Abstract
Linker histones bind to the nucleosomes and linker DNA of chromatin fibers, causing changes in linker DNA structure and stabilization of higher order folded and oligomeric chromatin structures. Linker histones affect chromatin structure acting primarily through their ~100 residue C-terminal domain (CTD). We have previously shown that the ability of the linker histone H1° to alter chromatin structure was localized to two discontinuous 24-/25-residue CTD regions (Lu, X., and Hansen, J. C. (2004) J Biol Chem 279, 8701–8707). To determine the biochemical basis for these results, we have characterized chromatin model systems assembled with endogenous mouse somatic H1 isoforms, or recombinant H1° CTD mutants in which the primary sequence has been scrambled, the amino acid composition mutated, or the location of various CTD regions swapped. Our results indicate that specific amino acid composition plays a fundamental role in molecular recognition and function by the H1 CTD. Additionally, these experiments support a new molecular model for CTD function, and provide a biochemical basis for the redundancy observed in H1 isoform knockout experiments in vivo.
Linker histones (e.g., H1, H5) are chromatin architectural proteins found in all eukaryotes (1, 2). They are abundant, with a stoichiometry of ~0.8 total linker histones per nucleosome in most tissues (3 and references therein). Linker histones are modularly structured proteins that have an ~35 residue unstructured N-terminal domain (NTD)1, a central globular winged helix domain, and an ~100 residue unstructured C-terminal domain (CTD) (4). Linker histones bind to chromatin fibers through interaction of the globular domain with nucleosomal sites(s) (1, 2, 5), and the CTD with linker DNA (6, 7). Higher eukaryotes have at least six somatic linker histone isoforms, which differ primarily in their CTD primary sequences (1, 8). The H1 isoform CTDs do, however, share a very similar and characteristic amino acid composition (9). At the molecular level, little is known about the actions of the isoforms. Linker histones are multifunctional, with roles in chromatin condensation (1, 2, 10, 11), nucleosome spacing (12, 13), specific gene expression (13) and references therein), DNA methylation (13) and other nuclear processes. In addition to chromatin, linker histones bind to many nuclear proteins, e.g., DFF40/CAD (14), BAF (15), Purα/YB1 (16). Thus, a key unanswered question related to linker histone function is how these small, simply constructed proteins can specifically recognize and interact with so many different macromolecular partners.
The relationships between linker histones, nucleosomal arrays, and chromatin fiber structure are well documented (1, 2, 10). In vitro, nucleosomal arrays are in salt-dependent equilibrium between unfolded, folded, and oligomeric conformational states (10, 11). Binding of linker histones to nucleosomal arrays affects chromatin structure in at least three distinct ways: the linker DNA between nucleosomes assumes an apposed stem/loop motif (7, 17, 18). Secondly, the folded conformational states of linker histone-bound chromatin fibers are stabilized relative to the folding seen with nucleosomal arrays alone, i.e., linker histones shift the equilibrium in favor of the folded chromatin fiber structures. Finally, Mg2+-dependent chromatin fiber oligomerization is also stabilized. Linker histones that lack their long unstructured CTD can bind to chromatin fibers but cannot mediate formation of apposed linker DNA segments or stabilization of folded or oligomeric conformational states (7, 19). Thus, the H1 CTD is essential for maintaining the structure and stability of chromatin fibers. Recently, CTD truncation studies showed that the abilities of the CTD to alter linker DNA structure and affect chromatin fiber folding and oligomerization were localized to two discontinuous 24-/25-residue regions (H1° residues 98–122 and 147–170) (7). While these results ruled out one obvious model in which the entire CTD is needed to bind to DNA and neutralize negative charge, the molecular basis for this observation remains unresolved. One possibility is that each of these two regions contains structured primary sequence motifs that mediate function, e.g., stable α-helices. However, an alternative possibility is that the action of the two functional CTD regions is linked to intrinsic protein disorder (9).
The concept of intrinsic protein disorder has been explored extensively in recent years (see (20 and references therein). Intrinsically disordered protein domains mostly or completely lack classical secondary structure, but undergo disorder to order transitions concomitant with binding to their macromolecular targets (9, 21, 22). While much progress has been made predicting the prevalence of intrinsically disordered protein domains in eukaryotic proteomes (23), there is a relative paucity of experimental biochemical data that address the molecular mechanisms that underlie how intrinsically disordered regions function. Several groups have speculated that intrinsically disordered domains contain short primary sequence elements imbedded within the domain (24–26). In addition, interesting attributes related to amino acid composition have been observed, such as reduced levels of hydrophobic residues and increased levels of charged residues (21, 27). We have noted that the amino acid composition of the somatic linker histone CTDs is distinctive; it is very similar between isoforms but differs from that of other intrinsically disordered protein regions such as yeast prion domains (9). Based on these observations, we have hypothesized that the chromatin condensing functions of the CTD may be mediated by its specific amino acid composition (9). A potential role for amino acid composition is intriguing because dogma holds that protein function is linked to primary amino acid sequence rather than composition. To examine the role of primary sequence and specific amino acid composition, and to better understand how linker histone isoforms affect chromatin structure in vitro, here we have used sedimentation velocity and differential centrifugation to characterize the linker DNA structure and folded and oligomeric states of defined chromatin model systems bound to endogenous mouse somatic H1 isoforms and specific recombinant H1 CTD mutants. Our results support a mechanism for H1 CTD function that is based on its amino acid composition and intrinsic protein disorder rather than classical modes of molecular recognition. Further, these data provide a molecular explanation for how the linker histone isoform CTDs can have redundant functions in chromatin condensation in vivo.
EXPERIMENTAL PROCEDURES
Materials
208-12 DNA (28–30) and chicken erythrocyte core histone octamers (31) were purified as described. All mouse linker histone H1° mutants were created based on the wild type H1° plasmid, pET-H1°-11d (32), using standard methods (33). Plasmids were sequenced to confirm the desired mutations were present. Wild type and mutant H1° proteins were expressed and purified in E. coli as described (7). Mouse endogenous linker histone variants were prepared as described (14). The following procedure was used to select the sequences of the two H1° mutants containing randomized sequences in their subdomain 1. Ten randomized sequences were first generated at web site, http://www.cellbiol.com/scripts/randomizer/sequence_randomizer.html, using wild type subdomain 1 as input. Program Excel (Microsoft, Seattle, WA) was then used to generate 2 random numbers between 1 and 10 and the two sequences corresponding to these two numbers were selected and named H1°RanS1#1 and H1°RanS1#2 (see Table 1 for sequences).
Table 1. Primary sequences of linker histone H1° CTD regions 1–4 and specific CTD mutants.
The amino acid differences between CTD region 1 and the S1TD mutant are shown in bold underline.
| Region | Residues | Sequence |
|---|---|---|
| 4 | 171–194 | SKPKKAKTVKPKAKSSAKRASKKK |
| 3 | 147–170 | KKKPAATPKKAKKPKVVKVKPVKA |
| 2 | 123–146 | AAKPKKAASKAPSKKPKATPVKKA |
| 1 | 98–122 | GDEPKRSVAFKKTKKEVKKVATPKK |
| RanS1#1 | -- | KTRKVDKPEPKKFKGEAVSKKAVTK |
| RanS1#2 | -- | SVKGAVEKVKAEKPDKPTKTKFKRK |
| S1TD | -- | GDEPKRSNAFKKPKKENKKNATPKK |
Reconstitution of nucleosomal arrays and linker histone-containing chromatin fibers
Nucleosomal arrays in which approximately 50% of the sample was saturated with nucleosomes were assembled from 208–12 DNA and chicken histone octamers as described previously (7). Briefly, histone octamers and DNA were mixed in 10 mM Tris, 0.25 mM EDTA, pH 7.8 (TE) containing 2 M NaCl at a molar ratio of ~1.1. The mixture was subsequently dialyzed against TE buffer containing 1 M, 0.75 M, and 2.5 mM NaCl, respectively, for at least 6 hr each. The mixture was then dialyzed against fresh TE buffer containing 2.5 mM NaCl (TEN) overnight. To assemble linker histone-containing chromatin model systems, the parent nucleosomal arrays were first brought to 50 mM NaCl by addition of appropriate amounts of 0.5 M NaCl. Linker histone was added to the solution at a ratio of ~1.3 moles linker histone per mol 208 bp repeat, and the solution dialyzed against TEN overnight (7, 18). The integrity of the reconstituted nucleosomal arrays and linker histone-containing chromatin fibers was confirmed by sedimentation velocity in TEN.
Sedimentation velocity
Sedimentation velocity experiments were performed in a Beckman XLA or XLI ultracentrifuge equipped with scanner optics as described (7, 18, 34). Scans were analyzed by the modified method of van Holde and Weischet (35, 36) using Ultrascan data analysis software (35). This method yields the integral distribution of sedimentation coefficients of the chromatin sample, G(s), plotted as boundary fraction versus the diffusion corrected sedimentation coefficient (35, 36).
Differential centrifugation
The assay for chromatin fiber oligomerization was performed as described (7, 18). Chromatin model systems (A260 1.2–1.4) were mixed with an equal volume of 2× MgCl2 solution to achieve the desired final salt concentration. After incubation for 5 minutes at room temperature, samples were centrifuged for 5 minutes at 16,000g in a microcentrifuge at room temperature. The supernatant was then removed and the A260 measured. The percentage of the sample remaining in the supernatant was plotted against MgCl2 to yield the oligomerization curve (7, 18). Each data point in the plot reflected the mean of 2–5 experiments. The oligomerization curve was used to obtain the Mg50, defined as the MgCl2 concentration at which 50% of the sample had oligomerized and pelleted during centrifugation.
RESULTS
When saturated with 12 nucleosomes per DNA, the model nucleosomal arrays used in our experiments sediment at 28–29 S in Mg2+-free TEN buffer (10, 18). As a control, the sample nucleosomal arrays used in Figures 1–5 were characterized by sedimentation velocity in TEN buffer and the boundaries analyzed to yield the diffusion corrected integral distribution of sedimentation coefficients, G(s) (35, 36). The data shown in Figure 1A indicated that roughly half of the sample sedimented at 28–29 S (boundary fraction 50–90%) and hence was saturated with 12 nucleosomes per DNA. The remainder of the sample sedimented at 26–27 S, and thus contained 10–11 nucleosomes per DNA, i.e., they were slightly “subsaturated”. This is typical of array preparations used in previous studies (7, 18, 29, 31).
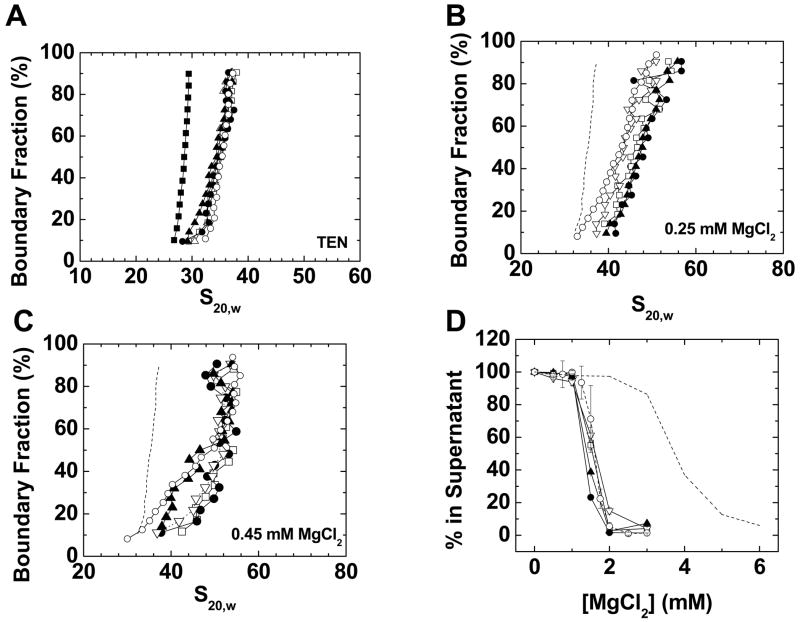
Figure 1. Characterization of model chromatin fibers bound to endogenous mouse somatic linker histone isoforms.
A) Sedimentation coefficient distributions in low salt TEN buffer of 208–12 nucleosomal arrays (dashed line) assembled with endogenous mouse H1° (○), H1S-1(●), H1S-1(□), H1S-3 (▲), and H1S-4 (▽). B) G(s) plots of 208–12 chromatin fibers bound to wild type H1° (○), H1S-1(●), H1S-1(□), H1S-3 (▲), or H1S-4 (▽) in 0. 25 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. C) G(s) plots of 208–12 fibers bound to wild type H1° (○), H1S-1(●), H1S-1(□), H1S-3 (▲), or H1S-4 (▽) in 0. 45 mM MgCl2. D) Oligomerization as a function of MgCl2. Shown are data for 208–12 nucleosomal arrays (dashed line) and 208–12 chromatin fibers bound to wild type H1° (○), H1S-1(●), H1S-1(□), H1S-3 (▲), or H1S-4 (▽). All plots are representative of results seen with 3–4 different chromatin preparations.
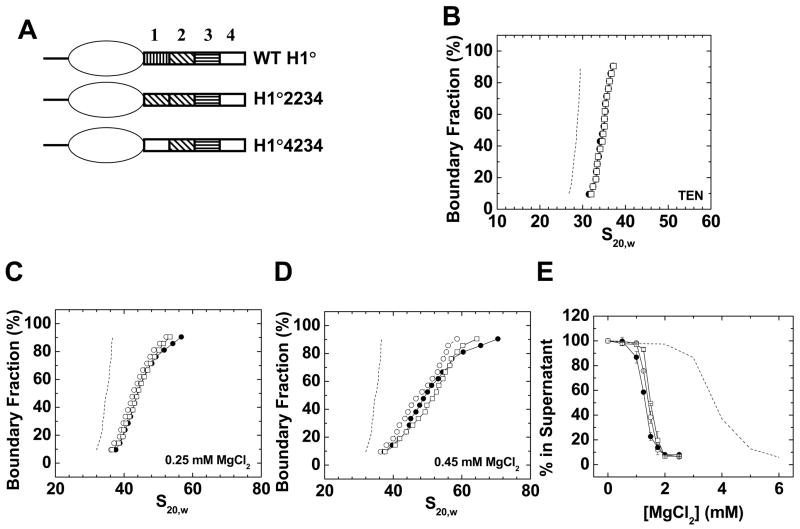
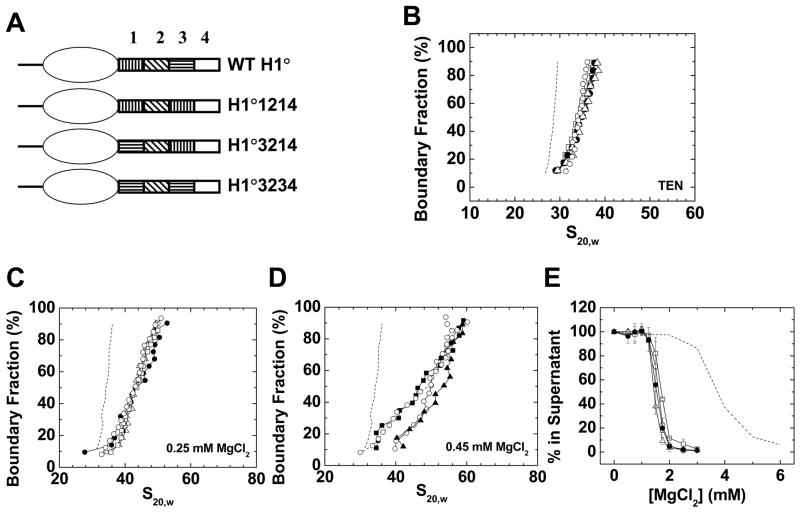
Figure 5. Replacement of H1° CTD region 1 with regions 2 or 4.
A) Schematic diagrams of wild type H1°, H1°2234, and H1°4234. B) Sedimentation coefficient distributions in low salt TEN buffer of 208–12 nucleosomal arrays (dashed line) assembled with wild type H1° (○), H1°2234 (●), or H1°4234 (□). C) Sedimentation velocity analysis. G(s) plots of 208–12 chromatin fibers bound to wild type H1° (○), H1°2234 (●), or H1°4234 (□) in 0. 25 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. D) G(s) plots of 208–12 fibers bound to wild type H1° (○), H1°2234 (●), or H1°4234 (□) in 0. 45 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. E) Oligomerization as a function of MgCl2. Shown are data for 208–12 nucleosomal arrays (dashed line) and 208–12 chromatin fibers bound to wild type H1° (○), H1°2234 (●), or H1°4234 (□). All plots are representative of results seen with 3–4 different chromatin preparations.
Effects of endogenous mouse linker histone isoforms on chromatin fiber structure
The CTDs of the somatic linker histone isoforms have divergent primary sequences (1, 8), but a very similar amino acid composition (9). Hence, to test the relative importance of primary sequence versus composition we first characterized model systems assembled with five purified endogenous mouse H1 isoforms (H1°, H1S-1, H1S-2, H1S-3, H1S-4; see Supplementary Figure 1 for correlation of isoform nomenclatures). The experiments described below in each case assay three H1 CTD-mediated functions related to chromatin condensation: apposition of linker DNA structure, stabilization of the intrinsic folding of nucleosomal arrays (4, 7, 18), and stabilization of nucleosomal array oligomerization (7, 18).
Previous studies have shown that saturated 12-mer nucleosomal arrays bound to one linker histone per nucleosome sediment at ~36 S in TEN (7, 18). This increase in sedimentation coefficient from 29 to ~36 S is due to both an increase in mass from linker histone binding to the nucleosomal arrays and the formation of the apposed stem-loop motif, the latter of which shortens the linker DNA and thus decreases the frictional coefficient of the fibers (7, 18). When mixed with the nucleosomal arrays at a ratio 1.3 moles linker histone per mole nucleosome, the sedimentation coefficient distributions of all isoform samples fibers increased to 30-38 S. The G(s) profiles in figure 1B indicate that approximately 40% of each isoform (boundary fraction 50–90%) sedimented at 36–38 S and hence consisted of chromatin fibers saturated with both one nucleosome per 208 bp repeat and one H1 per nucleosome (7, 18). The remainder of each sample was subsaturated and lacked stoichiometric amounts of core histones, linker histones, or both. The data in Figure 1A demonstrate that all isoforms could bind to linker DNA and are consistent with each isoform equally inducing formation of the apposed linker DNA stem/loop motif.
Our observation of similar relative binding affinities of the somatic isoforms differs from that reported previously by several investigators (37, 38). However, these differences are likely due to the experimental conditions chosen to study binding. Talasz et al. (38) used gel electrophoretic mobility shift assays and nanomolar protein and nucleosome concentrations to conclude that there were ~8-fold differences (2–16 nM) in binding affinities of the somatic isoforms for 210 bp mononucleosomes. Suau and colleagues (37) found 20-fold differences in relative binding affinities. In contrast to the these studies, binding to nucleosomal arrays in our work occurred at micromolar concentrations of linker histones and nucleosomal arrays and in low salt TEN buffer, i.e., conditions designed to saturate the model nucleosomal arrays with stably bound H1. Under these conditions we would not expect to see relatively small differences in binding affinities in the nanomolar range.
Chromatin folding is assayed by sedimentation velocity in the presence of MgCl2, and is evidenced by an increase in sedimentation coefficient beyond 36 S (7, 10, 18, 39). Folding of 12-mer chromatin fibers is complete when a stable 55 S structure is achieved (7, 10, 18). To examine the effects of H1 proteins on stabilization of chromatin fiber folding, experiments were performed in 0.25 mM MgCl2, which induces only partial folding, and 0.45 mM MgCl2, which induces nearly complete folding of saturated chromatin fibers that contain stoichiometric amounts of bound linker histone. We purposely did not work at higher MgCl2 concentrations to avoid induction of oligomerization. In 0.25 mM MgCl2, small differences in the extent of folding were observed, with H1S-1 = H1S-2 = H1S-3 > H1° = H1S-4 (Fig. 1B). However, by 0.45 mM MgCl2, all isoforms were capable of stabilizing the model chromatin fibers in 50-55 S structures, as indicated by the data between boundary fractions 50–90% of the G(s) plots (Fig. 1C). These data indicate that all H1 isoforms can stabilize chromatin fibers in extensively folded states. The differential effects of the isoforms in condensing saturated fibers in 0.25 mM MgCl2 suggests that small isoform-dependent differences in the stability of the folded structures may exist.
Mg2+-dependent oligomerization is assayed by a differential centrifugation protocol that measures reversible, cooperative self-association of individual fibers (10, 40). Figure 1D shows the characteristic sigmoidal curves that were obtained for model chromatin fibers bound to each H1 isoform. Although very subtle differences in Mg50 were observed, all isoforms led to essentially the same pronounced decrease in Mg50 relative to the parent nucleosomal arrays.
When taken together, a major conclusion from the data in Figure 1 is that all three chromatin condensing functions mediated by the H1 CTD are comparable for the different isoforms, despite divergent isoform CTD primary sequences. It follows from these results that amino acid composition may be a more important determinant of CTD action than the specific order of the amino acids.
Randomization of the primary sequence of residues 98–122 (CTD region 1) does not abolish function
To directly test the role of amino acid composition in CTD function, in the remainder of the studies we focused attention on the “functional” and “nonfunctional” CTD regions identified in previous CTD truncation experiments (7) (see Fig. 2A). Residues 98–122 (CTD region 1) have been shown to be required for formation of the stem-loop linker DNA motif, and stabilization of fiber folding and oligomerization. Residues 147–170 (CTD region 3) were found to assist in stabilizing folded chromatin fiber structures (7). Regions 2 (residues 123–146) and 4 (171–194) were nonfunctional, i.e., they could be deleted without any noticeable effects on H1 function (7). In the experiments described below, recombinant technology was used to scramble the primary sequence, mutate the amino acid composition, and switch the position of the functional and nonfunctional CTD regions of recombinant H1°.
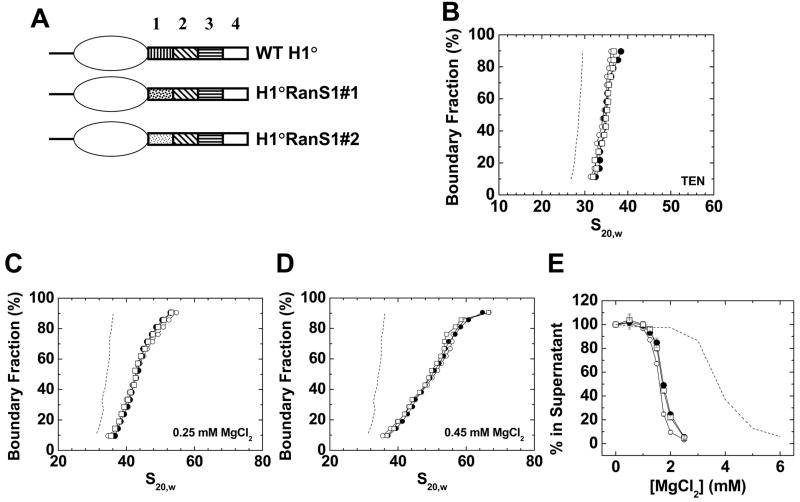
Figure 2. Randomization of the primary sequence of CTD region 1.
A) Schematic diagrams of wild type H1°, H1°RanS1#1, and H1°RanS1#2. B) Sedimentation coefficient distributions in TEN buffer of 208–12 nucleosomal arrays (dashed line) assembled with wild type H1° (○), H1°RanS1#1 (●), or H1°RanS1#2 (□). B) G(s) plots of 208–12 chromatin fibers bound to wild type H1° (○), H1°RanS1#1 (●), or H1°RanS1#2 (□) in 0. 25 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. D) G(s) plots of 208–12 fibers bound to wild type H1° (○), H1°RanS1#1 (●), or H1°RanS1#2 (□) in 0. 45 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. E) Oligomerization as a function of MgCl2. Shown are data for 208–12 nucleosomal arrays (dashed line) and 208–12 chromatin fibers bound to wild type H1° (○), H1°RanS1#1 (●), or H1°RanS1#2 (□). All plots are representative of results seen with 3–4 different chromatin preparations.
Figure 2A schematically illustrates two different mutants that were constructed in which the primary sequence of region 1 was randomized while the amino acid composition was kept constant. Table 1 shows that the two mutants have no primary sequence homology with wild type region 1. Sedimentation velocity experiments in TEN indicated that both randomized region 1 mutants behaved the same at binding to linker DNA and inducing stem-loop formation (Fig. 2B). We also observed no differences in stabilization of folded chromatin fibers in 0.25 mM (Fig. 2C) and 0.45 mM MgCl2 (Fig. 2D), and of Mg2+-dependent oligomerization (Fig. 2E). The similar behavior of the two randomized mutants in Figure 2 also supports a mechanism of CTD function that does not involve primary sequence but is dependent on specific amino acid composition.
Mutation of the amino acid composition of H1° residues 98–122 and 147–170 (CTD regions 1 and 3) affects chromatin folding
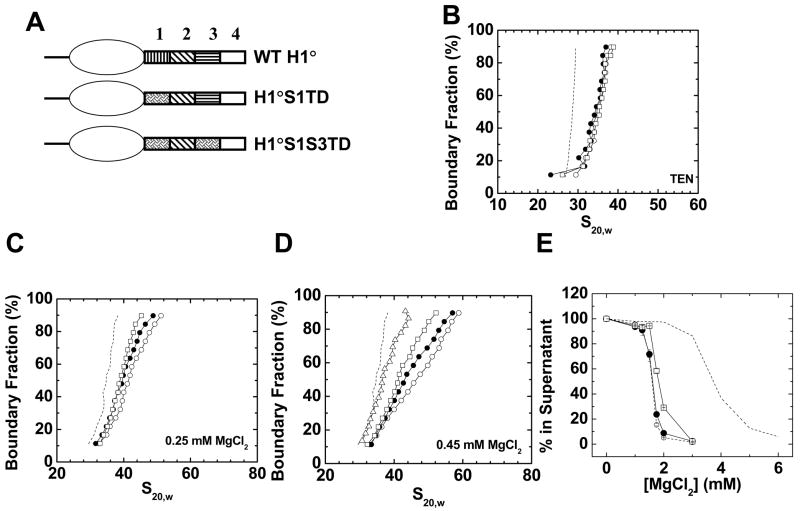
Given that scrambling the primary sequence did not abolish function, we attempted to disrupt H1 effects on chromatin structure by altering the specific amino acid composition of either CTD region 1 alone (H1°S1TD) or CTD regions 1 and 3 together (H1°S1S3TD) (Fig. 3A). Three Val residues were replaced with Asn residues, which normally are absent from the linker histone CTD (9), and a Pro was inserted in place of a Thr. (Table 1). Val is the only nonpolar amino acid present in significant quantities in the linker histone CTDs (9). The lysine content was kept constant to keep the high positive charge density of the mutants the same. Results indicated that both mutants behaved very similarly to wild type in terms of formation of the 36 S structure in low salt (Fig. 3B) and salt-dependent oligomerization (Fig. 3E). However, the H1°S1TD mutant stabilized folded chromatin fiber structures to a lesser degree in both 0.25 and 0.45 mM MgCl2, and the H1°S1S3TD mutant was even less effective in both salts (Figs. 3C, D). Specifically, in 0.45 mM MgCl2, the mutant fibers folded roughly half-way between that of the wild type fibers and H1-bound fibers completely lacking their C-terminal domains (Fig. 3D). Thus, at least for the case of chromatin fiber folding, altering the amino acid content of CTD regions 1 and 3 led to reproducible disruption of CTD function. This partial disruption could be due to alteration of the amino acid composition per se, or to point mutation(s) that disrupt the ability of CTD region 1 to form a key secondary structure element upon binding to DNA (see Discussion).
Figure 3. Mutagenesis of the amino acid composition of CTD region 1.
A) Schematic diagrams of wild type H1°, H1°S1TD, and H1°S1S3TD. B) Sedimentation coefficient distributions in TEN buffer of 208–12 nucleosomal arrays (dashed line) assembled with wild type H1° (○), H1°S1TD (●), or H1°S1S3TD (□). B) G(s) plots of 208–12 chromatin fibers bound to wild type H1° (○), H1°S1TD (●), or H1°S1S3TD (□) in 0. 25 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. D) G(s) plots of 208–12 fibers bound to wild type H1° (○), H1 CTD “tailless” (△), H1°S1TD (●), or H1°S1S3TD (□) in 0. 45 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. E) Oligomerization as a function of MgCl2. Shown are data for 208–12 nucleosomal arrays (dashed line) and 208–12 chromatin fibers bound to wild type H1° (○), H1°S1TD (●), or H1°S1S3TD (□). All plots are representative of results seen with 3–4 different chromatin preparations.
All CTD regions are functionally interchangeable given proper positioning relative to the globular domain
In the final set of experiments, a series of mouse H1° CTD mutants were constructed in which either the order, or position relative to the globular domain, of the various functional and nonfunctional CTD regions were altered (Fig. 4A, 5A). The primary sequences of the four CTD regions are listed in Table 1, and show that there is little sequence conservation between the regions. Table 2 shows the amino acid composition of the four regions, which is much more similar. Relative to wild type, H1 proteins were constructed that either exchanged the positions of regions 1 and 3 (H1°3214), replaced region 3 with a second copy of region 1 (H1°1214), or replaced region 1 with a second copy of region 3 (H1°3234) (Fig. 4A). Because the positions of key functional regions had been moved or replaced with a different region, one might expect the mutants to be nonfunctional. However, given the results from Figures 1–3, and the shared similarity in amino acid composition of the two functional regions, our prediction was that the mutants would behave the same as wild type H1°.
Figure 4. Exchanging and swapping H1° CTD regions 1 and 3.
A) Schematic diagrams of wild type H1°, H1°1214, H1°3214 and H1°3234. B) Sedimentation coefficient distributions in TEN buffer of 208–12 nucleosomal arrays (▪) and 208–12 chromatin fibers bound to wild type H1° (○), H1°1214 (●), H1°3214 (△), or H1°3234 (□). C) G(s) plots of 208–12 fibers bound to wild type H1° (○), H1°1214 (●), H1°3214 (△), or H1°3234 (□) in 0. 25 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. D) G(s) plots of 208–12 fibers bound to wild type H1° (○), H1°1214 (●), H1°3214 (△), or H1°3234 (□) in 0. 45 mM MgCl2. The G(s) plot of H1°-bound chromatin fibers in low salt (dashed line; data from panel 1A) is shown for reference. E) Oligomerization as a function of MgCl2. Shown are data for 208–12 nucleosomal arrays (dashed line) assembled with wild type H1° (○), H1°1214 (●), H1°3214 (△), or H1°3234 (□). All plots are representative of results seen with 3–4 different chromatin preparations.
Table 2.
Amino acid composition of the H1° regions.
| Region | Lys1 | Ala | Pro | Gly | Arg | Glu | Asp | Asn | Gln | Ser | Thr | Cys | Met | His | Val | Ile | Leu | Phe | Trp | Tyr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 36 | 8 | 8 | 4 | 4 | 8 | 4 | 0 | 0 | 4 | 8 | 0 | 0 | 0 | 12 | 0 | 0 | 4 | 0 | 0 |
| 2 | 38 | 29 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| 3 | 46 | 17 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 |
| 4 | 46 | 17 | 8 | 0 | 4 | 0 | 0 | 0 | 0 | 17 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
Each amino acid residue is expressed as percent of the total composition.
Figure 4B shows that the G(s) profiles in TEN of 208–12 model systems bound to wild type H1° and all three CTD region 1/3 swap mutants were identical within error. When the same samples were examined for folding in 0.25 (Fig. 3C) and 0.45 mM (Fig. 3D) MgCl2, there was no appreciable difference in the G(s) plots of the saturated fraction (boundary fraction = 50–90%) at both salt concentrations. Hence the stabilization of folding also was unaffected by the mutations. The differences in the subsaturated portion of the samples were most likely due to differences in the amount of bound H1. Figure 4E shows that all four samples exhibited nearly identical oligomerization behavior and Mg50 values. Collectively, the data in Figure 4 indicate that subdomains 1 and 3 were interchangeable for nucleosomal array binding, linker DNA stem/loop motif formation, and stabilization of condensed chromatin fiber structures, despite having different primary sequences. An even more telling experiment examined mutants in which CTD region 1 was replaced with either nonfunctional CTD region 2 (H1°2234) or region 4 (H1°4234) (Fig. 5A). These mutants bound to nucleosomal arrays and formed the ~36 S fiber structure the same as wild type (Fig. 5B). Similarly, the extent of folding in 0.25 mM (Fig. 5C) and 0.45 mM MgCl2 (Fig. 5D) was nearly identical to wild type (if anything the mutants were slightly more folded at the higher salt concentration). As shown in Figure 5E, the oligomerization curves of both mutants also were essentially indistinguishable from wild type. Thus, both of the CTD regions identified as nonfunctional in truncation experiments could replace native CTD region 1 if moved to the appropriate position along the H1° polypeptide chain.
DISCUSSION
The H1 CTD is an intrinsically disordered domain
Intrinsically disordered regions of proteins lack secondary structure, but nevertheless are functional (see 9, 20, 22, 41 and references therein). In particular, they often are involved in binding and macromolecular recognition. Further, intrinsically disordered regions often undergo a disorder to order transition that is coupled to recognition of their binding partners (9, 22, 41). Early experiments showed that the H1 CTD is unstructured in solution (4, 42, 43). Despite lacking native structure, the H1 CTD can bind DNA (43–45), nucleosomal arrays (7, 18, 19), and specific proteins (14–16). When H1 CTD peptides bind to DNA they assume α-helical structure (43, 44). By these criteria, the H1 CTD meets the definition of an intrinsically disordered protein domain.
Amino acid composition as a determinant of molecular recognition and H1 CTD function
In a previous study we characterized H1° mutants in which 24 or 25-residue segments were successively deleted from the C-terminus of the CTD (7). We found that deletion of regions 1 (residues 98–122) and 3 (residues 147–170) together led to loss of apposed linker DNA structure and destabilization of condensed chromatin fibers, while regions 2 (residues 123–146) and 4 residues (171–194) could be deleted without affect (7). At the time we termed regions 1–4 “subdomains”. A straightforward explanation for these results is that there are stable structured primary sequence element(s) in CTD regions 1 and 3 that mediate macromolecular recognition and function. In this case, scrambling the primary sequence should disrupt H1 function. However, in both mutants tested we observed that the primary sequence of CTD region 1 could be scrambled without influencing the ability of this CTD region to bind to linker DNA and stabilize condensed chromatin fibers (Fig. 2). Thus, the chromatin condensing functions of the H1 CTD do not appear to be dependent on CTD primary sequence.
One factor that was held constant in the scrambling experiments was amino acid composition. The amino acid composition of the H1 CTDs is distinctive and almost constant between isoforms. Approximately 40% of the residues are Lys and 20–30% Ala. The other resides are mainly Ser/Thr, Pro, Val, and some Gly (9). For all practical purposes, 13 of the common amino acids are absent from the isoform CTDs, including the bulky hydrophobic residues. In addition to the sequence scrambling experiments, three additional results support a role for specific amino acid composition as a mechanistic determinant of H1 CTD action. First, all somatic linker histone isoforms functioned nearly identically. The isoforms have divergent primary sequences (1, 8) but have maintained a very similar amino acid composition (9). Second, replacement of three Val and one Thr residues in CTD region 1 with Asn and Pro, which led to an altered amino acid composition depleted of nonpolar residues, partially disrupted H1 effects on chromatin fiber folding (Fig. 3C, D). Finally, CTD regions 2, 3 and 4 all could replace functional region 1 if they were placed in the proper position along the polypeptide chain (Figs. 4, 5). None of the four regions has primary sequence homology (Table 1) but their amino acid composition is more similar (Table 2). Collectively, these results are consistent with a mechanism in which specific amino acid composition mediates H1 CTD function in linker DNA binding and chromatin fiber condensation.
Amino acid composition has recently been shown to determine the function of the prion domains of the yeast proteins, Ura1p and Sup35p (9, 46, 47). Interestingly, the amino acid composition of the prion domains is Asn and Gln rich and differs fundamentally from that of the linker histone CTDs. The prion domains are intrinsically disordered, but form parallel in-register beta sheet structures concomitant with molecular recognition and amyloid fiber assembly (48). The prion domain results and the H1 CTD work reported here indicate that different specific amino acid compositions can dictate formation of different secondary structures when intrinsically disordered domains interact with their targets. While little is known about the protein chemistry that underlies the connections between amino acid composition, intrinsic protein disorder, and molecular recognition, the widespread prevalence of intrinsically disordered domains in eukaryotic proteomes (23, 49) suggests that mechanisms of macromolecular recognition based on amino acid composition may be common.
A new working model for H1 CTD action
Many events are involved in H1 regulation of chromatin structure in vitro, including (but not limited to) binding to chromatin fiber targets, formation of apposed linker DNA, stabilization of fiber folding, and stabilization of fiber oligomerization. Because of its importance in regulating genome structure and integrity, the molecular mechanism of H1 function has been studied for nearly 30 years. Based on its uniform high positive charge density (~4 Lys per 10 residues along all somatic H1 CTDs; see 50), the H1 CTD initially was proposed to function through an electrostatic mechanism in which the CTD was bound to linker DNA and screened negative charge, thereby facilitating close approach of nucleosomes (50, 51). Inherent in this proposal was that the entire CTD was engaged with linker DNA. However, as described extensively above, only two discontinuous regions (CTD regions 1 and 3 of 25 and 24 residues in length, respectively) are involved in mediating linker DNA apposition and stabilization of chromatin fiber condensation (7).
How does one explain this result given the functional interchangeability of the four CTD subdomains identified in the present studies (Figs. 4, 5)? In other words, if all regions can substitute for one another, why are some regions “functional” while others are “nonfunctional” in the intact protein? We hypothesize that this reflects the importance of the position of the CTD regions relative to another structural and functional reference point in the protein, in this case the centrally located globular domain. The globular domain binds to nucleosomes, although the details of this interaction remain unresolved (5). In our working model, binding of the globular domain helps positions CTD regions 1 and 3 to interact with linker DNA and possibly other chromatin components as well. The initial interaction can be purely electrostatic (7), but the specific amino acid composition of the CTD allows formation of one or more short α-helices in these regions upon DNA binding that mediate CTD function in chromatin. We note that there is one Pro per seven residues on average in the H1 CTDs, and that Pro residues often are found at the beginning of α-helices (52), as well as being helix breakers. The finding that the amino acid composition mutants only affected chromatin folding (Fig. 3) indicates that the three CTD-mediated functions may involve different specific disorder to order transitions. In this regard, several different α-helices may form in any given stretch of 24 CTD residues.
If only two discontinuous 24 residue CTD regions and a 73 residue long CTD are needed to stabilize condensed chromatin fibers (7), why are all the H1 isoform CTDs nearly 100 residues in length and maintain a consistent amino acid composition throughout the entire domain? The answer, at least in part, is that the H1 CTD can mediate interactions with specific proteins as well as chromatin. In the case of the apoptotic nuclease, DFF40/CAD, it has been shown that the mouse somatic H1 isoforms bound to and activated the enzyme equally well. Further, any CTD peptide greater than 47 residues in length were capable of activating the enzyme (14). However, for the H1-DFF40/CAD interaction, CTD regions 2, 3, and 4 (but not CTD region 1) were needed to mediate the protein-protein interaction in truncation experiments. Based on these results, we hypothesize that H1-DFF40/CAD binding also is mediated by the specific amino acid composition of the H1 CTD, but that a different specific pattern of α-helices form when the 72 C-terminal most CTD residues interact with the DFF40/CAD surface than when the CTD binds to chromatin. By acting through a mechanism involving intrinsic disorder and amino acid composition, the linker histone CTD is likely to be involved in competitive interactions with both the chromatin fiber and many other nuclear proteins. This “molecular plasticity” in binding may explain much of the multifunctionality of the H1 proteins.
Redundant functions of the linker histone isoforms in chromatin condensation
Skoultchi and colleagues have investigated linker histone isoform function in vivo by knocking out the H1 isoform genes singly (12), in pairs (12), and in triple combinations (13). They found that isoforms could be knocked out singly and in pairs without noticeable effects on chromatin structure or biological phenotype, and that expression of the other isoforms compensated for the knocked out isoforms to maintain total wild type H1 stoichiometry (3, 12). Their data indicated that the functions of the isoforms were redundant in vivo at the level of maintaining large-scale genome structure and organization. The results of our in vitro experiments in high Mg2+ showing similar condensation abilities of five of the mouse H1 isoforms (Figs. 1D, E) support the in vivo data. We hypothesize that the characteristic amino acid composition of the H1 isoform CTDs leads to redundant functions of the H1 isoforms in chromatin condensation when the isoforms are stably bound to chromatin fibers and chromosomal domains, acting through the mechanisms described above. Simultaneously, isoform-specific differences in other molecular properties, e.g., chromatin binding affinity, protein-protein interactions, post-translational modifications, can lead to isoform-specific effects on gene expression and other biological processes involving linker histones.
Supplementary Material
Supplementary Figure 1. shows the correlations between three isoform nomenclatures. This material is available free of charge via the Internet at http//:pubs.acs.org
Acknowledgments
We thank Drs. Steven McBryant, Heather Szerlong, and Robert Woody for critically reading the manuscript.
ABBREVIATIONS
- NTD
N-terminal domain
- CTD
C-terminal domain
Footnotes
This work was supported by NIH grant GM45916 to JCH.
References
- 1.van Holde KE. Chromatin. Springer-Verlag; New York: 1988. [Google Scholar]
- 2.Wolffe A. Chromatin : structure and function. 3. Academic Press; San Diego: 1998. [Google Scholar]
- 3.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 4.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 5.Thomas JO. Histone H1: location and role. Curr Opin Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 6.Hendzel MJ, Lever MA, Crawford E, Th'ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 7.Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J Biol Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 8.Ponte I, Vidal-Taboada JM, Suau P. Evolution of the vertebrate H1 histone class: evidence for the functional differentiation of the subtypes. Molecular Biology and Evolution. 1998;15:702–708. doi: 10.1093/oxfordjournals.molbev.a025973. [DOI] [PubMed] [Google Scholar]
- 9.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 10.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 11.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 12.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Molecular and Cellular Biology. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 14.Widlak P, Kalinowska M, Parseghian MH, Lu X, Hansen JC, Garrard WT. The histone H1 C-terminal domain binds to the apoptotic nuclease, DNA fragmentation factor (DFF40/CAD) and stimulates DNA cleavage. Biochemistry. 2005;44:7871–7878. doi: 10.1021/bi050100n. [DOI] [PubMed] [Google Scholar]
- 15.Montes de Oca R, Lee KK, Wilson KL. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J Biol Chem. 2005;280:42252–42262. doi: 10.1074/jbc.M509917200. [DOI] [PubMed] [Google Scholar]
- 16.Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, Johnson EM, Brock HW, An W. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 19.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 20.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends in Biochemical Sciences. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 22.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. Journal of Molecular Graphics & Modelling. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 23.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 25.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. Journal of Proteome Research. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–584. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 28.Georgel P, Demeler B, Terpening C, Paule MR, van Holde KE. Binding of the RNA polymerase I transcription complex to its promoter can modify positioning of downstream nucleosomes assembled in vitro. J Biol Chem. 1993;268:1947–1954. [PubMed] [Google Scholar]
- 29.Gordon F, Luger K, Hansen JC. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J Biol Chem. 2005;280:33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 30.Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 31.Hansen JC, Ausio J, Stanik VH, van Holde KE. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 32.Wellman SE, Song Y, Su D, Mamoon NM. Purification of mouse H1 histones expressed in Escherichia coli. Biotechnol Appl Biochem. 1997;26(Pt 2):117–123. [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning : a laboratory manual. 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1989. [Google Scholar]
- 34.Carruthers LM, Tse C, Walker KP, 3rd, Hansen JC. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 1999;304:19–35. doi: 10.1016/s0076-6879(99)04004-5. [DOI] [PubMed] [Google Scholar]
- 35.Demeler B, van Holde KE. Sedimentation velocity analysis of highly heterogeneous systems. Anal Biochem. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 36.van Holde KE, Weischet WO. Boundary Analysis of Sedimentation Velocity Experiments with Monodisperse and Paucidisperse Solutes. Biopolymers. 1978;17:1387–1403. [Google Scholar]
- 37.Orrego M, Ponte I, Roque A, Buschati N, Mora X, Suau P. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biology. 2007;5:22. doi: 10.1186/1741-7007-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talasz H, Sapojnikova N, Helliger W, Lindner H, Puschendorf B. In vitro binding of H1 histone subtypes to nucleosomal organized mouse mammary tumor virus long terminal repeat promotor. J Biol Chem. 1998;273:32236–32243. doi: 10.1074/jbc.273.48.32236. [DOI] [PubMed] [Google Scholar]
- 39.Tse C, Hansen JC. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 41.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartman PG, Chapman GE, Moss T, Bradbury EM. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The three structural regions of the histone H1 molecule. Eur J Biochem. 1977;77:45–51. doi: 10.1111/j.1432-1033.1977.tb11639.x. [DOI] [PubMed] [Google Scholar]
- 43.Vila R, Ponte I, Collado M, Arrondo JL, Suau P. Induction of secondary structure in a COOH-terminal peptide of histone H1 by interaction with the DNA: an infrared spectroscopy study. J Biol Chem. 2001;276:30898–30903. doi: 10.1074/jbc.M104189200. [DOI] [PubMed] [Google Scholar]
- 44.Clark DJ, Hill CS, Martin SR, Thomas JO. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988;7:69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mamoon NM, Song Y, Wellman SE. Binding of histone H1 to DNA is described by an allosteric model. Biopolymers. 2005;77:9–17. doi: 10.1002/bip.20172. [DOI] [PubMed] [Google Scholar]
- 46.Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Molecular and Cellular Biology. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shewmaker F, Ross ED, Tycko R, Wickner RB. Amyloids of shuffled prion domains that form prions have a parallel in-register beta-sheet structure. Biochemistry. 2008;47:4000–4007. doi: 10.1021/bi7024589. [DOI] [PubMed] [Google Scholar]
- 49.Dosztanyi Z, Chen J, Dunker AK, Simon I, Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. Journal of Proteome Research. 2006;5:2985–2995. doi: 10.1021/pr060171o. [DOI] [PubMed] [Google Scholar]
- 50.Subirana JA. Analysis of the charge distribution in the C-terminal region of histone H1 as related to its interaction with DNA. Biopolymers. 1990;29:1351–1357. doi: 10.1002/bip.360291003. [DOI] [PubMed] [Google Scholar]
- 51.Clark DJ, Kimura T. Electrostatic mechanism of chromatin folding. J Mol Biol. 1990;211:883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- 52.Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science (New York, NY) 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. shows the correlations between three isoform nomenclatures. This material is available free of charge via the Internet at http//:pubs.acs.org