Abstract
Many studies of alcohol adaptation in Drosophila melanogaster have focused on the Adh polymorphism, yet the metabolic elimination of alcohol should involve many enzymes and pathways. Here we evaluate the effects of glycerol-3-phosphate dehydrogenase (Gpdh) and cytosolic malate dehydrogenase (Mdh1) genotype activity on adult tolerance to ethanol. We have created a set of P-element-excision-derived Gpdh, Mdh1, and Adh alleles that generate a range of activity phenotypes from full to zero activity. Comparisons of paired Gpdh genotypes possessing 10 and 60% normal activity and 66 and 100% normal activity show significant effects where higher activity increases tolerance. Mdh1 null allele homozygotes show reductions in tolerance. We use piggyBac FLP–FRT site-specific recombination to create deletions and duplications of Gpdh. Duplications show an increase of 50% in activity and an increase of adult tolerance to ethanol exposure. These studies show that the molecular polymorphism associated with GPDH activity could be maintained in natural populations by selection related to adaptation to alcohols. Finally, we examine the interactions between activity genotypes for Gpdh, Mdh1, and Adh. We find no significant interlocus interactions. Observations on Mdh1 in both Gpdh and Adh backgrounds demonstrate significant increases in ethanol tolerance with partial reductions (50%) in cytosolic MDH activity. This observation strongly suggests the operation of pyruvate–malate and, in particular, pyruvate–citrate cycling in adaptation to alcohol exposure. We propose that an understanding of the evolution of tolerance to alcohols will require a system-level approach, rather than a focus on single enzymes.
THE genus Drosophila has an evolutionary history of exposure to alcohols, and it is believed that the adaptation to alcohols has facilitated the cosmopolitan spread of Drosophila melanogaster to temperate environments (Geer et al. 1993). Both larval and adult fruit flies feed on yeast, and this ecological niche exposes them to toxic fermentation products, including alcohols. In particular, it is believed that the high tolerance of D. melanogaster to alcohols is an evolved phenotype because other members of the melanogaster subgroup, such as D. simulans, show lower tolerance and avoid alcohol exposure (McKenzie and Parsons 1972; David and Bocquet 1975). In contrast, D. melanogaster utilizes ethanol as a carbon source and adult tolerance is highest in temperate climates (Cohan and Graf 1985), suggesting either increasing exposure to, or increased utilization of, alcohols in these regions. As a complex quantitative phenotype, both larval and adult alcohol tolerances show significant genetic variance (Cohan and Graf 1985; Cohan and Hoffmann 1986). Over several decades, this example of adaptation to a novel niche, one constituting both a resource and an environmental stress, has become a paradigm in evolutionary genetics.
The power of joining genetics and molecular analysis has made Drosophila an established model in studies of alcohol metabolism and tolerance. The induction of behaviors that are similar to those in humans is well noted, as are the parallels with alcohol metabolism in mammals (Scholz et al. 2000). In particular, there are two facets of alcohol tolerance that have been studied using Drosophila as a model. The first addressed short-term acquisition of tolerance, measured as a shift in knockdown time following a period of ethanol exposure (Scholz et al. 2000, 2005). The second (the focus of this study) is the metabolic elimination of alcohol and its relationship to tolerance and survival (Geer et al. 1993). Most of this second focus has centered on the relationship of biochemical variation in the alcohol dehydrogenase gene (Adh) to tolerance in both adults and larvae (Geer et al. 1993). Such studies have led to the textbook story of the Adh allozyme polymorphism (Freeman and Herron 2004; Futuyma 2005). However, the study of ADH has followed a path set down more by historical precedence than by design. ADH was the first enzyme system in Drosophila in which histochemical staining was used to detect electrophoretic variants (Johnson and Denniston 1964), and Adh was one of the first Drosophila genes cloned in the late 1970s (Kreitman 1983). Unfortunately, this precedence of Adh has directed interest away from the study of the development of metabolic tolerance to ethanol as a larger-scale problem involving many genes and pathways. The rapid elimination of ingested alcohols and its metabolic products is a system-wide challenge and must involve downstream pathways and metabolic networks, with possible interactions—all kept in redox balance.
In Drosophila, other genes and pathways have been implicated in ethanol tolerance (Van der Zel et al. 1991; Pecsenye and Saura 1998; Montooth et al. 2006; Morozova et al. 2006, 2007). For example, it was shown the next enzyme downstream, aldehyde dehydrogenase (Aldh), also plays a role in the subsequent metabolism of acetaldehyde to acetate in D. melanogaster larvae (Fry and Saweikis 2006; Fry et al. 2008). Glycerol-3-phosphate dehydrogenase (Gpdh) is another gene implicated in ethanol tolerance (Geer et al. 1993); a common allozyme polymorphism is found in natural populations. The derived GpdhS allele possesses increased GPDH activity and is more common in temperate latitudes (Oakeshott et al. 1982, 1984; Sezgin et al. 2004). Furthermore, ADH and GPDH activity levels are coordinately induced in larvae exposed to alcohols (Geer et al. 1983; Lissemore et al. 1990). In population cage experiments, allozyme polymorphisms for both genes, as well as cytosolic malate dehydrogenase (Mdh1), responded to ethanol exposure over time (Cavener and Clegg 1978). These observations all imply that these other enzymes may play roles in adaptation to alcohols.
The hypothesis that Gpdh and Mdh1 are involved in ethanol tolerance has not been directly tested using partial or full knockout alleles in rigidly controlled genetic backgrounds. To test this hypothesis, we use sets of P-element-excision alleles of the Gpdh (Merritt et al. 2006) and Mdh1 genes to determine if reductions in GPDH and cytosolic MDH activity influence adult tolerance to alcohol. Furthermore, since in natural populations the higher-activity GpdhS allele geographically covaries with the higher activity AdhF allele, we also examine the effect of increases in GPDH activity by creating Gpdh gene duplications using piggyBac transposon insertions and the FLP–FRT site-specific recombination system (Parks et al. 2004). Finally, we explore the possibility of gene interactions among Gpdh, Adh, and Mdh1 and their effect on ethanol tolerance.
MATERIALS AND METHODS
Lines:
The Gpdh lines are described in Merritt et al. (2006). They consist of three alleles derived from mobilization of the KG02555 P-element insertion: GpdhΔ9.2, GpdhΔ24.1, and GpdhΔ10.2, with 0, 21, and 100% activities relative to normal. The progenitor allele in this line is the GpdhF allele. Their white-marked X chromosomes are derived from Bloomington stock 2475, w*;T(2;3)apXa/Cy;TM3, Sb1, and the third chromosome backgrounds are replaced by using marker-assisted introgression in inbred line w;CyO/Tft;VT83.
The Mdh1 alleles are created using excision of the EY08761 P-element insertion in gene CG5362. This insertion site lies inside the 5′-UTR, 12 bases upstream of the start codon. Mdh1Δ18.1 has lost the mini-white construct, but retains >5 kb of the P element. In wild-type flies, ∼15% of the crude MDH activity is cytosolic, while the remainder represents mitochondrial MDH2 leakage during homogenization (Hay and Armstrong 1976). The loss of cytosolic MDH enzyme activity in Mdh1Δ18.1 is clearly seen after electrophoresis and allozyme staining (data not shown). Mdh1Δ10.5 is a precise excision and recovers full gene activity. The X chromosome is the white-marker chromosome from Bloomington stock 2475, and the third chromosome is from VT46.
The Adh test alleles are derived from mobilization of the KG05345 P element that is inserted in exon 3. AdhΔ25 is a partial excision that retains a small piece of the P element in exon 3 and possesses no ADH activity. AdhΔ17 is a precise excision and possesses activity equal to a normal Fast Adh allele. The X chromosome is from Bloomington line 2475, and the third chromosome is replaced by that from inbred line w;CyO/Tft;VT83.
Lines VT46 and VT83 are derived from inbred lines collected in 1997 in Whiting, Vermont. Line w;6326;6326.1 is a derivative of Bloomington stock 6326 that has the X chromosome from Bloomington stock 2475.
P elements were excised in male flies using standard dysgenic crosses (Merritt et al. 2006). Excision chromosomes (indicated by flies with white eyes) were isolated using the balancer chromosome CyO. Approximately 80–100 excision lines were sampled for each dysgenic cross. Relative allele function was determined by direct spectrophotometric assay of crude mass-adjusted enzyme activity (see below). Interline crosses were used to create heterozygotes and to test additivity in allele combinations in the event that transvection effects were present (Merritt et al. 2005). PCR and sequencing with flanking primers were used to determine molecular changes in the gene. All full-activity alleles were confirmed to have sequences consistent with the “precise” excision or gene conversion to a normal sequence. Reduced activity alleles possessed a spectrum of molecular changes from deletion of entire exons to retention of large pieces of the original P element. None of the alleles show single residue changes in amino acid sequence and thus catalytic function. Paired test genotypes differ only in the gene of interest.
A deletion-duplication series of Gpdh alleles was created using FRT–FLP-driven recombination (Parks et al. 2004) between piggyBac transposon insertions f00109 and e03988. These insertions are ∼40 kb apart and, upon FLPase-induced FRT recombination, will delete eight genes or duplicate seven genes. None of these other genes has an obvious relationship to ethanol tolerance. Eighty potentially recombinant lines were collected and screened by eye color, viability, PCR products, and GPDH activity. Chromosomes were genetically extracted using the CyO balancer chromosome and a subset of 40 chromosomes, which included 6 lethals, were further screened using PCR primer combinations and sequencing designed to detect hybrid piggyBac elements resulting from recombination between FRT sites. Five lethals were deletions. Three lines were duplications, including one lethal. The recovery rate for both deletions and duplications was ∼10%. All second chromosomes had the X and third chromosome backgrounds replaced using line w; 6326.6326.1. The progenitor allele in this line is GpdhS and is already in the 6326 second chromosome.
Enzyme activity measurements:
Flies were homogenized in grinding buffer (0.01 m KH2PO4, 1.0 mm EDTA, pH 7.4) at a “concentration” of five individuals pooled in 1 ml of grinding buffer and centrifuged at 13,000 rpm for 5 min at 4° to pellet all solids. The supernatant was recovered and transferred to a 96-well plate and used in all enzymatic assays. Enzyme activity assays were carried out on a Molecular Designs SpectraMax 384 Plus 96-well plate spectrophotometer using 10 μl of fly extract and 100 μl of assay buffer, and optical density was measured every 9 sec for 3 min. All activity assays were conducted at 25°. In all experiments, each of 10 replicate samples were assayed twice and the average was used as an estimate of each genotype activity. Enzyme activity is expressed as nanomolars of NAD+ reduced/min/fly (see Merritt et al. 2006). The assay buffers for the three enzymes assayed in this study were as follows: GPDH (0.1 m glycine NaOH, 2.5 mm NAD+, 15 mm α-glycerol-3-phosphate, pH 7.4), ADH (0.1 m Tris–HCL, 4.0 mm NAD+, 0.8 m ethanol, pH 8.6), and MDH (0.1 m Tris–HCL, 4.0 mm NAD+, 40.0 mm malate, pH 8.0). Initial values for appropriate pH, substrate, and cofactor concentrations for the reactions were taken from the literature and modified to give maximum enzyme activity.
Crosses to set up test genotypes:
All flies were reared on standard cornmeal media in 200-ml plastic flasks. In two sets of experiments, test genotypes were created with alleles GpdhΔ9.2, GpdhΔ24.1, and GpdhΔ10.2. In experiment 1, GpdhΔ9.2 and GpdhΔ10.2 males were mated with GpdhΔ24.1 females producing genotypes with 15 and 60% activities relative to a 10.2/10.2 genotype. The 10.2/10.2 genotype possesses activity that is ∼12% higher than the average GPDH activity of the 10 wild second chromosome lines assayed in Merritt et al. (2006). In experiment 2, GpdhΔ9.2 and GpdhΔ10.2 males (50 each) were separately mated with w; 6326;6326.1 females (100 each), producing genotypes with 66 and 100% relative GPDH activities. The 6326/6326 genotype possesses GPDH activity that is 25% higher than 10.2/10.2 and possesses the GpdhS allele. Densities were standardized in each bottle. Emerging males were collected from multiple replicate bottles, pooled by genotype, aged 4–6 days, and used in the assay. GpdhΔ9.2 homozygous genotypes were not tested because the homozygous null GPDH genotypes possess very low viability (Merritt et al. 2006).
For Mdh1 testcrosses in experiment 3, alleles 18.1 and 10.5 were combined to create 0, 50, and 100% normal MDH activity genotypes using the same rearing and collection methods as for Gpdh. All Mdh1 genotypes bear the EY P-element progenitor second chromosome and the white-marked X and VT46 third chromosomes.
For Adh testcrosses in experiment 4, the AdhΔ17 and AdhΔ25 alleles were combined to create 0, 50, and 100% normal ADH activity genotypes. All Adh genotypes bear the KG progenitor second chromosome and the white-marked X and VT83 third chromosomes.
In the Gpdh duplication series crosses for experiment 5, the Gpdh pB10 and GpdhpB23 alleles were used as representative single-copy and duplicate alleles and combined to produce three genotypes: GpdhpB10/GpdhpB10, GpdhpB10/GpdhpB23, and GpdhpB23/GpdhpB23 with 100, 125, and 150% relative GPDH activities. These lines all bear the same white-marked X chromosome (as the previous lines) and the 6326.1 second and third chromosomes.
In experiment 6, Mdh118.1 and Mdh110.5 males (50 each) were separately mated with AdhΔ17 and AdhΔ25 homozygous females to create four MDH:ADH genotypes with predicted 50:50, 50:100, 100:50, and 100:100 normal activity genotypes. In experiment 7, Mdh118.1 and Mdh110.5 homozygous females (50 each) were separately mated with CyO/GpdhΔ9.2 and GpdhΔ10.2 males to create four MDH:GPDH genotypes with predicted 50:50, 50:100, 100:50, and 100:100 normal activity genotypes. In experiment 8, CyO/GpdhΔ9.2 and GpdhΔ10.2 males (50 each) were separately mated with AdhΔ17 and AdhΔ25 homozygous females to create four GPDH:ADH activity genotypes with 50:50, 50:100, 100:50, and 100:100 normal activity genotypes. Enzyme assays of emerging flies indicate that these activity ratios are present as expected.
Basic tolerance assay:
The ethanol tolerance assay included replicated vials each with 10 adult males aged 5–7 days. A standard-sized cotton ball was pressed into the bottom of each vial and saturated with 2.5 ml of a solution of 2% sucrose and 15% ethanol. Vials were checked at two 24-hr intervals, recording the number of dead flies appearing over the 2-day interval. If <10% average mortality was observed, counts were extended another day. No significant mortality was ever observed for control flies (maintained on 2% sucrose over a 3-day period).
Statistics:
Statistical analysis was carried out on arc–sine transformed measures of the percentage surviving. Single-nested ANOVAS (single-locus tests), two-way ANOVAs (for di-locus interactions), and Tukey's honestly significant difference multiple comparison tests (Tukey's HSD test) were conducted using the JMP software package (release 5.0.1a, SAS Institute).
RESULTS
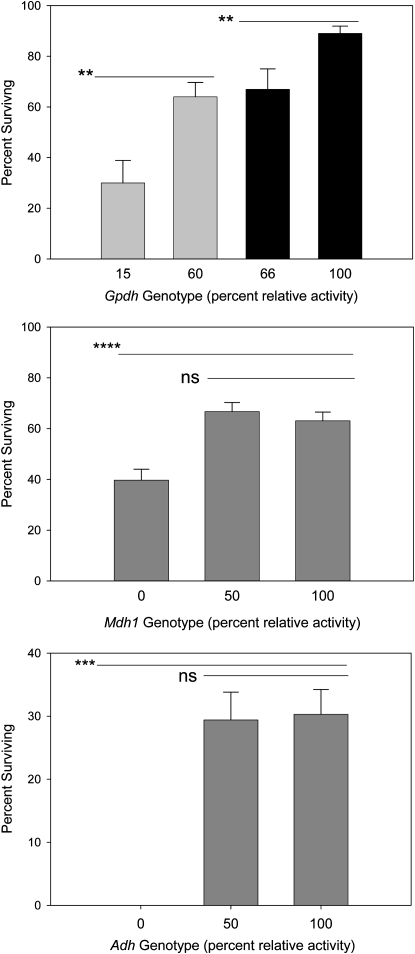
Experiments 1 and 2 compare the effects of GPDH activity reduction on ethanol tolerance. In each test comparison, there is a paired reference genotype with GPDH activity that scales within the normal range (Merritt et al. 2006). The first experiment (Figure 1A, shaded bars) contrasted Gpdh genotypes with 15% (9.2/24.1) and 60% (10.2/24.1) activity relative to a 10.2/10.2 genotype. Clearly, what is “normal” activity is arbitrary here because GPDH activity varies across wild alleles and backgrounds. The 10.2/10.2 genotype has 12% higher activity than the average for the 10 wild second chromosome lines reported in Merritt et al. (2006). Over the 48 hr of ethanol exposure, the survival rate of the low-activity genotype is less than one-half of the high-activity genotype (F1,36 = 11.83, P < 0.0015). Experiment 2 (Figure 1A, solid bars) compared genotypes constructed by crossing 10.2 and 9.2 males with 6326 females. The 10.2/6326 reference genotype possesses GPDH activity again in the normal range (∼12% higher than 10.2/10.2) and the 6326 line possesses the GpdhS allele. The 9.2/6326 genotype has a relative activity that is 66% of 10.2/6326 (Figure 1A) There is a highly significant difference in ethanol tolerance (F1,31 = 7.78, P < 0.009). These two experiments show a reduction in tolerance with lower GPDH activity.
Figure 1.—
Genotype-specific adult (male) survivorship after 48 hr of exposure to a 15% ethanol, 2% sucrose solution. (A) Experiments 1 and 2 using genotypes that possess 10 and 60% normal GPDH activity (shaded bars)—Gpdh9.2/24.1 (n = 17) and Gpdh24.1/10.2 (n = 20)—and (solid) genotypes Gpdh 9.2/6326 (n = 13) and 10.2/6326 (n = 20) possessing 66 and 100% normal activity. (B) Experiment 3 using Mdh1 genotypes 18.1/18.1 (n = 33), 18.1/10.5 (n = 39), and 10.5/10.5 (n = 36), representing 0, 50, and 100% normal MDH activities. The homozygous null genotype has a significantly lower survival rate (P < 0.0001). (C) Experiment 4 using three Adh genotypes: 25/25 (n = 29), 17/25 (n = 33), and 17/17 (n = 35) possessing 0, 50, and 100% normal ADH activity. Error bars represent ±1 SE. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Combining the Mdh1Δ18.1 and Mdh1Δ10.5 alleles, we created genotypes with 0, 50%, and full cytosolic MDH activities in experiment 3. There was a highly significant effect of Mdh1 genotype on ethanol tolerance (Figure 1B; F2,105 = 14.69, P < 0.0001). This was attributed to the full homozygous null Mdh1 genotype, 18.1/18.1, which possessed significantly reduced tolerance relative to the 50 and 100% activity genotypes.
Experiment 4 using the AdhΔ25 (null) and AdhΔ17 alleles found a highly significant effect with the homozygous null genotypes showing significantly lower tolerance (Figure 1C; F = 21.35, P < 0.001). However, there was no significant difference in tolerance between the 50 and 100% activity genotypes after 48 hr.
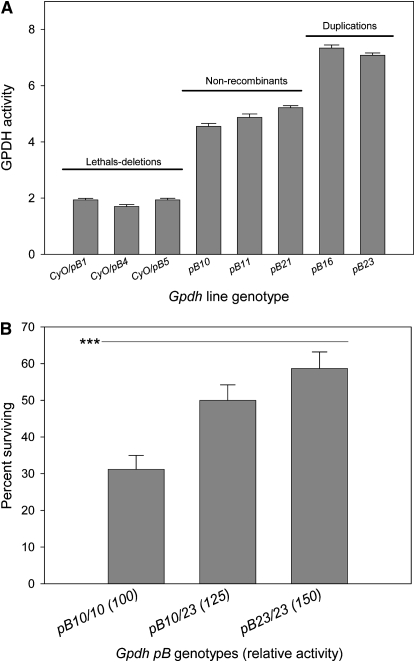
In experiment 5, using piggyBac FRT–FLP-facilitated recombination, we duplicated an 8-kb region spanning the Gpdh gene and placed these alleles in isogenic X and third chromosome backgrounds. The progenitor chromosomes for the piggyBac insertions are the 6326 line. The GPDH activities of the final duplication-deletion Gpdh allele sets are shown in Figure 2A. The duplicated alleles, pB16 and pB23, possess a 50% activity increase over single-copy alleles. When tested for ethanol tolerance (Figure 2B) using alleles pB10 and pB23, we observed a highly significant effect of elevated GPDH activity (F2,83 = 7.45, P < 0.001).
Figure 2.—
Gpdh gene duplication and deletion by piggyBac FLP–FRT site-specific recombination and effects on ethanol tolerance. (A) The GPDH activities as ΔOD units for seven piggyBac (pB) alleles recovered in the FRT–FLP recombination crosses and confirmed by diagnostic PCR and direct sequencing. All deletions are semilethal and balanced over CyO. (B) Experiment 5 showing the percentage of survival of three Gpdh genotypes possessing 100% (n = 34), 125% (n = 28), and 150% (n = 24) relative GPDH activity to normal. Among genotypes, differences are statistically significant by ANOVA on arc–sine transformed values (F2,83 = 7.44, ***P < 0.001). Error bars represent ±1 SE.
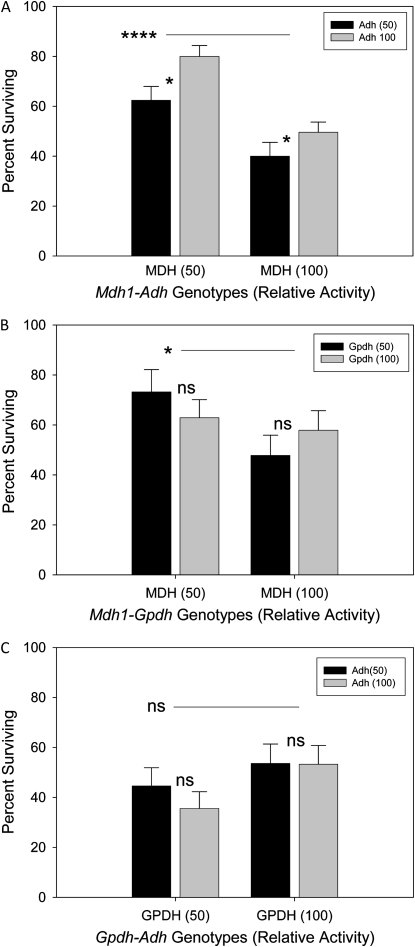
Experiments 6, 7, and 8 address interactions in di-locus combinations that yield full and half-full activity genotypes (Figure 3, A–C). With respect to tolerance, there were no significant interactions between genotypes in any experiment. Interestingly, there are highly significant main effects of the Mdh1 genotype (Mdh118.1 and Mdh110.5) in both Adh (Figure 3A; F1,73 = 21.9, P < 0.0001) and Gpdh (Figure 3B; F1,10 = 8.0, P < 0.038) backgrounds. The lower-activity Mdh1 genotype has significantly higher tolerance. This is suggested in experiment 3 as well (see Figure 1B). Adh genotypes showed significant genotype effects with the higher-activity AdhΔ17 allele possessing increased tolerance in combination with both Mdh1 genotypes (Figure 3A; F1,73 = 6.00, P < 0.017), but was not significant in combination with the Gpdh genotypes (Figure 3C). GpdhΔ9.2 and GpdhΔ10.2 genotypes were not significant in either background (Figure 3C), although the differences in the Adh background (F1,110 = 2.71, P < 0.102) are consistent with higher ethanol tolerance associated with the high-activity Gpdh genotype in both tests.
Figure 3.—
The ethanol tolerance as percentage of survival of Mdh1, Adh, and Gpdh di-locus genotypes. No experiments found statistically significant interactions. (A) Experiment 6 using Mdh1 and Adh genotypes with ratios of 50:50 (n = 15), 50:100 (n = 23), 100:50 (n = 16), and 100:100 (n = 26) normal activities. There are significant Mdh genotype effects (****P < 0.0001) and significant Adh genotype effects (*P < 0.017). (B) Experiment 7 using Mdh1 and Gpdh genotypes with ratios of 50:50 (n = 22), 50:100 (n = 34), 100:50 (n = 27), and 100:100 (n = 29) normal activities. There are significant Mdh genotype effects (*P < 0.038). (C) Experiment 8 using the Gpdh and Adh genotypes with ratios of 50:50 (n = 28), 50:100 (n = 34), 100:50 (n = 25), and 100:100 (n = 27) normal activities. Neither Gpdh nor Adh genotype effects were significant. The expected relative activities of genotypes are shown inside parentheses. Error bars represent ±1 SE.
In summary, in the five experiments that assessed Gpdh genotype effects all showed increased tolerance with increasing activity and three were statistically significant. In the three experiments in which the Mdh1 genotype effects were tested, all showed increasing tolerance with a 50% reduction in cytosolic MDH activity and two were statistically significant. While there are no statistically significant interactions in the strictest sense, these studies raise the possibility of the impact of genetic background on tolerance.
DISCUSSION
When adult flies possess no alcohol dehydrogenase activity, exposure to ethanol vapors results in knockdown within minutes, as often reported and again confirmed in our experiment. This sensitivity to ethanol exposure emphasizes the need for rapid elimination of ethanol, which requires not only the functioning of the initial ADH and ALDH steps and the downstream elimination of products, but also cofactor regeneration. Because both initial steps in ethanol breakdown consume NAD and produce NADH (two moles of NADH for each mole of ethanol), an essential consideration in ethanol metabolism is the maintenance of the redox potential in the cell. In Drosophila larvae exposed to dietary ethanol, there is a notable shift in the NADH:NAD ratio (Geer et al. 1983) and in adults a drop in NAD levels during 24 hr of exposure (McElfresh and McDonald 1983). In mammals, the maintenance of cellular redox balance is a central challenge in mammalian alcohol detoxification as well (Berry et al. 1994), and an important mechanism for the restoration of the redox balance in mammals is the malate–aspartate shuttle. In insects, where the glycerol phosphate shuttle is believed to have a major role in transferring NADH equivalents into the mitochondria, GPDH should be important in an analogous fashion.
In a series of early cage experiments on allozyme polymorphisms for Adh, Gpdh, and Mdh, Cavener and Clegg (1978) replicated selection under ethanol exposure in supplemented food in experiments that ran >50 generations. Both Adh and Gpdh showed repeatable responses, indicating selection favoring the AdhF and GpdhS allozyme alleles in the ethanol-exposed cage populations. Control populations showed no effective response, and cage populations removed from ethanol selection (relaxed) ceased allele changes. Mdh1 allozyme frequencies did not immediately respond to alcohol exposure and were followed less closely, but at generation 57 both ethanol-exposed populations were fixed for the MdhS allele, while controls were still polymorphic. This allele is most common in natural populations, typically <97.5% (Hay and Armstrong 1976). These studies suggest a participation of not just Adh, but Gpdh and possibly Mdh1 in the adaptation to alcohols in natural populations. However, as typical of cage experiments and as noted by the investigators, lines were started with small samples of wild chromosomes, and the initial linkage disequilibrium associated with this sampling potentially confounds interpretation. These demographic effects can be avoided by direct manipulation of enzyme levels as we have done here.
Our results show that after >48 hr of ethanol exposure adult male ethanol tolerance can depend on the activity levels of GPDH. We have also shown that changes in tolerance can be affected by both decreases and increases in GPDH activity relative to expected “normal” levels. Demonstrating this latter observation is important because in natural populations the derived GpdhS electrophoretic allele consistently shows 20% higher activity than the GpdhF allele (Miller et al. 1975; Laurie-Ahlberg and Bewley 1983; Bewley et al. 1984; Kang et al. 1998). Furthermore, the GpdhS allele frequency increases with latitude (Miller et al. 1975; Oakeshott et al. 1982; Sezgin et al. 2004), consistent with the hypothesis that increased activity is associated with increased tolerance in temperate climates. The intrapopulation sequence variation for Gpdh shows features associated with historical balancing selection and high levels of silent polymorphism relative to the associated interspecific divergence (Takano et al. 1993; Kreitman and Akashi 1995), similar to the situation at Adh (Hudson et al. 1987, but see Begun et al. 1999).
Because the relationship between metabolic flux and enzyme activity is often expected to be hyperbolic (Hartl et al. 1985; Dykhuizen et al. 1987), it does not necessarily follow that increases in activity above normal would also show enhanced tolerance. To test this hypothesis, we have used the piggyBac transposon and FLP–FRT recombination to produce a duplication of the Gpdh region and show that a 50% increase in GPDH activity (as seen for the derived GpdhS allele) causes increased tolerance to ethanol. While this method was introduced by Parks et al. (2004) to create site-specific deletions, our study is the first reported use of this genetic tool to increase gene function through duplication.
It is unclear if increased tolerance associated with Gpdh activity results from better maintaining the redox balance, increased triglyceride accumulation, or both. In a microarray study of genes induced under exposure to ethanol in adults (Morozova et al. 2006), both Gpdh and its mitochondrial shuttle partner, Gpo-1, show strong induction of transcripts. In larvae, GPDH is also strongly induced under dietary ethanol along with the accumulation of triglycerides (Geer et al. 1983; Lissemore et al. 1990). However, while the wild-type larval NADH:NAD ratio increases under ethanol exposure (0.22–0.36), this shift was not significantly different in Gpdh null genotypes (Geer et al. 1983). On sucrose control diets, both null Gpdh and wild-type larvae possess equal cofactor concentrations. However, under ethanol exposure, wild-type larvae see a 22% increase in total cofactor concentrations, but Gpdh null larvae experience a 24% drop. Therefore, on ethanol diets Gpdh null genotypes possess only 61% of the combined cofactor concentration of wild-type larvae. If the same phenomenon exists in adults, then the gain of tolerance with increased GPDH activity may derive from increased concentrations of both NADH and NAD, and not from the redox balance.
The different genotype-specific effects across experiments suggest a dependency on genetic background. Within each experiment, line constructions vary only in the targeted genes, but between experiments genotypes have different genetic backgrounds (the unique progenitor chromosomes of the KG, EY, and piggyBac elements), and the different outcomes certainly raise the possibility of genomewide interactions. In experiments 1–3, the higher-activity Gpdh genotypes always possess greater tolerance. In the interaction experiments, where Mdh1 and Adh genotypic backgrounds are varied, the Gpdh main effects are again in the same direction but nonsignificant. They certainly suggest further potential interactions. There are likely to be numerous other genes capable of participating in ethanol tolerance (Morozova et al. 2006, 2007), and genetic variation in these could contribute to background effects and differences between experiments.
The role of variation in cytosolic MDH activity in conferring ethanol tolerance appears complex. Complete loss of activity results in reduced ethanol tolerance (homozygous null Mdh1 genotypes are still normal in viability and fecundity), but genotypes with half-normal MDH activity clearly show significant increases in tolerance. Therefore, partial reduction of cytosolic MDH must enhance the elimination or metabolism of ethanol. In mammals, the malate–aspartate shuttle is responsible for the transfer of NADH equivalents into the mitochondria and the maintenance of redox balance, but its action in Drosophila is unknown. The cytosolic and mitochondrial glutamate–oxalacetate transaminases necessary for the shuttle are abundant, as is the aspartate–glutamate carrier (Aralar1). If this shuttle is present in flies, then a reduction in tolerance due to the complete loss of cytosolic MDH activity is understandable. However, the increased tolerance with partial reductions in activity is not expected and requires a different explanation. One alternative hypothesis is that Mdh1 plays a less direct role and the associated shuttle is an energy-state signal in Drosophila, triggering top–down responses as it clearly does in insulin secretion in the pancreatic β-cells (Rubi et al. 2004).
There is good evidence that potential malate–pyruvate and pyruvate–citrate cycles (Farfari et al. 2000; Guay et al. 2007) are strongly induced after adult response to ethanol. This is because Morozova et al. (2006) noted strong increases (nearly twofold) in transcription response for cytosolic malic enzyme (Men), but especially phosphoenopyruvate carboxykinase (Pepck) and the pyruvate carboxylase gene (CG1516). These cycles would act in the metabolic elimination of ethanol-derived acetyl-CoA as mitochondrial effluxes of malate and citrate. The cytosolic citrate is converted into oxalacetate and malonyl-CoA by ATP-citrate lyase, and oxalacetate is returned by PEPCK to gluconeogenesis and used in triglyceride formation. Malonyl-CoA is directed toward lipid synthesis (Freriksen et al. 1994). However, the efflux of citrate by the tricarboxylate carrier requires an exchange of malate; therefore, it is apparent that reduction of the pyruvate–malate shuttle could increase tolerance if the major detoxification pathway uses lipid and triglyceride synthesis. Since MDH1 metabolically bridges PEPCK and MEN, activity variation in it could control their relative roles in ethanol metabolism.
The association of increased alcohol tolerance with low cytosolic MDH activity certainly reflects a potential for natural selection to act on Mdh1 activity levels in natural populations, but, unlike Gpdh and Adh, there is no DNA sequence-based evidence that the Mdh1 gene is responding to positive or balancing selection in D. melanogaster. There is no common amino acid polymorphism and no evidence for clines in SNP sites inside the Mdh1 gene (Sezgin et al. 2004). This does not rule out regulatory variation polymorphism affecting enzyme levels, but this remains to be investigated. All population genetic evidence points to simple purifying selection. It is possible that general negative pleiotropic fitness effects associated with reduced MDH activity prevent its participation in naturally occurring variation in ethanol tolerance.
In natural populations of Drosophila, alcohol tolerance is a complex genetic trait and genetic variation for Adh has been shown to only partly contribute to the final phenotype (Cohan and Graf 1985). Clearly, genetic variation in many other enzymes should be important in the metabolic elimination of alcohols. Adh, Aldh, Gpdh, and Mdh1 possess different levels and patterns of intraspecific polymorphism and interspecific divergence, and this emphasizes the distinction between (1) identifying pathways of potential detoxification and (2) finding points of genetic variation realized as an adaptive response. The former distinction depends on the individual idiosyncratic properties of the enzymes as well as their context in a system of pathways. However, the impact of natural selection acting on genes in adaptive response to alcohol-based fitness reduction will also depend on the pleiotropic effects on other fitness components and their trade-offs in nonalcohol environments. Nevertheless, a complete understanding of the evolution of tolerance to alcohols will require a large-scale or system-level approach, rather than a focus on single enzymes.
Acknowledgments
We thank John True and two anonymous reviewers for constructive comments on the earlier version of the manuscript. We thank the Cell Biology Department of the Harvard Medical School and Exelixis for maintaining and providing support for the piggyBac insertion collection and for stocks maintained by the Bloomington Stock Center. Some of the project sequencing was carried out using the Molecular Evolution of Adaptation and Diversity Laboratory created through an equipment grant from the National Science Foundation (Multi-User Equipment and Instrumentation Program, DBI-0400829). This study was supported by U. S. Public Health Service grant GM-45247 to W.F.E. This is contribution no. 1140 from the Graduate Program in Ecology and Evolution, State University of New York, Stony Brook, New York.
References
- Begun, D. J., A. J. Betancourt, C. H. Langley and W. Stephan, 1999. Is the fast/slow allozyme variation at the Adh locus of Drosophila melanogaster an ancient balanced polymorphism? Mol. Biol. Evol. 16 1816–1819. [DOI] [PubMed] [Google Scholar]
- Berry, M. N., R. B. Gregory, A. R. Grivell, J. W. Phillips and A. Schon, 1994. The capacity of reducing-equivalent shuttles limits glycolysis during ethanol oxidation. Eur. J. Biochem. 225 557–564. [DOI] [PubMed] [Google Scholar]
- Bewley, G. C., D. W. Niesel and J. R. Wilkins, 1984. Purification and characterization of the naturally occurring allelic variants of sn-glycerol-3-phosphate dehydrogenase in Drosophila melanogaster. Comp. Biochem. Physiol. B 79 23–32. [DOI] [PubMed] [Google Scholar]
- Cavener, D. R., and M. T. Clegg, 1978. Dynamics of correlated genetic systems. IV. Multilocus effects of ethanol stress environments. Genetics 90 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan, F. M., and J.-D. Graf, 1985. Latitudinal cline in Drosophila melanogaster for knockdown resistance to ethanol fumes and for rates of response to selection for further resistance. Evolution 39 278–293. [DOI] [PubMed] [Google Scholar]
- Cohan, F. M., and A. A. Hoffmann, 1986. Genetic divergence under uniform selection. II. Different responses to selection for knockdown resistance to ethanol among Drosophila melanogaster populations and their replicate lines. Genetics 114 145–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, J. R., and C. Bocquet, 1975. Similarities and differences in latitudinal adaptation of two Drosophila sibling species. Nature 257 588–590. [DOI] [PubMed] [Google Scholar]
- Dykhuizen, D. E., A. M. Dean and D. L. Hartl, 1987. Metabolic flux and fitness. Genetics 115 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfari, S., V. Schulz, B. Corkey and M. Prentki, 2000. Glucose-regulated anaplerosis and cataplerosis in pancreatic beta-cells: possible implication of a pyruvate/citrate shuttle in insulin secretion. Diabetes 49 718–726. [DOI] [PubMed] [Google Scholar]
- Freeman, S., and J. C. Herron, 2004. Evolutionary Analysis, Ed. 3. Pearson-Prentice Hall, Upper Saddle River, NJ.
- Freriksen, A., D. Seykens and P. W. H. Heinstra, 1994. Differences between larval and adult Drosophila in metabolic degradation of ethanol. Evolution 48 504–508. [DOI] [PubMed] [Google Scholar]
- Fry, J. D., and M. Saweikis, 2006. Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet. Res. 87 87–92. [DOI] [PubMed] [Google Scholar]
- Fry, J. D., K. Donlon and M. Saweikis, 2008. A worldwide polymorphism in aldehyde dehydrogenase in Drosophila melanogaster: evidence for selection mediated by dietary ethanol. Evolution 62 66–75. [DOI] [PubMed] [Google Scholar]
- Futuyma, D. J., 2005. Evolution. Sinauer, Sunderland, MA.
- Geer, B. W., S. W. McKechnie and M. L. Langevin, 1983. Regulation of sn-glycerol-3-phosphate dehydrogenase in Drosophila melanogaster larvae by dietary ethanol and sucrose. J. Nutr. 113 1632–1642. [DOI] [PubMed] [Google Scholar]
- Geer, B. W., P. W. Heinstra and S. W. McKechnie, 1993. The biological basis of ethanol tolerance in Drosophila. Comp. Biochem. Physiol. 105B: 203–229. [DOI] [PubMed]
- Guay, C., S. R. M. Madiraju, A. Aumais, E. Joly and M. Prentki, 2007. A role for ATP-citrate lyase, malic enzyme, and pyruvate/citrate cycling in glucose-induced insulin secretion. J. Biol. Chem. 282 35657–35665. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., D. E. Dykhuizen and A. M. Dean, 1985. Limits of adaptation: the evolution of selective neutrality. Genetics 111 655–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, R. E., and F. B. Armstrong, 1976. Biochemical characterization of allelic forms of soluble malate dehydrogenase of Drosophila melanogaster. Insect Biochem. 6 367–376. [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, F. M., and C. Denniston, 1964. Genetic variation of alcohol dehydrogenase in Drosophila melanogaster. Nature 204 906. [DOI] [PubMed] [Google Scholar]
- Kang, S. J., S. H. Lee and K. S. Park, 1998. DNA polymorphisms at alpha-Gpdh locus of Drosophila melanogaster in Korean population. Genes Genet. Syst. 73 227–235. [DOI] [PubMed] [Google Scholar]
- Kreitman, M., 1983. Nuleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304 412–417. [DOI] [PubMed] [Google Scholar]
- Kreitman, M., and H. Akashi, 1995. Molecular evidence for natural selection. Annu. Rev. Ecol. Syst. 26 403–422. [Google Scholar]
- Laurie-Ahlberg, C. C., and G. C. Bewley, 1983. Naturally occurring genetic variation affecting the expression of sn-glycerol-3-phosphate dehydrogenase in Drosophila melanogaster. Biochem. Genet. 21 943–961. [DOI] [PubMed] [Google Scholar]
- Lissemore, J. L., C. A. Baumgardner, B. W. Geer and D. T. Sullivan, 1990. Effect of dietary carbohydrates and ethanol on expression of genes encoding sn-glycerol-3-phosphate dehydrogenase, aldolase, and phosphoglycerate kinase in Drosophila larvae. Biochem. Genet. 28 615–630. [DOI] [PubMed] [Google Scholar]
- McElfresh, K. C., and J. F. McDonald, 1983. The effect of alcohol stress on nicotinamide adenine dinucleotide (NAD+) levels in Drosophila. Biochem. Genet. 21 365–374. [DOI] [PubMed] [Google Scholar]
- McKenzie, J. A., and P. A. Parsons, 1972. Alcohol tolerance: an ecological parameter in the relative success of Drosophila melanogaster and D. simulans. Oecologia 10 373–388. [DOI] [PubMed] [Google Scholar]
- Merritt, T. J., D. Duvernell and W. F. Eanes, 2005. Natural and synthetic alleles provide complementary insights into the nature of selection acting on the Men polymorphism of Drosophila melanogaster. Genetics 171 1707–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, T. J., E. Sezgin, C. T. Zhu and W. F. Eanes, 2006. Triglyceride pools, flight and activity variation at the Gpdh locus in Drosophila melanogaster. Genetics 172 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S., R. W. Pearcy and E. Berger, 1975. Polymorphism at the alpha-glycerophosphate dehydrogenase locus in Drosophila melanogaster. I. Properties of adult allozymes. Biochem. Genet. 13 175–188. [DOI] [PubMed] [Google Scholar]
- Montooth, K. L., K. T. Siebenthall and A. G. Clark, 2006. Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster. J. Exp. Biol. 209 3837–3850. [DOI] [PubMed] [Google Scholar]
- Morozova, T. V., R. R. Anholt and T. F. Mackay, 2006. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome Biol. 7: R95. [DOI] [PMC free article] [PubMed]
- Morozova, T. V., R. R. Anholt and T. F. Mackay, 2007. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome Biol. 8: R231. [DOI] [PMC free article] [PubMed]
- Oakeshott, J. G., J. B. Gibson, P. R. Anderson, W. R. Knibb, D. G. Anderson et al., 1982. Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution 36 86–96. [DOI] [PubMed] [Google Scholar]
- Oakeshott, J. G., S. W. McKechnie and G. K. Chambers, 1984. Population genetics of the metabolically related Adh, Gpdh, and Tpi polymorphisms in Drosophila melanogaster. I. Geographic variation in Gpdh and Tpi allele frequencies in different continents. Genetica 63 21–29. [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36 288–292. [DOI] [PubMed] [Google Scholar]
- Pecsenye, K., and A. Saura, 1998. Interaction between the Adh and Odh loci in response to ethanol in Drosophila melanogaster. Biochem. Genet. 36 147–170. [DOI] [PubMed] [Google Scholar]
- Rubi, B., A. del Arco, C. Bartley, J. Satrustegui and P. Maechler, 2004. The malate-aspartate NADH shuttle member Aralar1 determines glucose metabolic fate, mitochondrial activity, and insulin secretion in beta cells. J. Biol. Chem. 279 55659–55666. [DOI] [PubMed] [Google Scholar]
- Scholz, H., J. Ramond, C. M. Singh and U. Heberlein, 2000. Functional ethanol tolerance in Drosophila. Neuron 28 261–271. [DOI] [PubMed] [Google Scholar]
- Scholz, H., M. Franz and U. Heberlein, 2005. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature 436 845–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin, E., D. D. Duvernell, L. M. Matzkin, Y. Duan, C. T. Zhu et al., 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, T. S., S. Kusakabe and T. Mukai, 1993. DNA polymorphism and the origin of protein polymorphism at the Gpdh locus of Drosophila melanogaster, pp. 179–190 in Mechanisms of Molecular Evolution, edited by N. Takahata and A. G. Clark. Japan Scientific Societies Press and Sinauer Associates, Tokyo/New York.
- Van der Zel, A., R. Dadoo, B. W. Geer and P. W. H. Heinstra, 1991. The involvement of catalase in alcohol metabolism in Drosophila melanogaster larvae. Arch. Biochem. Biophys. 287 121–127. [DOI] [PubMed] [Google Scholar]