Abstract
Background
Upon induction of DNA double-strand breaks (DSBs), Mre11 and Rad50 proteins of the Mre11 DNA repair complex accumulate at the sites of DSBs and form discrete nuclear foci. Precision in scoring of Mre11/Rad50-containing foci depends upon detection of those foci, some of which have a fluorescence staining intensity that is too close to the fluorescence staining intensity of the remaining Mre11 and Rad50 proteins that have not been incorporated into foci.
Methods
Human U-1 melanoma cells in exponential growth were irradiated with various doses of X-rays (0-12 Gy) to induce the formation of repair foci. Four hours after irradiation, cells were simultaneously labeled for Mre11 and Rad50 proteins, using a two-color immunofluorescence staining technique. Laser scanning confocal microscopy was used to collect the composite images of randomly selected cell nuclei. Intensity correlation analysis (ICA) of equally intense fluorescence signals from Mre11 and Rad50 proteins was performed to obtain the regions with correlated pixels.
Results
ICA permitted enhanced detection of low level fluorescence of Mre11/Rad50 foci (“hidden” foci) that can be barely detected upon imaging of only one protein. For example, while imaging of only one protein (either Mre11 or Rad50) in the nucleus of a 6 Gy-irradiated cell revealed 9 foci, imaging of two proteins with ICA revealed 11 foci. ICA permitted an evaluation of the dose-dependence of nuclear foci in cells irradiated with various doses of X-rays, with focus formation increasing up to a dose of 6 Gy.
Conclusions
Our data accumulated using two-color immunofluorescence staining of Mre11 and Rad50 proteins and ICA of these two target proteins provide a basis for enhanced detection and accuracy in the scoring of DNA repair foci.
Keywords: DNA double-strand breaks, DNA repair nuclear foci, Mre11 and Rad50 proteins, immunofluorescence, intensity correlation analysis, correlated pixels
INTRODUCTION
DNA damage may be caused by a variety of intrinsic and extrinsic factors, including exposure to ionizing radiation (IR). There are many different types of DNA lesions induced by IR in mammalian cells, but the DNA double-strand break (DSB), if misrepaired or left unrejoined, can cause transformation or cell death (1). Normally, DSBs are recognized and then repaired via the non-homologous DNA end-joining and homologous recombination pathways. The human Mre11 complex, composed of Mre11 (85 kDa), Rad50 (153 kDa), and Nbs1 (95 kDa) proteins, is believed to be a central player in the cellular response to DSBs, including DNA damage checkpoint activation and repair, and telomere maintenance (2-4). The complex is highly conserved and consists of a single Nbs1 molecule bound to a heterotetramer of Mre11 and Rad50 which is in turn comprised of a dimer of each protein (5). Mre11 stably associates with Rad50 (6), which can then bind to the third component, Nbs1. While Mre11 and Rad50 are involved in DNA binding and end processing, Nbs1 regulates the activities of the complex and may actually serve to initiate DNA damage-induced checkpoint response (7). Mre11, Rad50, and Nbs1 are essential genes: cell lines having deficiency or deletion mutations in each of these genes are hypersensitive to IR, presumably due to problems with DSB repair (8-10).
Normally, the human Mre11 complex is uniformly distributed throughout the cell nucleus. Efficient repair of DSBs appears to require the formation of nuclear foci comprised of Mre11, Rad50, and Nbs1 (10-12). Treatment of cells with DSB-inducing agents, such as IR, results in a rapid association between Mre11 and damaged DNA (13). The Mre11 complex likely functions as a primary detector of DNA damage (14). Discrete and bright nuclear foci stained for either Nbs1, Mre11 or Rad50 have been reported to appear several hours after irradiation of human fibroblasts with X- or γ-rays (≥ 1 Gy) (10-12,15,16). Formation of nuclear foci is dose-dependent, and foci remain until DSB repair is complete (11,15,16).
Nuclear repair foci, including foci containing the Mre11 complex or phosphorylated histone H2AX (γ-H2AX) (17) have often been used as an indicator of the DNA damage response, as their presence has been correlated with sites of DSBs. Foci are usually visualized by using indirect immunofluorescence. Over the past ten years, laser scanning confocal microscopy (LSCM) has been successfully used for obtaining high-resolution optical images of nuclear foci containing various proteins involved in different stages of the DNA repair. LSCM allows evaluation of the 3D distribution of nuclear foci with improved spatial resolution compared to epifluorescence microscopy. Both quantitative and qualitative information pertaining to localization and composition of nuclear foci may be obtained through the use of commercial image analysis software packages and individually developed computational image analysis algorithms which run on a personal computer (18). This permits fast and consistent analyses of foci observed in confocal images, generated under objectively defined criteria. However, there are some obvious limitations to such an analytical approach. First, upon increasing the number of foci per nucleus, foci reach a certain degree of overlap that reduces precision in focus scoring. Second, all foci should be bright enough to be included in scoring. Staining intensities of foci may vary from very bright to very dim, depending upon how much the protein of interest is expressed within a focus, and to what extent the primary antibody binds this protein. Third, dynamic changes in the density of chromatin at the sites of DNA DSBs (19) can be a factor that affects antibody staining of the protein residing in a focus. Dim foci sometimes may not be distinguishable from nuclear proteins that may still be uniformly distributed in nuclei and not incorporated into foci, and these foci may erroneously be excluded from scoring.
Here we describe the use of an analytical approach that permits enhanced detection of dim foci composed of Mre11 complex proteins, specifically Mre11 and Rad50. Using a two-color immunofluorescence technique to simultaneously label Mre11 and Rad50 proteins, LSCM was used to collect composite images of randomly selected cell nuclei of unirradiated and irradiated cells. Nuclear foci were analyzed on the basis of imaging of correlated pixels obtained by merging of equally intense fluorescence signals from Mre11 and Rad50 proteins. This approach was used to examine the dependence of focus formation on radiation dose.
MATERIALS AND METHODS
Cell Culture and Irradiation
Human U-1 melanoma cells were cultured in monolayer in McCoy's 5A medium with L-glutamine (Mediatech, Herndon, VA, USA) and 10% iron-supplemented bovine calf serum (BCS; Hyclone, Logan, UT, USA). Cells were maintained at 37°C in a humidified environment of 5% CO2 and 95% air. Forty thousand asynchronous cells were plated into each well of Lab-Tek II® 4-well tissue culture chamber slides (Nalge Nunc International, Naperville, IL, USA) and grown for 2 days until 80-90% confluent. Prior to irradiation, the culture medium in each well was replaced with pre-warmed 37°C fresh medium. The chamber slides were placed on ice for 5 min and cells were then irradiated using a 160 kVp Faxitron X-ray machine (Wheeling, IL, USA). Cells were irradiated with doses of 0, 3, 6, and 12 Gy (dose rate: 2.44 Gy/min). After irradiation, the culture medium in each well was replaced with pre-warmed 37°C fresh medium and cells were then incubated for 4 h in a CO2-incubator.
Immunofluorescence staining
After treatment, cells were washed twice in PBS (pH 7.4). Fixation and permeabilization of cells were performed as described by Scully et al (20). Briefly, cells were fixed for 10 min in PBS-buffered solution containing 3% formaldehyde/2% sucrose, and then permeabilized for 5 min in Triton buffer (0.5% Triton X-100 in 20 mM HEPES, pH 7.4; 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) on ice. Prior to immunofluorescence staining, cells were incubated in blocking solution (10% fetal bovine serum in PBS) for 1 h at 4°C. Cells were washed thrice with PBS and incubated overnight at 4°C with primary antibodies: anti-hRad50 mouse monoclonal antibody, dilution 1:1000 (Upstate, Lake Placid, NY, USA; clone 2C6; catalogue number: 05-525), and anti-hMre11 rabbit polyclonal antibody, dilution 1:100 (Calbiochem, La Jolla, CA, USA; catalogue number: PC388). Cells were then washed thrice with PBS and incubated at 37°C for 1 h with the corresponding secondary antibodies: Alexa Fluor 594 goat anti-mouse antibody, dilution 1:400 (Molecular Probes, Eugene, OR, USA; catalogue number: A11032), and Alexa Fluor 488 goat anti-rabbit antibody, dilution 1:200 (Molecular Probes; catalogue number: A11034). All antisera were diluted in 1% BSA in PBS. After staining, cells were washed four times with PBS, and then mounted in ProLong Gold antifade reagent (Molecular Probes) followed by examination of nuclear foci using LSCM. Boundaries of cell nuclei were clearly seen under these staining conditions. DNA counterstaining with DAPI showed the same nuclear boundaries (data not shown). Since co-localization between Nbs1 and Mre11/Rad50 foci was not routinely observed in our cells, this protein was not included in our studies.
LSCM
A Zeiss LSM-510Meta confocal microscope (Carl Zeiss, Jena, Germany) equipped with a C-Apochromat 63x/1.2 water immersion objective was used for examining nuclear foci. Argon (488 nm) and helium-neon (543 nm) lasers were used to excite Alexa Fluor 488 and Alexa Fluor 594, respectively. The fluorescence emitted by Alexa Fluor 488 was detected through a 505-530 nm band-pass filter. The fluorescence emitted by Alexa Fluor 594 was detected through a 560 long-pass filter. To minimize crosstalk between channels, the multi-track mode was used, which allows sequential collection of red fluorescence (from Rad50) and green fluorescence (from Mre11). Ten to eleven fields were randomly chosen from each sample, and from these, 2-3 representative cell nuclei were then chosen for analysis. Composite images of each nucleus were collected using LSM software (Carl Zeiss, version 4.0 SP2). These images consisted of a confocal series (z-series) of up to 18 sequential scans (0.5 µm step size). Scan speed and zoom were set on 6 and 3, respectively. To ensure the acquisition of high quality images, the pinhole size for each channel was reduced to the extent that guaranteed a superior spatial resolution and synchronous collection of green and red fluorescence signals through the entire stack of optical sections. Green and red fluorescence signals were collected at about equally intensive gray levels. That is, amplifier and gain settings were adjusted such that green and red fluorescence intensity was nearly equal. All settings (including amplifier settings and laser power) were the same, while performing the image acquisition within each set of experiments.
Image processing and analysis
The images were processed and analyzed with ImageJ software (version 1.37c) (21), modified at the Wright Cell Imaging Facility (WCIF; Toronto Western Research Institute, Canada) (website http://www.uhnresearch.ca/wcif). To visualize the association between Mre11 and Rad50 proteins within the nucleus, co-localization analysis was performed in accordance with instructions provided by WCIF. Briefly, the fluorescence intensities from these two target proteins were subjected to intensity correlation analysis (ICA) described by Li et al. (22).1 First, background subtraction was performed on each slice of the stack of images obtained from both photomultiplier tubes (PMTs) (red and green fluorescence). A region of interest was then defined by manually drawing the nuclear boundary on one of the images of the stack (of the middle section of the cell). Image correlation analysis was then performed by combining two stacks of images (combining green and red fluorescence images). The resulting combined images display positive PDM [product of the differences of the mean (22)] values representative of the regions with best-correlated pixels. The regions with best-correlated pixels that represent foci were further analyzed in order to obtain quantitative data (number of foci, their size in voxels, and their intensity in arbitrary units). To perform this analysis, the images with positive PDM values were converted from 32-bit to 8-bit format, and then they were subjected to “connexity analysis” (that runs under “3D objects counter” function) to score positive foci using a 10-voxel size threshold. That is, positive foci were determined based on whether they had > 10 voxels.
RESULTS
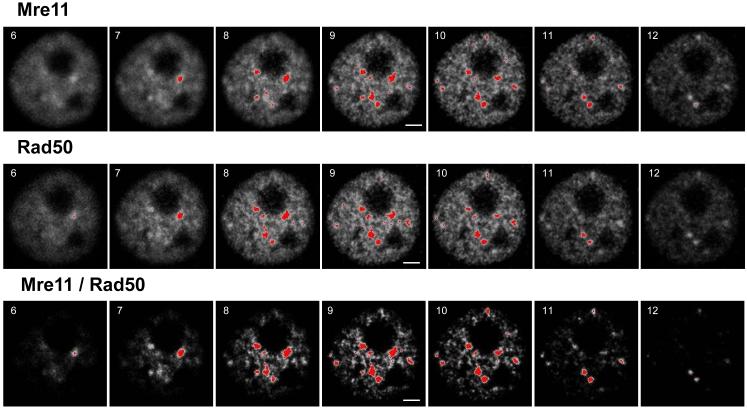
The major advantage for using a double target imaging approach with ICA over the conventional imaging approach for focus scoring, which involves imaging of only one protein, is that the former approach allows for more accurate detection and quantification of foci. Selected optical sections of a cell nucleus containing IR-induced Mre11/Rad50 foci are shown in Figure 1. The fluorescence intensities (in gray values) and the staining patterns of both Mre11 and Rad50 proteins were similar (top and middle panels, respectively, Fig. 1). To prevent the interference of background levels of nuclear Mre11 and Rad50 (that is, Mre11 and Rad50 that are not part of Mre11 complexes or foci) in the focus analysis, a threshold was set. Since the fluorescence intensities of both target proteins were almost equal (data not shown), the threshold was set at the same level for each protein. Correlation analysis was performed for these individual intensities, and positive PDM values indicative of best correlated pixels are shown in images in the bottom panel of Fig. 1. In these images, foci look more discrete and discernible due to exclusion of uncorrelated pixels. Derivation of positive PDM values enabled enhancement of foci that were weakly detected in either or both of the individual sets of images (top or middle panel, Fig. 1).
Figure 1.
Composite image of the nucleus of one of the human U-1 melanoma cells that were irradiated (6 Gy of X-rays), then incubated at 37 °C for 4 h, fixed/permeabilized, and immunostained for Mre11 and Rad50 proteins, as described in Materials and Methods. Each slice has the number in its left-top corner, which indicates the sequence within z-series collected from top to bottom. Fluorescence intensities are shown in 8-bit gray format. Both Mre11 and Rad50 are abundant proteins in the nucleus, but are absent in nucleoli (top and middle panels, respectively). The foci are shown in red with a threshold boundary set at 175 for both sets of images. The bottom panel shows the 8-bit images of positive PDM values (that mainly represent the enhanced foci) derived from image correlation analysis between these two sets of images (top panel vs. middle panel). After the enhancement of foci, the threshold boundary was readjusted to 220 to reduce their size (threshold level value of 175 that was adjusted for “Mre11” and “Rad50” sets of images is too low for “Mre11/Rad50 sets of images, since it results in the remarkable enlargement of foci leading to their fusion). Scale bars = 3 µm.
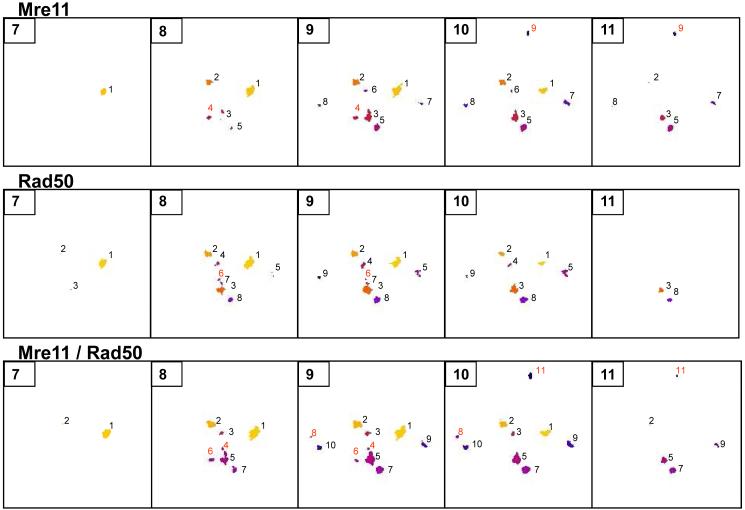
Foci shown in Figure 1 were analyzed using the “3D objects counter” function of ImageJ. A three-dimensional analysis of Mre11 and Rad50 foci is shown in Figure 2. Foci are enumerated as numbered pseudo-colored spots (Fig. 2). This analysis was performed in order to track and quantify the objects of interest (foci) through the entire stack of images. While the total number of foci in both individual Mre11 and Rad50 sets of images was 9 when integrated over all slices (top and middle panels, respectively, Fig. 2), the total number of foci in the merged Mre11/Rad50 set of images after ICA was 11 (bottom panel, Fig. 2). The set of images obtained for Mre11 showed two additional foci compared to the set of images obtained for Rad50 (numbered as 4 and 9, slices 8-11, top panel, in red). Also, there was one Rad50 focus which was not present in the set of images obtained for Mre11 (numbered as 6, slices 8 and 9, middle panel, in red). However, all these missing foci were present in the merged Mre11/Rad50 set of images (numbered as 4, 6, and 11, bottom panel, in red). The merged images also show the one focus (numbered as 8, in red) that was missing in both individual sets of images of Mre11 and Rad50. A listing of foci identified from individual and merged scans of nucleus from cell irradiated with 6 Gy of X-rays is shown in Table 1.
Figure 2.
3D objects analysis. Three sets of images shown in Figure 1 (“Mre11”, “Rad50”, and “Mre11/Rad50” sets of images) were subjected to this analysis. The spots represent foci. Each spot has its own number and color that allow tracking of foci through the entire stack of images. Numbers shown in red indicate the spots missing in one of the two sets of images (“Mre11” or “Rad50”), or in both sets of images (“Mre11” and “Rad50”). A 10-voxel size threshold was set to exclude the objects having sizes ≤ 10 voxels.
Table 1.
Identification of foci based on spot numbers shown in Figure 2
| Focus ID | Mre11 Spot# | Rad50 Spot# | Mre11/Rad50 Spot# |
|---|---|---|---|
| A | 1 | 1 | 1 |
| B | 2 | 2 | 2 |
| C | 6 | 4 | 3 |
| D | missing | 6 | 4 |
| E | 3 | 3 and 7 | 5 |
| F | 4 | missing | 6 |
| G | 5 | 8 | 7 |
| H | missing | missing | 8 |
| I | 7 | 5 | 9 |
| J | 8 | 9 | 10 |
| K | 9 | missing | 11 |
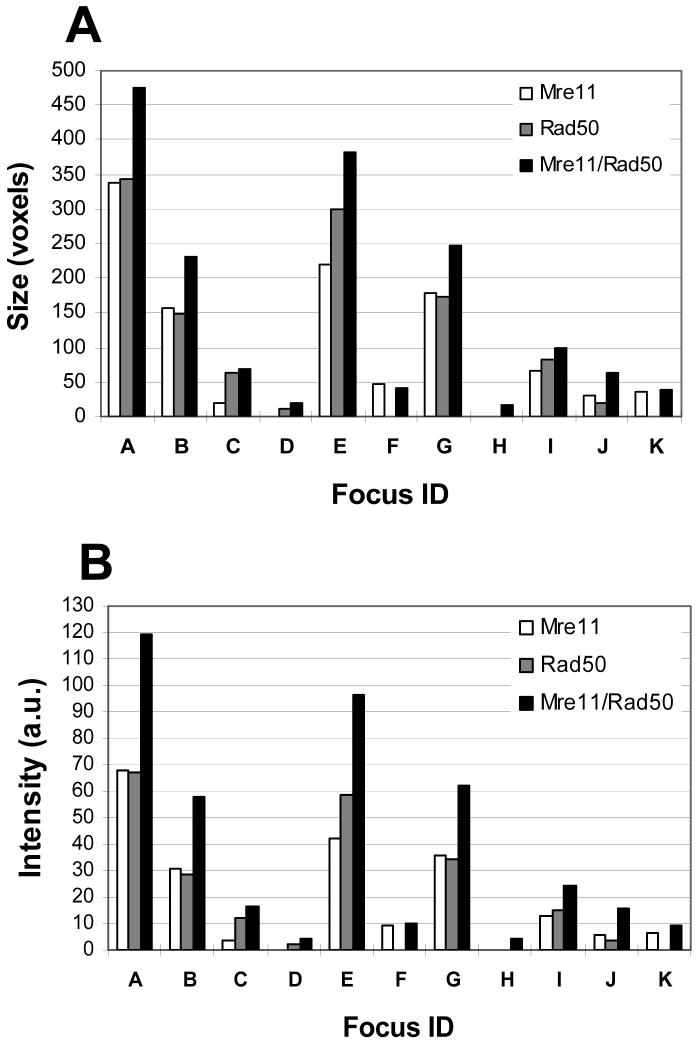
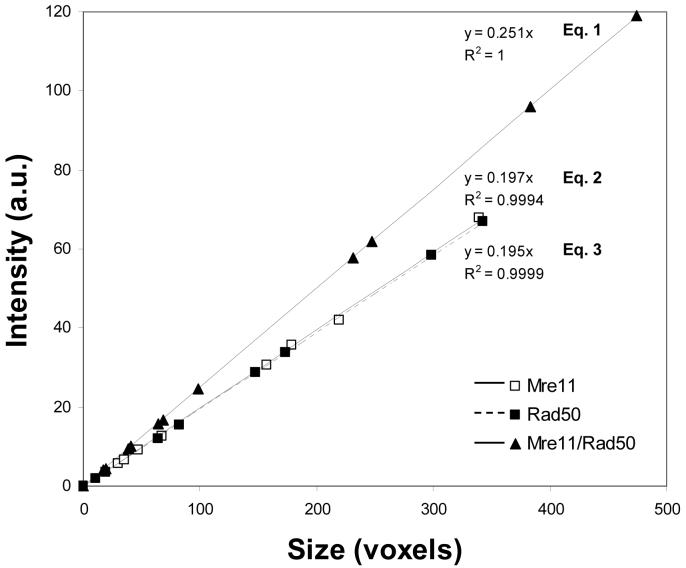
A comparative analysis of the sizes and fluorescence intensities of foci from the Mre11 and Rad50, and merged Mre11/Rad50 sets of images is shown in Figures 3A and 3B, respectively. In the merged Mre11/Rad50 set of images, there was an increase in fluorescence intensities of nearly all foci compared to the individual Mre11 or Rad50 set of images, and this was accompanied by an increase in size of the foci. Although the size of the object and its total intensity may not necessarily correlate, this analysis showed that the intensity of the focus is directly proportional to its size. Correlation between the sizes and the intensities of foci listed in Table 1 are shown in Figure 4. This correlation was perfect for each set of images (R2∼ 1.0). Coincidence of the trend lines of the size-intensity relationship for foci in the individual Mre11 and Rad50 sets of images is indicative of the fact that foci of equal sizes had equal fluorescence intensities. A markedly elevated slope for the trend line of the size-intensity relationship for foci in the merged Mre11/Rad50 set of images indicates that foci are more intense than foci in either of the individual (e.g., Mre11 or Rad50) set of images. In order to determine the contribution of fluorescence intensity in the enhancement of foci independently from their size, we obtained the equations of the size-intensity relationship for foci in each set of images. All three equations, shown in Figure 4, are linear, and each of them has its own slope. By dividing the slope value of Equation 1 by the slope value of either Equation 2 or 3, we can obtain the values of the enhancement factor as follows: 0.251 / 0.197 = 1.27 (Mre11/Rad50 set of images vs. Mre11 set of images) and 0.251 / 0.195 = 1.29 (Mre11/Rad50 set of images vs. Rad50 set of images). These values assume that if a focus in the merged Mre11/Rad50 set of images has exactly the same size as a focus in the separate Mre11 and Rad50 sets of images, then this focus is 1.27-fold brighter than a focus in the Mre11 set of images, or 1.29-fold brighter than a focus in the Rad50 set of images.
Figure 3.
Comparison of the sizes (A) and intensities (B) of foci in the “Mre11”, “Rad50”, and “Mre11/Rad50” sets of images, subjected to 3D objects analysis. In the “Mre11/Rad50” set of images, foci were larger and more intense than foci in both “Mre11” and “Rad50” sets of images. Sizes are shown in voxels. Intensities are shown in arbitrary units (a.u.). The abscissa (focus ID) corresponds to individual foci described in Table 1.
Figure 4.
Size-intensity relationship for foci in the “Mre11”, “Rad50”, and “Mre11/Rad50” sets of images, subjected to 3D objects analysis. Sizes are shown in voxels. Intensities are shown in arbitrary units (a.u.).
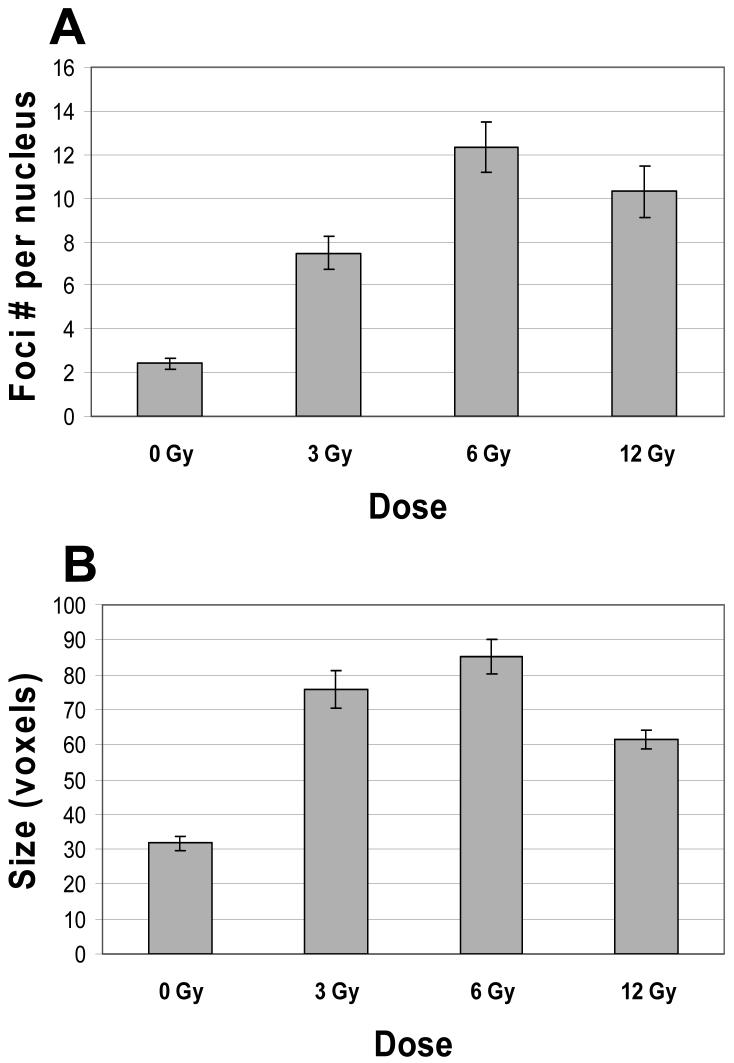
This double target imaging approach with ICA was also used to examine the induction of Mre11/Rad50 foci in the nuclei of cells irradiated with various doses of X-rays (0-12 Gy). Although nuclear foci were detected in both unirradiated control (∼ 60%) and irradiated cells (∼ 95%), 70-90% of irradiated cells had nuclei with large foci (≥1 large focus per nucleus having ≥ 40 voxels), while only 20% of control cells had nuclei with large foci.2 Since IR-induced foci are heterogeneous in size, all foci (including small foci having 11-39 voxels) were included in an analysis of the average number of foci per nucleus. The average number of foci per nucleus was calculated as follows: F = N1/N0, where F is the average number of foci per nucleus; N1 is the total number of foci; N0 is the total number of nuclei examined. The F values for control cells (0 Gy) and cells irradiated with 3, 6, and 12 Gy, are shown in Figure 5A. The number of foci per cell nucleus increased with dose up to 6 Gy. A 12 Gy exposure did not cause a further increase of focus formation compared to 6 Gy-irradiated cells (Fig 5A). Analysis of the average sizes of nuclear foci showed that the nuclear foci in control (unirradiated) cells were much smaller than those in irradiated cells (Fig. 5B). Interestingly, the nuclear foci in 3 Gy- and 6 Gy-irradiated cells were larger than those in 12 Gy-irradiated cells (Fig. 5B). Background staining of Mre11 and Rad50 protein decreased in irradiated cells, although no dose dependency was observed (data not shown).
Figure 5.
(A): Induction of nuclear focus formation in human U-1 melanoma cells that were irradiated with 0 (unirradiated control), 3, 6, and 12 Gy of X-rays, then incubated at 37 °C for 4 h, fixed/permeabilized, and immunostained for Mre11 and Rad50 proteins, as described in Materials and Methods. As expected, 6 Gy and 12 Gy exposures induced more foci per nucleus than a 3 Gy exposure did. However, a 12 Gy exposure did not induce more foci per nucleus than a 6 Gy exposure. The differences between F values for 3 Gy-irradiated cells and F values for control (0 Gy) and 6 Gy-irradiated cells were statistically significant (P < 0.05). The differences between F values for 12 Gy-irradiated cells and F values for 3 Gy- and 6 Gy-irradiated cells were statistically insignificant (P > 0.05). The total numbers of analyzed nuclei of control cells (0 Gy), and 3 Gy-, 6 Gy-, and 12 Gy-irradiated cells, were 156, 79, 77, and 93, respectively. (B): The average size of nuclear foci in control cells (0 Gy), and in 3 Gy-, 6 Gy-, and 12 Gy-irradiated cells (shown in voxels). All doses caused an increase in the size of foci. The average size of nuclear foci in irradiated cells was in 1.9-2.7 times larger than the average size of those in control cells. The difference between the average size of nuclear foci in 12 Gy-irradiated cells and the average sizes of foci in 3 Gy- and 6 Gy-irradiated cells was statistically significant ( P < 0.05). Data presented are the mean ± standard error of the mean of five independent experiments.
DISCUSSION
By means of double target imaging of nuclear foci stained for Mre11 and Rad50 proteins, and ICA of fluorescence signals from these proteins, we were able to identify those foci that were obscured due to the presence of Mre11 and Rad50 proteins in the nucleus that were not associated with foci. Unmasking of these “hidden foci” allowed for a more accurate focus detection and analysis. The conventional approach for focus analysis based on a single target imaging technique, when only one protein of the Mre11 complex (Mre11 or Rad50) is chosen for imaging of nuclear foci, was less accurate because of the high degree of uncertainty in the detection of “hidden foci”. For example, after the enhancement of foci by staining nuclei for Mre11 and Rad50 proteins and subsequent ICA of fluorescence signals from these proteins, we obtained 11 foci (“Mre11/Rad50” set of images, Fig. 2), compared to 9 foci obtained from images of a single target (either “Mre11” or “Rad50” set of images, Fig. 2). This enhancement of detection of foci of 22% was representative of other samples we analyzed. Single-color analysis is compared to two-color analysis in Table 2, which shows the magnitude of enhancement of focus detection in a group of cells irradiated with 6 Gy.
Table 2.
Numbers of foci in the group of nuclei (n = 13) of 6 Gy-irradiated cells before and after enhancement using ICA
| Nucleus | Mre11 foci No. before enhancement | Rad50 foci No. before enhancement | Mre11/Rad50 foci No. after enhancement |
|---|---|---|---|
| Overall enhancement for Mre11 foci: 26 % | |||
| Overall enhancement for Rad50 foci: 29 % | |||
| 1. | 4 | 3 | 5 |
| 2. | 12 | 6 | 11 |
| 3. | 13 | 13 | 16 |
| 4. | 9 | 9 | 11 |
| 5. | 15 | 17 | 19 |
| 6. | 12 | 10 | 12 |
| 7. | 4 | 7 | 12 |
| 8. | 18 | 16 | 20 |
| 9. | 6 | 8 | 8 |
| 10. | 9 | 13 | 12 |
| 11. | 11 | 16 | 17 |
| 12. | 8 | 3 | 9 |
| 13. | 7 | 5 | 10 |
| Mean ± SEM | 9.85 ± 1.16% | 9.69 ± 1.37% | 12.46 ± 1.22% |
Although the threshold adjustment for quantification of foci is subjective, it should be noted that with background staining of very low intensity, the number of the objects (e.g., foci) to be scored is affected very little within a broad range of threshold values. In the images with positive PDM values (“Mre11/Rad50” set of images, Fig. 1), there was a considerable reduction of background staining intensity (which is high in either “Mre11” or “Rad50” sets of images, Fig. 1). Thus, the analytical approach for focus analysis proposed in this study allows more flexibility in threshold adjustments, and better determination of the number of foci. The enhancing factor (see Results) can be used as a criterion for threshold adjustments to quantify foci obtained by using this approach. For example, if a threshold level value of 175 is chosen to score foci in both individual Mre11 and Rad50 sets of images, by multiplying this value by the enhancing factor of 1.28, we obtain the threshold level value (i.e., 222), that can be set for quantifying foci in the merged Mre11/Rad50 set of images. In the present work, the threshold settings were kept constant while performing the comparative analysis of foci in randomly selected nuclei of control cells and irradiated cells.
To date, there is only limited information on the quantitative aspects of IR-induced formation of Mre11/Rad50/Nbs1 nuclear foci in mammalian cells. A few laboratories have used epifluorescence microscopy and manually (i.e., by eye) scored Mre11 and/or Rad50 foci in human fibroblasts irradiated with various doses (1-12 Gy) of X- or γ-rays (11,15,16). In these studies, the number of nuclei of cells irradiated with doses greater than 1 Gy that exhibited focus formation was similar to that observed in our study (if one takes into account only the nuclei that have large foci). However, in our study, we observed that ∼ 20% of unirradiated cells have large nuclear foci. This is about 2-fold more than the number of focus-containing unirradiated human fibroblasts. The increased number of nuclear foci in melanoma cells may be attributed to an increased number of DNA DSBs compared to normal fibroblasts. Malignant melanoma cells have been shown to express elevated levels of γ-H2AX foci (23), which is indicative of the presence of DSBs. Inactivation of the retinoblastoma tumor suppressor (pRb) gene that occurs in many tumor cells lines, can deregulate activity of E2F1 transcription factor (24), a mechanism which links to induction of DSBs (25). Malignant melanoma cells have also been shown to have deregulated E2F transcriptional activity (26). Interestingly, E2F1, which is the most abundant of E2F family members in malignant melanoma cells (26), when it is overexpressed in normal human dermal fibroblasts, leads to formation of nuclear foci similar to IR-induced foci that result from DSBs (27). These foci were positively stained for Mre11, Nbs1, γ-H2AX, and 53BP1 proteins, and all of the proteins co-localized (27). F values obtained for melanoma cells (Fig. 5A) were not much different from F values obtained for primary human fibroblasts (11,15,16). The only major difference between our findings and those findings described by Maser et al. (11), van Veelen at al. (15), and Digweed et al. (16) is that we did not observe a further increase in the number of nuclear foci when the radiation dose was increased from 6 Gy to 12 Gy (Fig. 5A). This finding is perplexing in light of the findings that IR induces DSBs in a dose-dependent, linear fashion (28,29). At the present time, one can only speculate about why there is a discrepancy between the number of DSBs and foci for a given dose. There is a lack of information in the literature about time-dependent effects on Mre11/Rad50/Nbs1 focus formation after irradiation of cells with various doses of IR. However, perhaps the discrepancy is due to a saturation effect, whereby the number of nuclear foci remains unchanged, despite of progressive induction of DSBs [35-40 DSBs/cell/Gy (28,30)] with increasing dose. Therefore, the number of foci per nucleus would remain unchanged or could drop. Indeed, there is a slight drop in F value of 12 Gy-irradiated cells, compared to F value of 6 Gy-irradiated cells (Fig. 5A). Since the experimental results revealed much less foci per nucleus than theoretically predicted (31), this supports our hypothesis. While an excessive amount of DSBs in close proximity could result in the coalescence of smaller foci into larger ones, this cannot explain the data for 12 Gy irradiated cells, where the size of the foci is smaller than foci in 3 Gy- or 6 Gy-irradiated cells (Fig. 5B). It is possible that with an increased amount of damage, the amount of each protein per focus decreases because there is a finite number of copies of each particular protein. However, chromatin density at the sites of DSBs can influence antibody staining; thus it is also possible that chromatin density changes are dose-dependent over a finite dose range, leading to a non-linear relationship between dose and number of foci per nucleus (19). Localized changes in chromatin density within different chromatin domains in a nucleus could be one of the reasons why one focus is brighter than another within the same nucleus. This could also explain the saturation phenomenon observed at higher radiation doses.
In summary, correlation analysis of images obtained using LSCM permitted enhanced detection of DNA-damage induced Mre11/Rad50 foci, and more accurate scoring of foci. This tool may be helpful for researchers who rely on scoring of nuclear foci in their studies.
ACKNOWLEDGEMENTS
This work was supported by NIH grant CA108582 (JRD). The authors wish to acknowledge support from the Indiana Center for Biological Microscopy.
Footnotes
ICA assumes that if two proteins are parts of the same complex then their staining intensities should vary in synchrony, whereas if they are on different complexes or structures they will exhibit asynchronous staining.
Large foci had the range of diameters of 0.6-2.0 µm, while the range of diameters of small foci was 0.3-0.59 µm.
LITERATURE CITED
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E, et al. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- 3.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assenmacher N, Hopfner K-P. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 5.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 6.Dolganov GM, Maser RS, Novikov A, Tosto L, Chong S, Bressan DA, Petrini JHJ. Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J-H, Ghirlando R, Bhaskara V, Hoffmeyer MR, Gu J, Paull TT. Regulation of Mre11/Rad50 by Nbs1: effects on nucleotide-dependent DNA binding and association with ataxia-telangiectasia-like disorder mutant complexes. J Biol Chem. 2003;278:45171–45181. doi: 10.1074/jbc.M308705200. [DOI] [PubMed] [Google Scholar]
- 8.Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JHJ. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci U S A. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, et al. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol. 2001;21:2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maser RS, Monsen KJ, Nelms BE, Petrini JHJ. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd., Hays L, Morgan WF, Petrini JHJ. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 13.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JHJ. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 14.Petrini JHJ. The mammalian Mre11-Rad50-Nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am J Hum Genet. 1999;64:1264–1269. doi: 10.1086/302391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Veelen LR, Cervelli T, van de Rakt MWMM, Theil AF, Essers J, Kanaar R. Analysis of ionizing radiation-induced foci of DNA damage repair proteins. Mutat Res. 2005;574:22–33. doi: 10.1016/j.mrfmmm.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Digweed M, Demuth I, Rothe S, Scholz R, Jordan A, Grötzinger C, Schindler D, Grompe M, Sperling K. SV40 large T-antigen disturbs the formation of nuclear DNA-repair foci containing MRE11. Oncogene. 2002;21:4873–4878. doi: 10.1038/sj.onc.1205616. [DOI] [PubMed] [Google Scholar]
- 17.Sedelnikova OA, Rogakou EP, Panyutin IG, Bonner WM. Quantitative detection of 125IdU-induced DNA double-strand breaks with γ-H2AX antibody. Radiat Res. 2002;158:486–492. doi: 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 18.Böcker W, Iliakis G. Computational methods for analysis of foci: validation for radiation-induced γ-H2AX foci in human cells. Radiat Res. 2006;165:113–124. doi: 10.1667/rr3486.1. [DOI] [PubMed] [Google Scholar]
- 19.Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 20.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 21.Rasband WS. ImageJ. U. S. National Institutes of Health; Bethesda, Maryland, USA: pp. 1997–2007. http://rsb.info.nih.gov/ij/ [Google Scholar]
- 22.Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin 1, Gα0, and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warters RL, Adamson PJ, Pond CD, Leachman SA. Melanoma cells express elevated levels of phosphorylated histone H2AX foci. J Invest Dermatol. 2005;124:807–817. doi: 10.1111/j.0022-202X.2005.23674.x. [DOI] [PubMed] [Google Scholar]
- 24.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 25.Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene. 2006;25:746–755. doi: 10.1038/sj.onc.1209103. [DOI] [PubMed] [Google Scholar]
- 26.Halaban R, Cheng E, Smicun Y, Germino J. Deregulated E2F transcriptional activity in autonomously growing melanoma cells. J Exp Med. 2000;191:1005–1015. doi: 10.1084/jem.191.6.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frame FM, Rogoff HA, Pickering MT, Cress WD, Kowalik TF. E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene. 2006;25:3258–3266. doi: 10.1038/sj.onc.1209352. [DOI] [PubMed] [Google Scholar]
- 28.Nevaldine B, Longo JA, King GA, Vilenchik M, Sagerman RH, Hahn PJ. Induction and repair of DNA double-strand breaks. Radiat Res. 1993;133:370–374. [PubMed] [Google Scholar]
- 29.Löbrich M, Rydberg B, Cooper PK. Repair of x-ray-induced DNA double-strand breaks in specific Not I restriction fragments in human fibroblasts: joining of correct and incorrect ends. Proc Natl Acad Sci U S A. 1995;92:12050–12054. doi: 10.1073/pnas.92.26.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Löbrich M, Ikpeme S, Kiefer J. Measurement of DNA double-strand breaks in mammalian cells by pulsed-field gel electrophoresis: a new approach using rarely cutting restriction enzymes. Radiat Res. 1994;138:186–192. [PubMed] [Google Scholar]
- 31.Costes SV, Ponomarev A, Chen JL, Nguyen D, Cucinotta FA, Barcellos-Hoff MH. Image-based modeling reveals dynamic redistribution of DNA damage into nuclear subdomains. PLoS Comput Biol. 2007;3:1477–1488. doi: 10.1371/journal.pcbi.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]