Abstract
Fluorescence recovery after photobleaching was used to study the diffusion of two integral membrane proteins, bacteriorhodopsin and beta2-adrenergic receptor, in lipidic cubic phase (LCP). We found that the diffusion properties within the LCP matrix strongly depend on the protein construct and applied screening conditions. Common precipitants often induce restriction on diffusion of proteins in LCP and thereby impede their chances for crystallization. A high protein mobile fraction and a fast diffusion rate correlate very well with known crystallization conditions. Using this knowledge, one can now pre-screen precipitant conditions with microgram quantities of material to rule out conditions that are not conducive to diffusion, nucleation, and crystal growth. The results of this assay will narrow membrane protein crystallization space by identifying suitable protein constructs, stabilizing compounds and precipitant conditions amenable to in meso crystallization. Crystallization pre-screening will significantly increase the chances of obtaining initial crystal hits, expediting efforts in generating high-resolution structures of challenging membrane protein targets.
1. Introduction
Integral membrane proteins (IMPs) and their complexes are involved in a number of important cellular and physiological processes. They communicate signals and transport chemicals across the membrane and thus are the central family of proteins for signal transduction. The most successful technique for elucidating three-dimensional structures is X-ray crystallography, which requires growing well ordered protein crystals. In spite of their biological importance, IMPs constitute only ~500 (~150 unique) out of almost 50,000 structural entries (i.e., approx. 1%) in the PDB. This disparity in numbers reflects the difficulties in producing X-ray-diffraction-quality-grade crystals (current success rates, determined as the number of crystal structures relative to the number of generated clones, are <3% for all proteins and nearly zero for membrane proteins (http://targetdb.pdb.org/statistics/TargetStatistics.html)). Technological improvements to increase this abysmal success rate are urgently needed. Currently, the expected success rate of determining a structure goes to almost 80% once diffraction grade crystals become available, placing the bottleneck clearly on the crystallization step of the structure determination process.1
The traditional approach to membrane protein crystallization involves extracting the protein with an appropriate detergent followed by purification and vapor diffusion crystallization of the protein/detergent complexes (PDCs). Often the protein is not stable in detergent micelles that are conducive to crystallization, resulting in protein aggregates or poorly ordered crystals that do not diffract. An alternative approach, utilizes lipidic cubic phase (LCP) for crystallizing membrane proteins.2 The initial success of the LCP (or in meso) technique was in elucidating details of bacteriorhodopsin (bR) photocycle by providing high-resolution structures of photocycle intermediates and various bacteriorhodopsin mutants.3 After years of technology development aimed toward working with extremely small volumes of non-colored and less stable proteins, another notable success came recently with the publication of the high-resolution structure of human beta2-adrenergic receptor (β2AR),4,5 a G-protein coupled receptor in complex with a diffusible ligand. Currently there are a total 53 structures of 9 different membrane proteins in PDB attributed to the in meso technique (http://www.mpdb.ul.ie).6
Crystallization trials are characterized by the need to process large numbers of samples in a vast multidimensional crystallization space while searching for crystal nucleation and growth. With the in meso method the screening is complicated by the need to include the host lipid identity and identities of any lipid-like additives and their concentrations as variables to be explored and optimized. Therefore careful biochemical and biophysical characterization of the target protein constructs and suitable pre-crystallization assays are critical to narrow down the crystallization space and increase chances for successful crystallization.
For membrane proteins the in meso crystallization technique has number of attractive characteristics.7 Prior to crystallization, proteins are reconstituted into lipid membranes, a presumably more native environment than detergent micelles. Crystals growing in meso are type I8 with extensive hydrophobic as well as hydrophilic contacts between protein molecules. Such crystals typically have lower solvent content and higher order in comparison to their counterparts grown in detergent solutions, resulting in stronger diffraction. In meso crystallization is especially well suited for proteins with mostly intramembrane and small extramembrane components, which resist crystallization in detergent micelles.
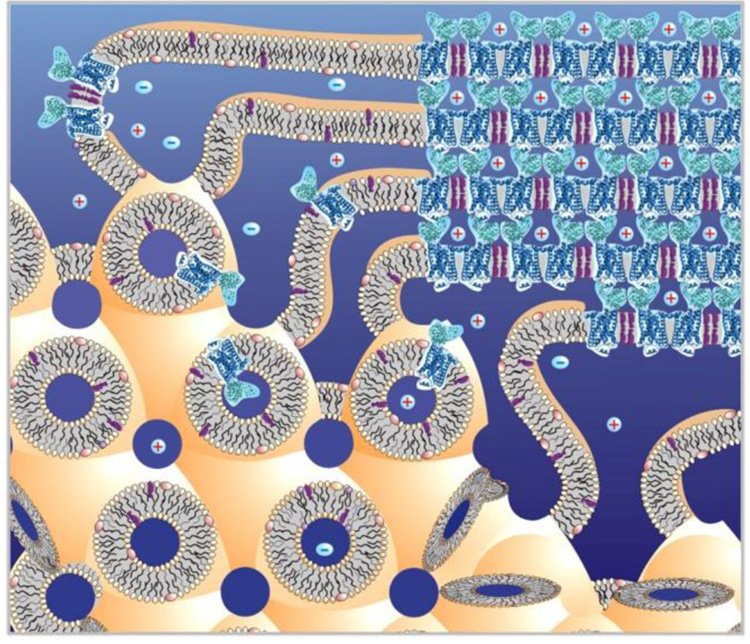
As with any emerging technique, a number of problems with in meso crystallization have been discovered. Challenges related to reconstitution of proteins into lipidic mesophases and to handling small volumes of viscous and sticky lipidic mesophases have largely been solved,9–13 rendering high-throughput in meso crystallization a reality. The underlying mechanism of in meso crystallization has been approached from different angles,7,14–16 however, a clear understanding of all events accompanying nucleation and crystal growth are still lacking. One of the prerequisites for successful crystallization in lipidic mesophases is the ability of membrane proteins to freely diffuse within the three dimensional cubic phase structure formed by a single connected lipid bilayer (see Figure 1 for a cartoon on how crystal growth is envisioned). The lipidic cubic phase spatial arrangement imposes certain limitations on the size of membrane proteins that will diffuse within the system unaffected by the curvature imposed on the lipid bilayer and able to pass unimpeded through the narrow solvent channels. The cubic-Pn3m phase of monoolein, the most commonly used for crystallization host lipid, has a lattice parameter of 110 Å and a solvent channel diameter of 50 Å.17 We have previously shown that the cubic phase must be induced to swell in order to crystallize the light-harvesting complex 2 (LH2), a 123 kDa protein.18 We speculate that the swelling relaxes spatial constrains increasing mobility of large proteins within lipidic cubic phase. Indeed, photosynthetic reaction center from Rhodobacter sphaeroides, 101 kDa,19 outer membrane vitamin B12 transporter BtuB, 66 kDa,20 and beta2-adrenergic receptor – T4L lysozyme fusion protein (β2AR-T4L), 55 kDa,4 were all crystallized from swollen lipidic mesophases. Recently Photosynthetic core complex, RC-LH1, 440 kDa, from Blastochloris viridis was also shown to crystallize in a swollen lipidic sponge phase.21
Figure 1.
Cartoon of the in meso crystallization process. Membrane protein (blue, β2AR-T4L, PDB ID 2RH1) is embedded in the lipid bilayer of the lipidic cubic phase (low left corner). Upon addition of a precipitant the crystal nucleates. The lipidic cubic phase transforms into a multilamellar phase in the vicinity of the growing crystal and serves as a portal connecting the bulk cubic phase to the growing crystal16 (upper right corner). Membrane proteins are expected to diffuse in 3-dimensions within the single lipid bilayer and approach the crystal through the lamellar phase portal. If proteins form micro-aggregates (such as shown in the middle-bottom part of the cubic phase),their diffusion through narrow channels in LCP will be highly restricted and no crystals will grow.
Since lipidic mesophases are not static, rigid structures, it is impossible to predict from their structural parameters whether a particular protein will or will not diffuse and what effect the spatial constraints will have on the diffusion rate of the protein. Therefore, we adapted the Fluorescence Recovery after Photobleaching (FRAP) technique to study the long-range translational diffusion of amphiphilic and soluble macromolecules. We examined the effects of shrinking and swelling the cubic phase by common precipitant agents on the diffusion rates of these macromolecules as well as the inclusion of various salts and buffers. Our results indicate that diffusion of membrane proteins in LCP depends not only on the size of the protein and structural parameters of the cubic phase but also on the identity of the protein and the screening conditions. Therefore, we propose to use FRAP in LCP as a pre-crystallization assay to select a suitable range of conditions for in meso crystallization of differening protein constructs and host lipids and eliminate conditions that are not compatible with protein diffusion from subsequent crystallization screens.
2. Experimental Section
2.1. Bacteriorhodopsin expression and purification
Wild-type Bacteriorhodopsin (bR) was solubilized with 1.2 %w/v octylglucoside (OG) from purple membranes isolated from Halobacterium salinarum (strain S9) using established protocols.22–24 Protein was concentrated to 7 mg/ml, and stored at −80 °C prior to labeling.
2.2. Beta2-adrenergic receptor expression and purification
Two thermally stabilized constructs of the human beta2 – adrenergic receptor were used in this study. The first construct (β2AR(E122W)) included stabilizing mutation E122W,25 a C-terminal truncation at residue 348, and the removal of the third glycosylation site,5,25 and deletion of residues 245 to 259 in the third intracellular loop (ICL3). The second construct (β2AR(E122W)-T4L) in addition to modifications of the first construct contained fused T4 lysozyme replacing ICL3 between transmembrane helices 5 and 6.5,25 Both constructs had FLAG tag at the N-terminus and 10xHis tag at the C-terminus. Both proteins were expressed in Spodoptera frugiperda (Sf9) insect cells infected by high-titer recombinant baculovirus (prepared using standard protocol in the Bac-to-Bac system, Invitrogen), and purified as described previously.25 After extensive membrane washes, the target proteins were solubilized from the membranes with 0.5 %w/v dodecylmaltoside (DDM)/ 0.1 %w/v cholesteryl hemi-succinate (CHS) in the presence of 1 mM timolol. After solubilization, proteins were further purified in the presence of 1 mM timolol using both Cobalt charged TALON IMAC resin and Nickel charged IMAC column. Purity and monodispersity were checked by SDS-PAGE and analytical size exclusion chromatography (SEC) using SepaxNanofilm SEC-250 (Sepax) on a Dionex Ultimate HPLC system. The mobile phase for SEC characterization was 20 mM HEPES pH 7.5, 150 mM NaCl, 2 %v/v glycerol, 0.05 %w/v DDM. The ligand was exchanged for 100 µM carazolol and the protein was deglycosylated by incubating with PNGaseF (NEB) at 4 °C overnight on the second IMAC column. PNGaseF was washed out using ?? column volumes of 20 mM Hepes pH 7.5, 150 mM NaCl, 0.05 %w/v DDM, 0.01 %w/v CHS, µ100 M carazolol, and the protein was eluted in a minimal volume of the same buffer supplemented with 200 mM imidazole.
2.3. Protein labeling
Protein samples were labeled with 5,5′-disulfato-1′-ethyl-3,3,3′,3′-tetramethylindocarbocyanine (Cy3) dye. The Cy3 dye was selected because of its relatively hydrophilic properties. Two commercially available Cy3 protein conjugation carriers were used: Cy3 mono-maleimide reacting with free sulfhydral groups of cysteine residues, and N-hydroxylsuccinimidyl (NHS) ester reacting with free amino groups (e.g. N-terminus, lysine residues). Using thiol-reactive probe requires availability of free cysteine residues exposed at the protein surface while amino-reactive labeling is more universal. The amino-reactive conjugation was plagued by an inadvertent labeling of phosphatidylethanolamine (PE) lipids, which are abundant in mammalian and insect cells and co-purified with the protein. Separation of the protein sample from the labeled lipids involves extensive washing and multiple SEC steps which could affect the stability of the protein and should be avoided if possible. Therefore, β2AR(E122W) containing surface exposed Cys264 was successfully labeled with Cy3-monomaleimide. The β2AR(E122W)-T4L construct, however, was unstable after labeling with Cy3-monomaleimide and instead was labeled with Cy3-mono NHS ester. Trace amounts of lipids were detected in this sample after extensive purification. Bacteriorhodopsin, lacking cysteines, was labeled with Cy3-Mono NHS ester. The purple membranes do not have free amine containing lipids,26 therefore lipid labeling was not a problem in this case. Specific details of the protein labeling are described in the following two subsections.
2.3.1. Labeling of bacteriorhodopsin
The Cy3-mono NHS ester (G.E. Healthcare) stock solution was prepared by adding 200 µl of dimethylformamide (DMF, Sigma) to a vial containing 1 mg of the dye to achieve the final concentration of 5 mg/mL. 25 µg of Cy3-mono NHS ester was incubated with 500 µg of bR in 500 µL of 25 mM Na/K phosphate pH 7.1, 1.2 %w/v OG at 4 °C for 2–4 hours in dark. Size-exclusion column (Superdex 75, GE Healthcare) equilibrated with 1.2 %w/v OG, 25 mM Na/K phosphate pH 5.5 was used immediately to separate labeled protein from the free dye. Fractions containing protein were collected and concentrated to 5 mg/mL using a Vivaspin concentrator with 50 kDa molecular weight cutoff. No labeled lipid or free dye was detected by thin layer chromatography (see section 2.5. Thin layer chromatography (TLC).). To estimate the protein labeling percentage sample absorptions at 280 nm and 550 nm were recorded. Cy3 has negligible absorption at 280 nm, while both bR and Cy3 absorb at 550 nm. Absorption ratio between 280 nm and 550 nm for unlabeled bR was found to be 1.8 and this ratio was used to extract individual contributions from bR and Cy3 in total absorption at 550 nm. Then using extinction coefficients of bR (58,000 M−1cm−1 at 550 nm)2 and Cy3 (150,000M−1cm−1 at 550 nm) the labeling percentage was estimated as 1.5%.
2.3.2. Labeling of β2AR(E122W) and β2AR(E122W)-T4L
β2AR(E122W) was labeled before solubilization, then purified as previously described. Stock of Cy3-mono maleimide was made the same way as the stock of Cy3-NHS ester. Membranes containing 0.5–1mg of β2AR(E122W) were treated with 300 µg of Cy3-mono maleimide (G.E. Healthcare) in 10 mM HEPES (pH7.5), 20 mM KCl, 10 mM MgCl2, at 4 °C for 2–4 hours in dark. After purification, no labeled lipids or free dye were detected by TLC. Labeling percentage was estimated to be 40 % using the theoretical extinction coefficients for β2AR(E122W) (93,445 M−1cm−1, at 280 nm) and Cy3 (150,000 M−1cm−1, at 550 nm), the final concentration of β2AR(E122W) was about 5 mg/mL.
Purified β2AR(E122W)-T4L was labeled by Cy3-mono NHS ester following a protocol similar to that of bR, however, instead of the SEC step, β2AR(E122W)-T4L was bound to Ni2+ charged resin and washed extensively with a 50 mM Hepes pH 7.5, 150 mM NaCl, 0.05 %w/v DDM, 0.01 %w/v CHS, 20 mM imidazole, 1 mM timolol buffer. Final protein concentration was 6.3 mg/mL and labeling percentage was 2%.
2.4. Lipid labeling
5 mg of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE, Avanti Polar Lipids) was dissolved in100 µL chloroform in a glass amber vial. 25 µg of Cy3-Mono NHS ester in DMF was added to the vial and incubated for 2–4 hours in the dark at room temperature. After reaction the lipid was separated from the free unreacted dye using selective solvent extraction. For this purpose 1 mL water was added to the vial and vortex mixed. After brief centrifugation at 4,000 × g the aqueous layer was discarded. This process was repeated several times until there was no detectable 550 nm absorption in the aqueous layer. Chloroform was evaporated by a stream of dry nitrogen. Residual traces of the solvent were removed under a vacuum (50 mTorr) for 4 hours. No free dye was detected in the final sample by TLC.
2.5. Thin-layer chromatography (TLC)
5 cm × 10 cm High Performance Thin-Layer Chromatography plates (EMD Chemicals, Germany) were used to detect labeled lipid and free dye. Prior to experiments, TLC plates were pre-run in pure methanol (HPLC grade, Fisher). After drying, 0.3–2 µl of 50 µg/mL Cy3 dye, labeled DOPE and labeled proteins were spotted 1.2 cm above the edge of the plate. After warming the plate at 60 °C to remove the traces of solvents and absorbed moisture, the plate was run in the chloroform : methanol : water = 65 / 35 / 4 solvent system. After drying the solvent, the plate was examined under a fluorescent microscope described below.
2.6. Fluorescence recovery after photobleaching
2.6.1. Sample preparation
Labeled protein was mixed with molten monoolein (1-oleoyl-rac-glycerol, Sigma) in 2/3 volume ratio using a syringe lipid mixer10 to form lipidic cubic phase (LCP). Samples were set up similar to the manual in meso crystallization setup in glass sandwich plates11 containing 50 µm thick transfer tape (3M, 9482PC), defining sample thickness, with 7 mm diameter punched holes forming individual wells. 70 nL of protein laden cubic phase and 700 nL of screening solutions were dispensed in each well and sealed with a 0.18 mm thick glass cover slip. All samples were incubated at room temperature (21–23 °C) overnight (12–15 hrs) prior to FRAP measurements. Most of the measurements, except for time-course series, were performed between 12 and 20 hrs after setup.
2.6.2. Instrument setup
FRAP experiments were performed on a Zeiss AxioImager-A1 customized microscope (Carl Zeiss, Germany) with an EC-plan 10x objective lens (NA=0.3, Zeiss). HBO100 illuminator with OSRAM Mercury Short Arc Photo Optic Lamp was used along with a custom filter set (excitation 543/22 nm, emission 575 – 640 nm) to observe fluorescence from the Cy3 labeled samples. Samples were bleached by a sequence of 10 – 20 short 5 ns pulses at 20 Hz repetition rate from a tunable dye-cell (set at 551 nm) MicroPoint laser system (Photonic Instruments) attached to the AxioImager microscope. Fluorescent images were captured by a CoolSnap HQ2, 14 Bit, cooled (−30 °C) CCD (1392 × 1040 pixels, 6.45 µm/pixel) monochrome FireWire camera (Photometrics). The acquisition area was set at 501 × 501 pixels to reduce the read-out and image transfer times. Fluorescent shutter, laser trigger, and image capture sequences were controlled with ImagePro (Media Cybernetics).
2.6.3. FRAP data collection
The image acquisition sequence consisted of three sections. The experiment started by taking five images to record pre-bleached fluorescence of the sample. Then the laser was triggered, immediately followed by a fast post-bleached sequence of 200 images (100 – 500 ms exposure time per image) streaming as fast as possible into computer memory. After saving the fast sequence, an additional 50 images were taken with various time delays between images (1 – 20 s). To reduce photobleaching from the fluorescent field illumination, the shutter was closed during the idle time between images. All images were acquired and processed using ImagePro. All experiments were conducted at room temperature (21–23 °C).
2.6.4. FRAP data analysis
For each frame, fluorescence intensity inside the bleached spot was integrated within a 6.3 µm diameter circular Region of Interests (ROI). Averaged integrated intensity of four 12.6 µm × 12.6 µm square ROI’s positioned near four corners of the image frames was used to correct for photobleaching from illuminating light during the image sequence acquisition. The correction was performed by dividing the value of the integrated intensity in the bleach spot ROI by the average integrated intensity of the four square ROI’s. Fractional fluorescence recovery curves F(t) were calculated using the following equation:27
| (Eq. 1) |
where f(t) is the corrected fluorescence intensity of the bleached spot, fo is the corrected fluorescence intensity of the bleached spot in the 1st image after bleaching, and f∞ is the average of corrected fluorescence intensity in the pre-bleached images.
The fractional fluorescence recovery curves were fitted with a 2-dimensional diffusion equation derived by Soumpasis:27
| (Eq. 2) |
where M is the mobile fraction of diffusing molecules, T is the characteristic diffusion time, t is the real time of each recorded frame, I0and I1are 0th and 1st orders modified Bessel functions.
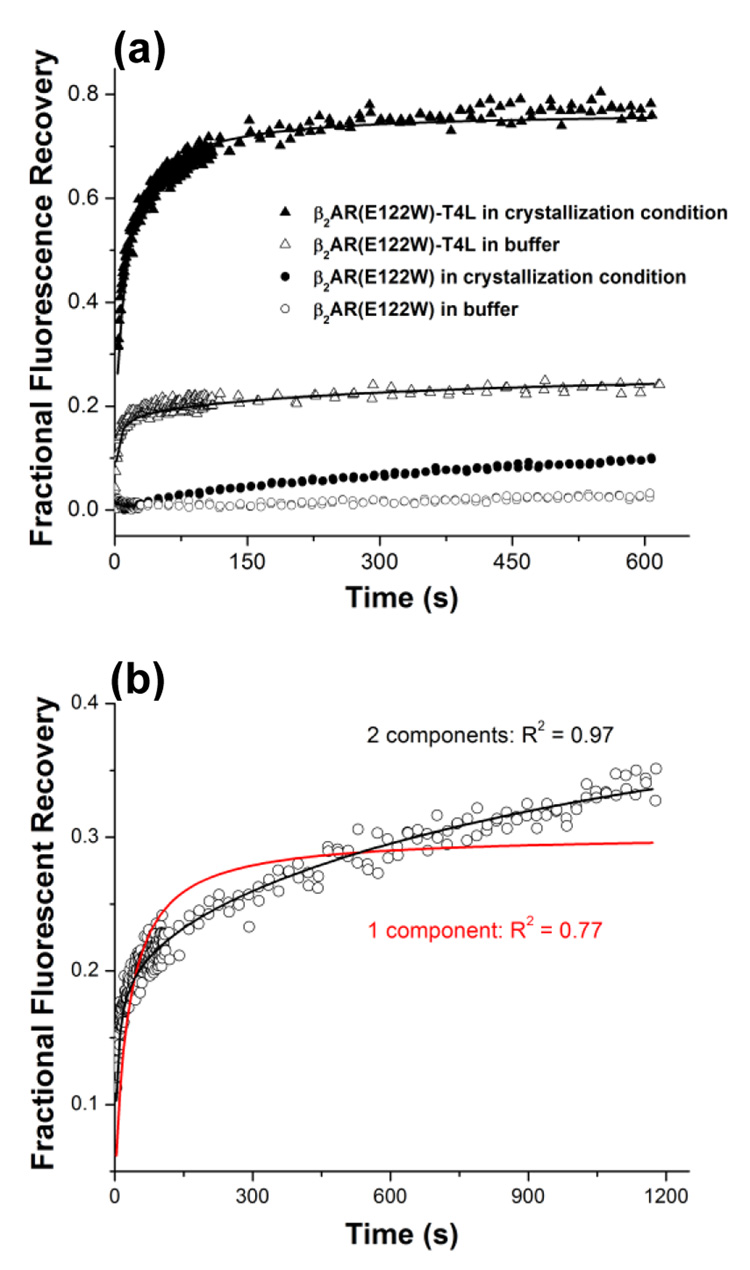
In some instances, the one component FRAP equation could not be used to adequately describe the fractional fluorescence recovery curves. In these cases the data were fitted with a two component FRAP equation and the rationale for using it is provided in the text:
| (Eq. 3) |
The size of the bleached spot was obtained by fitting the radially averagedspot profile with a Gaussian:
| (Eq. 4) |
The half width at half maximum (HWHM) of the Gaussian R = r (2ln2)0.5 was used as the measure of the bleached spot radius and the diffusion coefficient, D, was calculated as:28
| (Eq. 5) |
Non-linear curve fittings were done using Prism (GraphPad Software, San Diego).
3. Results
3.1. Validation of the FRAP data collection and analysis protocol
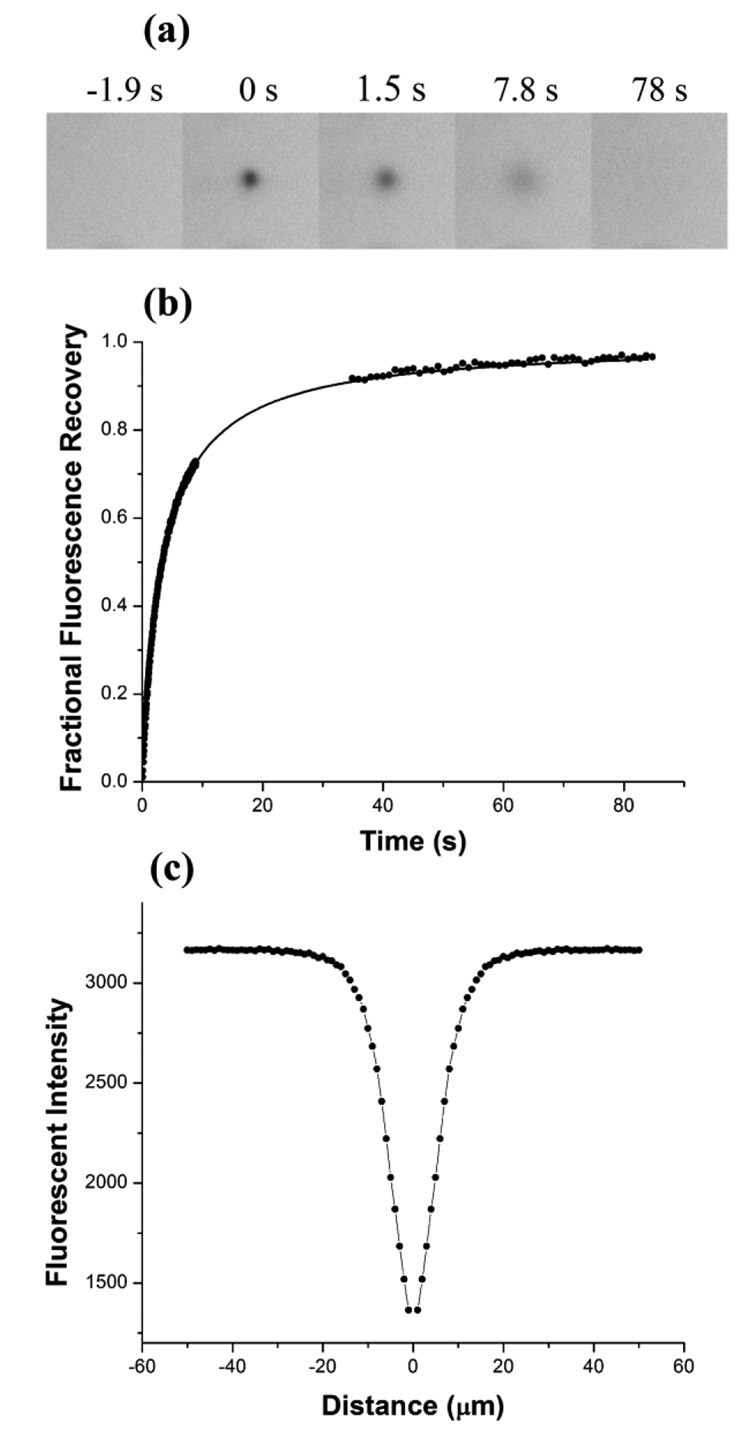
To validate the instrumental setup, experimental conditions and data analysis, we performed FRAP measurements on a standard well-defined sample of Rhodamine 6G in 90 %w/w glycerol solution. The sample was sandwiched between a microscope slide and a glass coverslip using a 50 µm spacer, defining the thickness of the sample. The FRAP experiment started with recording 5 pre-bleached images through a 10x objective with numerical aperture NA=0.3. Then 10 – 20 laser pulses at 20 Hz frequency were used to bleach a spot in the sample, immediately followed by recording a recovery image sequence. The recovery sequence consisted of two stages – the fast stage, during which 200 images were taken as quickly as possible (~50 ms per frame), and the slow stage, where 50 images were recorded with 1 s interval between the frames. A typical sequence of fluorescent images, normalized recovery curve and radially averaged profile of the bleached spot are shown in Figure 2. The recovery curve was fitted extremely well by a single component 2-D diffusion, Eq.2 (correlation coefficient R2 = 0.99), yielding 100 % mobile fraction and characteristic diffusion time of 3.67 s. The bleached spot profile was modeled by a Gaussian with HWHM 4.02 µm. Therefore, the diffusion rate of Rhodamine 6G in 90% glycerol was calculated using Eq. 5 as 1.10 ± 0.03 µm2/s. This value translates into diffusion coefficient of 242 µm2/s for Rhodamine 6G in water, correcting for 220 times difference between viscosity of 90 %w/w glycerol and water.29 This result is consistent with diffusion coefficient 280 ± 30 µm2/s determined for Rhodamine 6G in water.30
Figure 2.
FRAP measurements of the diffusion of Rhodamine 6G in 90 %w/w glycerol solution. (a) Fluorescence images recorded during the FRAP acquisition sequence. Time 0 s corresponds to the bleaching event. Images are cropped to 120 × 120 pixels area. (b) Fractional fluorescence recovery curve fitted with a one-component diffusion recovery equation. (c) Radially averaged profile of the bleached spot modeled with a Gaussian.
3.2. Diffusion of lipids in LCP
Free-amine group containing lipids, such as phosphatydilethanolamines, tend to co-purify with membrane proteins expressed in insect or mammalian cell lines and easily react with the succinimidyl dye derivatives during membrane proteins labeling. In most cases it is difficult to completely eliminate labeled lipids from the sample without affecting protein stability. In order to understand the factors influencing diffusion of lipids in LCP and to use these data to correct recovery curves from proteins contaminated with labeled lipids, we performed FRAP experiments on labeled lipids embedded in monoolein cubic phase. DOPE was labeled with Cy3 succinimidyl ester and separated from unreacted dye using selective extraction in chloroform. TLC analysis did not detect any free dye left in the resulted labeled lipid sample.
The fluorescence recovery curves for labeled lipids in LCP in all tested conditions were fitted with a single component diffusion equation (Eq.2). The diffusion coefficient of Cy3 labeled lipids in a regular monoolein cubic phase incubated with Bis-tris propane pH 7.0 buffer was found to be 2.4 µm2/s. Diffusion of labeled lipids was slightly slower in conditions with high PEG 400 concentrations (e.g. diffusion coefficient is 1.6 µm2/s in 0.1 M Bis tris propane pH 7, 32 %v/v PEG400, 0.1 M Na sulfate) and high salt concentrations (e.g. diffusion coefficient was 1.7 µm2/s in 1 M Na/K phosphate pH 5.6). Obtained values are in a good agreement with diffusion coefficients ~6 µm2/s measured by FRAP for single chain lipophilic probes in monoolein cubic phase at 22 °C,31,32 taking into account a strong dependence of diffusion on temperature and a factor of two difference in diffusion coefficients between single and double chain lipids in LCP.33
3.3. Diffusion of bacteriorhodopsin in LCP
Bacteriorhodopsin is a compact (Mw~27 kDa) integral membrane protein with 7 transmembrane helices. It functions as a light-driven proton pump in halobacteria, moving protons across the membrane out of the cell upon absorption of a photon by the retinal molecule covalently attached to Lys216. The proton gradient is further converted into chemical energy by ATP synthases. bR easily crystallizes in LCP composed of monoolein at protein concentrations above ~3 mg/mL using 2 – 3 M Na/K phosphate pH 5.6 as a precipitant.2
Bacteriorhodopsin absorbs light with maximum absorption at 550 nm, however it does not have strong fluoresce. Therefore, in order to perform FRAP measurements, we labeled the protein with a small fluorescent dye Cy3. Since bR does not contain cysteines we used Cy3 succinimidyl ester at pH 7.1 to preferentially label the N-terminus of the protein.
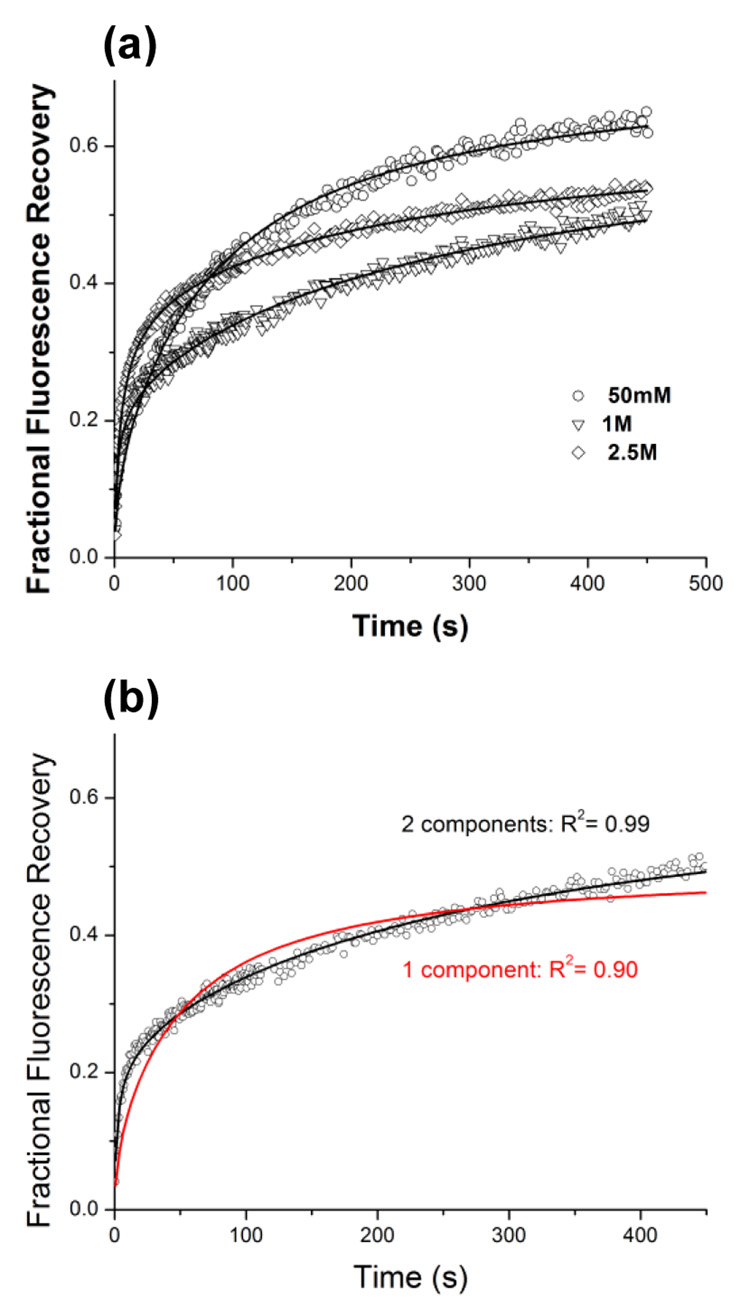
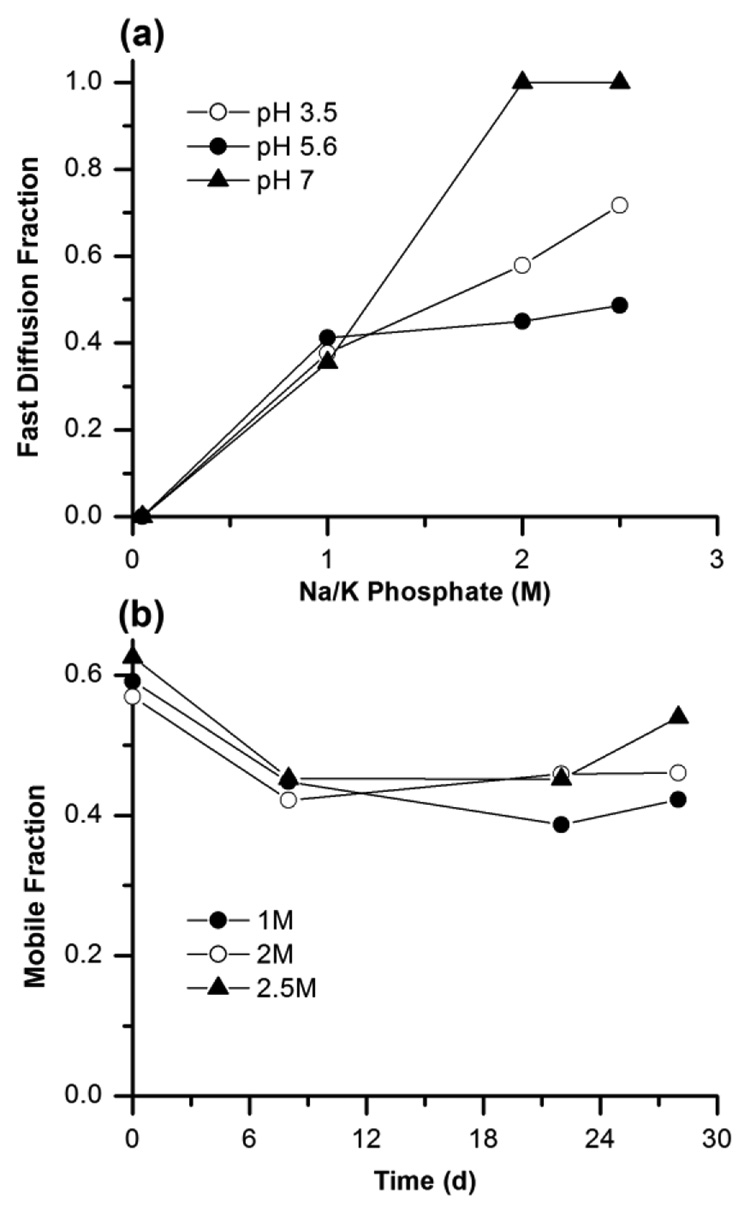
The fluorescence recovery curves for Cy3 labeled bR in LCP at different concentrations of Na/K phosphate pH 5.6 are shown in Figure 3a. At low concentration of salt (50 mM Na/K phosphate pH 5.6) diffusion of the protein is relatively slow and described well by a one-component diffusion with diffusion coefficient 6·10−2 µm2/s. When concentration of salt is increased the recovery curve could not be fitted with the one component diffusion equation, instead a two component equation (Eq.3) was necessary to describe the data (Figure 3b). This approach yielded two diffusion rates differing by at least an order of magnitude, implying the existence of two distinct oligomeric states for the bR in LCP. We interpret the fast diffusion component as bR monomers and the slow diffusion component as, trimers, in agreement with observation of monomeric and trimeric populations of bR after reconstitution of the solubilized protein in liposomes.31 The fraction of the fast diffusing population increases with salt concentration (Figure 4a). At high salt concentration (2.5 M Na/K phosphate pH 7) diffusion of the protein is fast and described well by the one-component diffusion with diffusion coefficient ~0.8 µm2/s and mobile fraction 60 %.
Figure 3.
(a) Fluorescence recovery curves for bacteriorhodopsin in LCP at different concentrations of Na/K phosphate pH 5.6. Recovery curve at 50 mM salt is fitted by a one-component equation (Eq.2), while data at both 1 and 2.5 M salt required using two-component equation (Eq. 3). (b) Example of one-component versus two-component curve fitting for bR in at 1 M Na/K phosphate pH 5.6
Figure 4.
(a) Dependence of the fraction of fast diffusing bR molecules on salt concentration at different pH. (b) Change in the total mobile fraction of bR in LCP over time at different concentrations of Na/K phosphate pH 5.6.
We followed diffusion properties of bR in LCP at different salt concentrations for period of a month (Figure 4b). After 7 days all the recovery curves can be fitted by fast one-component diffusion, indicating that all bR has monomerized in LCP with time. The total mobile fraction of the protein did not appreciably change with time suggesting high stability of bR in LCP in agreement with previous observations that bR crystals can continue to grow in LCP for months.12
3.4. Comparison between β2AR(E122W) and β2AR(E122W)-T4L
Beta2-adrenergic receptor is a member of G protein-coupled receptor (GPCR) family. These receptors reside primarily in the smooth muscles throughout the body, mostly in lungs and heart, and respond to the catecholamine hormones adrenaline and noradrenaline. In order to crystallize β2AR we have modified it by introducing a stabilizing mutation E122W,25 fusing T4 lysozyme between the fifth and sixth transmembrane helices4,5 and truncating the C-terminus at residue 348.25 Here we compare diffusion of β2AR(E122W) and β2AR(E122W)-T4L bound to an inverse agonist carazolol in LCP at different precipitant conditions.
Receptors were expressed in the insect Sf9 cells, membranes of which contain ~40% of phosphatidylethanolamine.32 To avoid labeling of lipids by succinimidyl ester we tried a thiol-reactive derivative of the dye (Cy3 maleimide). Both of the β2AR constructs contain dye accessible cysteine residues (Cys264), and both were labeled successfully. However β2AR(E122W)-T4L has largely aggregated upon labeling. A possible explanation is that Cys264 resides in close proximity to the interface between the receptor and T4 lysozyme, and attachment of Cy3 to this residue may affect the conformation and stability of the chimeric receptor. Therefore β2AR(E122W)-T4L was labeled with Cy3 succinimidyl ester at pH 7.1 and the free unreacted dye and most of the labeled lipids were removed by extensive washing. TLC analysis detected presence of some labeled lipids in the sample.
Fluorescence recovery curves for both variants of β2AR in LCP incubated with 100 mM Hepes pH 7.0, are shown in Figure 5a. In contrast to bR, no diffusion was detected for β2AR(E122W) construct. Increasing salt concentration did not change this behavior and β2AR(E122W) remained stationary in LCP (data not shown). For β2AR(E122W)-T4L reconstituted in LCP we observed very fast one-component fluorescence recovery with mobile fraction ~20 % and characteristic time 2 s. We attributed this recovery to the diffusion of labeled lipids, since we have determined that the sample did contain labeled lipids and the observed diffusion coefficient corresponds to that of labeled lipids alone in LCP. These data indicate that β2AR(E122W)-T4L also does not diffuse after reconstitution in LCP. Since β2AR(E122W)-T4L can be crystallized in LCP we performed FRAP measurements at variety conditions including ingredients from the successful crystallization screens in different combinations.
Figure 5.
(a) Fluorescence recovery curves recorded for β2AR(E122W) and β2AR(E122W)-T4L in LCP incubated with Bis tris propane pH 7.0 and with the crystallization conditions for β2AR(E122W)-T4L (0.1 M Bis tris propane pH 7.0, 25 %v/v PEG 400, 0.1 M Na sulfate, 5 %v/v 1,4-butanediol). (b) Two component fitting of the fluorescence recovery curve obtained for β2AR(E122W)-T4L in 0.1 M Bis tris propane pH 7.0, 15 %v/v PEG 400, 0.1 M Na sulfate, 5 %v/v 1,4-butanediol conditions. The fast component describes diffusion of labeled lipids and the slow component – diffusion of protein.
Successful crystallization conditions of β2AR(E122W)-T4L comprise 0.1 M Bis-tris propane pH 6.5 – 8.0, 25 – 35 %v/v PEG 400, 0.05 – 0.2 M Na sulfate and 2 – 5 %v/v 1,4-butanediol. Fluorescence recovery curves for β2AR(E122W)-T4L in LCP typically contained two components (Figure 5b). The fast component describes lipid motion and the slow component – protein diffusion. The curves were fitted by the two-component diffusion equation (Eq. 3). In order to increase the robustness and reliability of the fit, we fixed the characteristic diffusion time and the mobile fraction of the fast component using experimentally determined diffusion coefficients for lipids measured at the same conditions (see section 2.2. Diffusion of lipids in LCP) and mobile fraction of 20 % obtained from the measurements in Bis tris propane buffer pH 7.0, in which we found that only lipids but not proteins were moving.
To determine the effects of the individual ingridients from crystallization conditions on diffusion of β2AR(E122W)-T4L we started with 0.1 M Bis tris propane pH 7.0, 25 %v/v PEG 400, 0.1 M Na sulfate, 5 %v/v 1,4-butanediol and systematically varied concentrations of the main precipitant, salt, additive and pH of the buffer. The results are described in the following subsections. With β2AR(E122W) only very slow and limited diffusion with fluorescence recovery less than 10% after 600 s was observed at these conditions (Figure 5a).
3.4.1. Effect of PEG 400
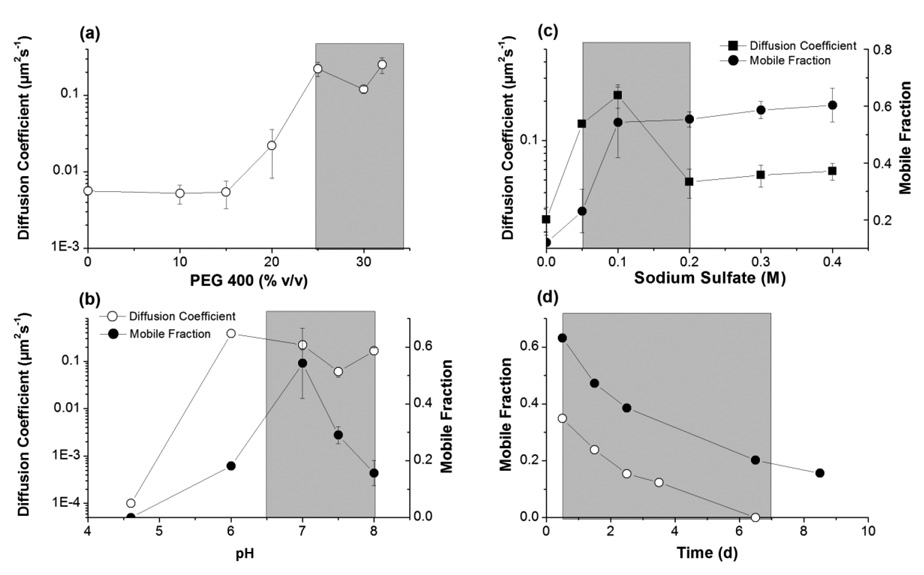
Addition of PEG 400 is known to swell the lipidic cubic phase.36 Typically the lattice parameter of the cubic Pn3m phase increases from 103 Å at 0 %v/v PEG 400 to 120 Å at 30 %v/v PEG 400. We observed that diffusion of β2AR(E122W)-T4L at low PEG 400 concentrations (below 20 %v/v) is extremely slow with diffusion coefficient ~3·10−3 µm2/s (Figure 6a). At PEG 400 concentration ~25 %v/v there is a sharp transition in protein mobility, at which the diffusion coefficient increases to 0.1 – 0.2 µm2/s. A possible explanation for this transition is that the diameter of the water channels in the regular lipidic cubic phase significantly restricts mobility of the receptor with attached T4 lysozyme. Swelling of the LCP with PEG 400 releases these constraints and allows the protein to move more freely. Notably, this transition in the protein mobility coincides with the PEG 400 concentration boundary of β2AR(E122W)-T4L crystallization (Figure 6a).
Figure 6.
Trends for diffusion coefficient and mobile fraction of β2AR(E122W)-T4L obtained by systematically varying concentration of PEG 400 (a), Na sulfate (b) and pH (c), starting from 0.1 M Bis tris propane pH 7.0, 25 %v/v PEG 400, 0.1 M Na sulfate, 5 %v/v 1,4-butanediol. (d) Time-course dependence of the β2AR(E122W)-T4L mobile fraction in 0.1 M Bis tris propane pH 7.0, 25 %v/v PEG 400, 0.2 M Na sulfate, 5 %v/v 1,4-butanediol (close circles) and in 0.1 Bis tris propane pH 8.0, 25 %v/v PEG 400, 0.1 M Na sulfate, 5 %v/v 1,4-butanediol (open circles). The shaded areas corresponds to concentrations of PEG 400 (a), Na sulfate (b) and pH (c) supporting β2AR(E122W)-T4L crystallization and to the time frame (d), during which the crystals continue to grow. No diffusion was detected in (c) at pH 4.6 and the diffusion coefficient was assigned an arbitrary low value of 10−4 µm2/s to represent a point on the log scale. All measurements were made at least in duplicates. The error bars are shown for data points measured for three or more times.
3.4.2. Effect of salt
Salts typically shrink the lipidic cubic phase in concentration dependant manner.14,15,37 However, the effect of low concentrations of salt (less than 0.1 – 0.2 M) on the lattice parameter of LCP is minimal. We found a bell shaped dependence of the protein diffusion coefficient on the Na sulfate concentration (Figure 6b). At 0 M Na sulfate diffusion of the protein was very slow (diffusion coefficient ~2·10−2 µm2/s). With increasing salt concentration the diffusion coefficient increased strongly reaching its maximum of 0.2 µm2/s at 0.1 M Na sulfate. Further increases in salt concentration resulted in a slow decrease in the diffusion coefficient.
Since we did not expect significant changes in the LCP structure between salt concentrations of 0 and 0.1 M, the most plausible explanation was that salt screens the charges and/or specifically bind to the protein surface, thereby changing the interactions between protein molecules, preventing their non-specific aggregation. Further increase in salt concentration may slightly decrease the lattice parameter of LCP, slowing down the protein diffusion rate. The best crystallization condition contained 0.1 M Na sulfate, corresponding to the fastest receptor mobility in LCP.
3.4.3. Effect of butanediol
1,4-butanediol swells the lipidic cubic phase at high concentration.18 Addition of 5 %v/v 1,4-butanediol does not appreciably change the lattice parameter of LCP. However we observed dramatic effect of low concentration of butanediol on the mobile fraction of the receptor. The mobile fraction increased from 10 % at 0 %v/v to 60 % at 5 %v/v of 1,4-butanediol, suggesting that butanediol stabilizes β2AR(E122W)-T4L and prevents its aggregation in LCP. 1,4-butanediol was not an essential component for crystallization, but was found to be one of the best additives significantly increasing the size of the crystals. Diffusion experiments, therefore, provide a possible explanation of the effect of this additive.
3.4.4. Effect of cholesterol
Addition of 10 %w/w cholesterol to monoolein has also substantially improved the quality of β2AR(E122W)-T4L crystals.4,25 Cholesterol is known for its ability to condense38 and rigidify39 lipid bilayers. The effect of 10% cholesterol on the lattice parameter of the fully hydrated monoolein cubic Pn3m phase is negligible.40 Nevertheless, cholesterol may modulate the response of the cubic phase lattice to addition of different ingredients from the crystallization cocktails. Diffusion of β2AR(E122W)-T4L in cholesterol-doped LCP at crystallization conditions was on average ~2 times slower in agreement with the rigidifying effect of cholesterol. The general trends of varying other ingredients on the diffusion properties remained unchanged in the presence of cholesterol, indicating that some specific effects of cholesterol on the protein rather than its effects on the lipid bilayer properties and protein diffusion are responsible for the improved crystal growth.
3.4.5. Effect of pH
Structural parameters of the lipidic Pn3m cubic phase are not affected by pH in the range 4 – 8.5.37 Accordingly, we did not find any significant differences in the diffusion coefficient of β2AR(E122W)-T4L in the pH range between 6 and 8 (Figure 6c). The mobile fraction, however, has a distinct maximum at pH 7.0 suggesting higher stability of the protein at neutral pH. The protein did not show any motion at pH 4.6, consistent with low stability and fast aggregation of the receptor in LCP at this pH.
3.4.5. Time-course of diffusion
Diffusion of β2AR(E122W)-T4L in LCP at crystallization conditions was monitored over a time course of several days (Figure 6d). The mobile fraction had gradually decreased from 62 % in the first day to less than 20 % after 6 days in 0.1 M Bis tris propane pH 7.0, 25 %v/v PEG 400, 0.2 M Na sulfate, 5 %v/v 1,4-butanediol, and from 36 % down to 0 % after 6 days in 0.1 M Tris pH 8.0, 25 %v/v PEG 400, 0.1 M Na sulfate, 5 %v/v 1,4-butanediol. This decay likely reflects relative instability of the receptor in LCP. Over time protein irreversibly aggregates losing its mobility. Remarkably, the characteristic decay time of the receptor mobility determined by FRAP coincides with the timeframe of crystallization. β2AR(E122W)-T4L crystals typically nucleate within 12–24 hrs and stop growing after 5–7 days.
4. Discussion
Crystal nucleation and growth require 3-dimensional protein diffusion. While diffusion in aqueous solutions is implicitly assumed, diffusion of protein in lipidic mesophases is not as trivial due to the microstructure of lipidic cubic phase which restricts diffusion of large proteins and protein aggregates. We have used FRAP to measure the mobility of bR and two constructs of β2AR (with and without fused T4 lysozyme) in lipidic cubic phase equilibrated with different screening solutions in an attempt to deconvolute the factors in known crystallization conditions.
We observed that bR diffuses well in LCP in all tested conditions. Depending on the condition, however, diffusion of bR can be described as one or two components, reflecting two distinct oligomeric states, likely monomers and trimers. At crystallization conditions, in high concentration of Na/K phosphate, bR moves in LCP predominately as a monomer with diffusion rate of 0.8 µm2/s.
To our surprise, a stabilized mutant of beta2-adrenergic receptor, β2AR(E122W), remained completely motionless in LCP. This receptor has similar architecture of 7 transmembrane helices as bR and molecular weight of 40 kDa and therefore should not be significantly restricted in mobility. Hence, the most plausible conclusion is that β2AR(E122W) quickly associates in LCP forming micro-aggregate complexes that are unable to diffuse. These aggregates, however, are too small to be visualized using light or fluorescent microscopy (the samples appear uniformly fluorescent under a microscope), and therefore FRAP measurements provide important feedback of events occurring with proteins embedded in LCP. A logical imperative in such cases when diffusion is completely lacking is that the protein will not crystallize.
We have previously crystallized beta2-adrenergic receptor fused with T4 lysozyme, β2AR(E122W)-T4L, in LCP.25 In the current study we determined how different ingredients from crystallization cocktail affect diffusion of β2AR(E122W)-T4L. We observed that crystallization conditions correlate with increased mobile fraction and faster diffusion of the studied proteins. Therefore fusion of T4 lysozyme to β2AR not only helps to establish crystal packing interactions, but also prevents non-specific aggregation and facilitates diffusion in LCP. Many screened conditions, however, resulted in complete suppression of the protein motion. Based on these results we believe that diffusion measurements by FRAP in representative precipitant conditions can serve as an excellent tool to pre-screen different membrane proteins before setting up crystallization trials in LCP.
The time dependence of the protein mobile fraction measured by FRAP provides an indication of protein stability in LCP. More stable proteins such as bacteriorhodopsin remains monomeric and diffuse well in LCP for a period of months, while the mobile fraction of less stable proteins such as β2AR(E122W)-T4L decays to almost zero within a week. The experimental data suggest that micro-aggregation of membrane proteins in LCP can be both reversible and irreversible. For example, immediately after embedding β2AR(E122W)-T4L in LCP, the protein does not diffuse. Mobility can be rescued by addition of salt, PEG400 and 1,4-butanediol. However, decrease in protein diffusion over time is likely observed due to irreversible micro-aggregation. There are several possible reasons for membrane protein instability and resulting aggregation in LCP, including lack of native lipids, bilayer curvature stress, lack of certain ions, or lack of other chaperon-like proteins. Effects of these and other factors will be investigated in future work employing the FRAP diffusion assay.
The LCP-FRAP pre-screening assay has number of attractive features. In contrast to crystallization experiments, most of which end up with a clear drop, FRAP measurements are quantitative, providing diffusion rates and mobile fractions, which can be used to guide and design subsequent crystallization experiments. Protein diffusion is not as sensitive as crystallization to the concentration of components. Therefore fewer conditions can be screened in a FRAP-based pre-crystallization assay while still obtaining a good indication of a protein’s behavior in LCP. The FRAP measurements described here requires microgram quantities of purified protein. About 10 µL of 1–5 mg/mL labeled protein (10–50 g) is sufficient to make ~200 samples. Samples can be set up using an in meso crystallization robot.12 We are currently adapting the assay for automation in a 96-well format. Automation will be an essential improvement as the measurement time required per sample can be quite extensive.
Finally, we would like to stress that diffusion is required, but not sufficient for crystallization. One needs to satisfy more stringent conditions to drive specific interactions between protein molecules to form crystal packing interactions. Therefore, the utility of the LCP-FRAP assay is to pre-screen different protein constructs in a variety of different classes of precipitant solutions and different host lipids, to efficiently eliminate those constructs, precipitant classes or host lipids which do not support protein diffusion in LCP from subsequent crystallization trials, thereby increasing the likelihood of finding the right crystallization conditions.
Acknowledgement
This work was supported in part by the NIH Roadmap Initiative grant P50 GM073197.
References
- 1.Abola EE, Stevens RC. The importance of target selection strategies in structural. In: Sussman JL, Silman I, editors. Structural Proteomics and its Impact on the Life Sciences. World Scientific Publishing Company; 2008. [Google Scholar]
- 2.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. U S A. 1996;93(25):14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landau EM, Pebay-Peyroula E, Neutze R. Structural and mechanistic insight from high resolution structures of archaeal rhodopsins. FEBS Lett. 2003;555(1):51–56. doi: 10.1016/s0014-5793(03)01082-2. [DOI] [PubMed] [Google Scholar]
- 4.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318(5854):1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 6.Raman P, Cherezov V, Caffrey M. The Membrane Protein Data Bank. Cell. Mol. Life Sci. 2006;63(1):36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caffrey M. Membrane protein crystallization. J. Struct. Biol. 2003;142(1):108–132. doi: 10.1016/s1047-8477(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 8.Deisenhofer J, Michel H. Nobel lecture. The photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis. Embo J. 1989;8(8):2149–2170. doi: 10.1002/j.1460-2075.1989.tb08338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherezov V, Caffrey M. A simple and inexpensive nanoliter-volume dispenser for highly viscous materials used in membrane protein crystallization. J. Appl. Cryst. 2005;38(2):398–400. [Google Scholar]
- 10.Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chem. Phys. Lipids. 1998;95(1):11–21. doi: 10.1016/s0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 11.Cherezov V, Caffrey M. Nano-volume plates with excellent optical properties for fast, inexpensive crystallization screening of membrane proteins. J. Appl. Cryst. 2003;36:1372–1377. [Google Scholar]
- 12.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr. D Biol. Crystallogr. 2004;60(Pt 10):1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 13.Cherezov V, Caffrey M. Picolitre-scale crystallization of membrane proteins. J. Appl. Cryst. 2006;39(4):604–606. [Google Scholar]
- 14.Nollert P, Qiu H, Caffrey M, Rosenbusch JP, Landau EM. Molecular mechanism for the crystallization of bacteriorhodopsin in lipidic cubic phases. FEBS Lett. 2001;504(3):179–186. doi: 10.1016/s0014-5793(01)02747-8. [DOI] [PubMed] [Google Scholar]
- 15.Efremov R, Shiryaeva G, Bueldt G, Islamov A, Kuklin A, Yaguzhinsky L, Fragneto-Cusani G, Gordeliy V. SANS investigations of the lipidic cubic phase behavior in course of bacteriorhodopsin crystallization. J. Cryst. Growth. 2005;275(1–2):e1453–e1459. [Google Scholar]
- 16.Cherezov V, Caffrey M. Membrane protein crystallization in lipidic mesophases. A mechanism study using X-ray microdiffraction. Faraday Discuss. 2007;136:195–212. doi: 10.1039/b618173b. [DOI] [PubMed] [Google Scholar]
- 17.Qiu H, Caffrey M. The phase diagram of the monoolein/water system: metastability and equilibrium aspects. Biomaterials. 2000;21(3):223–234. doi: 10.1016/s0142-9612(99)00126-x. [DOI] [PubMed] [Google Scholar]
- 18.Cherezov V, Clogston J, Papiz MZ, Caffrey M. Room to move: crystallizing membrane proteins in swollen lipidic mesophases. J. Mol. Biol. 2006;357(5):1605–1618. doi: 10.1016/j.jmb.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 19.Katona G, Andreasson U, Landau EM, Andreasson LE, Neutze R. Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.35A resolution. J. Mol. Biol. 2003;331(3):681–692. doi: 10.1016/s0022-2836(03)00751-4. [DOI] [PubMed] [Google Scholar]
- 20.Cherezov V, Yamashita E, Liu W, Zhalnina M, Cramer WA, Caffrey M. In meso structure of the cobalamin transporter, BtuB, at 1.95 A resolution. J. Mol. Biol. 2006;364(4):716–734. doi: 10.1016/j.jmb.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wohri AB, Johansson LC, Wadsten-Hindrichsen P, Wahlgren WY, Fischer G, Horsefield R, Katona G, Nyblom M, Oberg F, Young G, Cogdell RJ, Fraser NJ, Engstrom S, Neutze R. A lipidic-sponge phase screen for membrane protein crystallization. Structure. 2008;16(7):1003–1009. doi: 10.1016/j.str.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Oesterhelt D, Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31(Pt A):667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 23.Dencher NA, Heyn MP. Preparation and properties of monomeric bacteriorhodopsin. Methods Enzymol. 1982;88(Biomembranes Pt I):5–10. [Google Scholar]
- 24.Gordeliy VI, Schlesinger R, Efremov R, Buldt G, Heberle J. Crystallization in lipidic cubic phases: a case study with bacteriorhodopsin. Methods Mol. Biol. 2003;228:305–316. doi: 10.1385/1-59259-400-X:305. [DOI] [PubMed] [Google Scholar]
- 25.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A Specific Cholesterol Binding Site is Established by the 2.8 A Structure of the Human beta(2)-Adrenergic Receptor. Structure. 2008;16(6):897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renner C, Kessler B, Oesterhelt D. Lipid composition of integral purple membrane by 1H and 31P NMR. J. Lipid Res. 2005;46(8):1755–1764. doi: 10.1194/jlr.M500138-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Soumpasis DM. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 1983;41(1):95–97. doi: 10.1016/S0006-3495(83)84410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pucadyil TJ, Chattopadhyay A. Confocal fluorescence recovery after photobleaching of green fluorescent protein in solution. J. Fluoresc. 2006;16(1):87–94. doi: 10.1007/s10895-005-0019-y. [DOI] [PubMed] [Google Scholar]
- 29.Handbook of Chemistry and Physics. 54th edition. Cleveland: CRC Press; 1973. ed. [Google Scholar]
- 30.Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13(1):29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 31.Cribier S, Gulik A, Fellmann P, Vargas R, Devaux PF, Luzzati V. Cubic phases of lipid containing systems. A translational diffusion study by fluorescence recovery after photobleaching. J. Mol. Biol. 1993;229(2):517–525. doi: 10.1006/jmbi.1993.1051. [DOI] [PubMed] [Google Scholar]
- 32.Tsapis N, Reiss-Husson F, Ober R, Genest M, Hodges RS, Urbach W. Self diffusion and spectral modifications of a membrane protein, the Rubrivivax gelatinosus LH2 complex, incorporated into a monoolein cubic phase. Biophys. J. 2001;81(3):1613–1623. doi: 10.1016/S0006-3495(01)75815-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eriksson PO, Lindblom G. Lipid and water diffusion in bicontinuous cubic phases measured by NMR. Biophys J. 1993;64(1):129–136. doi: 10.1016/S0006-3495(93)81347-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gulik-Krzywicki T, Seigneuret M, Rigaud JL. Monomer-oligomer equilibrium of bacteriorhodopsin in reconstituted proteoliposomes. A freeze-fracture electron microscope study. J. Biol. Chem. 1987;262(32):15580–15588. [PubMed] [Google Scholar]
- 35.Marheineke K, Grunewald S, Christie W, Reilander H. Lipid composition of Spodoptera frugiperda (Sf9) and Trichoplusia ni (Tn) insect cells used for baculovirus infection. FEBS Lett. 1998;441(1):49–52. doi: 10.1016/s0014-5793(98)01523-3. [DOI] [PubMed] [Google Scholar]
- 36.Cherezov V, Liu W, Derrick JP, Luan B, Aksimentiev A, Katritch V, Caffrey M. In meso crystal structure and docking simulations suggest an alternative proteoglycan binding site in the OpcA outer membrane adhesin. Proteins. 2008;71(1):24–34. doi: 10.1002/prot.21841. [DOI] [PubMed] [Google Scholar]
- 37.Cherezov V, Fersi H, Caffrey M. Crystallization screens: compatibility with the lipidic cubic phase for in meso crystallization of membrane proteins. Biophys. J. 2001;81(1):225–242. doi: 10.1016/S0006-3495(01)75694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hung WC, Lee MT, Chen FY, Huang HW. The condensing effect of cholesterol in lipid bilayers. Biophys. J. 2007;92(11):3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrache HI, Harries D, Parsegian VA. Alteration of lipid membrane rigidity by cholesterol and its metabolic precursors. Macromol. Symp. 2005;219:39–50. [Google Scholar]
- 40.Cherezov V, Clogston J, Misquitta Y, Abdel-Gawad W, Caffrey M. Membrane protein crystallization in meso: lipid type-tailoring of the cubic phase. Biophys J. 2002;83(6):3393–3407. doi: 10.1016/S0006-3495(02)75339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]