Abstract
Missense mutations in the PTPN11 gene, which encodes the protein tyrosine phosphatase SHP-2, cause clinically similar but distinctive disorders, LEOPARD (LS) and Noonan (NS) syndromes. The LS is an autosomal dominant disorder with pleomorphic developmental abnormalities including lentigines, cardiac defects, short stature and deafness. Biochemical analyses indicated that LS alleles engender loss-of-function (LOF) effects, while NS mutations result in gain-of-function (GOF). These biochemical findings lead to an enigma that how PTPN11 mutations with opposite effects on function result in disorders that are so similar. To study the developmental effects of the commonest LS PTPN11 alleles (Y279C and T468M), we generated LS transgenic fruitflies using corkscrew (csw), the Drosophila orthologue of PTPN11. Ubiquitous expression of the LS csw mutant alleles resulted in ectopic wing veins and, for the Y279C allele, rough eyes with increased R7 photoreceptor numbers. These were GOF phenotypes mediated by increased RAS/MAPK signaling and requiring the LS mutant’s residual phosphatase activity. Our findings provide the first evidence that LS mutant alleles have GOF developmental effects despite reduced phosphatase activity, providing a rationale for how PTPN11 mutations with GOF and LOF produce similar but distinctive syndromes.
INTRODUCTION
LEOPARD syndrome (LS; OMIM# 151100) is an autosomal dominant genetic disorder named by Gorlin et al. as an acronym of its principal features: lentigines, electrocardiographic abnormalities, ocular hypertelorism, pulmonary valve stenosis, abnormal genitalia, retardation of growth and deafness (1). Particularly during infancy and early childhood when lentigines have not yet appeared, LS closely resembles Noonan syndrome (NS; OMIM# 163950), a more prevalent autosomal dominant disorder characterized by pleomorphic features that include congenital heart disease, facial dysmorphism, short stature, skeletal defects, cognitive deficits and hematological abnormalities (2,3). While the two disorders can be distinguished clinically, it was unclear whether they were genetically distinct. In 2001, we discovered that missense mutations in the PTPN11gene cause ∼50% of NS cases (4,5). Shortly thereafter, it was shown that specific PTPN11 missense defects cause roughly 90% of LS (6,7).
PTPN11 encodes SHP-2, which is an src homology-2 (SH2)-containing protein tyrosine phosphatase (PTP) that is highly conserved between species (8). SHP-2 consists of two SH2 domains (N-SH2 and C-SH2) in its N-terminal half and the catalytic PTP domain in the C-terminal half. SHP-2 is ubiquitously expressed and regulates signaling for several receptor tyrosine kinases (RTKs) such as EGFR and FGFR through the activation of the RAS/MAPK cascade, leading to cell proliferation, differentiation and migration. SHP-2 plays a positive role in signal transduction in most contexts.
SHP-2 has inactive and active conformations that are regulated through a molecular switching mechanism (9,10). In the inactive state, the backside of the N-SH2 domain forms a loop and is wedged into the PTP domain, blocking the catalytic site. When SHP-2 binds phosphotyrosyl-containing proteins at separate sites on the N- and C-SH2 domains, a conformational change releases the N-SH2 domain from the PTP domain, which makes the PTP catalytic cleft available.
Although the PTPN11 mutations observed in LS and NS are almost invariably missense mutations, the affected amino acids are distinct. Most NS mutations alter amino acid residues in the N-SH2 and PTP domains clustering at the interface between those domains. The effects of these NS mutations on signal transduction have been studied extensively, and it has been shown that these amino acid substitutions change the molecular switching of SHP-2 to favor the active state without affecting catalysis, resulting in gain-of-function (GOF) effects (11–15). In contrast, mutations in LS have been identified solely in the PTP domain and the two commonest LS alleles are located in the catalytic cleft. Recent biochemical studies indicated that amino acid alterations in LS result in significantly reduced phosphatase activity and are therefore deemed loss-of-function (LOF) defects (15–17). Moreover, expression of the LS-associated SHP-2 mutants in eukaryotic cells had dominant negative (DN) effects on downstream RAS signaling (17). These genetic and biochemical findings generated an enigma: how do PTPN11 mutations with opposite effects elicit overlapping phenotypes?
Previously, we generated transgenic Drosophila models of NS with mutations in corkscrew (csw), the fly orthologue of PTPN11 (14,18). Flies ubiquitously overexpressing the N308D allele, the commonest NS mutation, showed an ectopic wing vein phenotype due to increased RAS/MAPK activity as well as interactions with other signal transduction pathways (14). This provided the opportunity to examine the effects of the LS-associated mutations in the same developmental context. To study this, we generated transgenic Drosophila expressing the two commonest LS mutations in csw under control of UAS/GAL4 system (19). We report here that ubiquitous expression of those LS-causing alleles resulted in a GOF phenotype, which was similar to that of the NS transgenic flies. Moreover, we showed that the phenotype was dependent upon the residual phosphatase activity of the impaired LS mutant protein. Since transgenic overexpression of wild-type CSW engenders no phenotype, we concluded that dysregulated phosphatase activity of SHP-2, even at low level, is sufficient to cause the overlapping clinical features of LS and NS.
RESULTS
Generation of LS transgenic Drosophila models
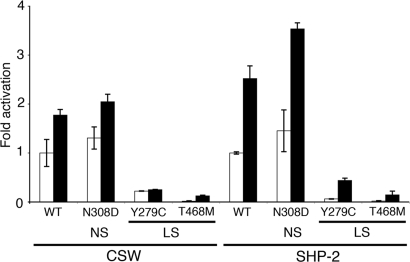
Since we intended to generate transgenic fruitflies expressing csw with the two commonest LS mutations, Y279C and T468M, we first sought to determine if those amino acid substitutions engendered similar LOF biochemical effects in SHP-2 and CSW. We expressed the wild-type and three mutant proteins in bacteria and assayed their phosphatase activities. The phosphatase activities of the Y279C and T468M CSW proteins were significantly reduced, comparable with what was observed for SHP-2 proteins with the same amino acid substitutions (Fig. 1). Throughout, we refer to amino acid changes based on human SHP-2; the actual CSW substitutions are shown in Table 1. Each csw allele was inserted into the pUASP vector and multiple independent transgenic lines were established for each construct. At least three independent lines of transgenic Drosophila, UAS-cswY279C, UAS-cswT468M, UAS-cswR465M and UAS-cswY279C/R465M (R465M, a mutant lacking phosphatase activity, called phosphatase-dead hereafter) (20), were obtained and stable stocks were established using balancer chromosomes. To compare the effects of LS alleles with wild-type CSW and the most common NS allele, N308D, we employed previously generated transgenic flies, UAS-cswWT and UAS-cswN308D (14).
Figure 1.
In vitro phosphatase activity assays of wild-type and mutant CSW and SHP-2 proteins. The LS mutant CSW proteins (Y279C and T468M) have reduced phosphatase activities. Activities were measured by phosphate release from a substrate, pNPP, without (white bars) and with (black bars) phosphotyrosyl peptide stimulation. Fold activation values are means ± SDs of at least three independent experiments and calculated from unstimulated wild-type CSW or SHP-2 for each group. NS, Noonan syndrome, LS, LEOPARD syndrome.
Table 1.
Transgenic csw alleles
| SHP-2 mutation | CSW mutation | Domain | Phenotype | Allele description |
|---|---|---|---|---|
| Y279C | Y258C | PTP | LS | UAS-cswY279C |
| T468M | T592M | PTP | LS | UAS-cswT468M |
| R465M | R589M | PTP | Phosphatase defective | UAS-cswR465M |
| Y279C/R465M | Y258C/R589M | PTP | LS/Phosphatase defective | UAS-cswY279C/R465M |
| N308D | N287D | PTP | NS | UAS-cswN308D |
LS, LEOPARD syndrome; NS, Noonan syndrome, PTP, protein tyrosine phosphatase.
RT–PCR (reverse transcription PCR) analyses were performed to evaluate the steady-state transgene expression levels. Transgenic csw expression driven by tubulin (tub)-GAL4 was stable and transcript levels in the wild-type and mutant csw transgenic fly lines were significantly higher than the endogenous csw expression level (Supplementary Material, Fig. S1 and Table S1). Although the T468M expression level was approximately double, we found this allele to be weaker (see later), not consistent with the possibility of expression dose effects.
Flies ubiquitously expressing LS alleles were viable and showed ectopic wing veins and rough eyes
To study the effects of the LS mutations during development, we overexpressed the transgenes ubiquitously using the tub-GAL4 driver. Transgenic males carrying the wild-type and mutant csw transgenes were crossed to tub-GAL4 females and the resulting offspring carrying both transgene and tub-GAL4 were studied. Adult flies expressing LS alleles, Y279C and T468M, appeared in the expected Mendelian ratio and were viable and fertile. No lethality, which was observed when some other csw alleles were overexpressed (14), was observed with the LS alleles.
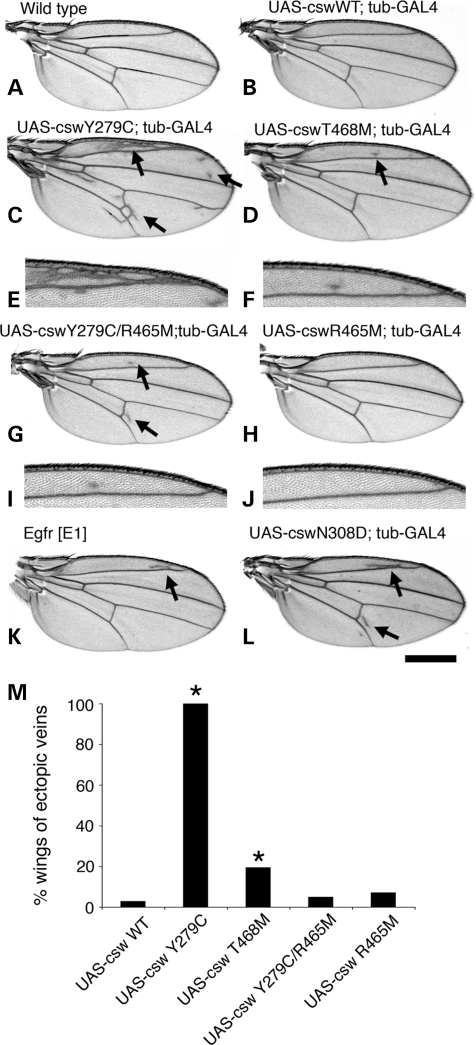
Next, we characterized phenotypic changes resulting from expression of the LS alleles. While the Y279C and T468M transgenes did not appear to alter early development, all of the Y279C and 20% of the T468M adult flies displayed ectopic wing veins. In contrast, the wings of flies expressing the wild-type csw transgene were normal except for the occasional presence of minimal ectopic veins (<3%) (Fig. 2). The wings of flies expressing LS mutant CSW exhibited ectopic veins mainly in the peripheral area of L2 and the posterior cross vein. Additionally, there were numerous small ectopic vein structures widely spreading over the entire area in the cswY279C transgene-expressing wings. When we compared the extent of ectopic vein formation between the alleles, the Y279C wings showed a significantly greater increase in ectopic veins than did the T468M (Fig. 2C–F and I), suggesting a strength difference between the mutations. This wing phenotype was the opposite of that observed in cswlf hypomorphic flies, which have interrupted wing veins that fail to reach the wing margin (21). The LS phenotype closely resembled those associated with the Egfr GOF allele, EgfrEl, (also known as Ellipse mutant) (22) and the NS N308D allele (Fig. 2K and L) (14). These findings indicated that the LS mutations behave as developmental GOF alleles rather than LOF or DN alleles as suggested from the in vitro phosphatase activity assays.
Figure 2.
Ubiquitous expression of LS csw alleles (Y279C and T468M) causes ectopic wing veins. Panels show adult wings from (A) wild type, (B) UAS-cswWT;tub-GAL4, (C and E) UAS-cswY279C;tub-GAL4, (D and F) UAS-cswT468M;tub-GAL4, (G and I) UAS-cswY279C/R465M;tub-GAL4, (H and J) UAS-cswR465M;tub-GAL4 (K) EgfrE1 and (L) UAS-cswN308D;tub-GAL4. (C–F) Ubiquitous expression of the LS alleles engenders ectopic veins, mainly in the peripheral areas of L2 and posterior cross vein (arrows), while wild-type csw transgene does not (B). (K and L) The LS wing phenotype resembles closely that observed with an Egfr gain-of-function allele, EgfrE1 and a NS allele, N308D. (G and I) Ubiquitous expression of a phosphatase-dead double mutant csw allele, Y279C/R465M. The Y279C ectopic wing veins were barely observed. Bar: 500 µm and 250 µm for the enlarged images (E, F, I and J). (M) The percentage of wings with ectopic wing veins. The csw transgenes described were crossed to tub-GAL4 driver. For each genotype, at least 40 wings were counted. Asterisks indicate statistical significance as compared to the UAS-cswWT expressing control (*P < 0.001).
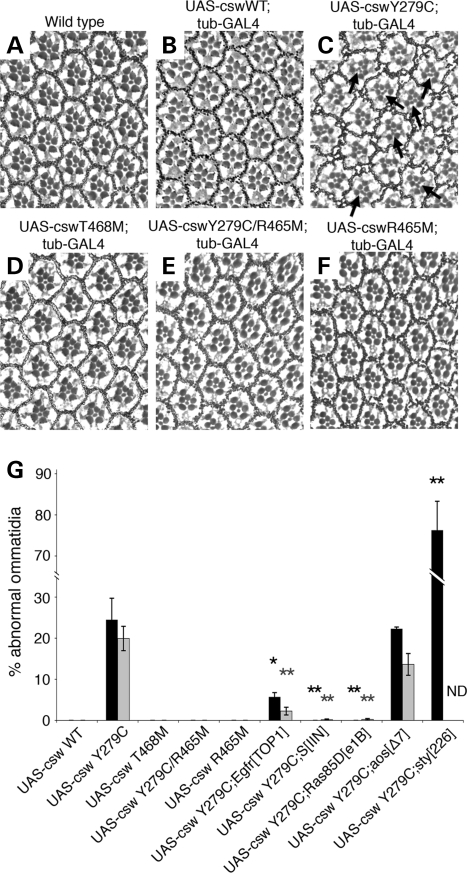
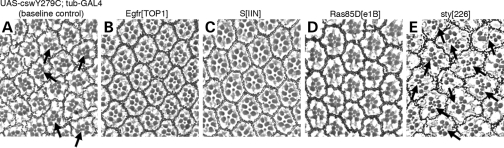
In addition to the wing phenotype, flies expressing the stronger mutant Y279C transgene exhibited a rough eye phenotype (Fig. 3). Although the size of the eye was not obviously changed, the arrangement of the ommatidia was altered. As shown in Figure 3C, the omatidia in the eyes of the Y279C-expressing flies were not symmetrically organized and microscopy of sections revealed the presence of multiple R7 photoreceptors in 24.5% of ommatidia (2–3/cell) (Fig. 3C and G). It is well known that cell fate specification of R7 is established through the RAS/RAF/MAPK pathway downsteam of the RTK, Sevenless, as well as through other receptors such as Notch and EGFR (23). This supernumerary R7 phenotype is a characteristic feature of flies with GOF sevenless, Ras and Raf alleles (24–26), suggesting that the Y279C allele was causing a GOF in RTK signaling. Moreover, these eyes displayed abnormalities of ommatidial rotation (20% of ommatidia) (Fig. 3C and G), for which EGFR signaling via MAPK has an important role (27). Of note, expression of neither the weaker T468M allele nor the wild csw transgene altered photoreceptor formation or ommatidial rotation.
Figure 3.
Images of sections of eyes. (A) wild type, (B) UAS-cswWT;tub-GAL4, (C) UAS-cswY279C;tub-GAL4, (D) UAS-cswT468M;tub-GAL4, (E) UAS-cswY279C/R465M;tub-GAL4 and (F) UAS-cswR465M;tub-GAL4. (C) Flies expressing the Y279C transgene exhibit a rough eye phenotype. Ommatidia were not symmetrically organized and their sections revealed the presence of multiple R7 photoreceptors (arrows) and abnormal rotations. (B and D) Expression of neither the weaker T468M allele nor the wild csw transgene alters photoreceptor formation or ommatidial rotation. (E) Flies expressing the phosphatase-dead double mutant, Y279C/R465M, do not display the Y279C eye phenotype. (G) The percentage of ommatidia with extra R7 photoreceptors (black bars) and misrotation (gray bars). All transgenes were overexpressed with tub-GAL4 and the additional mutant alleles of genes are heterozygous. Asterisks indicate statistical significance compared to the UAS-cswY279C expressing baseline control in order to evaluate genetic interaction effects (*P < 0.05; **P < 0.001). ND; not determined due to its phenotypic severity.
LS alleles have GOF effects through increased EGFR/RAS/MAPK signaling during development
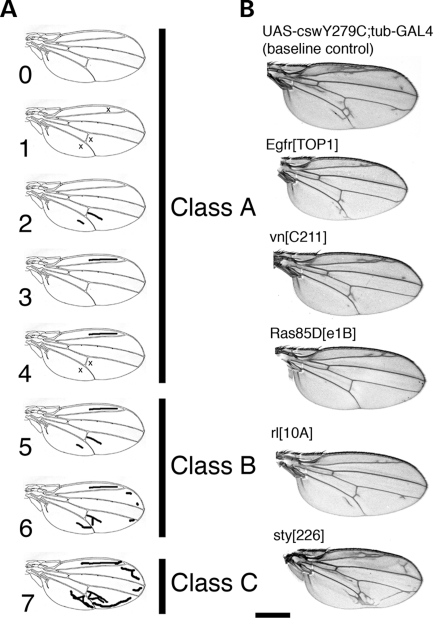
Since the wing phenotype observed in the LS transgenic flies closely resembled that of flies carrying an EGFR GOF mutation or the NS N308D csw GOF allele, it strongly suggested that the LS alleles increased EGFR signaling during wing vein formation and therefore had GOF effects in vivo. In the same way, formation of the rough eye phenotype due to increased numbers of R7 photoreceptors in the flies expressing Y279C csw implied increased activation of RTK/RAS/MAPK signaling during development. To test this, we generated flies expressing cswY279C ubiquitously as well as having LOF alleles for genes in the EGFR/RAS/MAPK signaling cascade. For the evaluation of wings, we used the wing classification system shown in Figure 4A (see Materials and Methods).
Figure 4.
Evaluation of the wing vein phenotype and wings of flies with ubiquitous Y279C CSW expression combined with mutant alleles for genes from EGFR/RAS/MAPK pathway. (A) Evaluation of the wing veins was performed according to the classification as illustrated. Each wing phenotype was assigned to one of the subclasses between 0 (no ectopic vein) and 7 (severe ectopic vein formation) according to the pattern of ectopic veins. Subclasses were grouped into class A, B or C as shown. Class B was designated as the baseline phenotype associated with the Y279C transgene. Assignment to classes A and C was considered based on suppression or enhancement, respectively. (B) Examples of ectopic wing vein phenotypes observed in flies of different genotypes. All flies had the UAS-cswY279C;tub-GAL4 genotype and were heterozygous for the indicated mutant alleles. LOF alleles for Egfr, vein, Ras85D, and rolled (EgfrTOP1, vnC211, Ras85De1B, and rl10A, respectively) suppressed the ectopic vein phenotype compared to the baseline Y279C CSW phenotype. In contrast, a LOF alleles for sprouty (sty226) enhanced the phenotype. Bar: 500 µm.
Crosses to LOF alleles of Ras85D, Son of sevenless (Sos) and rolled (rl, MAPK), which encode proteins in the RAS/MAPK pathway downstream of RTKs resulted in statistically significant suppression of ectopic wing vein formation (class A) (Table 2). Most wings showed no or mild ectopic vein formation in the anterior part of the wing (Fig. 4B, Table 2). In addition to intracellular positive regulators, LOF mutant alleles of Star (S), which is a positive extracellular regulator of EGFR signaling, and Egfr significantly suppressed the ectopic vein formation. LOF alleles of the Egfr ligands vein (vn) and spitz (spi) also caused minor, but insignificant, suppression of the ectopic vein formation (Fig. 4B). In contrast, crosses with LOF alleles of negative regulators of this signaling pathway, argos (aos), Gap1 and sprouty (sty), enhanced ectopic wing vein formation (Fig. 4B, Table 2).
Table 2.
Genetic interactions with the cswY279C ectopic wing vein phenotype
| Gene | % Class A wings (Suppression) | % Class B wings | % Class C wings (Enhancement) | P-value | Molecular features of genes |
|---|---|---|---|---|---|
| Control (w1118) | 8.8 | 91.2 | 0.0 | ||
| Egfrtop1 | 63.3 | 36.7 | 0.0 | <0.001 | Egfr |
| spi1 | 11.1 | 88.9 | 0.0 | 0.57 | Activating Egfr ligand |
| vnC221 | 0.0 | 100.0 | 0.0 | 0.06 | Activating Egfr ligand |
| aosΔ7 | 0.0 | 46.2 | 53.8 | <0.001 | Inhibitory Egfr ligand |
| SIIN | 93.8 | 6.3 | 0.0 | <0.001 | Egfr ligand processor |
| Sos34Ea-6 | 38.9 | 61.1 | 0.0 | <0.05 | Ras guanyl-nucleotide exchange factor |
| drke0A | 25.0 | 75.0 | 0.0 | 0.14 | SH3/SH2 adaptor protein |
| Ras85De1B | 96.7 | 3.3 | 0.0 | <0.001 | GTP binding protein |
| Gap1B2 | 0.0 | 0.0 | 100.0 | <0.001 | Ras GTPase activator |
| rl10A | 70.6 | 29.4 | 0.0 | <0.001 | MAPK |
| Sty226 | 0.0 | 0.0 | 100.0 | <0.001 | Egfr signaling antagonist |
n is at least 18 female wings.
Bold entries denote statistically significant suppression or enhancement of the ectopic wing vein phenotype.
We also evaluated the effects of these genetic interactions on the eye phenotype. LOF alleles of Egfr, Ras85D and S significantly suppressed the Y279C rough eye phenotype, normalizing the numbers of R7 and ommatidial rotation (Figs 3G and 5), while the S allele alone caused a rough eye phenotype (27,28). In contrast, a LOF allele of sty, a negative regulator of EGFR and other RTK signaling, enhanced the rough eye phenotype with the average number of R7 in each ommatidium increasing (>3 cells) (Figs 3G and 5E). Rotation defects could not be scored because the ommatidial alteration was too severe. These findings are consistent with the LS Y279C allele inducing its rough eye phenotype through increased EGFR signaling. Taken together, these genetic interaction studies documented that the Y279C transgene has GOF activity leading to increased EGFR signaling that then requires the RAS/MAPK cascade to induce the wing vein and eye phenotypes.
Figure 5.
Genetic interactions with the LS Y279C allele rough eye. Eye images of (A) UAS-cswY279C;tub-GAL4, (B) UAS-cswY279C;tub-GAL4;EgfrTOP1, (C) UAS-cswY279C;tub-GAL4;SIIN, (D) UAS-cswY279C;tub-GAL4;Ras85De1B, (E) UAS-cswY279C;tub-GAL4;sty226. (B–D) LOF alleles for Egfr, S, and Ras85D suppress the Y279C rough eye phenotype. Multiple R7 photoreceptors and abnormal rotation were not observed. (E) In contrast, a LOF allele of a negative regulator sprouty, sty226, enhanced the Y279C phenotype. The numbers of R7 cells are significantly increased (arrows).
Overexpression of LS CSW-induced MAPK activation
Kontaridis et al. showed that the Y279C and T468M SHP-2 mutants had DN effects, strongly inhibiting MAPK activation in transformed human embryonic kidney 293T cells (17). To determine whether similar DN effects occurred in the context of fly development, we performed western analysis using antibodies against dpERK and ERK.
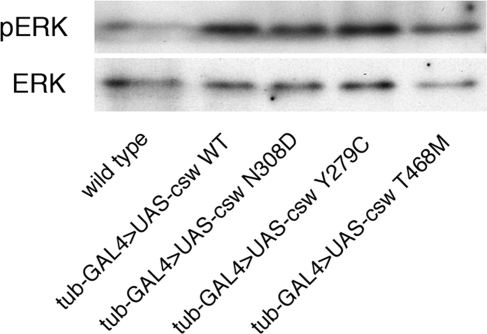
Contrary to the previous in vitro results, dpERK expression levels in the LS Y279C- and T468M-expressing larvae were significantly higher than that of wild-type larvae (tub-GAL4/+) and were similar to those of larvae expressing the wild-type and NS N308D transgenes (Fig. 6). ERK activation was not increased in the phosphatase-dead mutants, Y279C/R465M and R465M (Supplementary Material, Fig. S2). Immunohistochemical staining of fly embryos showed no differences in the pattern of ERK activation among these genotypes (data not shown) (29,30). These studies demonstrated that the LS alleles did not inhibit RAS/MAPK signaling during development.
Figure 6.
MAPK activation in ubiquitous LEOPARD and Noonan syndrome-transgene expressing larvae. Immunoblots of protein lysates from second instar larvae using antibodies against dpERK (activated MAPK) and ERK (all forms of MAPK). ERK is expressed at approximately equal levels in all fly lines. The expression of dpERK is increased in the wild-type and mutant transgenic larvae, as compared to the wild-type larvae showing a minimal level of dpERK.
PTP activity is necessary for the GOF effects of the LS alleles
As documented in the results presented thus far, the LS alleles engendered GOF effects during development and did not have DN effects on MAPK activation despite possessing reduced phosphatase activity. This raised the issue of whether the residual phosphatase activity of LS-associated CSW was necessary to induce the developmental phenotypes.
To address this question, we generated a double mutant transgenic fly line that eliminated any phosphatase activity in the Y279S CSW (Y279C/R465M; phosphatase-dead LS). As a control, single mutant transgenic flies expressing R465M CSW were also generated. Those transgenes were ubiquitously expressed using the tub-GAL4 driver. Of interest, ectopic wing vein formation was strongly suppressed in flies expressing the phosphatase-dead LS CSW (Fig. 2G, I and M). In addition, expression of the double mutant CSW protein did not result in the formation of supernumerary R7 photoreceptors and ommatidial rotation was normal (Fig. 3E and G). Ubiquitous expression of the R465M CSW single mutant did not result in any abnormal phenotype (Fig. 2H and J and 3F and G), documenting that this allele did not have DN effects on development per se.
DISCUSSION
CSW is the Drosophila orthologue of SHP-2 and works as a positive regulator of multiple RTK pathways (8,18,31). The amino acid sequence of SHP-2’s PTP domain is 63% identical to CSW, excluding an insertion of unknown consequence in the latter, and is 76% similar in the SH2 domains (18). Since the amino acids in the PTP domain altered by LS mutations in PTPN11 are conserved in the fly and the LS mutant CSW proteins showed reduced phosphatase activities in vitro, we hypothesized that transgenic Drosophila expressing mutant CSW would model the DN or LOF effects on signal transduction observed previously in cell culture. Moreover, we expected to observe phenotypes and epistatic interactions that differed from those observed in the transgenic Drosophila expressing the GOF alleles from NS. In fact, we observed the same ectopic wing vein phenotype in the LS fly models as described for the NS flies, and showed similar gene–gene interactions evidencing GOF in EGFR/RAS/MAPK signaling. In addition, we found a rough eye phenotype with supernumerary R7 photoreceptors, which had not been observed in the NS fly models but also behaved as a readout of increased RAS/MAPK signaling.
Although we demonstrated that the LS csw mutations act as GOF alleles during development, the mechanism for the increased EGFR signaling and GOF phenotypes remains unclear. The molecular function of SHP-2/CSW is complex but it is apparent that its PTP activity is important for normal function of these proteins (31). Here, we showed that the LS CSW mutants required their residual phosphatase activity in order to generate GOF effects in development. Our results would appear to be at odds with previous in vitro studies with LS SHP-2 mutants, which revealed that these proteins possessed minimal phosphatase activity and DN effects on signaling. There are, however, several considerations that make the current results comprehensible.
From a genetic perspective, it is worth noting that the LS-associated PTPN11 alleles are specific missense mutations. If their sole effect in transducing their phenotype was to reduce or eliminate SHP-2’s phosphatase activity, the existence of some haploinsufficient alleles such as nonsense mutations might be anticipated, but none has been observed (6,7,32). Similarly, loss of one Ptpn11 allele in mice results in no phenotype. Thus, simple LOF of SHP-2 seems inadequate to cause LS. While DN effects could be consistent with the genetic findings, our recent studies suggest that LS-causing Y279C and T468M mutations have a distinct perturbing effect in SHP-2 function (13). Moreover, our discovery that GOF RAF1 mutations cause NS and LS with hypertrophic cardiomyopathy renders that explanation improbable (33). Unlike the mutual exclusivity of PTPN11 mutations observed in NS and LS, one RAF1 missense defect has been observed in patients with both disorders and the other LS-associated RAF1 mutation clustered with one causing NS. The discovery of LS-associated RAF1 GOF alleles, which increase RAS/MAPK signaling, provides credence to our contention that biochemically impaired SHP-2 still engenders GOF developmental effects.
How might phosphatase-impaired SHP-2 mutants result in GOF developmental effects? While there is little question but that LS SHP-2 mutants are phosphatase impaired, previous characterizations of them, including our own, may have overestimated the degree of functional LOF. The reduced catalytic activities observed in vitro might not reflect the true enzymatic activity in vivo. The bona fide substrates for SHP-2 are still under investigation, and the affinity of the ones used in vitro may not accurately assess biologically relevant kinetics for the critical moieties (15,16,34). In cells, SHP-2 is recruited to complexes at the cell membrane through scaffolding proteins, which affects critical issues such as activation and substrate concentration (35). Biochemical assays in vitro, particularly when performed with SHP-2 prepared in bacteria and then isolated, fail to capture this complexity.
A second potentially relevant biochemical issue relates to the timing of SHP-2’s phosphatase activity. Wild-type SHP-2 is basally inactive due its molecular switch and become activated briefly through engagement of phosphotyrosyl residues at binding sites in the N- and C-SH2 domain. NS-associated GOF SHP-2 mutants with amino acid substitutions at the N-SH2/PTP interface generally have increased basal activity and remain active after stimulation for periods far in excess of normal. Of note, there are a few NS-associated PTPN11 mutations that alter the phosphotyrosyl binding sites, leaving the molecular switch and phosphatase domain intact. Their mutant SHP-2 proteins are basally quiescent, have normal peak phosphatase activity but show enhanced binding to phosphotyrosine-containing peptides, which promotes a prolonged activity following stimulation (12,13,15). Since these alleles cause NS, it is apparent that prolonged SHP-2 activity alone is sufficient to perturb development. Similarly, the LS-associated mutant SHP-2 proteins may have phosphatase activity that is prolonged. These LS mutants also exhibit enhanced association with a relevant docking protein, GAB1, after stimulation of cells with EGF (16,17). This suggests that LS-associated mutant SHP-2 proteins may remain in protein complexes and propagating signals at times when wild-type SHP-2 does not.
There is precedence for mutant proteins with impaired activity in the RAS/MAPK signaling pathway resulting in GOF phenotypic effect. BRAF is frequently mutated in cancer, particularly in melanoma (36). While most of the cancer-associated mutations increased the ability of BRAF to act as a MEK kinase, there are several mutations that result in kinase-impaired proteins. For some of the kinase-impaired mutants, increased signaling to MAPK occurs, which seems to result from increased activation of RAF1 in BRAF-RAF1 heterodimers. There are, however, a few BRAF mutants that are severely kinase impaired and do not induce increased MAPK activation. Their mechanism of action with respect to oncogenesis remains unknown. Analogously, RAF1 mutations in NS are generally GOF, often through loss of 14-3-3-mediated inactivation of RAF1 (33). Some RAF1 mutations, however, alter the activation loop in the kinase domain, which are analogous to the kinase-impaired BRAF mutations observed in cancer. Both of the activation loop RAF1 mutants characterized biochemically are kinase-impaired, but one increases MAPK activation while the other does not. While the activation loop mutations are not associated with hypertrophic cardiomyopathy like the other RAF1 defects are, they do engender the NS developmental features equivalently. As with the LS-associated PTPN11 mutations, the absence of genetic lesions to RAF1 that result in haploinsufficiency such as nonsense mutations suggests strongly that these activity-impaired mutant proteins are retaining functions necessary for inducing the genetic trait.
One limitation of the present study is that wing vein and eye phenotypes observed in fruitflies expressing the LS-associated CSW mutants may not model all aspects of LS pathogenesis. In a recent study using zebrafish, Shp-2 levels were reduced using morpholinos or overexpressed by injecting cRNAs encoding NS- or LS-associated Shp-2 proteins (37). Reduction of Shp-2 resulted in gastrulation defects due to perturbation of convergence and extension cell movements. These were attributable to perturbed signaling through RhoA, and no Ras/MAPK signaling defect was found. Overexpression of the NS and LS Shp-2 mutants also perturbed the convergence and extension cell movements, and craniofacial abnormalities and abnormal heart jogging movements were noted as well. No alteration in cell proliferation or specification was observed. Of particular interest, low-level expression of two different NS-associated or two different LS-associated Shp2 proteins resulted in synergistic phenotypes while expression of one NS and one LS Shp2 protein caused no phenotype. Since a mouse model of NS with a Ptpn11 mutation demonstrated increased Erk activation in several tissues during embryonic development, including the craniofacial region, as well as increased cellular proliferation in the endocardial cushions of the nascent cardiac atrioventricular valves (38), it is clear that the zebrafish models also failed to capture all aspects of the developmental pathogenesis of NS and LS. Nonetheless, the zebrafish model is important in demonstrating that some NS/LS developmental abnormalities probably do not arise through RAS/MAPK signaling perturbation and that NS- and LS-associated SHP2 proteins may alter signaling in non-synergistic ways while producing remarkably comparable phenotypes.
MATERIALS AND METHODS
Drosophila stocks
Unless otherwise specified, fly stocks are as described in FlyBase (http://www.flybase.net/), and were provided by the Bloomington Stock Center (http://flystocks.bio.indiana.edu/). Flies were cultured in a standard medium and crosses were performed at 25°C if not otherwise noted. As the wild-type control, w1118 or tub-GAL4/+ was used.
Generation of transgenic flies
Site-directed mutagenesis was performed to introduce two recurrent LS mutations into csw that correspond to Y279C and T468M mutations in PTPN11 using the Quick Change Site Directed Mutagenesis Kit (Stratagene). The cDNAs were sequenced to confirm the presence of the desired mutation. The wild-type and mutated csw cDNAs were inserted into a pUASP vector, and P-element-mediated transformation was performed to obtain LS transgenic flies (39). Likewise, other mutant transgenic flies were generated in the same manner. Multiple fly lines were generated for each construct, designated generally as UAS-cswtg and specific alleles as UAS-cswWT, UAS-cswY279C, UAS-cswT468M, UAS-cswR465M and UAS-cswY279C/R465M. Transgenic lines expressing similar levels of CSW protein using the same GAL4 driver were selected and used for further analyses.
Genetic analyses
For genetic interaction tests, a stable stock of each of the genotype UAS-cswY279C/CyO; tub-GAL4/TM2 was generated. This stock was then crossed to mutant flies with LOF alleles for genes of interest. We evaluated changes in vein patterning using the ectopic vein phenotype as the readout and assessed whether the presence of a mutant allele of for the gene of interest suppressed the ectopic vein. We used the classification system as previously described for wing vein grading and wings were categorized as subclassed 0–7 as shown in Figure 4A. Since the majority of the flies with the genotype of UAS-cswY279C/+; tub-GAL4/+ had wings with subclasses 5–6 ectopic wing veins, we designated these as the baseline phenotype of the cswY279C transgene, class B. If wings of progeny flies, carrying UAS-cswN308D; tub-GAL4 and a mutant allele, had no or small ectopic veins (subclasses 0–4), they were scored as class A (suppression) and if wings have longer and complex vein formation (subclass 7), they were scored as class C (enhancement). Fly eyes were observed using a light microscope. For statistical analysis, female wings were scored and χ2 tests were performed. Significant suppression or enhancement was declared with a threshold of P < 0.05.
Biochemical studies
Full-length human PTPN11 and Drosophila csw cDNAs were cloned into pGEX-4T-2 vector (Amersham Biosciences) and the LS Y279C and T468M mutations along with a NS allele, N308D, were introduced using the Quick Change Site Directed Mutagenesis Kit (Stratagene). Recombinant SHP-2 and CSW proteins were expressed in Escherichia coli (BL21) (Stratagene). GST-tagged expressed proteins were purified by chromatography using BugBuster GST-Bind purification kit (Novagen). In vitro phosphatase assays with 5 µg of purified recombinant SHP-2 and CSW proteins were performed using the PTP Assay Kit 2 (Upstate). Enzyme activities were measured after 30 min incubation at 37°C with pNPP as a substrate either in basal condition or with the activating PTP nonreceptor type substrate 1 (PTPNS1) bisphosphoryl tyrosine-based activation motif peptide (5 µm). Hydrolysis of pNPP was evaluated by measuring absorbance at 410 nm.
Immunoblotting, immunostaining and RT–PCR
For immunoblotting, protein lysates were obtained by homogenizing second instar larvae or adult flies in 100 or 200 µl of 2× Laemmli’s sample buffer (125 mm Tris–HCl, pH 6.8, 4% SDS, 30% glycerol, 1% β-mercaptoethanol), respectively. Total protein was separated with SDS–PAGE and transferred to PVDF membrane. Thirty microliters of protein lysates from 20 second instar larvae were used for the detections of dpERK and ERK levels. Anti-dpERK and ERK antibodies (Cell Signaling Technology) were used these assays. These immunoblotting were repeated three times for each assay and consistent results were obtained. For immunohistochemical detection of dp-ERK, developing embryos were collected, fixed and stained with mouse anti-dpERK (Sigma) (29,30) as described (14). Biotin-conjugated secondary antibody was employed and the signal was enhanced using the VECTASTAIN ABC and DAB substrate kit (Vector).
Total third instar larval RNAs were extracted using TRIZOL reagent (Invitrogen). To assess csw transgene expression, RT–PCR was performed using primer pairs for csw (F; 5′-GGTGACCCACATCAAAATCC-3′, R; 5′-GCCGACGTCGTACTTCTTGT-3′) and Rp49 (F; 5′-CAGCATACAGGCCCAAGAT-3′, R; 5′-GCACTCTGTTGTCGATACCC-3′) as a control (40). RT–PCR reactions were performed with One-Step RT–PCR System (Invitrogen) according to the manufacturer’s instructions using 500 ng of total RNA as template. Reactions (28 cycles) were performed at annealing temperatures of 56°C. No genomic DNA contamination was confirmed using an internal intron sequence of the Rp49 primers. For quantitative RT–PCR, reverse transcription was performed using Superscript II reverse transcriptase and oligo-dT primers (Invitrogen). Transcript levels were determined by real-time PCR using SYBR Green as a detection reagent and Rp49 as an internal control. These studies were performed in triplicate to obtain statistical significance.
Genotyping of embryos or larvae was performed using balancer chromosomes carrying either eve-lacZ or GFP markers.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the National Institutes of Health (HL71207, HD01294, HL074728 to B.D.G. and EY13256 to M.M.); March of Dimes (FY03-52, 6-FY07-286 to B.D.G.); Telethon-Italy grants (GGP07115 to M.T.); Programma di Collaborazione Italia-USA/malattie rare to M.T.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lizabeth Perkins for providing reagents, Bhaswti Pandit, Ruth Johnson, Audrey Au and Jiapeng Wang for their technical assistance and also the Bloomington Drosophila Stock Center at Indiana University for providing fly lines.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Gorlin R.J., Anderson R.C., Moller J.H. The Leopard (multiple lentigines) syndrome revisited. Birth Defects Orig. Artic. Ser. 1971;07:110–115. [PubMed] [Google Scholar]
- 2.Noonan J.A. Hypertelorism with Turner phenotype. A new syndrome with associated congenital heart disease. Am. J. Dis. Child. 1968;116:373–380. doi: 10.1001/archpedi.1968.02100020377005. [DOI] [PubMed] [Google Scholar]
- 3.Allanson J.E. Noonan syndrome. J. Med. Genet. 1987;24:9–13. doi: 10.1136/jmg.24.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S., et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 5.Tartaglia M., Kalidas K., Shaw A., Song X., Musat D.L., van der Burgt I., Brunner H.G., Bertola D.R., Crosby A., Ion A., et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am. J. Hum. Genet. 2002;70:1555–1563. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Digilio M.C., Conti E., Sarkozy A., Mingarelli R., Dottorini T., Marino B., Pizzuti A., Dallapiccola B. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am. J. Hum. Genet. 2002;71:389–394. doi: 10.1086/341528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Legius E., Schrander-Stumpel C., Schollen E., Pulles-Heintzberger C., Gewillig M., Fryns J.P. PTPN11 mutations in LEOPARD syndrome. J. Med. Genet. 2002;39:571–574. doi: 10.1136/jmg.39.8.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeman R.M., Jr, Plutzky J., Neel B.G. Identification of a human src homology 2-containing protein-tyrosine-phosphatase: a putative homolog of Drosophila corkscrew. Proc. Natl. Acad. Sci. USA. 1992;89:11239–11243. doi: 10.1073/pnas.89.23.11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barford D., Neel B.G. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 1998;6:249–254. doi: 10.1016/s0969-2126(98)00027-6. [DOI] [PubMed] [Google Scholar]
- 10.Hof P., Pluskey S., Dhe-Paganon S., Eck M.J., Shoelson S.E. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 11.Fragale A., Tartaglia M., Wu J., Gelb B.D. Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum. Mutat. 2004;23:267–277. doi: 10.1002/humu.20005. [DOI] [PubMed] [Google Scholar]
- 12.Keilhack H., David F.S., McGregor M., Cantley L.C., Neel B.G. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J. Biol. Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 13.Martinelli S., Torreri P., Tinti M., Stella L., Bocchinfuso G., Flex E., Grottesi A., Ceccarini M., Palleschi A., Cesareni G., et al. Diverse driving forces underlie the invariant occurrence of the T42A, E139D, I282V and T468M SHP2 amino acid substitutions causing Noonan and LEOPARD syndromes. Hum. Mol. Genet. 2008 doi: 10.1093/hmg/ddn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oishi K., Gaengel K., Krishnamoorthy S., Kamiya K., Kim I.K., Ying H., Weber U., Perkins L.A., Tartaglia M., Mlodzik M., et al. Transgenic Drosophila models of Noonan syndrome causing PTPN11 gain-of-function mutations. Hum. Mol. Genet. 2006;15:543–553. doi: 10.1093/hmg/ddi471. [DOI] [PubMed] [Google Scholar]
- 15.Tartaglia M., Martinelli S., Stella L., Bocchinfuso G., Flex E., Cordeddu V., Zampino G., Burgt I., Palleschi A., Petrucci T.C., et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am. J. Hum. Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanna N., Montagner A., Lee W.H., Miteva M., Vidal M., Vidaud M., Parfait B., Raynal P. Reduced phosphatase activity of SHP-2 in LEOPARD syndrome: consequences for PI3K binding on Gab1. FEBS Lett. 2006;580:2477–2482. doi: 10.1016/j.febslet.2006.03.088. [DOI] [PubMed] [Google Scholar]
- 17.Kontaridis M.I., Swanson K.D., David F.S., Barford D., Neel B.G. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J. Biol. Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 18.Perkins L.A., Larsen I., Perrimon N. Corkscrew encodes a putative protein tyrosine phosphatase that functions to transduce the terminal signal from the receptor tyrosine kinase torso. Cell. 1992;70:225–236. doi: 10.1016/0092-8674(92)90098-w. [DOI] [PubMed] [Google Scholar]
- 19.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Camb) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 20.Mohi M.G., Williams I.R., Dearolf C.R., Chan G., Kutok J.L., Cohen S., Morgan K., Boulton C., Shigematsu H., Keilhack H., et al. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer cell. 2005;7:179–191. doi: 10.1016/j.ccr.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Firth L., Manchester J., Lorenzen J.A., Baron M., Perkins L.A. Identification of genomic regions that interact with a viable allele of the Drosophila protein tyrosine phosphatase corkscrew. Genetics. 2000;156:733–748. doi: 10.1093/genetics/156.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker N.E., Rubin G.M. Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division, and cell death in eye imaginal discs. Dev. Biol. 1992;150:381–396. doi: 10.1016/0012-1606(92)90250-k. [DOI] [PubMed] [Google Scholar]
- 23.Nagaraj R., Banerjee U. The little R cell that could. Int. J. Dev. Biol. 2004;48:755–760. doi: 10.1387/ijdb.041881rn. [DOI] [PubMed] [Google Scholar]
- 24.Basler K., Christen B., Hafen E. Ligand-independent activation of the sevenless receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell. 1991;64:1069–1081. doi: 10.1016/0092-8674(91)90262-w. [DOI] [PubMed] [Google Scholar]
- 25.Rommel C., Radziwill G., Moelling K., Hafen E. Negative regulation of Raf activity by binding of 14-3-3 to the amino terminus of Raf in vivo. Mech. Dev. 1997;64:95–104. doi: 10.1016/s0925-4773(97)00052-x. [DOI] [PubMed] [Google Scholar]
- 26.Fortini M.E., Simon M.A., Rubin G.M. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- 27.Gaengel K., Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development (Camb) 2003;130:5413–5423. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- 28.Kolodkin A.L., Pickup A.T., Lin D.M., Goodman C.S., Banerjee U. Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila. Development (Camb) 1994;120:1731–1745. doi: 10.1242/dev.120.7.1731. [DOI] [PubMed] [Google Scholar]
- 29.Gabay L., Seger R., Shilo B.Z. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development (Camb) 1997;124:3535–3541. doi: 10.1242/dev.124.18.3535. [DOI] [PubMed] [Google Scholar]
- 30.Gabay L., Seger R., Shilo B.Z. In situ activation pattern of Drosophila EGF receptor pathway during development. Science (NY) 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 31.Allard J.D., Herbst R., Carroll P.M., Simon M.A. Mutational analysis of the SRC homology 2 domain protein-tyrosine phosphatase Corkscrew. J. Biol. Chem. 1998;273:13129–13135. doi: 10.1074/jbc.273.21.13129. [DOI] [PubMed] [Google Scholar]
- 32.Tartaglia M., Gelb B.D. Germ-line and somatic PTPN11 mutations in human disease. Eur. J. Med. Genet. 2005;48:81–96. doi: 10.1016/j.ejmg.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Pandit B., Sarkozy A., Pennacchio L.A., Carta C., Oishi K., Martinelli S., Pogna E.A., Schackwitz W., Ustaszewska A., Landstrom A., et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 34.Kalidas K., Shaw A.C., Crosby A.H., Newbury-Ecob R., Greenhalgh L., Temple I.K., Law C., Patel A., Patton M.A., Jeffery S. Genetic heterogeneity in LEOPARD syndrome: two families with no mutations in PTPN11. J. Hum. Genet. 2005;50:21–25. doi: 10.1007/s10038-004-0212-x. [DOI] [PubMed] [Google Scholar]
- 35.Neel B.G., Gu H., Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 36.Fecher L.A., Amaravadi R.K., Flaherty K.T. The MAPK pathway in melanoma. Curr. Opin. Oncol. 2008;20:183–189. doi: 10.1097/CCO.0b013e3282f5271c. [DOI] [PubMed] [Google Scholar]
- 37.Jopling C., van Geemen D., den Hertog J. Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 2007;3:e225. doi: 10.1371/journal.pgen.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Araki T., Mohi M.G., Ismat F.A., Bronson R.T., Williams I.R., Kutok J.L., Yang W., Pao L.I., Gilliland D.G., Epstein J.A., et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat. Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 39.Spradling A.C., Rubin G.M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science (NY) 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 40.Borghese L., Fletcher G., Mathieu J., Atzberger A., Eades W.C., Cagan R.L., Rorth P. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev. Cell. 2006;10:497–508. doi: 10.1016/j.devcel.2006.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.