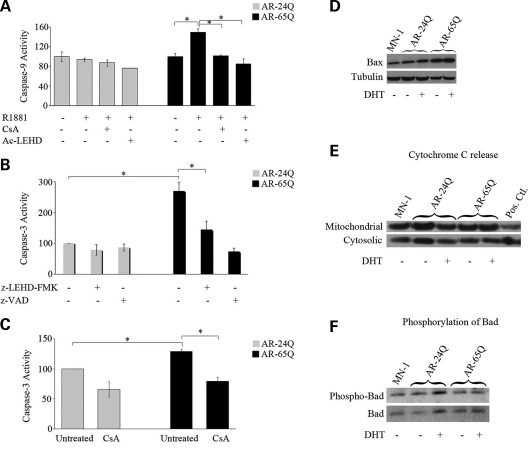
Previous studies have shown that the expression of mutant AR results in the activation of caspase 3 in motor neuron-like cells (MN-1) (31–34). To investigate whether caspase 9, a mediator of the mitochondrial caspase pathway and activator of caspase 3, is also involved in SBMA pathogenesis, we used MN-1 cells that stably express either normal (AR-24Q) or mutant AR (AR-65Q) (35). We found that the ligand exposure increases caspase 9 activity by 50 ± 12% in AR-65Q cells compared to AR-24Q cells (P ≤ 0.05) (Fig. 1A). Opening of the mitochondrial transition pore (MTP) is involved in the activation of caspase 9, and CsA inhibits opening of the MTP (36). Therefore, we tested whether the caspase 9 activation observed in MN-1 cells expressing mutant AR could be decreased by CsA. We found that the ligand-induced increase in caspase 9 activity in AR-65Q cells is decreased by 32 ± 8% by treatment of the cells with CsA (P ≤ 0.05) (Fig. 1A). As a positive control for caspase 9 inhibition, we used the specific caspase 9 inhibitor Ac-LEHD-CHO. Next, we asked whether caspase 9 is upstream of caspase 3 in MN-1 cells expressing mutant AR. Although in this cell model of SBMA caspase-3 activation is not ligand-dependent, it is polyglutamine-dependent (Fig. 1B), which is consistent with a previous report (31). Treatment of cells with another caspase 9 inhibitor z-LEHD-FMK blocked the increase in caspase 3 activity by 47%, indicating that the activation of caspase 3 is at least in part due to caspase 9 activation (Fig. 1B). To further test the hypothesis that the mitochondrial caspase pathway is involved in SBMA, we examined whether inhibiting the MTP abrogates caspase 3 activation using a fluorescent-based assay (MitoCasp). As a positive control for this assay, we used staurosporine, an apoptotic inducer (Supplementary Material, Fig. S1). We found that the increase of 29 ± 4% (P ≤ 0.01) in caspase 3 activity observed in the AR-65Q cells relative to the AR-24Q cells was blocked by treating the cells with CsA (39 ± 7%; P ≤ 0.01) (Fig. 1C).
Figure 1.
Mutant AR activates the mitochondrial caspase pathway. (A) MN-1 cells expressing the normal and mutant AR were exposed to vehicle alone or androgen ligand (R1881), with and without caspase 9 inhibitor (Ac-LEHD) or CsA. The AR-65Q cells had an increase in caspase 9 activity in the presence of R1881. CsA and the caspase 9 inhibitor block this increase. (B) Caspase 3 activity was measured with a fluorometric assay in the AR-24Q and AR-65Q MN-1 cells. These cells were treated with vehicle alone, caspase 9 inhibitor (z-LEHD-FMK) or pan-caspase inhibitor (z-VAD). AR-65Q cells showed a ligand-independent increase in caspase 3 activity. This increase is partially blocked by z-LEHD-FMK. (C) We measured caspase 3 activity with another fluorometric assay in MN-1 cells expressing AR-24Q and AR-65Q cells. The cells were treated with either vehicle or CsA. There was an increase in caspase 3 activity in cells expressing the mutant AR. This increase is ligand-independent and partially blocked with CsA. The caspase activities are indicated as percent activity relative to untreated AR-24Q (assigned as 100%) and normalized to total protein content. (D) Immunoblotting of MN-1 total cellular lysates shows increased Bax relative to tubulin in AR-65Q cells. The first lane shows vehicle-treated parental MN-1 cells. (E) Immunoblotting of MN-1 cells for cytochrome C in mitochondrial and cytosolic fractions show no change in cytochrome C release in AR-65Q cells relative to parental MN-1 or AR-24Q cells. The last lane shows HeLa cells treated with staurosporine as a positive control. (F) Immunoblotting of MN-1 total cell lysates for phosphorylated Bad and total Bad. Quantitation showed no change in the ratio of phosphorylated and total Bad in AR-65Q cells relative to AR-24Q cells. The graphs (A–C) show data representative of three experiments each done in triplicate with the error bars indicating standard error of mean (SEM); *P ≤ 0.05.
Alterations in the pro-apoptotic proteins, Bax and Bad and subcellular localization of cytochrome C are associated with the caspase 9/caspase 3 cascade (37–39). Therefore, we tested whether the expression of these markers of mitochondria-dependent apoptosis is altered in MN-1 cells expressing mutant AR. We found that the expression of expanded polyglutamine AR in MN-1 cells increased the levels of Bax protein by 15 ± 3% (P ≤ 0.05) (Fig. 1D). We did not detect any change in cytochrome C release (Fig. 1E). As control for Bax levels and release of cytochrome C, we used lysates of HeLa cells treated with staurosporine (data not shown). It has previously been shown that the pro-apoptotic protein Bad when phosphorylated remains inactive and sequestered in the cytoplasm by the transcription factor, 14-3-3. Upon the activation of the calcineurin pathway, Bad is dephosphorylated and translocates to the mitochondria where it inactivates the anti-apoptotic protein, Bcl-xL and then activates caspase 9 (39–41). To test whether Bad is activated in MN-1 cells expressing mutant AR, we did immunoblotting in MN-1 total cell extracts for phospho-Bad (Ser 136) and total Bad (Fig. 1F). We found no difference in phosphorylated Bad in AR-65Q cells compared to AR-24Q cells. Taken together, these results indicate that mutant AR increases Bax levels and activates the mitochondrial caspase pathway independently of cytochrome C release and dephosphorylation of Bad.
Mitochondrial mass and number are reduced in cells expressing mutant AR
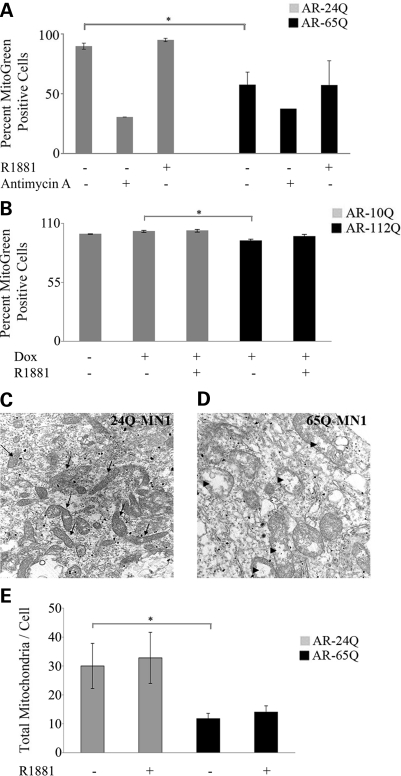
We next sought to determine whether cells expressing mutant AR have mitochondrial abnormalities. We tested this in both stable MN-1 cells and inducible PC12 cells expressing either normal or mutant AR (Fig. 2A and B). We used a mitochondria specific fluorescent dye, MitoTracker Green FM, to examine mitochondrial mass by flow cytometry. This dye localizes to mitochondria independent of the functional state. We found that the fluorescent signal was decreased by 43 ± 11% (P ≤ 0.01) and 6 ± 1% (P ≤ 0.0001) relative to normal cells in mutant MN-1 cells and PC12 cells, respectively. There was no further decrease in percent mitogreen positive cells upon the addition of ligands (Fig. 2A–B). As a positive control, we used antimycin A, an inhibitor of complex III of the electron transport chain, and we found that it decreases mitochondrial mass by greater than 60% in both AR-24Q and AR-65Q cells (Fig. 2A).
Figure 2.
Reduced mitochondrial mass and number in cells expressing mutant AR. (A) AR-24Q and AR-65Q MN-1 cells and (B) differentiated AR-10Q and AR-112Q PC12 cells left uninduced or induced to express AR were treated with vehicle or the mitochondrial toxin, antimycin A for 48 h. The cells were then stained with a fluorescent marker and mitochondrial mass was analyzed by flow cytometry. AR-65Q (A) and AR-112Q cells (B) have ligand-independent decrease in mitochondrial mass. The data in both panels were normalized to the respective vehicle treated normal cells (set as 100%). Shown are the mean ± standard error of three experiments; *P ≤ 0.05. (C and D) Representative high magnification (10 000×) electron micrographs of AR-24Q (C) and AR-65Q MN-1 (D) cells exposed to vehicle for 24 h. The micrographs show more mitochondria (arrows in B) in AR-24Q compared to AR-65Q cells. The AR-65Q cells have vesiculated cristae (arrowheads in D). (E) The total number of mitochondria was counted in 10–12 cells per sample. The error bars indicate SEM; *P ≤ 0.05.
We asked whether the expression of mutant AR in MN-1 cells affects mitochondrial morphology and number. We tested this in MN-1 cells expressing AR-24Q (Fig. 2C) and AR-65Q (Fig. 2D) by electron microscopy. We found that the morphology of mitochondria in mutant MN-1 cells is altered, as the mitochondria in the AR-65Q cells have vesiculated cristae (Fig. 2D; arrowheads), whereas there are numerous electron dense branched cristae in mitochondria of the AR-24Q cells (Fig. 2D; arrows). Quantitative assessment showed a 61 ± 6% (P ≤ 0.01) decrease in total mitochondrial number in AR-65Q cells compared to the AR-24Q cells (Fig. 2E). This decrease was not exacerbated by exposure of the cells to ligand. Together these data indicate a polyglutamine-dependent decrease in mitochondrial mass and number.
Cells expressing mutant AR have decreased mitochondrial membrane potential
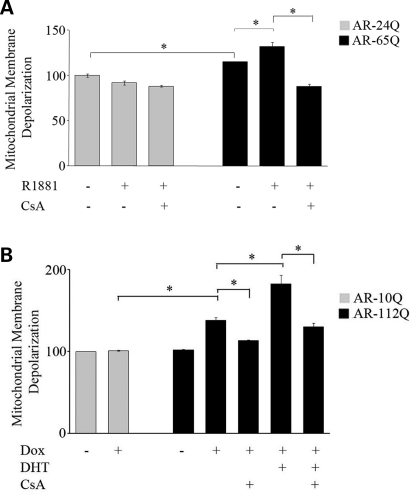
Abnormalities in mitochondrial membrane potential (MtMP) are associated with mitochondrial dysfunction through alteration in bioenergetic function (42,43). To investigate whether the activation of the mitochondrial caspase pathway and reduced number of mitochondria observed in the AR-65Q cells are associated with mitochondrial dysfunction, we examined the MtMP in MN-1 cells. We used the ratiometric cationic dye JC1, which localizes to the mitochondria, and emits a fluorescent signal that varies with the membrane potential (44,45). To capture the heterogeneity of responses across the cell population, we assessed the membrane potential using a fluorescent-activated cell sorter (FACS) (45). We observed an increase (15 ± 5%; P ≤ 0.05) in mitochondrial membrane depolarization (increased ratio of green to red fluorescence) in the AR-65Q cells compared to AR-24Q cells (Fig. 3A) and a 38 ± 3% (P ≤ 0.05) increase in the PC12 cells expressing mutant AR (Fig. 3B). This depolarization was further increased in the presence of the ligand in mutant MN-1 (32 ± 5%; P ≤ 0.05) and PC12 cells (46 ± 15%; P ≤ 0.01). CsA reversed the ligand-dependent depolarization in both MN-1 and PC12 cells, suggesting involvement of the MTP in this process (Fig. 3A and B). Assessment of the MtMP using TMRM, another mitochondria specific potentiometric dye, yielded results consistent with the FACS analysis using JC1 (data not shown). Together, these results suggest that mutant AR causes membrane changes indicative of mitochondrial dysfunction in an expanded polyglutamine-dependent and ligand-dependent manner.
Figure 3.
Mutant AR is associated with decreased MtMP. (A) AR-24Q and AR-65Q MN-1 cells and (B) differentiated AR-10Q and AR-112Q PC12 cells induced to express AR were exposed to vehicle alone, and ligand (R1881 or DHT) or CsA or both. Using flow cytometry, MtMP was measured with a fluorescent indicator of MtMP and expressed relative to the vehicle-treated cells. Depolarization was significantly greater in both AR-65Q MN-1 (A) and AR-112Q PC12 cells (B) compared to the respective normal AR expressing cells. The depolarization is significantly increased by the addition of ligand to the mutant cells. In addition, CsA blocks this depolarization. The histograms represent five (A) and two (B) independent experiments each done in triplicate, with the error bars showing the SEM; *P ≤ 0.05.
Increased reactive oxygen species in SBMA cell models
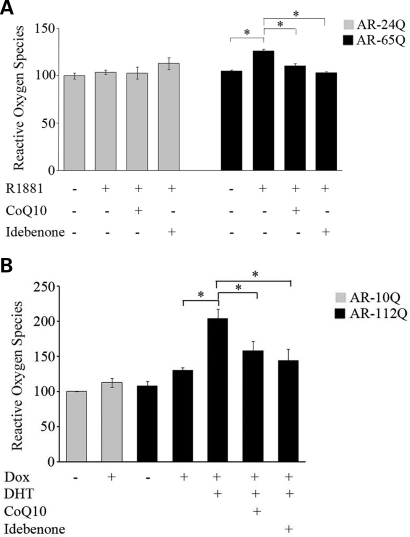
One of the downstream effects of mitochondrial dysfunction is oxidative stress through the generation of ROS (46,47). We assessed ROS using dihydrorhodamine 123 (DHR123), an oxidation-sensitive fluorometric dye that localizes to mitochondria. Exposure of AR-65Q cells to R1881 increased ROS levels by 20 ± 2% (P ≤ 0.01) (Fig. 4A). Treatment of these cells with the antioxidants co-enzyme Q10 (CoQ10) and idebenone blocked this increase in ROS by 13 ± 2% (P ≤ 0.01) and 18 ± 1% (P ≤ 0.01), respectively. Similarly we observed a 57 ± 13% (P ≤ 0.01) increase in ROS in mutant PC12 cells exposed to ligand (Fig. 4B). CoQ10 and idebenone blocked this increase in the mutant PC12 cells by 23 ± 13% (P ≤ 0.05) and 30 ± 17% (P ≤ 0.05), respectively (Fig. 4B).
Figure 4.
Mutant AR increases ROS (A) AR-24Q and AR-65Q MN-1 cells and (B) differentiated AR-10Q and AR-112Q PC12 cells induced to express AR were exposed to vehicle alone, ligand (R1881 or DHT) and CoQ10 or idebenone. ROS levels were measured with a fluorometric assay and are indicated relative to vehicle-treated AR-24Q cells (A) or vehicle-treated cells induced to express AR-10Q (B) (assigned as 100%). In both mutant MN-1 (A) and PC12 (B) cells, ROS levels increased with ligand and co-enzyme Q10 and idebenone blocked this increase. The graph shows representative results of one of the three independent experiments in (A), and the average of three experiments in (B), which were each done in triplicate. The error bars indicate the SEM; *P ≤ 0.05.
Mutant AR alters the expression of genes important for mitochondrial function
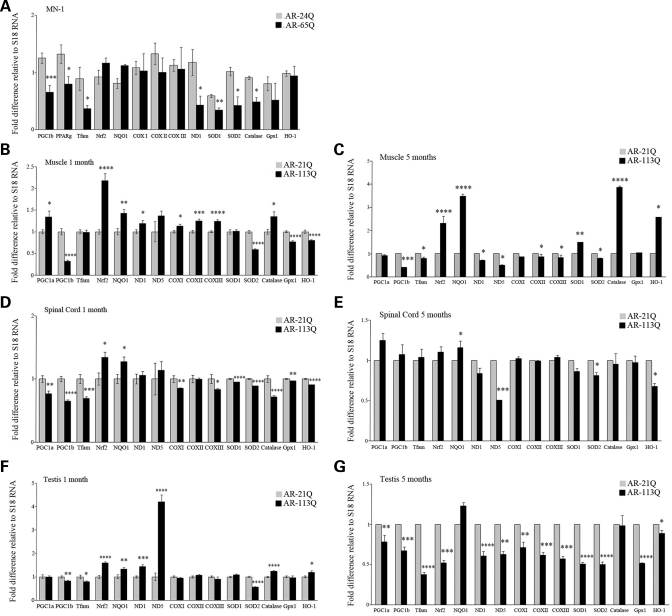
We next investigated whether mitochondrial dysfunction is associated with altered expression of genes related to mitochondrial function in AR-24Q and AR-65Q MN-1 cells when exposed to ligand (Fig. 5A). PGC-1 is a key regulator of mitochondrial biogenesis and function (26,48,49). We found that PGC-1β mRNA was decreased by 48 ± 10% (P ≤ 0.001) in the presence of ligand in cells expressing AR-65Q relative to cells expressing AR-24Q (Fig. 5A). Peroxisome proliferator-activated receptor-γ (PPARγ) and mitochondrial transcription factor A (Tfam) are two nuclear-encoded genes regulated by PGC-1. The expression levels of PPARγ and Tfam were downregulated by 40 ± 10 and 59 ± 7%, respectively, in the ligand-exposed AR-65Q cells (Fig. 5A). Tfam in turn regulates transcription of genes encoded by mitochondrial DNA, including subunits of complexes of the electron transport chain. We examined mRNA levels for the mitochondrial proteins NADH dehydrogenase 1 and 5 (ND1 and ND5), which are subunits of complex I, and cytochrome c oxidase I, II and III (COX I, II and III), which are subunits of complex IV. ND1 mRNA levels were significantly decreased (64 ± 13%; P ≤ 0.05) in AR-65Q cells exposed to ligand, while expression of ND5, COX I, II and III was unaltered (Fig. 5A). To further assess whether the transcription of genes involved in redox balance is affected in the ligand-exposed AR-65Q cells, we examined the transcript levels of antioxidant genes. We quantified the mRNA levels of SOD2 and other cellular antioxidants [SOD1, catalase and glutathione peroxidase (Gpx1)] and found that transcript levels of SOD1, SOD2 and catalase were reduced by 42 ± 6, 58 ± 17 and 46 ± 7%, respectively (P ≤ 0.05) (Fig. 5A). Gpx1 mRNA levels were unchanged. In addition, we found that ligand treatment did not alter transcript levels of NF-E2-related factor-2 (Nrf2), a regulator of antioxidant genes, and its targets, NAD(P)H:quinine oxidoreductase 1 (NQO1) and heme-oxygenase 1 (HO-1) (Fig. 5A). We verified that the decrease in SOD2 transcription translates into protein changes by immunocytochemistry (Supplementary Material, Fig. S2A). We found a decrease in SOD2 staining (FITC-labeled) in ligand-treated mutant MN-1 cells. As a mitochondrial marker, we used Hsp60, and we found that there was a decrease, consistent with reduced mitochondrial number or mass (Supplementary Material, Fig. S2A).
Figure 5.
Mutant AR alters expression of genes important for mitochondrial function. Total RNA from MN-1 cells (A), and hind limb muscle (B and C), spinal cord (D and E) and testis (F and G) of mice expressing AR-21Q, and AR-113Q was analyzed by quantitative RT–PCR as described in Materials and Methods. All transcript levels were normalized to the loading control, S18. (A) Gene expression of R1881-exposed MN-1 cells is shown relative to the respective vehicle treated controls (assigned as 1). (B–G) Transcript levels of pre-manifesting (B, D, F) and manifesting (C, E, G) AR-113Q mouse muscle (B, C), spinal cord (D, E) and testis (F, G) are shown relative to transcript levels in AR-21Q mice (assigned as 1). All experiments were done in triplicate, with the error bars indicating SEM; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001.
We then examined the expression of these genes in muscles, testis and spinal cord from a knock-in mouse model of SBMA (Fig. 5B–G). We analyzed mRNA levels in pre-manifesting 1 month old (Fig. 5B, D and F) and weakness-manifesting 5 month old (Fig. 5C, E and G) mutant AR mice (AR-113Q), and compared the results to mice expressing non-pathological repeat length AR (normal AR mice; AR-21Q).
Notably, we found that in pre-manifesting mice, PGC-1β mRNA was decreased by 68 ± 3% (P ≤ 1 × 10−6), 36 ± 2% (P ≤ 1 × 10−6) and 17 ± 3% (P ≤ 0.01) in hind limb muscle (Fig. 5B), spinal cord (Fig. 5D) and testis (Fig. 5F), respectively. The levels of PGC-1β mRNA remained reduced in muscle and testis of 5 month old mice (Fig. 5C and G). Moreover, we found that in 1 month old mutant mice, SOD2 mRNA was reduced in muscle, spinal cord and testis by 42 ± 2% (P ≤ 1 × 10−10; Fig. 5B), 11 ± 1% (P ≤ 1 × 10−8; Fig. 5D) and 44 ± 1% (P ≤ 1 × 10−8; Fig. 5F), respectively. In 5 month old mutant mice, the SOD2 transcript levels remained decreased by 20 ± 2% (P ≤ 0.05), 19 ± 3% (P ≤ 0.05) and 50 ± 3% (P ≤ 1 × 10−5) in muscle, spinal cord and testis, respectively (Figs 5C, E and G). We examined SOD2 protein levels in immunoblots of muscle and spinal cord tissue lysates of pre-manifesting (1 month) and manifesting (3 month) mice. We found decreased levels of SOD2 in both muscle (Supplementary Material, Fig. S2B) and spinal cord of 3 month old mice (Supplementary Material, Fig. S2C).
In addition, we found that the mRNA levels of Tfam were reduced in spinal cord (31 ± 3%; P ≤ 0.001) and testis (20 ± 2%; P ≤ 0.05) of mice at the pre-manifesting stage. ND1 and ND5 mRNA levels were increased in all three tissues in the 1 month old mice (Fig. 5B, D and F), but decreased as the disease progressed (Fig. 5C, E and G). The expression of Nrf2 and NQO1 was increased in the tissues of 1 month old mice (Fig. 5B, D and F). There was little or no change in PGC-1α, COX I, COX II, COX III, SOD1, Gpx1 and HO-1 in muscle (Fig. 5B and C) and spinal cord (Fig. 5D and E) of 1 and 5 month old mutant mice. In the testis of 5 month old mutant mice, all the analyzed genes had significant reduction in mRNA levels, with the exception of NQO1 and catalase, which were unaltered (Fig. 5G).
Altered subcellular distribution of androgen receptor
We hypothesized that the mitochondrial abnormalities found in cells expressing mutant AR may be due in part to direct association of mutant AR with mitochondria. To test this, we performed subcellular fractionation of MN-1 (Fig. 6A–C) and PC12 cells (Fig. 6D–E). We found that AR with normal (24Q) and expanded (65Q) polyglutamine localized to the cytosolic (Fig. 6A), mitochondrial (Fig. 6B) and nuclear fractions (Fig. 6C). To confirm the efficacy of the fractionation and purity of the lysates, we probed for tubulin, porin and HDAC1 as markers of cytosol, mitochondria and nucleus, respectively. As further confirmation of the purity of these fractions, we detected SOD2 and complex IV only in the mitochondrial fractions.
Figure 6.
Altered subcellular distribution of androgen receptor. Cytosolic (A and D), mitochondrial (B and E) and nuclear (C) fractions were probed for AR in vehicle-treated and R1881-exposed parental, AR-24Q and AR-65Q MN-1 cells (A–C) and in differentiated vehicle-treated PC12 cells (D and E). To confirm the purity of the fractions, we used antibodies for tubulin (cytosol), porin (mitochondria) and HDAC1 (nucleus). HDAC1 and tubulin were not detected in the mitochondrial fractions, and porin was not detected in the cytosolic and nuclear fractions (latter not shown). Heart mitochondrial lysate and knock-in mouse testis tissue lysates were used as negative and positive controls for AR. The parental samples, which do not express AR, were also a negative control. Sample assignments in the gels (A–C) are: lane 1, parental vehicle-treated MN-1; lane 2, vehicle-treated AR-24Q; lane 3, AR-24Q+R1881; Lane 4, vehicle-treated AR-65Q; lane 5, AR-65Q+R1881; lane 6, heart mitochondrial lysate; lane 7, knock-in mouse testis tissue lysate. (D and E) Lane 1, vehicle-treated Dox-induced parental cells; lane 2, vehicle-treated uninduced AR-10Q; lane 3, vehicle-treated Dox-induced AR-10Q; lane 4, Dox-induced AR-10Q+R1881; lane 5, Dox-induced AR-10Q+DHT; lane 6, vehicle-treated uninduced AR-112Q; lane 7, vehicle-treated Dox-induced AR-112Q; lane 8, Dox-induced AR-112Q+R1881; lane 9, Dox-induced AR-112Q+DHT; lane 10, human heart mitochondrial lysate. (F–H) Electron dense immunogold particles indicate AR in AR-24Q (F) and AR-65Q (G) MN-1 cells exposed to vehicle alone. 10 000× electron micrographs (F and G) show mitochondria with (arrows) or without (arrowheads) associated AR in AR-24Q (F) and AR-65Q cells (G). (H) The percentage of mitochondria with associated AR is higher in AR-65Q cells. The histogram represents averages from 10 to 12 cells, with the error bars indicating SEM; *P ≤ 0.05.
To examine the exact localization of AR in mitochondria, we did ultrastructural studies using transmission electron microscopy (Fig. 6F and G). Immunogold labeling for AR in MN-1 cells showed that AR localized within the organelle as well as along the membrane. Quantitative assessment showed an increased number of AR-positive mitochondria (56 ± 3%; P ≤ 0.001) in the AR-65Q MN-1 cells compared to AR-24Q cells (Fig. 6H). Thus, the results from both subcellular fractionation and ultrastructural studies indicate that AR associates with mitochondria.
Attenuation of cell death in MN-1 cells with mitochondrial modulators
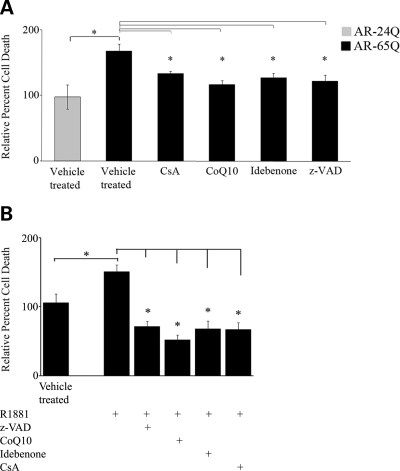
We then investigated whether CsA and antioxidants rescue mutant AR-induced cell death, using loss of membrane integrity as a measure of cell viability. We observed that the increase of 61 ± 11% (P ≤ 0.01) caused by the expression of mutant AR in MN-1 cells was mitigated by CsA (21 ± 4%; P ≤ 0.01), CoQ10 (31 ± 6%; P ≤ 0.01), idebenone (24 ± 6%; P ≤ 0.01) and the pan-caspase inhibitor, z-VAD (26 ± 9%; P ≤ 0.01) (Fig. 7A). Although ligand-dependent toxicity has been found in other models of SBMA, we found that exposure of the cells stably expressing AR-65Q to ligands did not exacerbate cell death (data not shown). However, when mutant AR expression was induced in PC12 cells, ligand exposure increased cell toxicity by 43 ± 10% (P ≤ 0.01) (Fig. 7B). The ligand-dependent toxicity was reversed by z-VAD (53 ± 7%; P ≤ 0.001), CoQ10 (65 ± 7%; P ≤ 0.001), idebenone (55 ± 11%; P ≤ 0.001) and CsA (56 ± 10%; P ≤ 0.001). Staurosporine, an apoptotic inducer, increased cell death by 3-fold (data not shown).
Figure 7.
Cell death in AR-65Q MN-1 cells and AR-112Q PC12 cells is attenuated by treatment with mitochondrial modulators. (A) AR-24Q and AR-65Q MN-1 cells were analyzed for cell toxicity by propidium iodide staining and FACS. Relative to the AR-24Q cells (assigned as 100%), the vehicle-treated AR-65Q cells had increased toxicity. This increase was attenuated by treatment with CsA, CoQ10, idebenone and z-VAD. The histogram represents two experiments each done in triplicate, with error bars indicating the SEM; *P ≤ 0.05. (B) Differentiated PC12 cells induced to express AR-10Q and AR-112Q were exposed to vehicle control, R1881, the pan-caspase inhibitor, z-VAD, CoQ10, idebenone or CsA were analyzed for cell toxicity by propidium iodide staining and FACS. Vehicle-treated AR-112Q cells did not show toxicity relative to the vehicle-treated AR-10Q cells (assigned as 100%). PC12 cells expressing AR-112Q cells exposed to R1881 had increased cell toxicity. This increase was attenuated by treatment with CsA, CoQ10, idebenone and z-VAD. The histogram represents three experiments each done in triplicate, with error bars indicating the SEM; *P ≤ 0.001.
In conclusion, we show that mitochondrial number and function are altered in cell models of SBMA. Treatment of the cells with CsA and antioxidants rescues cells from the toxicity induced by mutant AR. Therefore, we propose that mitochondrial dysfunction is at least in part responsible for the toxicity of mutant AR in SBMA.
DISCUSSION
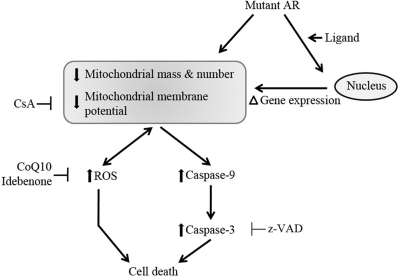
Mitochondrial abnormalities and oxidative stress-mediated neuronal toxicity have been implicated in many neurodegenerative diseases, including ALS and HD (16,18,19,50,51). Here we studied mitochondrial dysfunction in SBMA. We found that the expression of mutant AR is associated with activation of the mitochondrial caspase pathway, decreased MtMP, elevated ROS levels and increased cell death. These changes were mitigated with CsA and antioxidants. We also report consistent decreases in PGC-1β mRNA and in SOD2 mRNA and protein. Altered gene expression in a knock-in mouse model of SBMA was evident in pre-manifesting mice, suggesting that mitochondrial abnormalities and redox imbalance occur early in the disease mechanism. Interestingly, we found AR associated with the mitochondria in MN-1 and PC12 cells. Taken together, the evidence supports both indirect and direct effects of mutant AR on mitochondrial function in SBMA (Fig. 8).
Figure 8.
Mechanism of mitochondrial dysfunction in SBMA. Mitochondrial mass, number and membrane potential are likely affected by mutant AR indirectly through ligand-dependent alterations in mitochondrial gene expression and directly through association of AR with mitochondria. The decrease in MtMP is blocked by CsA. Downstream from the mitochondrial dysfunction, mutant AR increases ROS levels, which can reversibly result in further deleterious effects on the mitochondrial membrane. The antioxidants CoQ10 and idebenone block the increase in ROS levels. Mitochondrial dysfunction increases caspase 9 activity, which in turn increases caspase 3 activity. Elevated ROS levels and caspase 3 activation result in cell death. Activation of the caspases is mitigated by CsA, and cell death is attenuated by CsA, CoQ10, idebenone and the caspase inhibitor, z-VAD. Thus, regardless of the cause of mitochondrial dysfunction, downstream events may be targets for therapeutic intervention, for example with CsA and antioxidants.
SBMA is characterized by both a loss of normal AR function and a toxic gain of function. It is well established that there is transcriptional dysregulation due to partial loss of AR function and to ligand-dependent toxicity (9,10,12–14). The effects of mutant AR on mitochondrial function that we found were both ligand-dependent and -independent. Although previous studies identified gene expression changes in the presence of mutant AR (52), transcriptional effects on either nuclear-encoded mitochondrial genes or transcription of the mitochondrial DNA have not previously been reported. It is likely that mutant AR alters the expression of mitochondrial genes, and this in turn may affect mitochondrial turnover and respiratory function. The nuclear co-activator, PGC-1, is known to regulate mitochondrial biogenesis and function (23,49,53). PGC-1α knock-out mice have impaired energy homeostasis, hyperactivity, striatal degeneration and behavioral changes characteristic of movement disorders, including HD (54). PGC-1α and β regulate oxidant-detoxifying enzymes and also the expression of mitochondrial transcription factor A (Tfam), a direct regulator of mitochondrial DNA replication and transcription (53,55). We could not detect the expression of PGC-1α in MN-1 cells, but transcript levels of its isoform PGC-1β and Tfam were significantly decreased, and this could account for the reduced mitochondrial number and membrane potential that we observed. In addition, expression of cellular antioxidants, such as SOD2, was altered in MN-1 cells, consistent with increased oxidative stress. In pre-manifesting (1 month old) SBMA knock-in mice, we found alterations in transcripts that regulate mitochondrial biogenesis (PGC-1α, PGC-1β and Tfam), metabolism (subunits of complex I and IV that are encoded by the mitochondrial DNA) and antioxidants. This indicates that mitochondrial alterations and oxidative imbalance are early and important events in SBMA. It is also noteworthy that the transcript levels of ND1 and ND5 are decreased concurrent with decreased PGC-1β and Tfam in the manifesting (5 month old) SBMA mice. This suggests that the metabolic demand increases in the tissues of this mouse model as the disease progresses. Also, consistent with the previously reported pathological findings (56,57), the greatest changes occurred in muscle and testis early in the disease and persisted as the disease progressed. Future investigations are warranted to examine whether mutant AR alters bioenergetics and whether it does so by indirectly or directly affecting the activity of the electron transport chain and ATP levels.
Previous studies have shown the localization of steroid and thyroid hormone receptors to mitochondria (58–61). Hormone response elements in the mitochondrial genome and metabolic effects of glucocorticoids and estrogens suggest a direct action of these hormones on mitochondria (62). We are unaware of previous reports of normal or mutated AR localized to neuronal mitochondria. We found mitochondria-associated AR in MN-1 cells, which have constitutive expression of AR, and also in the inducible PC12 cells that express normal and mutant AR at comparable levels. This supports the conclusion that the association is not simply due to clonal variation or differences in the relative levels of AR expression. Although the association of AR with mitochondria that we found is not limited to the mutant form, it is likely that the presence of the expanded polyglutamine-containing AR directly confers a toxic gain of function resulting in mitochondrial abnormalities and oxidative imbalance. These results suggest that the mitochondrial defects may be due to both transcriptional events mediated by the nucleus and a direct toxic effect of the mutant AR on the mitochondria. The mutant proteins that cause HD and spinocerebellar ataxia 3 have also been reported to associate with mitochondria (20,63) suggesting that the mitochondrial association of the mutant proteins in polyglutamine expansion diseases may point towards a common pathogenic mechanism. Our finding that AR localizes to mitochondria in both MN-1 and PC12 cells may have important implications in understanding normal AR function and its role in disease.
It has not previously been well documented that mutant AR affects mitochondrial number and function. However, it has been reported in other neurological disorders that excitotoxicity and other insults cause mitochondria to undergo vesiculation of the cristae, vacuolation and fragmentation (51,64–66). Although in our study we cannot definitively demonstrate fragmentation, our results do indicate that the expanded polyglutamine in AR alters mitochondrial number and morphology of the cristae. The reduced number of mitochondria in PC12 cells induced to express mutant AR indicates that the decrease in MN-1 cells is not simply an adaptive change related to constitutive expression of the mutant protein. Mitochondrial number can be affected by differences in cellular proliferation or passage numbers of the clones. However, the different experimental conditions did not induce any variation in cellular proliferation, and we controlled for possible differences due to different passage numbers of the different clones. The vesiculation and vacuolation that we observed in the AR-65Q cells could be due to mitochondrial toxicity or vice versa. The mitochondrial membrane depolarization and the response to CsA treatment in the in vitro models of SBMA are further evidence for the role of mitochondrial dysfunction and the MTP in SBMA. The protective effect of CsA suggests but does not prove the involvement of the MTP, because CsA also has other effects, such as calcineurin inhibition (67). The effects we found on MtMP were both ligand-dependent and -independent. In light of these results, the effects of the mutant AR on mitochondrial mass, number and membrane potential may be due either to indirect nuclear-encoded transcriptional effects on mitochondria or to direct toxic interactions of the expanded polyglutamine with mitochondria. However, these are not mutually exclusive, as the disease mechanism could involve a combination of both effects.
Regardless of the cause of mitochondrial dysfunction, we found that the effects of the mutant AR include increased ROS, increased Bax protein levels, activation of caspases 9 and 3 and increased cell death. The mitochondrial abnormalities described in this study may be an important modifier of the pathogenic process, where the ligand-dependent effect of the mutant AR on transcription is likely to be exacerbated by a direct effect on mitochondria. The mitochondrial dysfunction may then tip the cell towards the activation of the caspase cascade and death. Alterations in oxidative phosphorylation may result in the generation of ROS and elevated oxidative stress (21,68). Also, increased ROS generation may further damage mitochondria. In vivo deletion of SOD2 in mice results in mitochondrial pathology and shortens lifespan (69). In another study, partial reduction of SOD2 in hAPP transgenic mice accelerates the onset of β amyloid-induced behavioral impairment and significantly worsens Alzheimer's disease pathology (70). We did not see a general decrease in transcript levels of cellular antioxidants, but the mitochondrial antioxidant, SOD2, was specifically and consistently reduced in all three affected tissues, indicating the importance of mitochondria-associated oxidative stress in SBMA. This oxidative imbalance can further result in mitochondrial membrane changes that initiate the intrinsic caspase cascade involving activation of caspases 9 and 3. Our results support cytochrome C release- and Bad-independent activation of caspase 9, consistent with previous reports of cytochrome C independent activation of caspase 9 (71–74). In addition our findings that the increase in Bax and caspase 3 activation is polyglutamine-dependent while caspase 9 activation is ligand-dependent also suggest involvement of other pathways in the activation of caspase 9. Post-translational modifications such as phosphorylation or the release of Smac/Diablo can mediate caspase 9 activation. Further studies of these pathways in cell culture and mouse tissues may be warranted. The finding that CsA mitigates MtMP changes and the activation of caspases 9 and 3 indicates that the expression of the mutant AR is associated with mitochondrial dysfunction, which can be reversed by pharmacological intervention. Importantly, treating these cells with antioxidants was effective not only in mitigating the increase in ROS but also in blocking cell death caused by the expression of mutant AR. While cell death in the MN-1 cells was dependent on mutant polyglutamine but not ligand, the toxicity of mutant AR in PC12 cells was ligand-dependent. With constitutive expression of AR-65Q, the mutant MN-1 clone may have adapted to cope with ligand-dependent effects. These in vitro studies warrant follow-up studies in tissue mitochondria and pre-clinical therapeutic studies of antioxidants in SBMA mouse models.
While we do not differentiate between primary and secondary roles of mitochondrial defects in SBMA pathogenesis, we propose that mitochondrial dysfunction and dysregulation of the antioxidant defense are important in the disease mechanism. Given the central role of mitochondrial integrity in bioenergetics and cell death pathways, improvement of mitochondrial function is worth considering as a therapeutic approach to SBMA.
MATERIALS AND METHODS
Reagents
All cell culture reagents were purchased from Invitrogen (Gaithersburg, MD) unless otherwise mentioned. The non-aromatizable synthetic ligand R1881 and the natural ligand dihydrotestosterone were purchased from Sigma-Aldrich (St Louis, MO, USA). 5,5′,6,6′-tetrachloro-1,1,3,3-tetraethylbenzimidazolcarbocyanine iodide (JC-1), tetramethyl rhodamine methyl ester (TMRM), MitoTracker Green FM and DHR 123 were obtained from Molecular Probes (Gaithersburg, MD, USA). The other compounds used in the study were nerve growth factor (NGF; BD Biosciences), doxycycline (Dox) (Sigma), CsA (Sigma), co-enzyme Q10 (CoQ10) (Sigma), idebenone (Santhera Pharmaceuticals, Switzerland), purified caspase 9 enzyme and caspase 9 inhibitor (Ac-LEHD-CHO; BIOMOL), caspase 9 inhibitor (z-LEHD-FMK; Alexis Biologicals), pan-caspase inhibitor (z-VAD-FMK; BD Pharmingen), staurosporine (STS; Calbiochem), p-trifluoromethoxy carbonyl cyanide phenyl hydrazone (FCCP; Sigma), antimycin A (Sigma), hydrogen peroxide (Fisher Scientific) and propidium iodide (PI; Sigma).
Cell culture
Stably transfected mouse spinal cord motor neuron-derived hybrid (MN-1) cells and stably transfected inducible rat pheochromacytoma (PC12) cells of SBMA were cultured as previously described (15,35). The PC12 cells were provided by Dr Diane Merry (Thomas Jefferson University, PA, USA). MN-1 cells were maintained in normal growth medium (Dulbecco's Modified Eagle's Medium) with 10% fetal bovine serum (FBS; Atlanta biologicals, GA, USA), 2 mm l-glutamine and 100 units/ml penicillin/streptomycin at 37°C in 5% CO2. The MN-1 clones constitutively expressing either the full-length wild-type AR with 24 glutamines (AR-24Q) or the full-length mutant AR with 65 glutamines (AR-65Q) were grown in the same medium supplemented with 350 μg/ml G418. It has been demonstrated that in the absence of ligand, the mutant protein is expressed at lower levels than the wild-type protein (35). The steady-state levels of both forms of the protein are increased with the addition of ligands (52). We used DHT, the natural ligand and R1881, the synthetic non-metabolizable ligand for our experiments. We did not see any difference in the effects of DHT and R1881 at the concentrations used in this study. The parental PC12 cells were grown on rat tail collagen I [0.01% w/v in 0.1 M glacial acetic acid / 1X phosphate-buffered saline (PBS)] coated dishes. Cells were cultured in DMEM with 10% heat inactivated horse serum (HyClone, UT), 5% tetracycline-negative FBS (Tet- FBS; Clontech, CA, USA), 1 mm sodium pyruvate, 2 mm l-glutamine, 100 units/ml penicillin/streptomycin, 200 µg/ml hygromycin B (complete medium) at 37°C in 10% CO2. The AR-10Q and AR-112Q PC12 clones were maintained in the complete medium supplemented with 100 µg/ml G418. PC12 cells were differentiated with 200 ng/ml NGF for 1 day and all experiments were done while maintaining the cells in a differentiated state. The MN-1 cells were not differentiated. All experiments in MN-1 and PC12 cells were done in complete medium containing 10% charcoal dextran stripped (CDSFBS) (HyClone, UT, USA). The experiments with PC12 cells were done in complete medium but replacing the Tet- FBS with CDSFBS and in the presence of 200 ng/ml NGF. The cells were either left uninduced or induced with 10 ng/ml of Dox. Unless otherwise mentioned, cells were treated with 10 nm R1881, 10 nm DHT, 250 nm CsA, 200 nm CoQ10, 200 nm idebenone, 10 µm FCCP, 250 nm Ac-LEHD-CHO, 10 µm z-LEHD-FMK, 5 µm z-VAD for either 24 or 48 h. As positive controls for different assays, we used the mitochondrial toxin, antimycin A (200 or 500 ng/ml), hydrogen peroxide (500 nm) and the apoptotic inducer, staurosporine (1 µm) for 16–18 h and 4 h, respectively.
Mouse model
The generation and characterization of the knock-in mouse model of SBMA (AR-21Q and AR-113Q) has been previously described (56,57). Breeder mice were provided by Drs A.P. Lieberman and D. Robins (University of Michigan, MI, USA). The colonies of mice with the non-pathological repeat length of glutamines knocked in the AR (AR-21Q; normal) were maintained by mating homozygotes. Male mice with 113 glutamines knocked in the AR (AR-113Q; mutant) males are infertile and hence the mutant mice colonies were maintained by mating females heterozygous for the targeted allele with wild-type C57BL/6J males. All experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and were approved by the National Institute of Neurological Disorders and Stroke Animal Care Committee. Tail DNA genotyping was done using the REDExtract-N-AMP tissue PCR kit (Sigma). The primers used for the genotyping were 5′-ggaagtgcagttagggctggga and 5′-gacactgccttacacaactccttggc.
For biochemical studies, mice were anesthetized with isoflurane and sacrificed by cervical dislocation. Spinal cord, hind limb skeletal muscle and testes were dissected and snap frozen in liquid nitrogen. The tissues were stored at −80°C and thawed on ice immediately prior to RNA and protein extraction.
Caspase 9 and caspase 3 assays
Caspase 9 activity was assessed with the Caspase-Glo 9 assay as per the manufacturer's protocol (Promega). MN-1 cells were seeded in 24-well plates at 6 × 104 cells/well. Cells were exposed to vehicle alone or androgen ligands (R1881 or DHT), and treated with or without caspase 9 inhibitor (Ac-LEHD-CHO; Biomol) and CsA. After 48 h, cells were trypsinized and centrifuged at 800g for 5 min. The cell pellets were washed in 1X PBS and re-suspended in 200 µl of 1X PBS. Half of the cell suspension was transferred to a 96-well plate for the luminescence assay and the other half of the suspension was used to determine total protein concentration. Purified caspase 9 enzyme (100 units/ml) was used as a positive control, and the vehicle control was used as a blank for the assay. Caspase-Glo 9 reagent (100 µl) was added to each sample in the 96-well plate; the samples were incubated for 1 h at room temperature and luminescence measured with a Victor3 multi-well luminometer (Perkin Elmer, MA, USA). The luminescence is a direct measure of the caspase 9 activity and was normalized to total cellular protein. An equal volume of the Mammalian Protein Extraction Reagent (M-PER; Pierce Biotechnology) was added to the 100 µl cell suspension to lyse the cells, and following centrifugation at 800g for 7 min the lysate was used to determine the protein concentration by the MicroBCA Protein Assay method as per manufacturer's protocol (Pierce Biotechnology).
Caspase 3 activity was measured with two different assays. In the first, caspase 3 activity was determined by the ApoTarget fluorometric assay (Biosource International) according to manufacturer's instructions and a published protocol (31). Another caspase 9 inhibitor (z-LEHD-FMK) and pan-caspase inhibitor (z-VAD) were used at concentrations of 10 and 5 µm, respectively. Protein concentrations were estimated on parallel samples using the bicinchoninic acid protein assay method.
In an independent caspase 3 assay, the activity was measured with MitoCasp (Cell Technology, CA, USA) as per manufacturer's instructions. Briefly, cells grown in 24-well dishes and treated for 48 h were stained with the MitoCasp reagents for 1 h at 37°C. After PBS washes and centrifugation, the re-suspended pellets were transferred to a black-wall 96-well plate. The caspase 3 activity was determined with a fluorescent-based assay wherein carboxyfluorescein (FAM)-labeled fluoromethyl ketone (FMK)-peptide inhibitors bind to active caspases and fluoresce green (emission peaks at 510 nm). The fluorescence was measured in a Victor3 multi-well plate fluorometer and was normalized to total protein content as described earlier (see caspase 9 activity assay).
Mitochondrial mass
MN-1 and PC12 cells were seeded at 2.5 × 105 and 3 × 105 cells/well, respectively, in 6-well dishes. Following 48 h exposure to vehicle or androgen ligands, the cells were stained with 100 nm MitoTracker Green FM (MTG) in complete medium prepared using phenol-red free DMEM for 30 min at 37°C (75). After trypsinizing the adherent cells, they were centrifuged at 800g for 5 min at room temperature. The pellets were washed twice in PBS supplemented with 0.5% FCS and re-suspended in 400 µl PBS/0.5% FCS. Green fluorescent cells were analyzed by flow cytometry (Becton Dickinson FACS Calibur) using 488 nm laser excitation and 530/30 nm MTG emission and Cell Quest software (Becton Dickinson). A total of 100,000 cells were analyzed in each sample. Cell populations were gated by size (forward scatter) and granularity (side scatter) to exclude debris and cell clumps for all samples. An unstained negative control sample and a positive control sample of normal cells stained with the MTG were used to establish a 100% positive reference region. Experimental samples stained with MTG were then evaluated for the percent green fluorescent cells in the reference region.
Mitochondrial membrane potential (MtMP)
MtMP was determined as per published protocols (45,76). The lipophilic cation JC-1 was used to obtain ratiometric determinants of membrane potential using flow cytometry (Becton Dickinson FACS Calibur) and Cell Quest software (Becton Dickinson). Lower potential causes JC1 to remain in a monomeric form with low amounts in the mitochondria and fluoresce green. In contrast, when the mitochondrial membranes are energized, the dye forms J-aggregates, accumulates in the mitochondria and fluoresces red. MN-1 and PC12 cells were seeded at 2 × 105 and 3 × 105 cells/well, respectively, in 6-well dishes. The cells were exposed to vehicle alone or androgen ligands with or without 250 nm CsA for 48 h. As a positive control, cells were exposed to 10 µm FCCP for 24 h. Cells were then stained with 5 µm JC-1 in complete medium prepared with phenol-red free DMEM for 40 min at 37°C. Adherent cells were collected by trypsinization and centrifuged at 800g for 5 min at room temperature. The pellets were then washed twice in PBS supplemented with 0.5% FCS and re-suspended in 400 µl PBS/0.5% FCS before analysis by flow cytometry. A total of 50,000 cells were analyzed. The degree of mitochondrial membrane depolarization was calculated as the ratio of depolarized (green) to polarized (red) signals from the dot plots.
In an independent assay, MtMP was determined using the fluorochrome TMRM, which localizes to mitochondria and detects membrane potential. This was normalized to total number of mitochondria, as measured with MTG, which labels all mitochondria irrespective of their functional state (76). The ratio between TMRM and MTG fluorescence indicates MtMP normalized to the number of mitochondria. Briefly, cells in 6-well dishes were exposed to vehicle alone, or to androgen ligands (R1881 and DHT), or treated with 250 nm CsA for 48 h. As a positive control for membrane depolarization, cells were exposed to the mitochondrial uncoupler, FCCP (10 µm) for 24 h. They were then stained with 100 nm each of TMRM and MTG for 30 min at 37°C. Following trypsinization and centrifugation, the cells were washed twice with PBS to remove excess fluorochrome, re-suspended in 1X PBS and gently lysed with 1% (v/v) Triton X-100. The relative fluorescence for TMRM (excitation, 550 nm; emission, 590 nm) and MTG (excitation, 405 nm; emission, 515 nm) was measured using the Victor (3) multi-well plate fluorometer.
ROS assay
ROS levels were measured in cell lines using fluorescence-based assays. MN-1 and PC12 cells were seeded at 6 × 104 and 1 × 105 cells/well, respectively, in 24-well dishes. These cells were either exposed to vehicle or androgen ligands for 48 h. As positive controls, the cells were treated for 16 h each with 200 ng/ml antimycin A, and 500 nm hydrogen peroxide. They were also treated with 200 nm each of the antioxidants CoQ10 and idebenone. After 48 h of treatment, the cells were loaded with the probe, DHR 123 at a final concentration of 5 µm. DHR 123 is an oxidation-sensitive lipophilic dye that enters a cell, localizes to the mitochondria and fluoresces when oxidized by mitochondrial ROS to positively charged derivatives. Cell fluorescence with a peak emission of 530 nm was measured in lysed cells using a Victor3 multi-well plate fluorometer. The relative fluorescence units (RFU) were normalized to total protein content, which was determined as for the caspase assays described earlier.
RNA isolation and quantitative real-time polymerase chain reaction (qRT–PCR)
Total RNA was isolated from MN-1 cells (1 × 106) exposed to vehicle control or the androgen ligands (16–24 h) and from about 40–60 mg of mouse tissues as per a previously published protocol (77). Briefly, the RNA was extracted using TRIzol (Invitrogen) and further purified using the RNAeasy clean-up kit according to the manufacturer's protocol (QIAGEN). Isolated RNA was first reverse transcribed to cDNA using random hexamer primers with the High Capacity cDNA Archive kit (Applied Biosystems, CA, USA). The qRT–PCR reactions were performed in triplicate with SYBR Green PCR core reagents using the ABI Prism 7900 Sequence Detector System (Applied Biosystems). Each of the primer sets demonstrated amplification, with no amplification of the negative controls. Since SYBR Green can bind primer dimers and produce non-specific signal, melting curves were generated for all primer sets in all experiments to ascertain specificity of amplified products. Target gene primer pairs are listed in Supplementary Material, Table S1. cDNA transcribed with known concentrations of RNA was diluted at 2-fold series to generate a standard curve for each primer set according to the cycle of threshold value obtained from real-time PCR. Primer efficiencies were estimated from the standard curves. Based on the standard curves, we used 300 nm each of the forward and reverse primers and 3 ng of the template for all experiments. Transcript levels were normalized to the loading control, S18 rRNA. Relative gene expression was determined by the Pfaffl method (78). The parental MN-1 cells, vehicle treated AR-24Q and AR-65Q clones were set to 1. The transcript levels of ligand-exposed AR-24Q and AR-65Q cells were compared to the respective vehicle treated samples. In the analysis of the knock-in mice, the non-pathological (AR-21Q; normal) and pathological (AR-113Q; mutant) samples were normalized to the AR-113Q wild-type littermates. Gene expression in the AR-113Q mice was calculated relative to that of the AR-21Q mice, which was assigned as 1. For the study, we used 1 month and 5 month old mice, and all experiments were conducted at least twice with each condition in triplicate. The numbers of 1 month old (pre-manifesting) and 3–5 month old (manifesting) mice used in the study were as follows: AR-21Q 1 month (n = 10) and 3–5 month old (n = 7); AR-113Q mutant 1 month (n = 9) and 3–5 month old (n = 4); AR-113Q wild type littermates 1 month (n = 6) and 3–5 month old (n = 7).
Immunocytochemistry and confocal microscopy
MN-1 cells were seeded in two-chamber CC2-coated slides (Lab-Tek; NUNC) at 105 cells/well and exposed to vehicle control or R1881 for 48 h. They were then fixed in ice cold 4% paraformaldehyde at room temperature for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 15 min and blocked with 5% normal goat serum for 1 h. The samples were incubated with polyclonal antibodies to SOD2 (4 µg/ml; SOD-110, Stressgen) and Hsp60 (1:150; Santa Cruz Biotechnology). Following washes in 1X PBS, they were incubated with FITC and Texas red conjugated secondary antibodies (Jackson Immuno-Research Laboratories), washed and mounted using DAPI-containing Vectashield mounting media (Vector Labs). Deconvoluted images were acquired with a Zeiss LSM510 laser scanning confocal microscope.
Total cell protein extraction, sub-cellular fractionation and immunoblotting
MN-1 and PC12 cells were seeded in 10 cm dishes at 8 × 105 and 1 × 106 cells/dish, respectively. They were exposed to vehicle alone or the androgen ligands for 48 h. The cells were then trypsinized and centrifuged at 800g for 7 min, and the pellets were washed twice in cold 1X PBS. For total cell lysates, the pellets were re-suspended in 200 µl lysis buffer (0.01 M sodium phosphate, pH 7.2, 0.15 M NaCl, 0.1% sodium dodecyl sulfate, 1% sodium deoxycholate, 1% NP40 and 2 mm EDTA) with a protease inhibitor cocktail (Roche Diagnostics) and incubated on ice for 10 min. The lysates were then sonicated for 10 s and microfuged at 20 000g for 10 min at 4°C. The supernatant was then collected as the total cell lysate. Total tissue homogenate was prepared by homogenizing 50–70 mg of tissue in 500 µl lysis buffer and protein extracted following the protocol as described above.
Mitochondrial subcellular fractions were prepared from cells using the mitochondrial isolation reagents (Sigma) as per the manufacturer's protocol. Briefly, cells suspended in extraction buffer were homogenized in a Dounce homogenizer using 15 strokes. The samples were then microfuged at 1000g for 5 min at 4°C. The supernatant was transferred to a fresh tube and centrifuged at 3500g for 10 min at 4°C. The resulting supernatant was stored as the cytosolic fraction. The pellet was re-suspended in 100 µl CelLytic M cell lysis reagent (Sigma) and stored as the mitochondrial fraction. Nuclear fractionations were prepared using the NE-PER nuclear and cytoplasmic extraction reagents (Pierce Biotechnology) following the manufacturer's instructions. Total protein concentrations of all preparations were determined with the BCA protein assay kit (Pierce Biotechnology) according to the manufacturer's protocol.
For SDS–PAGE, protein lysates (100 µg) were denatured at 70°C in 4× sample buffer (Invitrogen) for 10 min, separated on 4–12% Tris-glycine acrylamide gels and electro-transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). All immunoblotting procedures were carried out in 5% non-fat dry milk in 1× Tris-buffered saline (TBS). The antibodies used are as follows: rabbit anti-AR (N-20; sc-816, Santa Cruz Biotechnology; 1:1000), mouse anti-cytochrome C (MSA06; Mitosciences; 1:1000), rabbit anti-Bax (sc-493; Santa Cruz Biotechnology; 1:500), rabbit anti-phospho Bad (9295S; Cell Signaling Technology; 1:500), rabbit anti-Bad (9292; Cell Signaling Technology; 1:500), rabbit anti-SOD2 (SOD-110; Stressgen; 1:500), mouse anti-Hsp60 (SPA-829; Stressgen; 1:500), mouse anti-OxPhos complex IV (A6403; Invitrogen; 2 µg/ml), mouse anti-porin (A31855; Invitrogen; 0.5 µg/ml), mouse anti-α-tubulin (T9026; Sigma; 1:1000) and mouse anti-HDAC1 (89910; Pierce; 1:1000). Immunodetection was done with horse radish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (1:15,000; Jackson Immunoresearch), and visualized with enhanced chemiluminescence reagent (Perkin-Elmer) following the manufacturer's instructions. Re-probing of the blots was done after stripping the blots with Re-blot Plus (Chemicon) according to manufacturer's protocol. Densitometric quantitation of the resultant bands was done with Image J software (NIH, Bethesda) and protein amounts were normalized to loading controls (tubulin for total lysates and cytosolic fractions, porin for mitochondria and HDAC1 for nuclear fractions). The values were normalized to the mean of the vehicle-treated MN-1 cells, uninduced normal PC12 cells or normal (AR-21Q) mice, which were each assigned as 1. Phosphorylated Bad was normalized to total Bad levels. Cytochrome C release was determined as the ratio of cytosolic to mitochondrial cytochrome C.
Immuno-electron microscopy
MN-1 cells were plated on Permanox chambered slides (Lab-Tek; Nunc) and exposed to vehicle control or the androgen ligands for 24 h. Cells for immunogold labeling were fixed with ice cold 4% paraformaldehyde in PBS for 1 h at room temperature followed by blocking and permeablization with 5% goat serum/0.1% saponin for 1 h. The cells were then incubated with anti-AR antibodies (1:100; N20, Santa Cruz Biotechnology) in 5% goat serum/0.1% saponin for 1.5 h at room temperature. After three 10 min washes in 1% goat serum/1X PBS and two 10 min washes in 2% dry milk, they were incubated with Nanogold-conjugated secondary antibody (1:250; Nanoprobes) for 1 h at room temperature. Following four 10 min washes in 2% dry milk/1X PBS the cells were fixed in 2% glutaraldehyde/1X PBS for 30 min. The slides were subjected to staining and silver enhancement as described previously (79). After dehydration, silver enhancement, embedding and sectioning, the samples were examined with an electron microscope (1200EX; JEOL). Electron micrographs of 12–15 cells were acquired and the mitochondria quantified by two independent researchers blinded to the source of the sample. The number of mitochondria per cell and the number of AR positive mitochondria were quantified. Immunogold particles appearing as electron dense dark spots within and in the peripheral areas (within 30 nm) of the mitochondria were counted as AR associated with the organelle. This distance was approximated based on the lengths of two ∼15 nm long IgG molecules separating the gold and the antigen (80).
Cell death assays
We used a flow cytometry-based cell death assay as previously published (81). MN-1 and PC12 cells seeded at 2 × 105 and 3 × 105 cells /well in six well dishes were exposed to the vehicle control or the androgen ligands (R1881 and DHT), 1 µm staurosporine, 500 ng/ml antimycin A, 200 nm each of CsA, CoQ10 and idebenone. Except for staurosporine (4 h) and antimycin (16 h) treatments, the rest of the treatments were for 48 h. The cells were then trypsinized, gently pelleted and re-suspended in 1 ml of 1X PBS. The suspensions were then stained with 1 µg/ml propidium iodide (PI; Sigma) immediately before flow cytometry (Becton Dickinson FACS Calibur). For each sample, 80 000 non-gated events were acquired. PI fluorescence was analyzed in the FL-3 channel in dot and density blot formats. The results were expressed as a percentage of PI-positive dead cells divided by the total number of cells.
Statistics
All experiments were conducted at least twice with each condition in triplicate. Comparisons between any two groups of data were done using the single-factorial analysis of variance (ANOVA). A P-value of ≤0.05 was considered statistically significant. Numerical data were expressed as averages ± the standard error of mean (SEM).
SUPPLEMENTARY MATERIAL
FUNDING
The work was supported by intramural research funds from the NINDS, and by Fondazione Telethon (GFP04005) and a Kennedy's Disease Association research grant (to M.P.). Funding to pay the open access charges was provided by the intramural research funds from the NINDS.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs Andrew Lieberman and Diane Robins for providing us with the knock-in mouse model and to Dr Diane Merry for the PC12 cell lines. We thank Martha Kirby in the National Human Genome Research Institute FACS Core Facility, Carolyn Smith with the National Institute of Neurological Disorders and Stroke (NINDS) imaging Core Facility and Susan Cheng and Virginia Crocker in the NINDS Electron Microscopy Core Facility. We thank the members of the Neurogenetics Branch, particularly Melanie Knight and Isabella Palazzolo for their helpful discussions. We also thank Santhera Pharmaceuticals for providing idebenone for this study.
Conflict of Interest statement. The authors declare no conflict of interest.
REFERENCES
- 1.La Spada A.R., Wilson E.M., Lubahn D.B., Harding A.E., Fischbeck K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 2.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Ann. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 3.Katsuno M., Adachi H., Waza M., Banno H., Suzuki K., Tanaka F., Doyu M., Sobue G. Pathogenesis, animal models and therapeutics in spinal and bulbar muscular atrophy (SBMA) Exp. Neurol. 2006;200:8–18. doi: 10.1016/j.expneurol.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 4.Brooks B.P., Fischbeck K.H. Spinal and bulbar muscular atrophy: a trinucleotide-repeat expansion neurodegenerative disease. Trends Neurosci. 1995;18:459–461. doi: 10.1016/0166-2236(95)94497-s. [DOI] [PubMed] [Google Scholar]
- 5.Wieacker P.F., Knoke I., Jakubiczka S. Clinical and molecular aspects of androgen receptor defects. Exp. Clin. Endocrinol. Diabetes. 1998;106:446–453. doi: 10.1055/s-0029-1212014. [DOI] [PubMed] [Google Scholar]
- 6.Poletti A., Negri-Cesi P., Martini L. Reflections on the diseases linked to mutations of the androgen receptor. Endocrine. 2005;28:243–262. doi: 10.1385/ENDO:28:3:243. [DOI] [PubMed] [Google Scholar]
- 7.Palazzolo I., Gliozzi A., Rusmini P., Sau D., Crippa V., Simonini F., Onesto E., Bolzoni E., Poletti A. The role of the polyglutamine tract in androgen receptor. J. Steroid Biochem. Mol. Biol. 2008;108:245–253. doi: 10.1016/j.jsbmb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Adachi H., Waza M., Katsuno M., Tanaka F., Doyu M., Sobue G. Pathogenesis and molecular targeted therapy of spinal and bulbar muscular atrophy. Neuropathol. Appl. Neurobiol. 2007;33:135–151. doi: 10.1111/j.1365-2990.2007.00830.x. [DOI] [PubMed] [Google Scholar]
- 9.Taylor J.P., Taye A.A., Campbell C., Kazemi-Esfarjani P., Fischbeck K.H., Min K.T. Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev. 2003;17:1463–1468. doi: 10.1101/gad.1087503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCampbell A., Taylor J.P., Taye A.A., Robitschek J., Li M., Walcott J., Merry D., Chai Y., Paulson H., Sobue G., et al. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 11.Katsuno M., Adachi H., Tanaka F., Sobue G. Spinal and bulbar muscular atrophy: ligand-dependent pathogenesis and therapeutic perspectives. J. Mol. Med. 2004;82:298–307. doi: 10.1007/s00109-004-0530-7. [DOI] [PubMed] [Google Scholar]
- 12.Katsuno M., Adachi H., Doyu M., Minamiyama M., Sang C., Kobayashi Y., Inukai A., Sobue G. Leuprorelin rescues polyglutamine-dependent phenotypes in a transgenic mouse model of spinal and bulbar muscular atrophy. Nat. Med. 2003;9:768–773. doi: 10.1038/nm878. [DOI] [PubMed] [Google Scholar]
- 13.Katsuno M., Adachi H., Kume A., Li M., Nakagomi Y., Niwa H., Sang C., Kobayashi Y., Doyu M., Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 14.Chevalier-Larsen E.S., O'Brien C.J., Wang H., Jenkins S.C., Holder L., Lieberman A.P., Merry D.E. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J. Neurosci. 2004;24:4778–4786. doi: 10.1523/JNEUROSCI.0808-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walcott J.L., Merry D.E. Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. J. Biol. Chem. 2002;277:50855–50859. doi: 10.1074/jbc.M209466200. [DOI] [PubMed] [Google Scholar]
- 16.Trushina E., McMurray C.T. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145:1233–1248. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- 17.Seong I.S., Ivanova E., Lee J.M., Choo Y.S., Fossale E., Anderson M., Gusella J.F., Laramie J.M., Myers R.H., Lesort M., et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 18.Hervias I., Beal M.F., Manfredi G. Mitochondrial dysfunction and amyotrophic lateral sclerosis. Muscle Nerve. 2006;33:598–608. doi: 10.1002/mus.20489. [DOI] [PubMed] [Google Scholar]
- 19.Browne S.E., Beal M.F. Oxidative damage in Huntington's disease pathogenesis. Antioxid. Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 20.Panov A.V., Gutekunst C.A., Leavitt B.R., Hayden M.R., Burke J.R., Strittmatter W.J., Greenamyre J.T. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat. Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 21.Di Prospero N.A., Fischbeck K.H. Therapeutics development for triplet repeat expansion diseases. Nat. Rev. Genet. 2005;6:756–765. doi: 10.1038/nrg1690. [DOI] [PubMed] [Google Scholar]
- 22.Spiegelman B.M. Transcriptional control of energy homeostasis through the PGC1 coactivators. Novartis Found. Symp. 2007;286:3–6. [PubMed] [Google Scholar]
- 23.Spiegelman B.M. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Found. Symp. 2007;287:60–63. [PubMed] [Google Scholar]
- 24.Puigserver P. Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha. Int. J. Obesity. 2005;29:S5–S9. doi: 10.1038/sj.ijo.0802905. [DOI] [PubMed] [Google Scholar]
- 25.Shi Q., Gibson G.E. Oxidative stress and transcriptional regulation in Alzheimer disease. Alz. Dis. Assoc. Disord. 2007;21:276–291. doi: 10.1097/WAD.0b013e31815721c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 27.Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 28.Weydt P., Pineda V.V., Torrence A.E., Libby R.T., Satterfield T.F., Lazarowski E.R., Gilbert M.L., Morton G.J., Bammler T.K., Strand A.D., et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Piccioni F., Pinton P., Simeoni S., Pozzi P., Fascio U., Vismara G., Martini L., Rizzuto R., Poletti A. Androgen receptor with elongated polyglutamine tract forms aggregates that alter axonal trafficking and mitochondrial distribution in motor neuronal processes. FASEB J. 2002;16:1418–1420. doi: 10.1096/fj.01-1035fje. [DOI] [PubMed] [Google Scholar]
- 30.Beauchemin A.M., Gottlieb B., Beitel L.K., Elhaji Y.A., Pinsky L., Trifiro M.A. Cytochrome c oxidase subunit Vb interacts with human androgen receptor: a potential mechanism for neurotoxicity in spinobulbar muscular atrophy. Brain Res. Bull. 2001;56:285–297. doi: 10.1016/s0361-9230(01)00583-4. [DOI] [PubMed] [Google Scholar]
- 31.Palazzolo I., Burnett B.G., Young J.E., Brenne P.L., La Spada A.R., Fischbeck K.H., Howell B.W., Pennuto M. Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Hum. Mol. Genet. 2007;16:1593–1603. doi: 10.1093/hmg/ddm109. [DOI] [PubMed] [Google Scholar]
- 32.Piccioni F., Roman B.R., Fischbeck K.H., Taylor J.P. A screen for drugs that protect against the cytotoxicity of polyglutamine-expanded androgen receptor. Hum. Mol. Genet. 2004;13:437–446. doi: 10.1093/hmg/ddh045. [DOI] [PubMed] [Google Scholar]
- 33.Ellerby L.M., Hackam A.S., Propp S.S., Ellerby H.M., Rabizadeh S., Cashman N.R., Trifiro M.A., Pinsky L., Wellington C.L., Salvesen G.S., et al. Kennedy's disease: caspase cleavage of the androgen receptor is a crucial event in cytotoxicity. J. Neurochem. 1999;72:185–195. doi: 10.1046/j.1471-4159.1999.0720185.x. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi Y., Miwa S., Merry D.E., Kume A., Mei L., Doyu M., Sobue G. Caspase-3 cleaves the expanded androgen receptor protein of spinal and bulbar muscular atrophy in a polyglutamine repeat length-dependent manner. Biochem. Biophys. Res. Commun. 1998;252:145–150. doi: 10.1006/bbrc.1998.9624. [DOI] [PubMed] [Google Scholar]
- 35.Brooks B.P., Paulson H.L., Merry D.E., Salazar-Grueso E.F., Brinkmann A.O., Wilson E.M., Fischbeck K.H. Characterization of an expanded glutamine repeat androgen receptor in a neuronal cell culture system. Neurobiol. Dis. 1997;3:313–323. doi: 10.1006/nbdi.1997.0126. [DOI] [PubMed] [Google Scholar]
- 36.Waldmeier P.C., Zimmermann K., Qian T., Tintelnot-Blomley M., Lemasters J.J. Cyclophilin D as a drug target. Curr. Med. Chem. 2003;10:1485–1506. doi: 10.2174/0929867033457160. [DOI] [PubMed] [Google Scholar]
- 37.Tang T.S., Slow E., Lupu V., Stavrovskaya I.G., Sugimori M., Llinas R., Kristal B.S., Hayden M.R., Bezprozvanny I. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington's disease. Proc. Natl Acad. Sci. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin Z., El-Deiry W.S. Overview of cell death signaling pathways. Cancer Biol. Ther. 2005;4:139–163. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- 39.Masters S.C., Yang H., Datta S.R., Greenberg M.E., Fu H. 14-3-3 inhibits Bad-induced cell death through interaction with serine-136. Mol. Pharmacol. 2001;60:1325–1331. doi: 10.1124/mol.60.6.1325. [DOI] [PubMed] [Google Scholar]
- 40.Datta S.R., Ranger A.M., Lin M.Z., Sturgill J.F., Ma Y.C., Cowan C.W., Dikkes P., Korsmeyer S.J., Greenberg M.E. Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev. Cell. 2002;3:631–643. doi: 10.1016/s1534-5807(02)00326-x. [DOI] [PubMed] [Google Scholar]
- 41.Humbert S., Bryson E.A., Cordelieres F.P., Connors N.C., Datta S.R., Finkbeiner S., Greenberg M.E., Saudou F. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev. Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 42.Iijima T. Mitochondrial membrane potential and ischemic neuronal death. Neurosci. Res. 2006;55:234–243. doi: 10.1016/j.neures.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Nicholls D.G. Mitochondrial membrane potential and aging. Aging Cell. 2004;3:35–40. doi: 10.1111/j.1474-9728.2003.00079.x. [DOI] [PubMed] [Google Scholar]
- 44.Iijima T., Mishima T., Akagawa K., Iwao Y. Mitochondrial hyperpolarization after transient oxygen-glucose deprivation and subsequent apoptosis in cultured rat hippocampal neurons. Brain Res. 2003;993:140–145. doi: 10.1016/j.brainres.2003.09.041. [DOI] [PubMed] [Google Scholar]
- 45.Fillon S., Lang F., Jendrossek V. Pseudomonas aeruginosa triggered apoptosis of human epithelial cells depends on the temperature during infection. Cell Physiol. Biochem. 2002;12:207–214. doi: 10.1159/000066280. [DOI] [PubMed] [Google Scholar]
- 46.Adam-Vizi V., Chinopoulos C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol. Sci. 2006;27:639–645. doi: 10.1016/j.tips.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Brookes P.S. Mitochondrial H(+) leak and ROS generation: an odd couple. Free Radic. Biol. Med. 2005;38:12–23. doi: 10.1016/j.freeradbiomed.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 48.Butow R.A., Bahassi E.M. Adaptive thermogenesis: orchestrating mitochondrial biogenesis. Curr. Biol. 1999;9:R767–R769. doi: 10.1016/S0960-9822(00)80008-1. [DOI] [PubMed] [Google Scholar]
- 49.Puigserver P., Spiegelman B.M. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 50.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 51.Kong J., Xu Z. Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. J. Neurosci. 1998;18:3241–3250. doi: 10.1523/JNEUROSCI.18-09-03241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lieberman A.P., Harmison G., Strand A.D., Olson J.M., Fischbeck K.H. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet. 2002;11:1967–1976. doi: 10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- 53.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 54.Lin J., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Lin J., Tarr P.T., Yang R., Rhee J., Puigserver P., Newgard C.B., Spiegelman B.M. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J. Biol. Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 56.Yu Z., Dadgar N., Albertelli M., Gruis K., Jordan C., Robins D.M., Lieberman A.P. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J. Clin. Invest. 2006;116:2663–2672. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu Z., Dadgar N., Albertelli M., Scheller A., Albin R.L., Robins D.M., Lieberman A.P. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 CAG knock-in mice reveal toxic effects of the mutant protein. Am. J. Pathol. 2006;168:195–204. doi: 10.2353/ajpath.2006.050619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Solakidi S., Psarra A.M., Nikolaropoulos S., Sekeris C.E. Estrogen receptors alpha and beta (ERalpha and ERbeta) and androgen receptor (AR) in human sperm: localization of ERbeta and AR in mitochondria of the midpiece. Hum. Reprod. 2005;20:3481–3487. doi: 10.1093/humrep/dei267. [DOI] [PubMed] [Google Scholar]
- 59.Psarra A.M., Bochaton-Piallat M.L., Gabbiani G., Sekeris C.E., Tsacopoulos M. Mitochondrial localization of glucocortocoid receptor in glial (Muller) cells in the salamander retina. Glia. 2003;41:38–49. doi: 10.1002/glia.10130. [DOI] [PubMed] [Google Scholar]
- 60.Psarra A.M., Sekeris C.E. Steroid and thyroid hormone receptors in mitochondria. IUBMB life. 2008;60:210–223. doi: 10.1002/iub.37. [DOI] [PubMed] [Google Scholar]
- 61.Demonacos C., Tsawdaroglou N.C., Djordjevic-Markovic R., Papalopoulou M., Galanopoulos V., Papadogeorgaki S., Sekeris C.E. Import of the glucocorticoid receptor into rat liver mitochondria in vivo and in vitro. J. Steroid. Biochem. Mol. Biol. 1993;46:401–413. doi: 10.1016/0960-0760(93)90231-k. [DOI] [PubMed] [Google Scholar]
- 62.Psarra A.M., Solakidi S., Sekeris C.E. The mitochondrion as a primary site of action of regulatory agents involved in neuroimmunomodulation. Ann. NY Acad. Sci. 2006;1088:12–22. doi: 10.1196/annals.1366.019. [DOI] [PubMed] [Google Scholar]
- 63.Pozzi C., Valtorta M., Tedeschi G., Galbusera E., Pastori V., Bigi A., Nonnis S., Grassi E., Fusi P. Study of subcellular localization and proteolysis of ataxin-3. Neurobiol. Dis. 2008;30:190–200. doi: 10.1016/j.nbd.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 64.Berman S.B., Watkins S.C., Hastings T.G. Quantitative biochemical and ultrastructural comparison of mitochondrial permeability transition in isolated brain and liver mitochondria: evidence for reduced sensitivity of brain mitochondria. Exp. Neurol. 2000;164:415–425. doi: 10.1006/exnr.2000.7438. [DOI] [PubMed] [Google Scholar]
- 65.Brocard J.B., Rintoul G.L., Reynolds I.J. New perspectives on mitochondrial morphology in cell function. Biol. Cell. 2003;95:239–242. doi: 10.1016/s0248-4900(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 66.Chang D.T., Reynolds I.J. Differences in mitochondrial movement and morphology in young and mature primary cortical neurons in culture. Neuroscience. 2006;141:727–736. doi: 10.1016/j.neuroscience.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 67.Ho S., Clipstone N., Timmermann L., Northrop J., Graef I., Fiorentino D., Nourse J., Crabtree G.R. The mechanism of action of cyclosporin A and FK506. Clin. Immunol. Immunopathol. 1996;80:S40–S45. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- 68.Dineley K.E., Richards L.L., Votyakova T.V., Reynolds I.J. Zinc causes loss of membrane potential and elevates reactive oxygen species in rat brain mitochondria. Mitochondrion. 2005;5:55–65. doi: 10.1016/j.mito.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 69.Melov S., Schneider J.A., Day B.J., Hinerfeld D., Coskun P., Mirra S.S., Crapo J.D., Wallace D.C. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat. Genet. 1998;18:159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 70.Esposito L., Raber J., Kekonius L., Yan F., Yu G.Q., Bien-Ly N., Puolivali J., Scearce-Levie K., Masliah E., Mucke L. Reduction in mitochondrial superoxide dismutase modulates Alzheimer's disease-like pathology and accelerates the onset of behavioral changes in human amyloid precursor protein transgenic mice. J. Neurosci. 2006;26:5167–5179. doi: 10.1523/JNEUROSCI.0482-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bitzer M., Armeanu S., Prinz F., Ungerechts G., Wybranietz W., Spiegel M., Bernlohr C., Cecconi F., Gregor M., Neubert W.J., et al. Caspase-8 and Apaf-1-independent caspase-9 activation in Sendai virus-infected cells. J. Biol. Chem. 2002;277:29817–29824. doi: 10.1074/jbc.M111898200. [DOI] [PubMed] [Google Scholar]
- 72.Gyrd-Hansen M., Farkas T., Fehrenbacher N., Bastholm L., Hoyer-Hansen M., Elling F., Wallach D., Flavell R., Kroemer G., Nylandsted J., et al. Apoptosome-independent activation of the lysosomal cell death pathway by caspase-9. Mol. Cell Biol. 2006;26:7880–7891. doi: 10.1128/MCB.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ho A.T., Li Q.H., Hakem R., Mak T.W., Zacksenhaus E. Coupling of caspase-9 to Apaf1 in response to loss of pRb or cytotoxic drugs is cell-type-specific. EMBO J. 2004;23:460–472. doi: 10.1038/sj.emboj.7600039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morishima N., Nakanishi K., Takenouchi H., Shibata T., Yasuhiko Y. An endoplasmic reticulum stress-specific caspase cascade in apoptosis. Cytochrome c-independent activation of caspase-9 by caspase-12. J. Biol. Chem. 2002;277:34287–34294. doi: 10.1074/jbc.M204973200. [DOI] [PubMed] [Google Scholar]
- 75.Kim G.J., Fiskum G.M., Morgan W.F. A role for mitochondrial dysfunction in perpetuating radiation-induced genomic instability. Cancer Res. 2006;66:10377–10383. doi: 10.1158/0008-5472.CAN-05-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Panduri V., Weitzman S.A., Chandel N.S., Kamp D.W. Mitochondrial-derived free radicals mediate asbestos-induced alveolar epithelial cell apoptosis. Am. J. Physiol. 2004;286:L1220–L1227. doi: 10.1152/ajplung.00371.2003. [DOI] [PubMed] [Google Scholar]
- 77.Avila A.M., Burnett B.G., Taye A.A., Gabanella F., Knight M.A., Hartenstein P., Cizman Z., Di Prospero N.A., Pellizzoni L., Fischbeck K.H., et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanner V.A., Ploug T., Tao-Cheng J.H. Subcellular localization of SV2 and other secretory vesicle components in PC12 cells by an efficient method of preembedding EM immunocytochemistry for cell cultures. J. Histochem. Cytochem. 1996;44:1481–1488. doi: 10.1177/44.12.8985140. [DOI] [PubMed] [Google Scholar]
- 80.Liu J., Lillo C., Jonsson P.A., Vande Velde C., Ward C.M., Miller T.M., Subramaniam J.R., Rothstein J.D., Marklund S., Andersen P.M., et al. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 81.Levy J.R., Sumner C.J., Caviston J.P., Tokito M.K., Ranganathan S., Ligon L.A., Wallace K.E., LaMonte B.H., Harmison G.G., Puls I., et al. A motor neuron disease-associated mutation in p150Glued perturbs dynactin function and induces protein aggregation. J. Cell Biol. 2006;172:733–745. doi: 10.1083/jcb.200511068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.