Abstract
Bacterial initiation factor 3 (IF3) is organized into N-domain and C-domains separated by a linker. Mitochondrial IF3 (IF3mt) has a similar domain organization although both domains have extensions not found in the bacterial factors. Constructs of the N- and C-domains of IF3mt with and without the connecting linker were prepared. The Kd’s for the binding of full length IF3mt and its C-domain with and without the linker to mitochondrial 28S subunits are 30, 60 and 95 nM respectively, indicating that much of the ribosome binding interactions are mediated by the C-domain. However, the N-domain binds to 28S subunits with only a 10-fold lower affinity than full length IF3mt. This observation indicates that the N-domain of IF3mt has significant contacts with the protein-rich small subunit of mammalian mitochondrial ribosomes. The linker also plays a role in modulating the interactions between the 28S subunit and the factor; it is not just a physical connector between the two domains. The presence of the two domains and the linker may optimize the overall affinity of IF3mt for the ribosome. These results are in sharp contrast to observations with E. coli IF3. Removal of the N-domain drastically reduces the activity of IF3mt in the dissociation of mitochondrial 55S ribosomes although the C-domain itself retains some activity. This residual activity depends significantly on the linker region. The N-domain alone has no effect on the dissociation of ribosomes. Full-length IF3mt reduces the binding of fMet-tRNA to the 28S subunit in the absence of mRNA. Both the C-terminal extension and the linker are required for this effect. IF3mt promotes the formation of a binary complex between IF2mt and fMet-tRNA that may play an important role in mitochondrial protein synthesis. Both domains play a role promoting the formation of this complex
Keywords: initiation complex, initiation factor 3, mitochondria, protein synthesis, ribosome
Introduction
Mitochondria have their own genomes which in mammals contains only 16,000 base pairs of DNA. This DNA encodes thirteen proteins which are synthesized within the organelle. All of these proteins are inserted into the inner membrane and are involved in oxidative phosphorylation. The synthesis and assembly of these polypeptides in the inner membrane is not well understood.
The mammalian mitochondrial translation system has a number of interesting features. These mitochondrial ribosomes (55S) have a higher protein content than bacterial ribosomes, while their rRNAs are considerably truncated. The 55S particle consists of a 28S small subunit and a 39S large subunit 1. Cryo-electron microscopy indicates that the core of the mitochondrial ribosome is decorated with many proteins that have no bacterial homologs 2. Therefore, the interactions of these ribosomes with mRNA, tRNA and translational factors are expected to show interesting differences compared to those occurring in bacterial systems.
Two initiation factors have been found in mammalian mitochondria. Initiation factor 2 (IF2mt) promotes the binding of fMet-tRNA to the ribosome. Recently Gaur et al. 3 provided evidence that mammalian IF2mt serves the functions not only of IF2 but also that of IF1 in mitochondria. Human IF3mt has been cloned and expressed in E. coli. It has both similarities and differences with its bacterial counterparts.
The activities of E. coli IF3 are well established 4,5. This factor plays several roles in initiation complex formation including functioning as an anti-association (or dissociating) agent for the ribosomal subunits, enhancing codon-anticodon interactions at the P-site and increasing the rate of dissociation of noncanonical 30S initiation complexes 6–10. Recent kinetic studies have shown that the joining of the 50S subunit to 30S initiation complexes and the dissociation of IF3 from the small subunit are strongly regulated by the codon used for initiation 11. This result is supported by earlier investigations 12.
E. coli IF3 consists of two domains (N-domain and C-domain) joined by a flexible linker (Figure 1). The isolated C-domain of E. coli IF3 binds to 30S subunits but with a 100-fold lower affinity than full length IF3 and carries out most of the functions of IF3 9. The isolated N-domain does not bind detectably to 30S subunits and has no known function 9 although it clearly enhances the interaction of the C-domain with the small subunit. Recent studies indicate that the N-domain may modulate the association and dissociation of IF3 from the 30S subunit through a fluctuating interaction with the neck region of the subunit 13. It has also been reported that the N-terminal domain may provide a proof reading function since mutation of Tyr75 or iodination of Tyr70 in the N-domain results in a factor that could not dissociate incorrect initiation complexes 14,15.
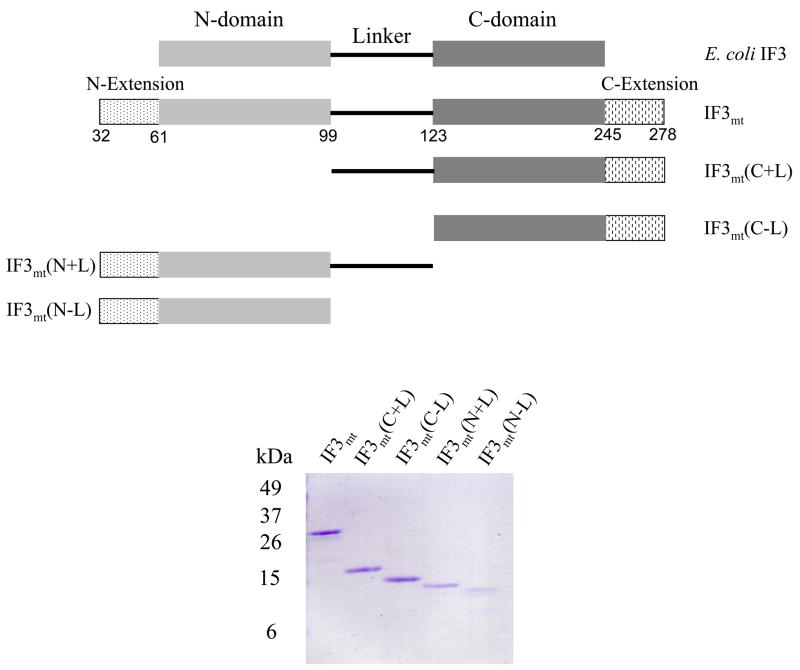
Figure 1. Domain organization of bacterial IF3 and human IF3mt and its truncated C- and N-domains with and without the linker.
(A) Pictorial representation of the domain structure of E. coli IF3 and human IF3mt and the four truncated derivatives used in this study. IF3mt and its C-terminal domain with the linker IF3mt(C+L) and without the linker IF3mt(C−L) and the N-terminal domain with the linker IF3mt(N+L) and without the linker IF3mt(N−L) region are shown. The numbers indicate the amino acid residues in IF3mt with the first residue of the signal sequence being designated as residue 1. The import signal was removed in the preparation of the full-length factor and the various constructs. (B) SDS-PAGE analysis of the IF3mt derivatives prepared following expression in E. coli. The amounts of the proteins used were IF3mt, 0.7 μg; IF3mt(C+L), 0.45 μg; IF3mt(C−L) 0.42 μg; IF3mt(N+L), 0.39 μg; and IF3mt(N−L), 0.23 μg. The gel was stained with Commassie Blue.
IF3mt has a central region with homology to the bacterial factors with additional N-terminal and C-terminal extensions (Figure 1A). The roles of IF3mt and its terminal extensions in initiation complex formation have been investigated 16–18. These studies suggest that the C-terminal extension reduces the affinity of the factor for 39S subunits, thus promoting proper subunit joining. In the present study we have examined the properties of the isolated N- and C-domains of IF3mt carrying the extensions on mitochondrial protein synthesis.
Results
Interactions of isolated N- and C-domains of IF3mt with small ribosomal subunits and the role of the linker region in this interaction
IF3mt has ~20–25 identity to bacterial IF3. Unlike bacterial IF3, mammalian IF3mt contains N-terminal and C-terminal extensions surrounding the homologous N- and C-domains (Figure 1A). To explore the role of the extended N- and C-domains and the linker of IF3mt in binding to 28S subunits and initiation complex formation, four derivatives were prepared (Figure 1A). Two His6-tagged derivatives were generated for the N-domain, one with the linker, IF3mt(N+L) and the other without the linker, IF3mt(N−L). Similarly two derivatives of the C-domain were generated, one with the linker, IF3mt(C+L) and the other without the linker, IF3mt(C−L) (Figure 1B).
The interaction of purified IF3mt(N+L), IF3mt(N−L), IF3mt(C+L), and IF3mt(C−L) with 28S subunits was determined by Microcon-100 centrifugation followed by Western blotting using antibodies against IF3mt 18. For quantification, calibration curves were generated for each domain because the antigenic responses of the various domains were quite different (Fig. 2 and Supplementary Figure S1). Full length IF3mt gives a strong antigenic response with less than 0.2 pmol of factor being readily detectable. Good responses are also observed with IF3mt(C+L) and IF3mt(N+L) using about 2.5-fold more factor. However, removal of the linker in the IF3mt(N−L) and IF3mt(C−L) derivatives reduces the antigenicity of the factor and 6-fold more protein is required for easy detection (Figure 2A). These observations suggest that the linker region contains major antigenic determinants.
Figure 2. Detection of IF3mt and its derivatives using antibodies in the presence and absence of 28S subunits.
The accessibility of IF3mt and its N- and C-domains to antibody in the presence of 28S subunits was studied using the dot blot method. (A) Representative dot blots are shown: left panel corresponds to protein alone and right panel corresponds to 28S+IF3mt. Reaction mixtures (100 μL) contained where indicated 5 pmol of 28S subunits and IF3mt (0.16, 0.25 and 0.41 pmol), IF3mt(C+L) (0.8, 1.64 and 2.5 pmol), IF3mt(C−L) (2.2, 4.4 and 6.6 pmol), IF3mt(N+L) (0.82, 1.7 and 2.5 pmol), and IF3mt(N−L) (2, 4 and 6 pmol). (B) Plot of the reduction in the antigenic response of the various IF3mt derivatives in the presence of 28S subunits. Standard curves were generated at different concentrations of protein in the absence of 28S subunits and fitted with an exponential raise equation [y=a(1− e−bx)] where y is the number of pixels, x is the concentration of IF3mt and 100% exposure to antibody is assumed in the linear part of the curve. In the presence of 28S subunits, plots of pixel intensity (28S+IF3mt) as a function of protein concentration are linear at lower concentrations of protein. Pixel values were determined at low concentrations of protein to maximize the amount of the protein bound to 28S subunits (IF3mt:28S ratios of 1:10 or less). The relative exposure of the epitopes of IF3mt and its derivatives to the antibody were calculated from the ratio of the pixels of IF3mt obtained in the presence of 28S subunits divided by the pixels observed in the presence of protein alone.
The binding of IF3mt or its derivatives to the 28S subunit leads to a strong quenching (about 80 %) of the antigenic response indicating that a number of the epitopes on the factor are hidden by interactions with the ribosome (Figure 2A and B). This quenching can be overcome by treatment of the samples with EDTA which damages the structure of the small subunit releasing the factor. The antigenicity of the C-domain with or without the linker is also reduced by binding to the small subunit although to a lower extent than observed with the full length factor. The epitopes in the N-domain with or without the linker are almost entirely obscured by binding to the 28S subunit indicating that this region of IF3mt makes significant contacts with the small subunit. Both the linker and the N-domain appear to be sequestered from the antibodies when this region of IF3mt is bound to the small subunit. This observation is surprising since the linker is quite exposed and can be cleaved by trypsin when E. coli IF3 is bound to the 30S subunit 19. The epitopes on all of these derivatives are exposed upon EDTA treatment of the samples allowing the antigenic detection of the factors in the experiments to quantitate their binding to the small subunit. Individual calibration curves were generated for each derivative in the presence of EDTA permitting accurate quantitation of the amount of factor bound to the small subunit.
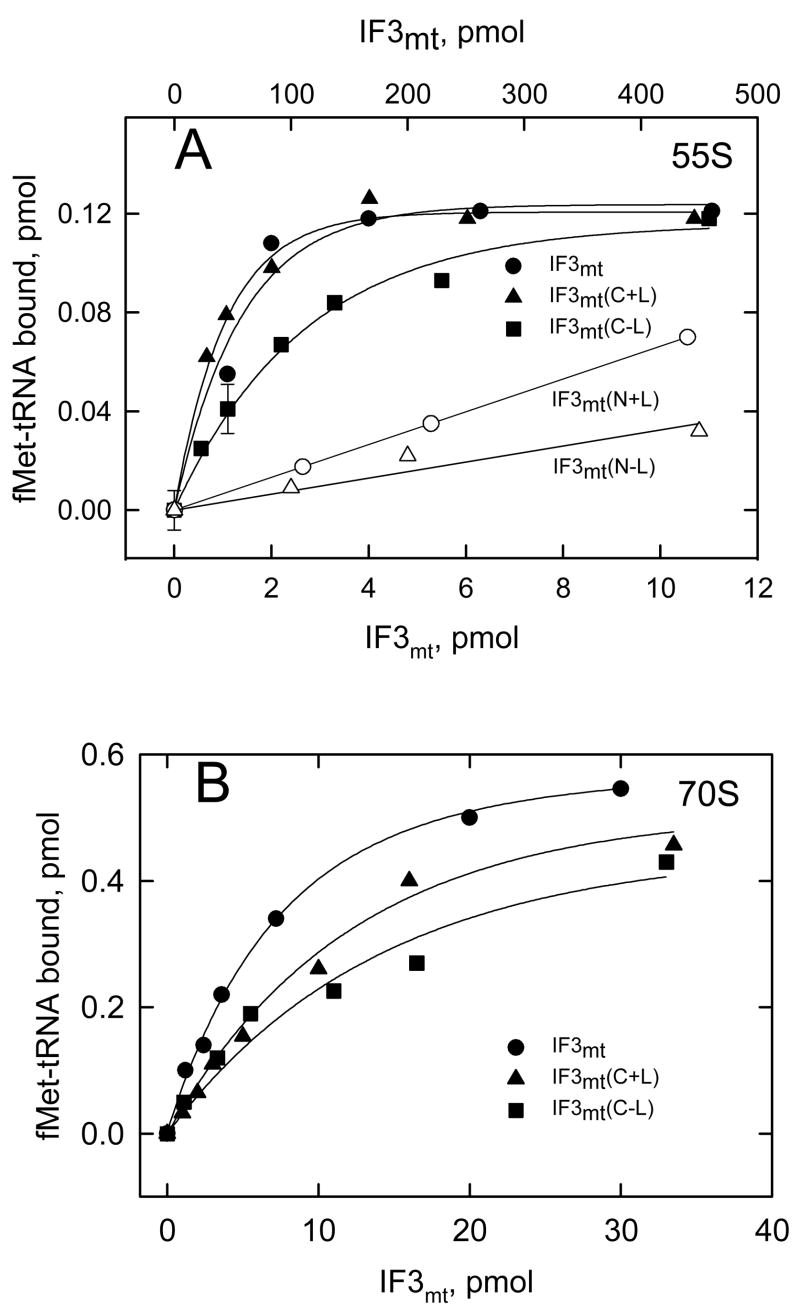
For determination of the apparent equilibrium dissociation constants, different amounts of the IF3mt domain derivatives were incubated with a fixed amount of 28S subunits, and the amounts of the derivatives bound to 28S subunits were calculated (Figure 3) permitting the documentation of the Kd values governing the interactions (Table I). The Kd values are 30, 60 and 95 nM for full length IF3mt, IF3mt(C+L), and IF3mt(C−L), respectively. Unlike the C-domain of E. coli IF3, the isolated C-domain of IF3mt with and without the linker binds to the small subunit strongly having only a 2- and 3-fold lower affinity than full length IF3mt for the 28S subunit.
Figure 3. Interaction of the C- and N-domains of IF3mt with small ribosomal subunits (28S).
(A) Amount of IF3mt(C+L) and IF3mt(C−L) bound to 28S subunits as a function of added protein. The amount of IF3mt or its derivatives bound to 28S subunits was determined by Microcon centrifugation followed by immunological detection using the standard curves for IF3mt(C+L) and IF3mt(C−L) (Supplement Figure S1). The apparent Kd values were estimated from the linear range of the calibration curves. (B) The amount of IF3mt(N+L) and IF3mt(N−L) bound to 28S subunits as a function of added IF3mt(N+L) and IF3mt(N−L). The amount of bound protein was determined as above.
Table 1.
The apparent equilibrium dissociation constant (Kd) for the interaction of IF3mt and its C- and N-domains to 28S subunits.
| IF3mt derivative used | Kd (nM) |
|---|---|
| IF3mt | 30 ± 4* |
| IF3mt(C+L) | 60 ± 5 |
| IF3mt(C−L) | 95 ± 13 |
| IF3mt(N+L) | 240 ± 40 |
| IF3mt(N−L) | 390 ± 60 |
value is taken from previous publication18. The apparent Kd was estimated from the following equation. Kd = [IF3mt]free[28S]free/[IF3mt·28S], where [IF3mt]free indicates the concentration of free IF3mt, [28S] indicates the free 28S subunits and [IF3mt·28S] indicates the concentration of IF3mt bound to 28S subunits.
In contrast to the N-domain of E. coli IF3, the N-domain of IF3mt binds to 28S subunits reasonably well, having only a 10-fold lower affinity than the full length factor for the small subunit (Table I). This result indicates that much of the ribosome binding interactions are mediated by the C-domain but that the N-domain clearly contributes to this interaction. These data also indicate that the linker region contributes only mildly to the tightness of the interaction between IF3mt and the small subunit. This observation is in contrast to observations made with chloroplast IF3 in which mutations of conserved lysine residues in the linker region had drastic effects (100-fold) on the binding of IF3chl to ribosomal subunits 20.
The results obtained for the binding of IF3mt and the IF3mt(N+L) derivative to 28S subunits obtained by Microcon centrifugation were compared to those obtained by sucrose density gradient centrifugation. For these experiments, IF3mt and IF3mt(N+L) bound to the small subunits were separated from unbound protein by centrifugation (Supplementary data, Figure S2). The Kd values obtained using the sucrose density gradient centrifugation method are 35 and 277 nM for IF3mt and IF3mt(N+L), respectively, which are very similar to those obtained using the Microcon method.
The experiments above were carried out on His-tagged derivatives of IF3mt. The interaction of IF3mt and its domains with 28S subunits could potentially be affected by the presence of the His-tag. To assess this possibility, untagged derivatives of IF3mt, IF3mt(N+L) and IF3mt(C+L) were prepared and tested for their activities in initiation complex formation on 55S ribosomes and for binding to 28S subunits (Supplementary data, Figure S4). The untagged derivatives had essentially the same activities as the His-tagged variants in stimulating initiation complex formation. Further, the Kd values for the binding of untagged IF3mt and its domains to 28S subunits are 40 nM, 70 nM and 232 nM for IF3mt, IF3mt(C+L) and IF3mt(N+L) respectively (Supplementary data, Figure S4). These values are essentially the same as those obtained for the His-tagged derivatives indicating that the tag was not affecting the properties of IF3mt or its derivatives.
Kinetics of the binding of IF3mt and its C-domain to 28S subunits using surface plasmon resonance (SPR)
The rates of association (kon) and dissociation (koff) of IF3mt and its C-domain from 28S subunits were determined by SPR. IF3mt and IF3mt(C+L) were active after the coupling reaction and were capable of binding to 28S subunits. However, the C-domain without the linker and the N-domain with or without the linker were inactive when coupled to the sensor chip and their binding to 28S subunits could not be studied using this method.
The kinetics of the binding of IF3mt(C+L) at different concentrations of 28S subunits are shown in Figure 4. The rate constants were determined as described previously 18 and the values are shown in Table II. The kon of the C-domain of IF3mt is 2-fold slower than that of the full-length factor; while koff is almost the same for both. This result indicates that the N-domain of IF3mt may play a small role in promoting the fast association of this factor with 28S subunits. The equilibrium dissociation constant was calculated from the ratio of koff/kon. These Kd alues agree with the values obtained using the Microcon assay. This observation suggests that there are no hidden intermediates during formation of the IF3mt·28S or IF3mt(C+L)·28S complexes.
Figure 4. The forward and reverse rate constants for the binding of IF3mt(C+L) to 28S subunits determined by SPR.
Sensorgram of IF3mt(C+L) binding to 28S subunits. The subunits were diluted to different concentrations (25, 50 and 75 nM) and 60 μL of this dilution was injected onto CM5 chips containing immobilized IF3mt(C+L) or an avidin control at a flow rate of 20 μL/min. The RU values shown are due to the binding of 28S subunits to IF3mt(C+L) (RU change due to IF3mt(C+L) – RU change due to avidin).
Table 2.
Kinetic rate constants (kon and koff) and calculated Kd values for the interaction of IF3mt and IF3mt(C+L) with the 28S subunit using SPR.
| Protein | kon (M−1s−1) | koff (s−1) | Kd (nM) |
|---|---|---|---|
| IF3mt * | (3.80 ± 0.2) × 105 | (1.35 ± 0.02) × 10−2 | 37 ± 9 |
| IF3mt(C+L) | (2.10 ± 0.1) × 105 | (0.92 ± 0.1) × 10−2 | 44 ± 8 |
Rate constants have been taken from previous results.18
Release of IF3mt(N+L) from 28S subunits in the presence of 39S subunits
The C-domain of IF3 is believed to bind to the platform of the small subunit in a region where there are significant contacts with the large subunit. Hence, it is logical that the C-domain would be displaced by the large subunit. However, it is less clear where the N-domain is positioned on the small subunit and whether the large subunit would promote its dissociation. To investigate this question, IF3mt(N+L) was bound to the small subunit. The 39S subunit was subsequently added and the amount of IF3mt(N+L) remaining associated with the ribosome was determined. No detectable IF3mt(N+L) was present in the resulting 55S ribosomes (data not shown) indicating that the N-domain is displaced by the large subunit.
Dissociation of 55S ribosomes by IF3mt and its N- and C-domain
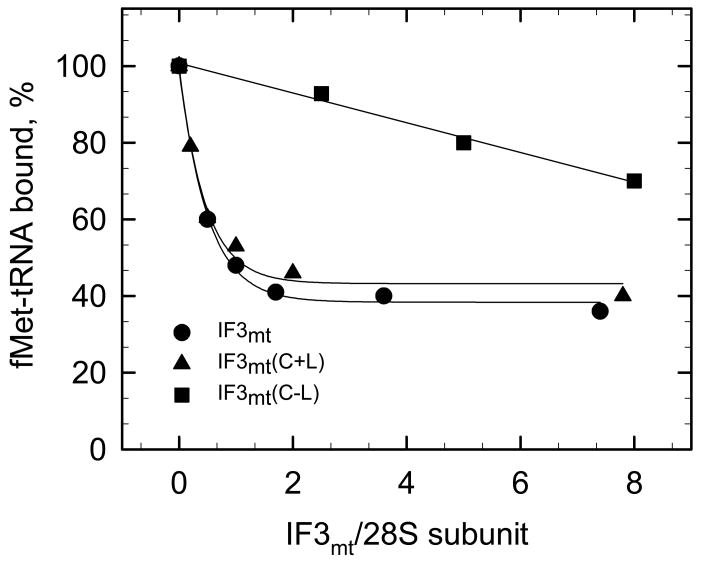
To explore the role of the N-and C-domains and the linker region of IF3mt on the dissociation of mitochondrial 55S ribosomes, the ribosomes were incubated with different concentrations of IF3mt or its derivatives, and the distribution of ribosomal particles was determined by sucrose density gradient centrifugation (Supplementary Material, Figure S3). In the absence of IF3mt, most of the ribosomes are present as 55S particles at 5 mM MgCl2. IF3mt had a significant effect on the fraction of the ribosomes that sedimented as 55S particles with about 50% dissociation observed at a ratio of three IF3mt/ribosome. The isolated N-domain with or without the linker region was inactive in promoting subunit dissociation even at very high concentrations (an IF3mt N-domain/55S ratio of 110:1). The C-domain with the linker had considerable activity in promoting subunit dissociation but was somewhat less active than the full length factor. An 18-fold excess of IF3mt(C+L) over 55S ribosomes was required to achieve 50% dissociation of the particles. This construct has only a 2-fold weaker binding to 28S subunits compared to intact IF3mt. Hence, it is less effective at subunit dissociation than one would predict based on the strength of its interaction with the small subunit. One possible explanation for this observation is that the C-domain alone is more readily displaced by the 39S subunit. The intact factor has contacts with the 28S subunit through both the N- and C-domains making it more difficult for the 39S subunit to displace it from the small subunit.
The linker region clearly contributes to the ability of the C-domain to promote subunit dissociation (Figure 5). A ratio of over 30 IF3mt(C−L) to ribosomes is required for 50% dissociation with this derivative. This value is also quite different from what one would expect based on the binding constant for the interaction of IF3mt(C−L) with 28S subunits which is only about 3-fold weaker than that of the intact factor. This observation again suggests that the linker region makes it more difficult for the 39S subunit to displace IF3mt from the 28S subunit and its removal leads to a more facile displacement of the C-domain by the large subunit.
Figure 5. Effect of IF3mt and its C-domain with and without the linker on the dissociation of mitochondrial 55S ribosomes.
Dissociation of 55S ribosomes (60 nM to 80 nM) was monitored in the presence of different protein concentrations of IF3mt or its C-domain derivatives. The concentrations of IF3mt were 0.18, 0.36, 0.54, 1.8 and 3.6 μM. The concentrations of IF3mt(C+L) were 0.37, 2.0, 4.0 μM and of IF3mt(C−L) were 0.37, 2.5 and 5.0 μM, respectively. Representative fractionation profiles of mitochondrial 55S ribosome dissociation in the absence and presence of full length IF3mt and its domains are shown in the Supplement (Figure S3). The area under the 55S peak in the absence of protein was set to 100% 55S ribosomes. The percentage of this value remaining was calculated in the presence of IF3mt or its derivatives and plotted as a function of the ratio of IF3mt to 55S ribosomes.
Effect of the N- and C-domains of IF3mt on initiation complex formation
The activities of the IF3mt and its N- and C-domains were examined by measuring their effect on fMet-tRNA binding to mitochondrial 55S and E. coli 70S ribosomes (Figure 6) in the presence of IF2mt and poly(A,U,G). IF3mt(C+L) is as active as full length IF3mt in promoting initiation complex formation on 55S ribosomes. This assay is believed to measure primarily the dissociation factor activity of IF3mt. IF3mt(C+L) is somewhat less active than the full length factor in promoting ribosome dissociation as measured by sucrose density gradient centrifugation. In the centrifugation assay, the apparent degree of dissociation reflects a competition between IF3mt binding and the reassociation of the 28S subunit with the 39S subunit leading to an apparent decrease in ribosome dissociation. In contrast, in the fMet-tRNA binding assay, the displacement of IF3mt by the 39S subunit leads to the 55S initiation complex which is measured in the assay. Thus, the high activity of IF3mt(C+L) in the fMet-tRNA binding assay most likely reflects its significant activity in subunit dissociation. IF3mt(C−L) is somewhat less active (about 30–40 %) in the fMet-tRNA binding assay. This reduced level of activity is likely to reflect the significantly lower activity of this construct in subunit dissociation. These observations suggest that the C-domain including the linker region is able to carry out the most basic function of IF3mt of promoting initiation complex formation beginning with 55S ribosomes. Basically similar results were obtained when E. coli 70S ribosomes were used in this assay in place of mitochondrial ribosomes (Figure 6B).
Figure 6. Effect of IF3mt and its C- and N-domain with and without the linker on initiation complex formation.
(A) [35S]fMet-tRNA binding to mitochondrial 55S particles (5 pmol, 50 nM) was tested in the presence of different concentrations of IF3mt and its C- and N-domain derivatives at a fixed amount of IF2mt (100 nM) using 10 μg poly(A,U,G) as the mRNA. A blank (about 0.18 pmol) representing the amount of label retained on the filter in the absence of IF3mt but in the presence of IF2mt has been subtracted from each value. The lower x-axis corresponds to full length IF3mt and its C-domain derivatives and the upper x-axis corresponds to the N-domain derivatives. (B) [35S]fMet-tRNA binding to E. coli 70S particles (20 pmol, 200 nM) was tested in the presence of different amounts of IF3mt and its C-domain derivatives at a fixed concentration of IF2mt (100 nM) using poly(A,U,G) as the mRNA. A blank representing the amount of label retained on the filter in the absence of IF3mt has been subtracted from each value (0.54 pmol). Representative errors are shown in panel A.
To test possible effects of the N-domain on fMet-tRNA binding to ribosomes, it was important to use significantly higher concentrations of the derivatives due to the lower binding constants observed with them (Table I). IF3mt(N+L) stimulates the binding of fMet-tRNA to 55S ribosomes in the presence of IF2mt slightly (Figure 6). This stimulation is enhanced about 2-fold by the presence of the linker in agreement with the higher activity of IF3mt(C+L) compared to IF3mt(C−L). The degree of stimulation observed with the N-domain derivatives is much lower than would be predicted from the reduction in the binding constant alone. It is in agreement with the undetectable ribosome dissociation activities of these derivatives (Figure S3). The slight increase in fMet-tRNA binding observed in the presence of the N-domain derivatives is intriguing. It may reflect an ability of the N-domain to enhance the reassociation of the 39S subunit stabilizing transient complexes formed or perhaps an enhancement of the binding of IF2mt to the small subunit.
Effect of IF3mt and its N- and C-domain on the binding of fMet-tRNA to 28S subunits in the absence of mRNA
In previous studies 17 it has been shown that the C-terminal extension on IF3mt plays an important role in reducing fMet-tRNA binding to 28S subunits in the absence of mRNA. It has no effect on fMet-tRNA binding to the small subunit in the presence of mRNA. The C-domain with and without the linker were tested for this affect (Figure 7). IF3mt(C+L) was as effective as the full length factor in reducing fMet-tRNA binding in the absence of mRNA. This effect is strongly influenced by the presence of the linker region since IF3mt(C−L) had little effect in this assay. These results, taken together with previous observations, suggest that both the C-terminal extension and the linker are important for this activity. The C-terminal extension is predicted to emerge from the C-terminal domain pointing toward the linker region in agreement with this idea 17.
Figure 7. Effect of IF3mt derivatives on the binding of [35S]fMet-tRNA to mitochondrial 28S subunits in the absence of mRNA.
(A) Reaction mixtures contained 160 nM IF2mt and increasing levels of IF3mt or its C-domain derivatives at a constant level of 28S subunits (50 nM). In the absence of IF3mt 0.1pmol of fMet-tRNA were bound to the 28S subunits by IF2mt in the reaction mixture. The estimated error of this experiment is ± 10%.
The effects of IF3mt(N+L) and IF3mt(N−L) on the IF2mt-dependent binding of fMet-tRNA to 28S subunits was also tested in the absence and presence of poly(A,U,G) or the AUG triplet. For these experiments, higher levels of IF3mt(N+L) and IF3mt(N−L) were used, reflecting their lower binding constants for the small subunit. The N-domain, both with or without the linker, had no effect on fMet-tRNA binding to the small subunit in the absence of mRNA and showed a very slight (~10%) stimulation in the presence of the AUG triplet (data not shown).
Effect of IF3mt and its domains on the formation of a complex between [35S]fMet-tRNA and IF2mt
Previous work 21 has shown that IF2mt, like E. coli IF2, can form a binary complex in solution with fMet-tRNA mediated primarily through the C-terminal domain of IF2. The Kd for the binding of fMet-tRNA to IF2mt is about 1 μM 21. The interaction between IF2mt and mitochondrial fMet-tRNA is specific and is 50-fold stronger than the binding of the unformylated mitochondrial initiator tRNA, Met-tRNA. Bovine IF2mt also has a high affinity for yeast cytoplasmic fMet-tRNA and E. coli fMet-tRNA 22. The effect of IF3mt on the interaction between IF2mt and fMet-tRNA was examined using a filter binding assay (Figure 8). Interestingly, IF3mt stimulated the formation of this complex nearly 10-fold perhaps promoting the formation of a ternary complex containing IF3mt:IF2mt:fMet-tRNA. Both the N- and C-domains of IF3mt are involved in the stimulation of fMet-tRNA binding to IF2mt (Figure 8) with the N-domain being somewhat more effective than the C-domain in this interaction. The effect of IF3mt on the interaction between IF2mt and Met-tRNA was also examined (data not shown). IF3mt as a very minor effect on IF2mt:Met-tRNA complex formation. This result indicates that the stimulation of fMet-tRNA binding to IF2mt by IF3mt is largely specific for the formylated initiator tRNA.
Figure 8.
Effect of IF3mt and its domains on the binding of [35S]fMet-tRNA to IF2mt[35S]fMet-tRNA binding to IF2mt (17 pmol, 170 nM) was tested in the presence of different concentrations of IF3mt and its C- and N-domain derivatives. The solid lines represent the binding observed in the presence of IF2mt alone or in the presence either IF3mt or its domains. The dashed lines represent label retained on the filter in the presence of IF3mt or its derivatives alone. This value has been subtracted from the data plotted in the presence of IF2mt to provide a clear estimate of the stimulatory effect of IF3mt on the interaction of IF2mt with fMet-tRNA.
Since IF3mt is a basic protein (pI 10.8), it was essential to ensure that the effect observed was not simply due to an electrostatic interaction between IF3mt and IF2mt (pI~ 6) or the acidic fMet-tRNA. Two controls were carried out. In the first, IF3mt alone was incubated with fMet-tRNA and its ability to retain the tRNA on the filter was tested. Like E. coli IF3 23,24, IF3mt had a measurable affinity for RNA in general. However, this binding was small compared to the stimulation of IF2mt:fMet-tRNA complex formation at the IF3mt concentrations used in the current experiments. None of the domain constructs had significant tRNA binding activity (Figure 8).
The second control was to test the effect of a non-specific basic protein on the formation of the IF2mt:fMet-tRNA complex. Neither avidin (pI 10.5) nor lysozyme (pI 10.7) had a significant effect on the formation of the IF2mt:fMet-tRNA complex indicating that the results obtained with IF3mt reflect a specific interaction between IF2mt and IF3mt. It is conceivable that this interaction takes place in solution prior to initiation factor binding to the small subunit. Alternatively, this interaction may reflect contacts between these factors on the surface of the 28S subunit.
Discussion
The IF3 from both bacteria and mitochondria is folded into distinct N- and C-domains separated by a linker of approximately 25 residues. Previous work 9 with E. coli IF3 indicates that the C-domain binds independently to the small ribosomal subunit and that this domain can carry out all the known in vitro activities of this factor. However, this domain binds 30S subunits with a 100-fold lower affinity than the intact factor. This result is in sharp contrast to those observed with IF3mt. The C-domain with the linker region of the human mitochondrial factor has a strong affinity for mitochondrial 28S subunits having a Kd only about 2-fold weaker than the full length factor. The C-domain of E. coli IF3 is believed to bind to the platform region of the small subunit physically blocking interaction with the large subunit 7. It makes contacts with helices H23, H24 and H45 blocking the formation of intersubunit bridges B2b, B2c and B7a. Examination of the truncated small subunit ribosomal RNA (12S) of mitochondrial ribosomes indicates that these helices have been preserved and that intersubunit bridges B2b, B2c and B7a represent 3 out of 6 conserved bridges between the subunits of the mitochondrial ribosome 2.
It is not immediately apparent why the independent C-domain of IF3mt binds so much better to the small subunit than does the bacterial factor. The C-domain of IF3mt does have an extension of about 30 amino acids following the region with homology to the bacterial factors. However, deletion of this region does not effect the binding of IF3mt to the 28S subunit 18. Rather, the C-terminal extension is thought to play an important role in promoting the dissociation of IF3mt from 28S subunits upon 39S subunit joining 18. The platform region of the small subunit where the C-domain binds is one of the more highly conserved regions between bacterial and mitochondrial ribosomes 2,25. However, the edges of the platform do contain proteins that are specific to the mitochondrial ribosome, and it is possible that IF3mt makes contacts with these proteins that are unavailable to the bacterial factor. It should also be noted that different experimental strategies were used in the E. coli and mitochondrial systems to determine the strength of the interaction. The binding of the C-domain of IF3mt was measured directly and independently while that of the E. coli factor was determined in competition with the full-length factor 9.
In E. coli IF3 the linker does not appear to contribute to the binding of this factor to 30S subunits or to have any functional activity other than to provide a physical connection between the N- and C-terminal domains 9. In contrast, the linker in IF3mt plays a functional role in binding to the small subunit, in the dissociation of 55S ribosomes and in initiation complex formation (Figure 5 and 6). This observation suggests that the linker has direct and important contacts with the surface of the small subunit and is not simply a physical connector between the two domains.
In the bacterial system, no binding of the independent N-domain to 30S subunits was detected 9, but NMR data indicates that this domain does interact with 30S subunits through a small number of residues in the full length factor 26. Again this observation differs from that made with the N-domain of IF3mt which binds to the 28S subunit with only a 10-fold lower affinity than the full length factor. There are considerable differences of opinion about the location of the binding site for the N-domain of IF3 7,13. One recent model suggests that this region of IF3 is located near A790 in helix 24 close to the P-site tRNA. This position places the N-domain close to the neck at the border between the platform and the body of the small subunit 13. This position is in agreement with the cross-linking of IF3 to ribosomal proteins S11, S13 and S19. While mammalian mitochondrial ribosomes have a homolog of S11, they do not have homologs of either S13 or S19 in the head of the small subunit. Rather, the head of the 28S subunit has several unidentified proteins specific to the mitochondrial ribosome. Interactions between the N-domain of IF3mt and these proteins could explain the significant binding of the N-domain to the small subunit observed with the mitochondrial factor.
One interesting suggestion for the role of the N-domain of IF3 proposed by Fabbretti et al. 13 is that a fluctuating interaction between the N-domain and the neck of the 30S subunit modulates the association and dissociation of the factor from the subunit. The significant binding observed with both the C-domain and the isolated N-domain of IF3mt argues that, in the mitochondrial system, this role may not be the major function of the N-domain. Kinetic analysis indicates that kon is reduced only about 2-fold when the N-domain is removed while the koff value is nearly unchanged. Thus, unlike E. coli IF3 the N-domain of IF3mt itself does not appear to have a strong role in modulating the interaction of the C-domain with the 28S subunit.
The role of the N-domain remains obscure. However, one possible hint for its effect lies in the stimulatory effect of IF3mt on the formation of the complex between IF2mt and fMet-tRNA. Both the N- and C-domains appear to play a role in this interaction. While this stimulation has been observed in solution, it may reflect an interaction that occurs on the surface of the small subunit in the 28S initiation complex. It should be noted that the N-domain has been positioned near A790 13 and that this residue interacts with the anticodon stem-loop of the P-site bound initiator tRNA 27.
Previous data also suggests that IF3 is positioned close to the P-site bound fMet-tRNA on the small subunit 7. The C-domain of IF3 is located close to the junction of the D-stem and the anticodon stem as well as being close to the anticodon itself. The N-domain including the linker has been positioned close to the acceptor stem and the D-arm on the opposite side of the tRNA seen with the C-domain. These data are quite compatible with the observations made here. Our data indicate that the C-domain helps promote the formation of the complex between IF2mt and that the N-domain, particularly with the linker, also facilitates the formation of this complex.
As indicated above, it seems unlikely that a major role of the N-domain of IF3mt is to modulate an interaction with the small subunit as suggested for the bacterial factor. Rather, it seems more likely that this region of IF3mt plays a role in promoting conformational changes in the small subunit prior to fMet-tRNA binding and that it may have a role in the kinetics of initiation complex formation. Since the majority of the mRNAs of mammalian mitochondria do not contain a 5′ untranslated leader region, it is also possible that the N-domain of IF3mt may play a role in ensuring the correct positioning of the 5′ start codon in the P-site of the ribosome. This possibility is currently under investigation.
Materials and Methods
Materials
High purity grade chemicals were purchased from Sigma-Aldrich or Fisher Scientific. A rabbit polyclonal primary antibody to the region of IF3mt homologous to the bacterial factors was made by Pacific Immunology Corporation. Secondary antibody, goat anti-rabbit IgG coupled to alkaline phosphatase, was purchased from Sigma-Aldrich. Protein free blocking agent was purchased from Pierce Technologies. Microcon-100 spin columns and pure nitrocellulose membrane filters were obtained from Millipore Corporation. Research grade CM5 sensor chips, 1-ethyl-3-(3-dimethylaminopropyl)- carbodiimide (EDC), N-hydroxysuccinimide (NHS) and 2-(2-pyridinyldithio)-ethaneamine hydrochloride were obtained from Biacore Company. Bovine mitochondria and mitochondrial ribosomes (55S), ribosomal subunits (28S and 39S), bovine IF2mt, yeast [35S]Met-tRNA and yeast [35S]fMet-tRNA were prepared as described 28–30.
Cloning of C- and N-domains of IF3mt
The C-domain with and without the linker was amplified by PCR using the mature IF3mt cDNA as template 16, using forward primers GGAATTCCATATGACAGGATTGCAGAT (for with linker), and GGAATTGGCCATATGAGAAAGGAACTGATTTTG (for without linker) and the reverse primer CCTTAACTCGAGCTGATGCAGAAC (for both). The N-domain with and without the linker were prepared using the forward primer GGAATTCATATGACAGCACCAGCACAG (for both) and the reverse primers CCTTAACTCGAGCAGGGTTGGTTTCCAGTTTTGGG (with linker) and CCTTAACTCGAGCTGATACTCTGCAGGTTC (without linker). These PCR products were purified and digested with NdeI and XhoI and cloned into the pET21 (c+) vector (Novagen), providing a six His-tag at the C-terminus of the expressed protein. Constructs carrying the IF3mt domains were transformed into E. coli DH5α and the nucleotide sequence of the inserted DNA was confirmed. The plasmids were subsequently transformed into E. coli BL21pArgU218 16.
For removal of the His-tag, IF3mt and its C- and N-domains with the linker were prepared carrying a TEV cleavage site between an N-terminal His-tag and the desired region of IF3mt. For these constructs PCR was used to amplify the mature IF3mt cDNA template 16 with forward primers GGAATTCATATGGAAAACCTGTATTTTCAGGGAACAGCACCAGCACAG (for full length and N-domain IF3mt), GGAATTCATATGGAAAACCTGTATTTTCAGGGAATGACAGGATTGCAGATC (for the C-domain) and the reverse primers CCTTAACTCGAGTTATTACTGATGCAGAAC (for full length and C-domain), and CCTTAACTCGAGTTATTACAGGGTTGGTCCAGTTTTGGG (for the N-domain). These PCR products were purified, digested with NdeI and XhoI and cloned into pET15b (Novagen), providing a six His-tag at the N-terminus followed by a TEV cleavage site proceeding the region of IF3mt to be studied. Constructs carrying IF3mt and its domains were transformed into XL10GOLD E. coli DH5α and the nucleotide sequence of the inserted DNA was confirmed. The plasmids were subsequently transformed into E. coli BL21RIL for expression.
Expression and purification of IF3mt and its C- and N-domain derivatives with and without the linker
The purification of the mature form of IF3mt and its domains was carried out as described previously 16,17 using Ni-NTA resins. IF3mt(C+L) and IF3mt(C−L) did not require further purification. However, the Ni-NTA preparations of IF3mt, IF3mt(N+L) and IF3mt(N−L) contained degradation products and were further purified by HPLC using a TSK-gel SP5-PW column 31. These derivatives expressed well although cells carrying the IF3mt(C−L) plasmid were slow to emerge from stationary phase.
The expressed N-terminally His-tagged proteins containing the TEV cleavage site were purified using Ni-NTA resins and did not require further purification. Proteins were digested with pro-TEV protease (Promega) in which the protease itself contains a His-tag following the manufacturer’s instructions). The protease and any uncut IF3mt remaining were removed using Ni-NTA resins. Untagged IF3mt, IF3mt(C+L) and IF3mt(N+L) protein were used to confirm the idea that the His-tag does not affect the interaction of IF3mt or its derivatives with 28S subunits.
Estimation of the exposure of IF3mt and its derivatives to antibodies when bound to 28S subunits
Reaction mixtures (100 μL) contained Binding Buffer (10 mM Tris-HCl, pH 7.6, 7.5 mM MgCl2, 40 mM KCl, 0.1 mM spermine and 1 mM dithiothreitol) with and without 28S subunits (5 pmol) with different amounts of IF3mt (0.16 to 0.66 pmol) and its derivatives (0.82 to 2.5 pmol for IF3mt(C+L) and IF3mt(N+L), and 2 to 6.5 pmol for IF3mt(C−L) and IF3mt(N−L)). The samples were incubated for 10 min at 25 °C. The reaction mixtures were applied to the dot blot apparatus and the IF3mt epitopes exposed to antibody in the absence and presence of 28S subunits were quantified colorimetrically as described previously 18.
Quantitation of the binding of IF3mt and its C- and N-domain to mitochondrial 28S subunits using Microcon centrifugation
The binding of IF3mt and its derivatives to 28S subunits was tested by Microcon centrifugation as described previously 18 using 50 nM 28S subunits (5 pmol) and different concentrations of IF3mt or its domain derivatives as indicated. The ribosome:protein mixtures were incubated for 20 min at 25 °C. Unbound proteins were separated from protein bound to 28S subunits using a Microcon spin filter device as described 18. The bound IF3mt and its derivatives were applied to a dot blot apparatus and the binding of IF3mt to 28S subunits was quantified colorimetrically using antibodies raised against IF3mt. As expected the presence of 28S subunits quenched the antibody-antigen response in the dot blot. This quenching effect was fully overcome by adding EDTA to a final concentration of 20 mM prior to carrying out the dot blot. The amount of bound protein was determined from individual calibration curves generated for each construct separately to account for the different antigenicity of the derivatives (Supplementary Materials, Figure 1).
To estimate the apparent equilibrium dissociation constant for the interaction of IF3mt and its domain derivatives with 28S subunits, it was necessary to determine the percentage of active molecules. These values were determined as described previously 32 and indicated that the IF3mt and its derivatives were almost 100 % active while the 28S subunits were 25–50 % active.
Binding of the N-domain of IF3mt to 28S subunits and its release in the presence of 39S subunits
The binding of IF3mt(N+L) to 28S subunits was performed at a fixed concentration of ribosomes (50 nM) and different concentrations of protein (2.2, 4.4, and 6.6 nM). The ribosomes alone and ribosome:protein mixtures were incubated for 20 min at 25 °C in Binding Buffer (100 μL). Subsequently 39S subunits (6.6 nM) were added to each reaction mixture (giving a final MgCl2 concentration of 8 mM) and incubated for an additional 5 min. This concentration of Mg2+ is sufficient to promote quantitative association of the subunits to 55S particles. Unbound proteins were separated from protein bound to 55S ribosomes using a Microcon spin filter device as described 18. EDTA (20 mM) was added to samples containing ribosomes and the amount of IF3mt(N+L) associated with the ribosomes was determined using the dot blot.
Binding of IF3mt IF3mt(C+L) to 28S subunits using surface plasmon resonance
The rate constants governing the association and dissociation of IF3mt and IF3mt(C+L) with 28S subunits were measured using a Biacore 2000 biosensor instrument located in the Univ. North Carolina Macromolecular Interactions Facility as described previously 18. To avoid crowding of the surface, the extent of immobilization was kept low (~200 RU). Samples (200 μL) of different concentrations of 28S subunits (25, 50 and 75 nM) were prepared in Running Buffer (20 mM HEPES-KOH, pH 7.6, 7.5 mM MgCl2 100 mM KCl, 1 mM DTT, 0.5% glycerol and 0.01% surfactant P20) and 60 μL of each solution was injected at a flow rate 20 μL/min. The RU change due to the binding of 28S subunits was obtained by subtracting the RU of the control surface (avidin bound at 200 RU) from the RU obtained from the surface containing IF3mt or IF3mt(C+L). Of the IF3mt derivatives, only IF3mt(C+L) retained activity when coupled to the surface. Rate constants for association and dissociation were calculated based on the approach outlined previously 18,33.
Dissociation of mitochondrial 55S ribosomes by IF3mt and its N- and C-domains
Mitochondrial ribosomes (60 nM) were incubated in the presence or absence of IF3mt or its domain derivatives at different concentrations as indicated in 100 μL of Gradient Buffer (25 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 40 mM KCl and 1 mM DTT) for 15 min at 37 °C. After incubation, reaction mixtures were placed on ice for 10 min and then layered onto a cold 4.8 mL 10–30% linear sucrose gradient prepared in Gradient Buffer. Gradients were centrifuged for 1 h and 45 min in a Beckman SW55 Ti rotor and fractionated using an ISCO gradient fractionator.
Initiation complex formation on mitochondrial 55S and E. coli 70S ribosomes
Stimulation of [35S]fMet-tRNA binding to either mitochondrial or E. coli ribosomes in the presence of IF3mt or its C- and N-domain derivatives was examined using filter binding assays as described previously 16,17. The amount of label retained on the filter in the absence of IF3mt has been subtracted from each value as indicated in the figure legends. Since high concentrations of IF3mt(N+L) and IF3mt(N−L) were used in several experiments, additional control experiments were performed to measure the retention of [35S]fMet-tRNA on the filter in the presence of IF2mt alone (no ribosomes added) with different concentrations of IF3mt(N+L) and IF3mt(N−L). The amount of [35S]fMet-tRNA bound on the filter due to IF3mt(N+L) or IF3mt(N−L) alone (0.18 pmol) has been subtracted from appropriate values.
Effect of IF3mt on the binding of fMet-tRNA to IF2mt
[35S]fMet-tRNA binding to IF2mt in the presence of IF3mt in solution was determined using the filter binding assay. Reaction mixtures (100 μL) contained IF2mt (17 pmol, 0.17 μM), [35S]fMet-tRNA (60 nM), 50 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 40 mM KCl, 1 mM DTT, 0.1 mM spermine, 0.25 mM GTP, 1.25 mM phosphoenolpyruvate, 0.9 U pyruvate kinase, and variable amounts of IF3mt or its N- and C-domains as indicated. The reactions were incubated for 10 min, then diluted with ice cold buffer (50 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 40 mM KCl) and filtered through nitrocellulose membranes with two buffer washes. The filters were dried for 8 min at 100 °C. The amount of [35S]fMet-tRNA retained as a complex with IF2mt and IF3mt was determined by scintillation counting.
Supplementary Material
The calibration curve for the dot blot analysis showing the image intensity of IF3mt(C+L). The insert shows the dot blot image analysis intensity of IF3mt(C−L). The immunological response of the C-domain with the linker is much stronger than without the linker. Similar calibration curves for the N-domain derivatives were generated (data not shown). The presence of 28S subunits quenched the immunological response in the dot blot but this quenching effect has been eliminated by adding 20 mM EDTA. These calibration curves were used to determine the amount of IF3mt bound to subunits. IF3mt and its truncated derivatives all had different responses to the IF3mt antibody and all the calibration curves were different.
Shown is a representative fractionation profile of 28S subunits. Dot blot images of the three relevant fractions are shown under the figure and the total pixels were determined for the binding of IF3mt(N+L) to 28S subunits from the difference between the combination of the pixels in the three dots of IF3mt(N+L) + 28S minus the combination of dots of the control pixels (28S subunit fractions alone). Reaction mixtures (100 μL) applied to the gradient contained 6 pmol (60 nM) 28S subunits and either no IF3mt(N+L) or 1.2 pmol IF3mt(N+L). The first row of the dot blot corresponds to the gradient run with 28S subunits alone and second row corresponds to the gradient containing both 28S subunits and IF3mt(N+L).
The fractional profiles are shown for 55S ribosomes alone, IF3mt/55S=7, IF3mt(C+L)/55S=7, IF3mt(C−L)/55S=31 and IF3mt(N+L)/55S=110. Gradients were run and analyzed as described previously18.
(A) Comparison of fMet-tRNA bound to 55S ribosomes (5 pmol, 50 nM) in the presence of His-tagged and untagged IF3mt (1 pmol, 10 nM) and IF3mt(C+L) (1 pmol, 10 nM) at a fixed amount of IF2mt (110 nM) using 10 μg poly(A,U,G) as the mRNA. Representative data from the linear part of the dose response curve is shown for each derivative. (B) Comparison of the binding of His-tagged and untagged IF3mt, IF3mt(C+L) and IF3mt(N+L) to 28S subunits as reflected in the pixel change upon immunological detection of the bound factor in the dot blot. The 28S subunits (4.5 pmol) were incubated with 0.5, 1.0 and 2 pmol of IF3mt, IF3mt(C+L) and IF3mt(N+L), respectively.
Supplementary
Supplementary information is available at the Journal online.
Acknowledgments
Funding was provided by the National Institutes of Health (Grant GM 32734) to LLS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O’Brien TW, Denslow ND, Faunce W, Anders J, Liu J, O’Brien B. Structure and function of mammalian mitochondrial ribosomes. In: Nierhaus K, Franceschi F, Subramanian A, Erdmann V, Wittmann-Liebold B, editors. The translational apparatus: Structure, function regulation and evolution. Plenum Press; New York: 1993. pp. 575–586. [Google Scholar]
- 2.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 3.Gaur R, Grasso D, Datta PP, Krishna PDV, Das G, Spencer A, Agrawal RK, Spremulli L, Varshney U. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol Cell. 2008;29:180–190. doi: 10.1016/j.molcel.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochem. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 5.Gualerzi CO, Brandi L, Caserta E, Teana A, Spurio R, Tomsic J, Pon CL. Translation initiation in bacteria. In: Garrett RA, Douthwaite SR, Liljas A, Matheson AT, Moore PB, Noller HF, editors. The Ribosome: Structure, function, antibiotics, and cellular interactions. ASM Press; Washington D.C.: 2000. pp. 477–494. [Google Scholar]
- 6.Dottavio-Martin D, Suttle DP, Ravel JM. The effects of initiation factors IF-1 and IF-3 on the dissociation of Escherichia coli 70 S ribosomes. FEBS Letters. 1979;97:105–110. doi: 10.1016/0014-5793(79)80062-9. [DOI] [PubMed] [Google Scholar]
- 7.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 8.La Teana A, Gualerzi CO, Brimacombe R. From stand-by to decoding site. Adjustment of the mRNA on the 30 S subunit under the influence of the initiation factors. RNA. 1995;1:772–782. [PMC free article] [PubMed] [Google Scholar]
- 9.Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO. Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J. 2001;20:4560–4569. doi: 10.1093/emboj/20.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol Cell. 2008;30:712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Teana A, Pon C, Gualerzi CO. Tanslation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc Natl Acad Sci USA. 1993;90:4161–4165. doi: 10.1073/pnas.90.9.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbretti A, Pon CL, Hennelly SP, Hill W, Lodmell J, Gualerzi CO. The real-time path of translation factor IF3 onto and off the ribosome. Molecular Cell. 2007;25:285–296. doi: 10.1016/j.molcel.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Bellis D, Liveris D, Goss D, Ringquist S, Schwartz I. Structure-function analysis of Escherichia coli translation initiation factor 3: Tyrosine 107 and lysine 110 are required for ribosome binding. Biochem. 1992;31:11984–11990. doi: 10.1021/bi00163a005. [DOI] [PubMed] [Google Scholar]
- 15.Bruhns J, Gualerzi CO. Structure--function relationship in Escherichia coli initiation factors: role of tyrosine residues in ribosomal binding and functional activity of IF-3. Biochem. 1980;19:1670–1676. doi: 10.1021/bi00549a023. [DOI] [PubMed] [Google Scholar]
- 16.Koc EC, Spremulli LL. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J Biol Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- 17.Bhargava K, Spremulli LL. Role of the N- and C-terminal extensions on the activity of mammalian mitochondrial translational initiation factor 3. Nuc Acids Res. 2005;33:7011–7018. doi: 10.1093/nar/gki1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haque ME, Grasso D, Spremulli LL. The interaction of mammalian mitochondrial translational initiation factor 3 with ribosomes: evolution of terminal extensions in IF3mt. Nuc Acids Res. 2008;36:589–597. doi: 10.1093/nar/gkm1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Cock E, Springer M, Dardel F. The interdomain linker of Escherichia coli initiation factor IF3: a possible trigger of translation initiation specificity. Mol Microbiol. 1999;32:193–202. doi: 10.1046/j.1365-2958.1999.01350.x. [DOI] [PubMed] [Google Scholar]
- 20.Yu NJ, Spremulli LL. Structural and mechanistic studies on chloroplast translational initiation factor 3 from Euglena gracilis. Biochem. 1997;36:14827–14835. doi: 10.1021/bi971185y. [DOI] [PubMed] [Google Scholar]
- 21.Spencer AC, Spremulli LL. Interaction of mitochondrial initiation factor 2 with mitochondrial (f)Met-tRNA. Nuc Acids Res. 2004;32:5464–5470. doi: 10.1093/nar/gkh886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao HX, Spremulli LL. Initiation of protein synthesis in animal mitochondria: Purification and characterization of translational initiation factor 2. J Biol Chem. 1991;266:20714–20719. [PubMed] [Google Scholar]
- 23.Wickstrom E, Laing LG. Physical studies of the interaction of Escherichia coli translational initiation factor 3 protein with ribosomal RNA. Meth Enzymol. 1988;164:238–58. 238–258. doi: 10.1016/s0076-6879(88)64046-8. [DOI] [PubMed] [Google Scholar]
- 24.Vermeer C, Boon J, Talens A, Bosch L. Binding of the initiation factor IF-3 to Escherichia coli ribosomes and MS2 RNA. Eur J Biochem. 1973;40:283–293. doi: 10.1111/j.1432-1033.1973.tb03196.x. [DOI] [PubMed] [Google Scholar]
- 25.Koc EC, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome: Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 26.Sette M, Spurio R, VanTilborg P, Gualerzi C, Boelens R. Identification of the ribosome binding sites of translation initiation factor IF3 by multidimensional heteronuclear NMR spectroscopy. RNA. 1999;5:82–92. doi: 10.1017/s1355838299981487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 28.Ma J, Spremulli LL. Expression, purification and mechanistic studies of bovine mitochondrial translational initiation factor 2. J Biol Chem. 1996;271:5805–5811. doi: 10.1074/jbc.271.10.5805. [DOI] [PubMed] [Google Scholar]
- 29.Graves M, Spremulli LL. Activity of Euglena gracilis chloroplast ribosomes with prokaryotic and eukaryotic initiation factors. Arch Biochem Biophys. 1983;222:192–199. doi: 10.1016/0003-9861(83)90516-7. [DOI] [PubMed] [Google Scholar]
- 30.Matthews DE, Hessler RA, Denslow ND, Edwards JS, O’Brien TW. Protein composition of the bovine mitochondrial ribosome. J Biol Chem. 1982;257:8788–8794. [PubMed] [Google Scholar]
- 31.Grasso DG, Christian BE, Spencer AC, Spremulli LL. Over-expression and purification of mitochondrial translational initiation factor 2 and initiation factor 3. (Lorsch, J., ed) Meth Enzymol. 2007;430:59–78. doi: 10.1016/S0076-6879(07)30004-9. [DOI] [PubMed] [Google Scholar]
- 32.Spencer AC, Spremulli LL. The interaction of mitochondrial translational initiation factor 2 with the small ribosomal subunit. Biochim Biophys Acta. 2005;1750:69–81. doi: 10.1016/j.bbapap.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 33.O’Shannessy DJ, Brigham-Burke M, Soneson KK, Hensley P, Brooks I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: use of nonlinear least squares analysis methods. Anal Biochem. 1993;212:457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The calibration curve for the dot blot analysis showing the image intensity of IF3mt(C+L). The insert shows the dot blot image analysis intensity of IF3mt(C−L). The immunological response of the C-domain with the linker is much stronger than without the linker. Similar calibration curves for the N-domain derivatives were generated (data not shown). The presence of 28S subunits quenched the immunological response in the dot blot but this quenching effect has been eliminated by adding 20 mM EDTA. These calibration curves were used to determine the amount of IF3mt bound to subunits. IF3mt and its truncated derivatives all had different responses to the IF3mt antibody and all the calibration curves were different.
Shown is a representative fractionation profile of 28S subunits. Dot blot images of the three relevant fractions are shown under the figure and the total pixels were determined for the binding of IF3mt(N+L) to 28S subunits from the difference between the combination of the pixels in the three dots of IF3mt(N+L) + 28S minus the combination of dots of the control pixels (28S subunit fractions alone). Reaction mixtures (100 μL) applied to the gradient contained 6 pmol (60 nM) 28S subunits and either no IF3mt(N+L) or 1.2 pmol IF3mt(N+L). The first row of the dot blot corresponds to the gradient run with 28S subunits alone and second row corresponds to the gradient containing both 28S subunits and IF3mt(N+L).
The fractional profiles are shown for 55S ribosomes alone, IF3mt/55S=7, IF3mt(C+L)/55S=7, IF3mt(C−L)/55S=31 and IF3mt(N+L)/55S=110. Gradients were run and analyzed as described previously18.
(A) Comparison of fMet-tRNA bound to 55S ribosomes (5 pmol, 50 nM) in the presence of His-tagged and untagged IF3mt (1 pmol, 10 nM) and IF3mt(C+L) (1 pmol, 10 nM) at a fixed amount of IF2mt (110 nM) using 10 μg poly(A,U,G) as the mRNA. Representative data from the linear part of the dose response curve is shown for each derivative. (B) Comparison of the binding of His-tagged and untagged IF3mt, IF3mt(C+L) and IF3mt(N+L) to 28S subunits as reflected in the pixel change upon immunological detection of the bound factor in the dot blot. The 28S subunits (4.5 pmol) were incubated with 0.5, 1.0 and 2 pmol of IF3mt, IF3mt(C+L) and IF3mt(N+L), respectively.
Supplementary
Supplementary information is available at the Journal online.