Abstract
The heterozygous Pro250Arg substitution mutation in fibroblast growth factor receptor 3 (FGFR3), which increases ligand-dependent signalling, is the most common genetic cause of craniosynostosis in humans and defines Muenke syndrome. Since FGF signalling plays dosage-sensitive roles in the differentiation of the auditory sensory epithelium, we evaluated hearing in a large group of Muenke syndrome subjects, as well as in the corresponding mouse model (Fgfr3P244R). The Muenke syndrome cohort showed significant, but incompletely penetrant, predominantly low-frequency sensorineural hearing loss, and the Fgfr3P244R mice showed dominant, fully penetrant hearing loss that was more severe than that in Muenke syndrome individuals, but had the same pattern of relative high-frequency sparing. The mouse hearing loss correlated with an alteration in the fate of supporting cells (Deiters'-to-pillar cells) along the entire length of the cochlear duct, with the most extreme abnormalities found at the apical or low-frequency end. In addition, there was excess outer hair cell development in the apical region. We conclude that low-frequency sensorineural hearing loss is a characteristic feature of Muenke syndrome and that the genetically equivalent mouse provides an excellent model that could be useful in testing hearing loss therapies aimed at manipulating the levels of FGF signalling in the inner ear.
INTRODUCTION
The auditory sensory epithelium, the organ of Corti, is located along the length of the basilar membrane in the spiral cochlear duct of the inner ear. In humans and mice, the hearing organ has four rows of sensory hair cells comprising a single row of inner hair cells, which transduce most of the auditory information to the brain, and three rows of outer hair cells, which play a critical role in amplifying and modulating the response of the inner hair cells to sound. Each type of hair cell has an associated support cell; phalangeal cells underlie inner hair cells, and Deiters' cells underlie outer hair cells. Another supporting cell type, the pillar cell, separates the rows of inner and outer hair cells along with their corresponding supporting cells. In normal ears, two pillar cells form a triangular space (the tunnel of Corti) and play a role in regulating the stiffness of the basilar membrane upon which the organ of Corti vibrates. The sensory tissue is relatively narrow at the base of the cochlear duct, where it emerges from the vestibular portion of the inner ear, and wider at the apical end of the cochlear duct, contributing to the basal-to-apical gradient of high-to-low frequency sensitivity to sound (1,2).
Fibroblast growth factor (FGF) signalling is required for the normal induction, patterning and morphogenesis of the inner ear epithelium, as well as for the differentiation of cell types within the organ of Corti (2–4). In particular, FGF20 and Fgfr1 (both expressed in the prospective sensory region) are required for sensory hair cell differentiation (5,6) and Fgf8 (expressed in developing inner hair cells) and Fgfr3 (expressed in developing pillar and Deiters' cells, and for a time in developing outer hair cells) are required for pillar cell differentiation (7–12). Over-activation of FGF signalling also affects cochlear sensory development. Electroporation of Fgf8 or application of FGF2 or FGF17 to cochlear cultures induces ectopic pillar cell development (10,13), and Spry2 null mutants, in which a negative regulator of FGF signalling expressed prominently in Deiters' cells is absent, show a Deiters' cell-to-pillar cell fate change resulting in excess pillar cell development (14). Interestingly, both Fgfr3 and Spry2 null cochleae also show supernumerary outer hair cell development (9,11,14). Both of these mouse mutants are hearing impaired, and the Spry2 null excess pillar cell and deafness phenotypes, but not the excess outer hair cell phenotype, are rescued by removing a single copy of Fgf8 (14). Thus, FGF8 is the likely ligand for fibroblast growth factor receptor 3 (FGFR3) in pillar cell differentiation, and the level of FGF signalling involved in the process appears to be regulated within a very tight range.
FGF receptor mutations that lead to increased FGF signalling are frequent in humans, with craniosynostosis (premature fusion of the cranial sutures) and short stature being the most common skeletal phenotypes (15,16). Muenke syndrome (OMIM 602849) is caused by a heterozygous 749C>G transversion in FGFR3, which changes proline 250 to arginine (17,18). This is the most common individual mutation that occurs in a craniosynostosis syndrome (19), and several reports indicate that it can be variably associated with sensorineural hearing loss (HL) (20–24). In in vitro assays of the FGFR3c spliceform, the proline to arginine substitution, located in the linker between the extracellular IgII and IgIII domains of the receptor, resulted in significantly enhanced binding to two of nine tested FGFs, FGF2 and FGF9 (25). These act as physiological ligands of FGFR3c (26), and Fgf9, in particular, is expressed in the developing cochlear duct (27). Collectively, these observations, together with the Spry2 null mouse phenotype, suggested that Muenke syndrome patients and a mouse model of the Muenke syndrome mutation (28) should undergo comprehensive auditory phenotyping.
We report here that a sample of 37 subjects with Muenke syndrome show significant, predominantly low-frequency hearing loss. Muenke model mice evaluated in four genetic backgrounds have dominant hearing loss that is more severe than that in Muenke syndrome patients, but have the same pattern of relative high-frequency sparing, with variation between strains with respect to the extent of sparing. Similarly to Spry2 null mice, the Muenke model mice show excess pillar cell development at the expense of Deiters' cell development along the entire length of the cochlear duct, with the most extreme abnormalities occurring apically, and excess outer hair cell development that is confined to the apical region. All phenotypes are intensified in homozygotes relative to heterozygotes. Surprisingly, we found that hearing loss was not rescued by removal of a single copy of either Fgf8 or Fgf9 or both. We conclude that low-frequency sensorineural hearing loss is a characteristic feature of Muenke syndrome and that the genetically equivalent mouse provides an excellent model of this aspect of the phenotype.
RESULTS
Muenke syndrome patients have mild low-frequency sensorineural hearing loss
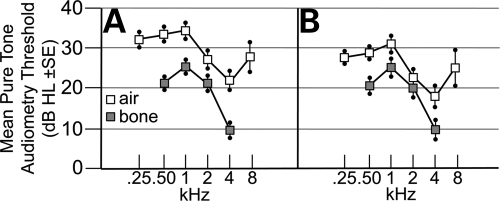
The largest previous study of hearing in Muenke syndrome combined data on 18 subjects tested at four different facilities (20). To determine the characteristics and penetrance of hearing loss in a large cohort of individuals with Muenke syndrome seen at a single facility, we measured air- and bone-conducted pure-tone audiometry thresholds in 37 heterozygotes for the FGFR3 P250R mutation (Fig. 1A). In the group as a whole, significant mild hearing loss was present, which was more marked at low frequencies (0.25–1 kHz; 32–35 dB HL [decibels hearing level]) than at high frequencies (2–8 kHz; 22–28 dB HL) (Fig. 1A). After excluding ears for which masked bone conduction could not be measured and/or there was evidence of middle ear disease (see Materials and Methods), we obtained a cleaned audiometric dataset for 41 ears from 30 different subjects (Fig. 1B). While the difference between the air and bone conduction was reduced at all frequencies compared with the original dataset, the mean thresholds measured for bone conduction remained virtually unchanged, ranging from 25 dB HL at 1 kHz to 10 dB HL at 4 kHz (Fig. 1B). This indicates that the hearing loss in Muenke syndrome is largely sensorineural. No significant sex difference was observed (data not shown). Estimation of the penetrance of significant sensorineural hearing loss (defined as ≥20 dB bone conduction) in this cleaned dataset confirmed that it is significantly more severe at lower frequencies, with 28 of 41 ears (68%) showing hearing loss at 1 kHz but only nine of 41 ears (22%) at 4 kHz (P = 4.8 × 10−5, Fisher's exact test, two-tailed). Nine subjects (22%) in our cohort have been fitted with hearing aids owing to the severity of their hearing loss.
Figure 1.
Individuals heterozygous for the Muenke syndrome mutation have low-frequency sensorineural hearing loss. (A) hearing loss measured in dB HL±SE and plotted as a function of frequency in kHz for 37 subjects with the FGFR3 P250R mutation. Results of air and bone conduction tests are plotted separately using white and grey symbols, respectively. (B) Cleaned dataset, comprising a total of 41 ears from 30 subjects for which paired sets of air and bone conduction data were available, and excluding ears showing evidence of compromise in middle ear function on tympanometry.
Muenke model mice are moderately hearing impaired, and hearing loss is more severe at low frequencies than at high frequencies
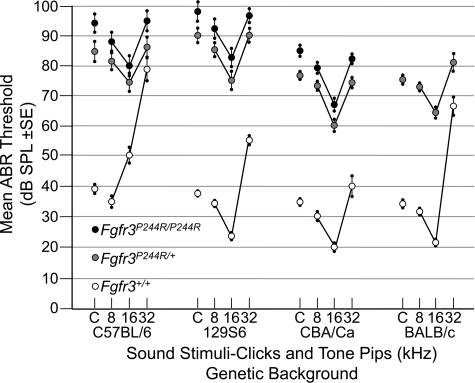
We have reported separately (28) the construction of a precise molecular mimic of the Muenke syndrome mutation in mice (Fgfr3P244R). Muenke model mice have craniofacial phenotypes that vary in penetrance and expressivity depending on genetic background, but do not include obvious middle ear anomalies. To assess hearing in this model, we measured auditory brainstem response (ABR) thresholds to click (broadband low frequency), 8, 16 and 32 kHz tone-pip stimuli in wild-type, heterozygous and homozygous Fgfr3P244R mice in four different genetic backgrounds (Fig. 2). As expected, wild-type mice in each background had a range of normal auditory thresholds (29,30), with the lowest thresholds occurring at 16 kHz in all strains, except in this particular C57BL/6 background (Fig. 2, open symbols). In contrast, all heterozygous mice, regardless of genetic background, had increased auditory thresholds (by 8–56 dB, depending on strain and stimulus) at all test points (Fig. 2, grey symbols) and homozygotes exhibited a further 5–9 dB decrement in hearing (16–62 dB above wild-type; Fig. 2, black symbols). BALB/c homozygotes could not be evaluated as we recovered too few live animals. Heterozygote and homozygote ABR thresholds were significantly elevated relative to wild-type at all test frequencies, and in all strain backgrounds (P < 0.0001) except at 32 kHz in the C57BL/6 background. In all cases, homozygous thresholds were greater than heterozygous thresholds, and in the case of the click response in all three backgrounds that could be evaluated, and at all frequencies tested in the CBA/Ca strain, the differences were statistically significant (0.0024 < P < 0.039). The small magnitudes not withstanding, these differences reflect 2–3-fold differences in hearing sensitivity. Unlike the craniofacial phenotypes (28), there was no effect of sex on the hearing loss phenotype (data not shown).
Figure 2.
Dominant hearing loss in Fgfr3P244R mice on four genetic backgrounds. ABR thresholds in decibels sound pressure level (dB SPL) ±SE were measured for click (C), 8, 16 and 32 kHz tone-pip stimuli in wild-type, heterozygous and homozygous mice inbred on four genetic backgrounds. Open circles indicate wild-type, grey circles indicate heterozygotes and black circles indicate homozygotes.
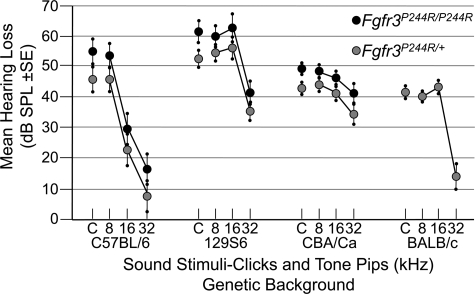
Hearing loss (average heterozygous or homozygous minus wild-type means) in all strains was less severe at 32 kHz than at lower frequencies (Fig. 3). These differences were highly significant (0.0001 < P < 0.0018) in the C57BL/6, 129S6 and BALB/c backgrounds, but less so in the CBA/Ca background (0.017 < P < 0.048), and not significant for the specific comparison of the CBA/Ca homozygotes at 32 and 16 kHz (P = 0.11). Furthermore, there was also relative sparing of hearing loss at 16 kHz in the C57BL/6 background, but this may be due to the relatively poor hearing of the wild-type strain at 16 kHz rather than to an effect of the mutation. These data show that the Muenke syndrome model has hearing loss that is more severe, but of a similar pattern (low frequency>high frequency) to that found in Muenke syndrome subjects (compare Figs 1 and 3). In addition, expressivity of the hearing loss phenotype varies with genetic background; it is more severe on the 129S6 and C57BL/6 backgrounds than on the CBA/Ca and BALB/c backgrounds. Finally, the hearing loss is Fgfr3P244R dosage sensitive; homozygotes are more severely affected than heterozygotes.
Figure 3.
Sparing of high-frequency hearing in Fgfr3P244R mice. Mean difference in dB SPL±SE between heterozygous or homozygous and wild-type mice is shown for each stimulus and genetic background. Symbols are the same as for Figure 2.
Muenke model mice have extra pillar cells and fewer Deiters' cells in the organ of Corti
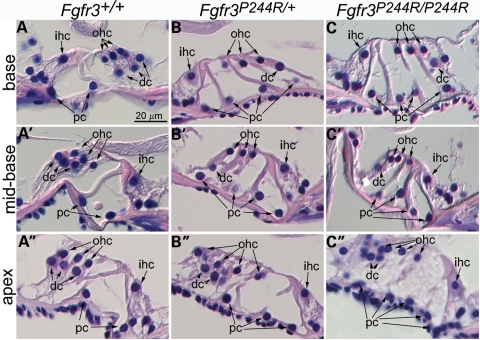
In mice, both inhibition of Fgfr3 function and excessive FGF signalling, mediated by the loss of Spry2, lead to hearing loss together with altered relative numbers of pillar and Deiters' cells (see Introduction); thus we suspected that deafness in the Muenke syndrome model animals might also be a consequence of aberrant organ of Corti development. Histological comparison of wild-type inner ears with those of heterozygous and homozygous animals in the 129S6 background suggests that this is indeed the case (Fig. 4). Cross-sections taken through the wild-type organ of Corti show three outer hair cells supported by three Deiters' cells at all basal to apical levels of the cochlea. Two pillar cells separate the outer hair cell/Deiters' cells from the inner hair cell (Fig. 4A–A′′). In contrast, heterozygous and homozygous Fgfr3P244R mutants showed three outer hair cells, but only one Deiters' cell and four pillar cells in basal and mid-basal cochlear cross-sections. In these mutants, inner hair cell numbers were normal (Fig. 4B, B′, C and C′). Apical cross-sections taken through the cochlear ducts of heterozygotes had the same cellular pattern, but occasionally showed a fourth outer hair cell, with or without an additional Deiters' cell (Fig. 4B′′). Apical cross-sections taken through the cochlear ducts of homozygotes sometimes showed as many as five pillar cells, one or two Deiters' cells and often showed four or more outer hair cells (Fig. 4C′′). These data demonstrate Fgfr3 dose-dependent abnormalities of organ of Corti differentiation that are more severe at apical (low frequency) than basal (high frequency) regions, consistent with the hearing loss in Muenke model mice.
Figure 4.
Extra pillar cells and fewer Deiters' cells at all levels in Muenke model cochlear ducts. H+E stained sections taken through basal (A–C), mid-basal (A′–C′) and apical (A′′–C′′) turns of the cochlear duct of 4–5-week-old wild-type (A and A′), heterozygous (B–B′′) and homozygous (C–C′′) 129S6-background mice. dc, Deiters' cells; ihc, inner hair cells; ohc, outer hair cells; pc, pillar cells.
Muenke model mice have extra outer hair cell in the apical region of the cochlear duct
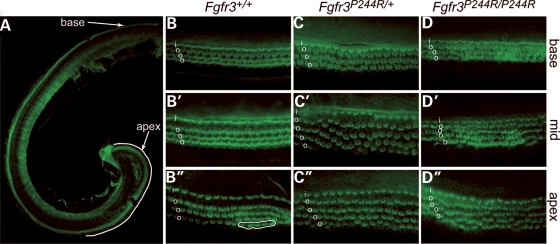
In addition to its effects on pillar and Deiters' cell differentiation, changes in signalling through FGFR3 also affect outer hair cell differentiation. Fgfr3 null mice show a continuous extra row of outer hair cells in the apical 60–67% of the cochlear duct (9,11), and Spry2 null mice have patchy extra outer hair cell rows that are more continuous at apical cochlear regions (14). To better assess outer hair cell development along the length of the Fgfr3P244R cochlear duct, we stained surface preparations of the P0–P5 129S6-derived organ of Corti with Alexa-488-phalloidin and visualized hair cell stereocilia (Fig. 5). Wild-type P3 epithelia had one row of inner hair cells and three rows of outer hair cells at basal, mid and apical levels of the cochlear duct (Fig. 5A, B, B′ and B′′). Only the most apical regions of wild-type epithelia showed small patches of extra outer hair cells (Fig. 5B′′). As expected from the adult histology, we found longer stretches of extra outer hair cells in both Fgfr3P244R heterozyotes and homozygotes. In heterozygotes, the extra outer hair cells were seen in the middle and apical domains of the cochlea and were discontinuous (Fig. 5C, C′ and C′′). In homozygotes, the extra outer hair cells formed a fairly continuous fourth row in approximately the apical 25% of the cochlear duct (Fig. 5D, D′ and D′′). Thus, signalling through FGFR3 induces apical outer hair cell development in a dose-dependent fashion.
Figure 5.
Extra outer hair cells at mid and basal regions of Muenke model cochlear ducts. Phalloidin-stained P3 cochlear surface preparations. (A) Whole cochlea illustrating the apical area (white line) in which the most complete rows of supernumerary hair cells could be found in Fgfr3P244R cochleae. Basal (B–D), mid (B′–D′) and apical (B′′–D′′) segments of wild-type (B–B′′), heterozygous (C–C′′) and homozygous (D–D′′) cochlear ducts. i, inner hair cells; o, outer hair cells. Circled region in B′′ shows the only extra outer hair cells found in an entire wild-type duct.
Reducing the copy number of Fgf8 or Fgf9 or both does not rescue Fgfr3p244r hearing loss
The hearing loss and cochlear cytoarchitecture found in Fgfr3P244R/+ mice appeared similar to that described for Spry2−/− mice, suggesting that reducing the levels of FGF8 signalling might rescue Fgfr3P244R/+ hearing loss, as is true for Spry2−/− mutants (14). We therefore crossed BALB/c-Fgfr3P244R/+ mice, which showed the mildest hearing loss, to Fgf8+/− mice and measured ABR thresholds in animals of each possible genotype. All Fgfr3P244R/+ animals in this hybrid background were hearing impaired, regardless of Fgf8 gene dosage (Supplementary Material, Fig. S1). Since the mutant FGFR3 P250R protein exhibits a large increase in affinity for FGF9 relative to the wild-type receptor (25), and Fgf9 is expressed in the cochlear duct, albeit rather far from the developing organ of Corti (27), we also tried reducing the dose of Fgf9 alone, or in combination with Fgf8. As with the Fgf8+/− allele alone, neither Fgf9+/− nor the combination of Fgf8+/− and Fgf9+/− was able to rescue Fgfr3P244R/+ hearing loss (Supplementary Material, Fig. S1).
DISCUSSION
Our data indicate that Fgfr3P244R/+ mice, a model of Muenke syndrome, have dominant, fully penetrant hearing loss that is more severe at low than at high frequencies. This is qualitatively similar, but more severe, than the hearing loss that we have documented in the largest audiological study of subjects with the Muenke syndrome mutation reported to date and is consistent with other recent work (20,22). These findings emphasize the need to ensure that all individuals heterozygous for the P250R mutation in FGFR3 are offered audiological testing and appropriate management for hearing loss. The differences in the absolute and relative magnitudes of high-frequency versus low-frequency hearing loss between the different mouse genetic backgrounds suggest that the variability in auditory phenotypes seen in the Muenke syndrome patients has, in part, a genetic basis.
Our results suggest that other individuals and mouse models bearing activating FGFR3 mutations might also present with sensorineural hearing loss. Indeed, some studies of achondroplasia patients (the great majority of whom are heterozygous for the FGFR3 G380R substitution) report sensorineural hearing loss (31–33), and the corresponding mouse model (Fgfr3G374R/+) could be tested for hearing (34). Many of the other common syndromes caused by FGFR3 activation are neonatal lethal in humans (15,16); however, the Fgfr3S365C/+ mouse strain, which models thanatophoric dysplasia type I, is viable (35) and could be evaluated for hearing loss and cochlear dysplasia. In humans, autosomal dominant non-syndromic low-frequency sensorineural hearing loss is relatively unusual. Some such individuals have mutations in DIAPH1 or WFS1 (36–39), but others are unexplained, and our data suggest that FGFR3 should be considered as a candidate for these other cases. Interestingly, a putative dominant negative mutation in FGFR3, R621H, appears associated with a reversed pattern of hearing loss to that documented for P250R, with hearing at higher frequencies more severely affected (40).
Our histological study of Fgfr3P244R cochleae revealed two patterning defects of the organ of Corti, one involving supporting cells and the other involving sensory cells. All cochlear levels showed at least two excess pillar cells at the expense of Deiters' cells, and this phenotype was most severe in the apical region, where three or more ectopic pillar cells formed at the expense of Deiter's cell. In addition, we found supernumerary outer hair cell development that was confined to the apical region. These cellular defects are similar to, but somewhat more severe, than those reported for hearing-impaired Spry2−/− mice, in which FGF signalling is increased by relief of negative regulation (14). Since Spry2−/− hearing loss and support cell phenotypes, but not the outer hair cell phenotype, are rescued when an Fgf8 null allele is introduced, it is likely that the support cell abnormalities seen in Fgfr3P244R mice are the major cause of their hearing loss. In addition, the increased severity of supporting cell mispatterning at the apical end of the Fgfr3P244R cochlear duct is consistent with the greater elevation of low-frequency relative to high-frequency ABR thresholds.
Both cytoarchitectural defects seen in Fgfr3P244R mice are confined to cells that normally express Fgfr3 during organ of Corti development. Fgfr3 expression initiates at E15.5 in the precursors of pillar, Deiters' and outer hair cells, turning on in a base-to-apex wave. Expression is subsequently restricted to pillar and Deiters' cells at P0, and finally to pillar cells only by P7 (9,14). Fgfr3 null or Fgf8 otic conditional null mice have defects in pillar cell differentiation (9–12), suggesting that FGF8, which is expressed from about E15.5 in the prospective inner hair cell, signals to the two closest FGFR3-expressing cells to differentiate as pillar cells. Presumably the FGF8 signal is too low to activate the FGFR3 receptors expressed by more lateral cells, which differentiate as Deiters' cells and then turn off Fgfr3. Spry2, which encodes an inhibitor of FGF signalling, is initially expressed similarly to Fgfr3 in prospective pillar cells, Deiters' cells and outer hair cells, but is restricted to Deiters' cells by P5. Thus, in its absence, FGF8-dependent signalling occurs normally in prospective pillar cells, but is likely increased in the prospective Deiters' cell most proximal to the inner hair cell, causing it to differentiate as a pillar cell. Therefore, removal of one copy of Fgf8 from the Spry2 null restores to some extent the balance of signalling back towards the wild-type differentiation pattern (14). In the Muenke model organ of Corti, we found that at least two prospective Deiters' cells adopt the pillar cell fate. This could reflect a higher level of signalling induced by the P244R mutation in FGFR3 relative to that induced by the wild-type ligand/receptor pair in the absence of Spry2. Indeed, in contrast to the Spry2 null, Muenke model hearing loss was not rescued by removal of a single copy of Fgf8. Thus, a further reduction in FGF8 and/or FGFR3 levels may be necessary to achieve rescue. Alternatively or in addition to FGF8, the mutant FGFR3 P244R may be activated by additional ligands. Removal of one copy of FGF9, which has a high affinity for FGFR3c in vitro, did not rescue the auditory phenotype, suggesting that it may not contribute significantly to mutant receptor activation in Deiters' cells. The other two FGFs in the FGF9 family are FGF16 and FGF20 (41), both of which are expressed in the cochlea (5,42). The latter, by virtue of its expression in the prosensory region (5), is a particularly attractive candidate for activation of signalling through FGFR3 P244R, though its expression may terminate too early to contribute to the excess pillar cell phenotype of Muenke model cochleae. A final consideration in seeking to explain the differences between the Muenke model and the Spry2 null cochlea is the relative timing of expression of inhibitors of FGF signalling, some of which are transcriptional targets of FGF signalling. SPRY2 acts as a feedback inhibitor in some situations (43), but this does not appear to be the case in the organ of Corti, where Spry2 is unaffected by the loss of Fgfr3 (9). Spry1 and Dusp6 (Mkp3) are also expressed in the developing organ of Corti (14,44), eventually appearing to be specific to pillar cells, but their responses to loss of Fgfr3 or Spry2 are unknown. Thus, a developmental study of the Muenke model, focusing on indicators and inhibitors of FGF signalling, as well as markers of cellular differentiation, should help to elucidate precisely which cells experience increased FGF signalling and when it occurs relative to Fgf gene expression.
It was notable that Fgfr3P244R cochleae developed supernumerary outer hair cells in the apical region. This phenotype was also more pronounced than in Spry2−/− mutants, in which excess outer hair cell development is patchy, but was less extensive than in Fgfr3−/− mutants, which have excess outer hair cell (and Deiters' cell) development along the apical 60–67% of the cochlea (9,11). How can both increases and decreases in FGF signalling cause additional outer hair cell development? Hayashi and colleagues proposed that Fgfr3−/− mice have extra outer hair cells because in the absence of cells expressing FGFR3 to bind the available FGF8, FGF8 is able to diffuse more laterally across the sensory epithelium to activate FGFR1 (9), which is normally stimulated by FGF20 to specify hair cell development (5,6). In addition, Shim and colleagues proposed that Spry2−/− mice have extra hair cells, not due to an increase in FGF8 signalling through FGFR3, but rather by increased signalling through FGFR1. Their observation that an Fgf8 null allele rescues the ectopic pillar cell phenotype of Spry2−/− mutants, but not the ectopic outer hair cell phenotype, supports this interpretation (14). To explain the excess outer hair cell phenotype in Meunke model mice, in which FGFR3, rather than FGFR1, activity is increased, we propose that the FGFR3 P244R mutant protein has an increased affinity for FGF20 (as it does for the structurally related FGF9) (25,41) and that this signalling event increases the extent of the prosensory domain that can differentiate as hair cells. As the base-to-apex wave of Fgf20 expression initiates 2 days before that of Fgfr3 and quickly recedes, it may be that the Fgfr3P244R apex is the only area that has abnormally high levels of FGF20-stimulated signalling, and thus is the only region that develops extra hair cells. It will be interesting to determine the timing of the appearance of the supernumerary hair cells relative to Fgf20 expression dynamics and to determine whether removal of one copy of Fgf20 will rescue the Fgfr3P244R outer hair cell phenotype.
In summary, we have shown that a mouse model of the Muenke syndrome mutation has hearing loss similar to that found in Muenke syndrome patients and that hearing loss correlates with abnormalities of organ of Corti differentiation, which are similar to those seen when a negative regulator of FGF signalling is removed. This mouse model should be useful in testing hearing loss therapies aimed at manipulating the levels of FGF signalling in the inner ear.
MATERIALS AND METHODS
Ascertainment and audiological assessment of subjects with Muenke syndrome
Audiological testing was performed on 41 individuals (including 14 affected parents) from 23 different families with Muenke syndrome presenting to the Oxford Craniofacial Unit. Heterozygosity for the 749C>G mutation was demonstrated by either NciI or BanI digestion, or DNA sequencing, of the human FGFR3 exon 7 amplimer (45).
Age-appropriate audiometry and tympanometry were performed on each patient according to British Society of Audiology recommended procedures. Audiometric tests were obtained in a double-walled soundproof test room (to ISO 8253-1 standards), using either a Grason–Stadler Inc 61 audiometer or a Kamplex AD27 audiometer (both machines comply with the BSEN 60645 part 1 specification for audiometers and are calibrated to ISO 389 series). Four children unable to perform pure-tone audiometry using headphones were excluded. For the remaining 37 subjects, separate measurements of air conduction in each ear (0.25–8 kHz), and unmasked bone conduction (0.5–4 kHz), were obtained (in one subject with normal hearing, bone conduction was not obtained but was assumed equal to air conduction in the better hearing ear). If the difference between bone conduction and air conduction for both ears was ≤10 dB at all frequencies for either ear, then the figure for bone conduction was attributed to both ears, and masking was not attempted (four subjects). Where this criterion was not met, additional measurements of masked bone conduction were obtained from 15 subjects (the others were unable to perform the masking procedure). For the cleaned dataset in the remaining 17 subjects, the unmasked bone conduction was compared with the air conduction in the better hearing ear.
Tympanometry was performed using either a GSI Tympstar or an Amplivox Otowave 102 hand-held portable tympanometer (both machines comply with the BSEN 60645 part 5 specification for tympanometers and are calibrated to ISO 389 series). Individual ears (n = 19) with flat tympanograms or markedly negative middle ear pressures (<150 daPa in children, <50 daPa in adults) were excluded from the cleaned dataset.
Mouse strains
All work with mice complied with protocols approved by the University of Utah Institutional Animal Care and Use Committee.
Fgfr3P244R/+ mice backcrossed to four inbred backgrounds (129S6, CBA/CaJ, C57BL/6 and BALB/c) were transferred to the University of Utah and genotyped as described by Twigg et al. (28). The auditory and histological studies reported here were carried out on subjects derived from heterozygous intercrosses of Muenke model animals in the seventh to ninth backcross generation. Similar to the results reported by Twigg et al., the penetrance of obvious craniofacial malformations in Utah-bred homozygotes was 75% in the C57BL/6 background, 64% in the 129S6 background and 36% in the CBA/Ca background. The only BALB/c homozygote that was recovered was affected. The penetrance of craniofacial malformations in heterozygotes on the C57BL/6, 129S6, CBA/Ca and BALB/c backgrounds was 11, 15, 0 and 5%, respectively.
Auditory rescue analyses were carried out on animals derived from crossing BALB/c-Fgfr3P244R/+ to Fgf8+/− animals originally obtained from Dr Anne Moon and maintained on a mixed genetic background (primarily C57BL/6, 129X1 and 129S1) (46). Fgfr3P244R/+; Fgf8+/− animals from the mixed genetic background were crossed to C57BL/6-Fgf9+/− animals obtained from Dr. David Ornitz to generate animals of all possible combinations of Fgfr3, Fgf8 and Fgf9 alleles (47).
Mouse ABR threshold measurements
To avoid the possibility of confounding age-related hearing loss with Fgfr3P244R-specific effects, all animals used for ABR testing were 4–5 weeks old. Mice were anesthetized using 0.02 ml/g Avertin. ABR thresholds for click (∼1–16 kHz, 47 µs duration, 29.3/s), 8, 16 and 32 kHz tone-pip stimuli (3 ms duration, 29.3/s, Exact Blackman envelope) were determined using high-frequency transducers controlled and analysed by SmartEP software (Intelligent Hearing Systems) as described (48). This protocol was chosen to enable comparisons with the mouse auditory screening project at The Jackson Laboratory (http://hearingimpairment.jax.org/screening.html) as well as with ABR tests of Spry2 and Fgfr3 null mice (8,11,14). Testing was carried out blind with respect to genotype, but in some cases, the genotype could be inferred based on craniofacial phenotypes (28). ABR results were obtained from the following numbers of inbred subjects: 129S6-Fgfr3+/+, 19; 129S6-Fgfr3P244R/+, 19; 129S6-Fgfr3P244R/P244R, 9; CBA/Ca-Fgfr3+/+, 11; CBA/Ca-Fgfr3P244R/+, 12; CBA/Ca-Fgfr3P244R/P244R, 13; BALB/c-Fgfr3+/+, 12; BALB/c-Fgfr3P244R/+, 13; C57BL/6-Fgfr3+/+, 11; C57BL/6-Fgfr3P244R/+, 12; C57BL/6-Fgfr3P244R/P244R, 13.
Statistics
The ABR data were analysed for differences between genotypes using a general linear model, controlling for strain background and stimuli. Differences between heterozygous or homozygous and wild-type means were calculated by least square means.
Inner ear histology and phalloidin staining of cochlear epithelia
For histology, adult mice were perfused with 4% PFA in PBS, the ears were removed and decalcified in 0.1 M EDTA/2% HCHO/25 mm HCl, then embedded in paraffin wax and sectioned in the plane of the modiolus at 5–10 µm. Sections were stained with haematoxylin and eosin.
For visualization of filamentous actin, cochleae were dissected in ice-cold PBS, rinsed in PBS containing 0.01% Triton X-100 (PBS/Triton) and then incubated for 15 min at 4°C with Alexa 488-phalloidin (Invitrogen) diluted 1:1000 in PBS/Triton. The labelled cochlea were rinsed in PBS/Triton and mounted in Fluoromount™ for observation.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (R01DC002043 to S.L.M.); the University of Utah (to S.L.M.) and Wellcome Trust Programme Grant (078666 to A.O.M.W).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Mr David Johnson for referral of families, Dr Tracy Lester for mutation analysis, Dr Andy Pain for case note analysis of audiograms in Muenke syndrome, Dr Jo Byren for clinical co-ordination, Jane Humphries, Jane Jones and Christina Kirkegard for audiometric testing, Dr Josh Yorgason for instruction in cochlear isolation, Dr Xiaoming Sheng for statistical analysis of the mouse ABR data, Dr Olivia Bermingham-McDonogh and Dr Toshi Hayashi for the phalloidin staining protocol, Dr Lisa Urness for assistance with phalloidin staining and Mark Hill for assistance with some of the Fgf9 ABR tests.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kelley M.W. Regulation of cell fate in the sensory epithelia of the inner ear. Nat. Rev. Neurosci. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 2.Shim K. The auditory sensory epithelium: the instrument of sound perception. Int. J. Biochem. Cell Biol. 2006;38:1827–1833. doi: 10.1016/j.biocel.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Schimmang T. Expression and functions of FGF ligands during early otic development. Int. J. Dev. Biol. 2007;51:473–481. doi: 10.1387/ijdb.072334ts. [DOI] [PubMed] [Google Scholar]
- 4.Wright T.J., Mansour S.L. FGF signaling in ear development and innervation. Curr. Top. Dev. Biol. 2003;57:225–259. doi: 10.1016/s0070-2153(03)57008-9. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi T., Ray C.A., Bermingham-McDonogh O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J. Neurosci. 2008;28:5991–5999. doi: 10.1523/JNEUROSCI.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirvola U., Ylikoski J., Trokovic R., Hebert J.M., McConnell S.K., Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 7.Bohne B.A., Harding G.W. Processing and analyzing the mouse temporal bone to identify gross, cellular and subcellular pathology. Hear. Res. 1997;109:34–45. doi: 10.1016/s0378-5955(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 8.Colvin J.S., Bohne B.A., Harding G.W., McEwen D.G., Ornitz D.M. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat. Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi T., Cunningham D., Bermingham-McDonogh O. Loss of FGFR3 leads to excess hair cell development in the mouse organ of Corti. Dev. Dyn. 2007;236:525–533. doi: 10.1002/dvdy.21026. [DOI] [PubMed] [Google Scholar]
- 10.Jacques B.E., Montcouquiol M.E., Layman E.M., Lewandoski M., Kelley M.W. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- 11.Puligilla C., Feng F., Ishikawa K., Bertuzzi S., Dabdoub A., Griffith A.J., Fritzsch B., Kelley M.W. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev. Dyn. 2007;236:1905–1917. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelarayan L.C., Vendrell V., Alvarez Y., Dominguez-Frutos E., Theil T., Alonso M.T., Maconochie M., Schimmang T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev. Biol. 2007;308:379–391. doi: 10.1016/j.ydbio.2007.05.033. [DOI] [PubMed] [Google Scholar]
- 13.Mueller K.L., Jacques B.E., Kelley M.W. Fibroblast growth factor signaling regulates pillar cell development in the organ of corti. J. Neurosci. 2002;22:9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shim K., Minowada G., Coling D.E., Martin G.R. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev. Cell. 2005;8:553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Ornitz D.M., Marie P.J. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- 16.Wilkie A.O.M. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 2005;16:187–203. doi: 10.1016/j.cytogfr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Bellus G.A., Gaudenz K., Zackai E.H., Clarke L.A., Szabo J., Francomano C.A., Muenke M. Identical mutations in three different fibroblast growth factor receptor genes in autosomal dominant craniosynostosis syndromes. Nat. Genet. 1996;14:174–176. doi: 10.1038/ng1096-174. [DOI] [PubMed] [Google Scholar]
- 18.Muenke M., Gripp K.W., McDonald-McGinn D.M., Gaudenz K., Whitaker L.A., Bartlett S.P., Markowitz R.I., Robin N.H., Nwokoro N., Mulvihill J.J., et al. A unique point mutation in the fibroblast growth factor receptor 3 gene (FGFR3) defines a new craniosynostosis syndrome. Am. J. Hum. Genet. 1997;60:555–564. [PMC free article] [PubMed] [Google Scholar]
- 19.Moloney D.M., Wall S.A., Ashworth G.J., Oldridge M., Glass I.A., Francomano C.A., Muenke M., Wilkie A.O.M. Prevalence of Pro250Arg mutation of fibroblast growth factor receptor 3 in coronal craniosynostosis. Lancet. 1997;349:1059–1062. doi: 10.1016/s0140-6736(96)09082-4. [DOI] [PubMed] [Google Scholar]
- 20.Doherty E.S., Lacbawan F., Hadley D.W., Brewer C., Zalewski C., Kim H.J., Solomon B., Rosenbaum K., Domingo D.L., Hart T.C., et al. Muenke syndrome (FGFR3-related craniosynostosis): expansion of the phenotype and review of the literature. Am. J. Med. Genet. A. 2007;143:3204–3215. doi: 10.1002/ajmg.a.32078. [DOI] [PubMed] [Google Scholar]
- 21.Hollway G.E., Suthers G.K., Battese K.M., Turner A.M., David D.J., Mulley J.C. Deafness due to Pro250Arg mutation of. FGFR3. Lancet. 1998;351:877–878. doi: 10.1016/S0140-6736(98)24012-8. [DOI] [PubMed] [Google Scholar]
- 22.Honnebier M.B., Cabiling D.S., Hetlinger M., McDonald-McGinn D.M., Zackai E.H., Bartlett S.P. The natural history of patients treated for FGFR3-associated (Muenke-type) craniosynostosis. Plast. Reconstr. Surg. 2008;121:919–931. doi: 10.1097/01.prs.0000299936.95276.24. [DOI] [PubMed] [Google Scholar]
- 23.Kress W., Schropp C., Lieb G., Petersen B., Busse-Ratzka M., Kunz J., Reinhart E., Schafer W.D., Sold J., Hoppe F., et al. Saethre-Chotzen syndrome caused by TWIST 1 gene mutations: functional differentiation from Muenke coronal synostosis syndrome. Eur. J. Hum. Genet. 2006;14:39–48. doi: 10.1038/sj.ejhg.5201507. [DOI] [PubMed] [Google Scholar]
- 24.Lowry R.B., Jabs E.W., Graham G.E., Gerritsen J., Fleming J. Syndrome of coronal craniosynostosis, Klippel-Feil anomaly, and sprengel shoulder with and without Pro250Arg mutation in the FGFR3 gene. Am. J. Med. Genet. 2001;104:112–119. doi: 10.1002/ajmg.10049. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahimi O.A., Zhang F., Eliseenkova A.V., Linhardt R.J., Mohammadi M. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum. Mol. Genet. 2004;13:69–78. doi: 10.1093/hmg/ddh011. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Ibrahimi O.A., Olsen S.K., Umemori H., Mohammadi M., Ornitz D.M. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pirvola U., Zhang X., Mantela J., Ornitz D.M., Ylikoski J. Fgf9 signaling regulates inner ear morphogenesis through epithelial-mesenchymal interactions. Dev. Biol. 2004;273:350–360. doi: 10.1016/j.ydbio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Twigg S.R.F., Healy C., Babbs C., Sharpe J.A., Wood W.G., Sharpe P.T., Morriss-Kay G.M., Wilkie A.O.M. Skeletal analysis of the Fgfr3P244R mouse, a genetic model for the Muenke craniosynostosis syndrome. Dev. Dyn. 2009 doi: 10.1002/dvdy.21790. in press. [DOI] [PubMed] [Google Scholar]
- 29.Johnson K.R., Zheng Q.Y., Noben-Trauth K. Strain background effects and genetic modifiers of hearing in mice. Brain Res. 2006;1091:79–88. doi: 10.1016/j.brainres.2006.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X., Jen P.H., Seburn K.L., Frankel W.N., Zheng Q.Y. Auditory brainstem responses in 10 inbred strains of mice. Brain Res. 2006;1091:16–26. doi: 10.1016/j.brainres.2006.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cobb S.R., Shohat M., Mehringer C.M., Lachman R. CT of the temporal bone in achondroplasia. AJNR Am. J. Neuroradiol. 1988;9:1195–1199. [PMC free article] [PubMed] [Google Scholar]
- 32.Glass L., Shapiro I., Hodge S.E., Bergstrom L., Rimoin D.L. Audiological findings of patients with achondroplasia. Int. J. Pediatr. Otorhinolaryngol. 1981;3:129–135. doi: 10.1016/0165-5876(81)90028-8. [DOI] [PubMed] [Google Scholar]
- 33.Shohat M., Flaum E., Cobb S.R., Lachman R., Rubin C., Ash C., Rimoin D.L. Hearing loss and temporal bone structure in achondroplasia. Am. J. Med. Genet. 1993;45:548–551. doi: 10.1002/ajmg.1320450504. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Spatz M.K., Kannan K., Hayk H., Avivi A., Gorivodsky M., Pines M., Yayon A., Lonai P., Givol D. A mouse model for achondroplasia produced by targeting fibroblast growth factor receptor 3. Proc. Natl Acad. Sci. USA. 1999;96:4455–4460. doi: 10.1073/pnas.96.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L., Li C., Qiao W., Xu X., Deng C. A Ser(365)–>Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum. Mol. Genet. 2001;10:457–465. doi: 10.1093/hmg/10.5.457. [DOI] [PubMed] [Google Scholar]
- 36.Fukuoka H., Kanda Y., Ohta S., Usami S. Mutations in the WFS1 gene are a frequent cause of autosomal dominant nonsyndromic low-frequency hearing loss in Japanese. J. Hum. Genet. 2007;52:510–515. doi: 10.1007/s10038-007-0144-3. [DOI] [PubMed] [Google Scholar]
- 37.Bespalova I.N., Van Camp G., Bom S.J., Brown D.J., Cryns K., DeWan A.T., Erson A.E., Flothmann K., Kunst H.P., Kurnool P., et al. Mutations in the Wolfram syndrome 1 gene (WFS1) are a common cause of low frequency sensorineural hearing loss. Hum. Mol. Genet. 2001;10:2501–2508. doi: 10.1093/hmg/10.22.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lalwani A.K., Jackler R.K., Sweetow R.W., Lynch E.D., Raventos H., Morrow J., King M.C., Leon P.E. Further characterization of the DFNA1 audiovestibular phenotype. Arch. Otolaryngol. Head Neck Surg. 1998;124:699–702. doi: 10.1001/archotol.124.6.699. [DOI] [PubMed] [Google Scholar]
- 39.Lynch E.D., Lee M.K., Morrow J.E., Welcsh P.L., Leon P.E., King M.C. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278:1315–1318. [PubMed] [Google Scholar]
- 40.Toydemir R.M., Brassington A.E., Bayrak-Toydemir P., Krakowiak P.A., Jorde L.B., Whitby F.G., Longo N., Viskochil D.H., Carey J.C., Bamshad M.J. A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. Am. J. Hum. Genet. 2006;79:935–941. doi: 10.1086/508433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh N., Ornitz D.M. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Hatch E.P., Urness L.D., Mansour S.L. Fgf16IRESCre mice: A tool to inactivate genes expressed in inner ear cristae and spiral prominence epithelium. Dev. Dyn. 2008 doi: 10.1002/dvdy.21681. September 4[Epub ahead of print] doi:10.1002/dvdy.21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mason J.M., Morrison D.J., Basson M.A., Licht J.D. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Urness L.D., Li C., Wang X., Mansour S.L. Expression of ERK signaling inhibitors Dusp6, Dusp7, and Dusp9 during mouse ear development. Dev. Dyn. 2008;237:163–169. doi: 10.1002/dvdy.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rannan-Eliya S.V., Taylor I.B., De Heer I.M., Van Den Ouweland A.M., Wall S.A., Wilkie A.O.M. Paternal origin of FGFR3 mutations in Muenke-type craniosynostosis. Hum. Genet. 2004;115:200–207. doi: 10.1007/s00439-004-1151-5. [DOI] [PubMed] [Google Scholar]
- 46.Moon A.M., Capecchi M.R. Fgf8 is required for outgrowth and patterning of the limbs. Nat. Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colvin J.S., Green R.P., Schmahl J., Capel B., Ornitz D.M. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 48.Li C., Scott D.A., Hatch E., Tian X., Mansour S.L. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.