Abstract
Amyotropic lateral sclerosis (ALS) is a neurodegenerative disease linked to misfolding and aggregation of the homodimeric enzyme superoxide dismutase (SOD1). In contrast to the precursors of other neurodegenerative diseases, SOD1 is a soluble and simple-to-study protein with immunoglobulin-like structure. Also, there are more than 120 ALS-provoking SOD1 mutations at the disposal for detailed elucidation of the disease-triggering factors at molecular level. In this article, we review recent progress in the characterization of the folding and assembly pathway of the SOD1 dimer and how this is affected by ALS-provoking mutations. Despite the diverse nature of these mutations, the results offer so far a surprising simplicity. The ALS-provoking mutations decrease either protein stability or net repulsive charge: the classical hallmarks for a disease mechanism triggered by association of non-native protein. In addition, the mutant data identifies immature SOD1 monomers as the species from which the cytotoxic pathway emerges, and point at compromised folding cooperativity as a key disease determinant. The relative ease by which these data can be obtained makes SOD1 a promising model for elucidating also the origin of other neurodegenerative diseases where the precursor proteins are structurally more elusive.
Amyotropic lateral sclerosis (ALS) is a neurodegenerative disease affecting primarily the upper and lower motor neurons in the spinal cord (Bruijn et al., 2004). A characteristic feature of ALS is the accumulation of granular, intracellular aggregates of the ubiquitous radical scavenger superoxide dismutase (SOD1) (Fig. 1). In common with other neurodegenerative disorders like Alzheimer’s disease and Parkinson’s disease, ALS is not caused by loss of function of the aggregating protein but seems rather to arise from a generic toxicity of misfolded proteins (Gurney et al., 1994; Ripps et al., 1995). The mechanism by which such misfolded proteins escape the cellular housekeeping system and provoke neurotoxicity is yet poorly understood (Balch et al., 2008; Dobson, 1999). A contributing factor is that the molecular identity of the toxic species has remained elusive. Partly, since they tend to be hidden in high backgrounds of aggregating material but also since the structures of the disease-provoking proteins are often intrinsically dynamic and hard to characterise (Dickson, 2003). From this perspective, ALS presents several simplifying features (Shaw and Valentine, 2007). The causative protein SOD1 is normally a perfectly soluble homo dimer with well defined tertiary structure, which is expressed and functional inside the neurons. Moreover, the first signs of injury to spinal cord arise before the accumulation of SOD1 inclusions in transgenic mice (Jonsson et al., 2006), indicating that the cytotoxic precursor does not always coexist with macroscopic aggregation (Zetterstrom et al., 2007). Finally, the familial form of ALS has been linked to more than 120 mutations in the SOD1 structure providing a unique tool for molecular analysis of the disease mechanism (Andersen et al., 2003) (Fig. 1). Studies of the structural behavior of SOD1 is thus not only important for understanding ALS but could also shed light on the molecular events underlying other misfolding diseases where the responsible proteins are less amenable to experimental analysis.
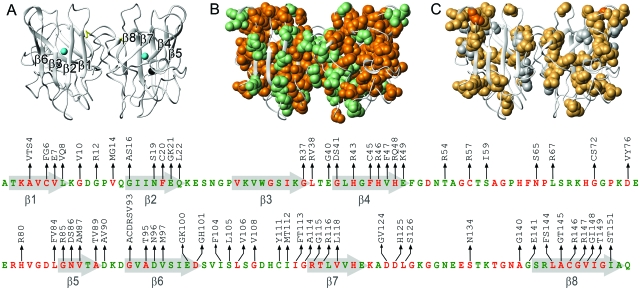
Figure 1. Structures of SOD1, pdb code: 1SPD (Deng et al., 1993), showing the distribution of conserved amino-acid positions and the ALS-associated mutations analyzed in this study.
(a) Ribbon structure showing the positions of the Zn (black) and Cu (blue) ions, and the 57–146 disulphide bond (yellow). (b) The distribution of ALS mutations in conserved (orange) and variable (green) sites. Below is the corresponding SOD1 amino-acid sequence showing the ALS mutations in conserved (orange) and variable (green) positions. (c) The distribution of ALS-associated SOD1 mutations that decrease (yellow) and increase (white) the proteins’ repulsive charge. In position G93 (orange) there are ALS mutations that both decrease (G93R) and increase (G93D) the net repulsive charge. Figure adapted from Sandelin et al. (2007).
To keep focus on the folding behavior of wildtype and mutant SOD1 we have been forced to leave out many of the interesting aspects of the ALS mechanism relating to pathology, cell cultures, and transgenic model organisms. As excellent reviews on these subjects we recommend (Bruijn et al., 2004; Shaw and Valentine, 2007).
SETTING THE CASE: MOLECULAR DETERMINANTS FOR PROTEIN AGGREGATION
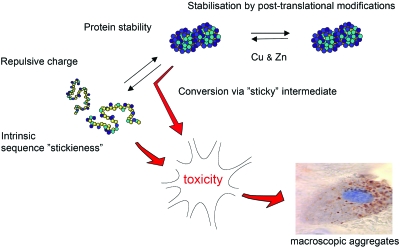
The molecular factors likely to influence protein aggregation in vivo are schematically outlined in Fig. 2, and can be summarized as follows: (1) Protein stability. The conformational accessibility of the precursor protein to aggregation is generally determined by protein stability. This describes the relative fractions of unfolded and folded protein and, thus, the effective concentration of aggregation-competent material. For peptide precursors that cannot adapt safe, folded structures the fraction of aggregation-competent, denatured material is 1, whereas for a globular protein it is typically much smaller at 10−2–10−8 (Glover et al., 1997). The molecular basis for protein stability is not only the cooperative action of residue-residue interactions (Fersht, 1999) but involves also the coordination of stabilizing cofactors (Wittung-Stafshede, 2004), competing interactions with membranes (Vande Velde et al., 2008), and other biological macromolecules (Dunker et al., 2002). (2) Net repulsive charge. To maintain solubility of the cellular interior most macromolecules carry a net repulsive charge that is negative under physiological conditions. Decreasing the repulsive charge is generally observed to increase the extent of protein aggregation. (3) Intrinsic “stickiness” of the polypeptide chain. In the simplest case, the intrinsic stickiness of an amino-acid sequence is determined by the local sequence hydrophobicity: the greasier the segment, the lower the solubility. There are also instances where relatively hydrophilic sequences are observed to assemble into highly ordered fibrillar aggregates, e.g., the polyglutamine repeats associated with Huntington’s disease (Michalik and Van Broeckhoven, 2003) and the yeast prion sup35 (Glover et al., 1997). Accordingly, the link between sequence signatures and aggregation propensity is sometimes intricate and seems to be different for different types of aggregate morphologies (Rousseau et al., 2006a). As a defence against protein aggregation, natural proteins seem to disperse their sequences with charges, so called aggregation gatekeepers, that prevent the hydrophobic segments from becoming too long (Otzen et al., 2000; Rousseau et al., 2006b; Thirumalai et al., 2003). Even so, most proteins require some degree of contiguous hydrophobicity for chaperone binding (Rousseau et al., 2006b), folding and structural stability, indicating that the coil solubility in vivo is a delicate balance that is easy to break. In the case of SOD1, there is also the possibility to form covalent crosslinks in the form of disulphide bridges since the protein contains four cysteines per monomer. (4) Sticky intermediates. It is further conceivable that aggregation is triggered at tertiary level by several residues coming together to form a “sticky patch.” An example of such aggregation precursors would be partly unfolded or misfolded intermediates with excessive exposure of hydrophobic surface area (Canet et al., 2002; Lynch et al., 2004; McParland et al., 2002; Nordlund and Oliveberg, 2006; Platt et al., 2005). Alternatively, incompletely folded structures can assembly through domain swap where native-like interactions appear across several copies of matching polypeptide chains (Liu et al., 2001). The susceptibility to domain swap is general and stems from the protein’s selective preference for native contact patterns (Oliveberg, 1998; Silow et al., 1999; Yang et al., 2004). Accordingly, domain swap has been put forward as a key mechanism in the evolution of multidomain structures (Bennett et al., 1994) and is also observed naturally in functional oligomerization (Wilczynska et al., 2003). As will be discussed below, the diverse set of ALS-associated SOD1 mutations seems to target the majority of these determinants for protein aggregation.
Figure 2. The molecular and thermodynamic factors controlling protein aggregation.
The simplistic scheme assumes that the toxic species is a precursor to the macroscopic SOD1 inclusions that accumulate in the neurons. Alternatively, the toxicity could arise from species that accumulate in parallel with SOD1 aggregation.
SOD1 STABILITY: A DELICATE SUM OF SEVERAL COMPONENTS
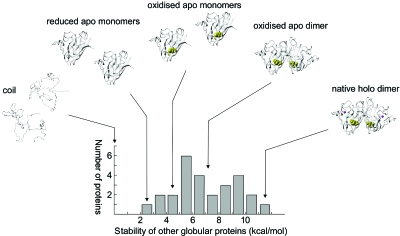
The native SOD1 structure contains several characteristic features that contribute to its stability: the intrinsic stability of the individual monomers, the energetic contribution of the C57-C146 disulphide bridge, the strength of the dimer interface, and the stabilising effect of coordinating the functional Cu and Zn ions (Fig. 3).
Figure 3. The species on the maturation pathway of SOD1 span the whole range of stabilities observed for other globular proteins.
It has been put forward that the low stability of the reduced apo monomer constitutes an Achille’s heel that makes the protein susceptible to neurotoxic misfolding (Lindberg et al., 2004). Stability data adapted from Jackson (1998). The protein folds as a three-state dimer where docking is preceded by the cooperative formation of the individual monomers, consistent with the relatively small dimer interface of SOD1 (Levy et al., 2004).
A simplifying factor in experimental analysis of SOD1 is that the individual monomers fold independent of the dimer interface and, thus, can be studied on their own. The analysis is further facilitated by the sidechain replacements F50E and G51E (Bertini et al., 1994) that selectively obstruct the dimer interface without significantly affecting the folding and stability of the monomeric species (Lindberg et al., 2004; Svensson et al., 2006). Like other immunoglobuline-like proteins (Clarke et al., 1999), the apo-SOD1 monomer acquires its structure in a highly cooperative two-state transition between the denatured (D) and native (N) conformations. The intermediate species in this transition are predominantly unstable forming a free-energy barrier that normally safeguards the structural integrity of the native state: the higher the barrier, the less susceptible is the protein to sample partly structured states (Fig. 4). For an ideal two-state protein the equilibrium constant for folding (KD∕N) is given by
| (1) |
where kf and ku are the rate constants for folding and unfolding, respectively (Fersht, 1999). Protein stability (ΔGD-N) is then defined as
| (2) |
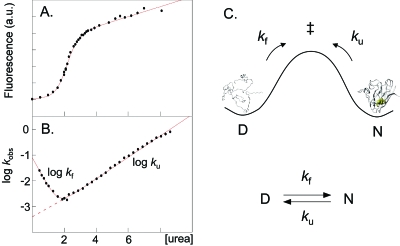
Figure 4. Equilibrium denaturation data (a) and chevron data (b) of the apo SOD1 monomer shows that it displays classical two-state transition between the denatured (D) and native (N) states.
Accordingly, the SOD1 stability can be described simply by the ratio of the unfolding and refolding rate constants (c).
An accurate method for determining the folding and unfolding kinetics and, thus, the protein stability is the chevron analysis that employs denaturants in the form of urea or GdmCl (Fersht, 1999). To obtain kf, the protein is first denatured in urea or GdmCl and then rapidly diluted into refolding conditions by stopped-flow mixing with physiological buffer. Vice versa, ku is obtained by mixing the folded protein into high concentrations of denaturant. The result is typically a v-shaped plot of log kf and log ku versus [urea] or [GdmCl] where the values of kf and ku in pure physiological buffer (log and log ) are obtained by linear extrapolation (Fig. 4)
| (3) |
For the reduced apo-SOD1 monomer, that represents the first step in the maturation pathway to the native homodimer, the values of kf and ku are about 10−1 and 10−3 s−1, respectively (Lindberg et al., 2004). In other words, in physiological buffer the protein flicks constantly between the denatured and folded conformations, spending on average 23 min in N and 10 s in D. The corresponding value of KD∕N is 1∕140 and ΔGD∕N=2.9 kcal∕mol. For comparison, other two-state proteins typically display stabilities in the range of 2–10 kcal∕mol (Glover et al., 1997). The stability of the apo-SOD1 monomer is thus relatively low. Addition of the native disulphide linkage between C57 and C146 offers small improvement, increasing the stability of the apo-SOD1 monomer to 4.5 kcal∕mol (Lindberg et al., 2004). A more substantial gain of stability is observed upon formation of the dimer interface, which contributes to an apparent first-order stability of 7 kcal∕mol at 4 μM of protein (Lindberg et al., 2004; Svensson et al., 2006). The levels of denatured and monomeric protein are roughly reduced by a factor of 300 at physiological protein concentrations, moving the apo-SOD1 dimer to a more normal stability regime (Fig. 3). Even so, the rate-limiting step in folding of homodimeric SOD1 remains the cooperative formation of the monomeric species. Dimerization is thus a downstream event that has no impact on the initial folding events; it seems just to trap the fully formed monomers. The largest individual contribution to SOD1 stability, however, seems to be the coordination of Cu and Zn: it even exceeds the intrinsic stability of the monomer framework. From equilibrium data the stability gain of metallation has been estimated to more than 10 kcal∕mol (Stroppolo et al., 2000), see also Lynch and Colon (2006); Potter and Valentine (2003), and preliminary results from monomeric protein yields even higher values, linked to theoretical lifetimes of several months for the holo monomer in pure buffer (unpublished). Regardless of the uncertainty in these numbers, it is clear that metallation of SOD1 efficiently locks the protein in a most stable and long-lived state that does not easily lose its structural integrity under in vitro conditions (Fig. 3).
However, the individual contributions to the SOD1 stability are not simply additive but are coupled through allosteric interactions that have important implications for how the maturation process advances. Most notably, the apo-SOD1 monomer is not able to dimerise at μM concentrations unless the C57-C146 disulphide linkage is formed (Furukawa et al., 2004; Lindberg et al., 2004). The structural basis for this redox regulation of the SOD1 assembly lies in the long loop IV between residues H48 and D83 that needs to be pinched to the β barrel to better fit the dimer interface (Hornberg et al., 2007). Consistently, dimerisation of reduced SOD1 can also be promoted by metallation that imposes similar restrictions to loop IV (Hornberg et al., 2007; Lindberg et al., 2004). From a disease perspective, this coupling means that a substantial fraction of the newly synthesized SOD1 molecules could reside as marginally stable monomers prior to disulphide oxidation. Although it is not yet clear how the disulphide oxidation is orchestrated in the reducing environment of the cytosol, several observations point at the involvement of the Cu-inserting chaperone CCS (Banci et al., 2006; Furukawa et al., 2004; Lamb et al., 2001). The activation of immature SOD1 into functional, metallated dimers would then occur in one concerted step, substantially limiting the number of intermediates in the maturation pathway. SOD1 would tend to polarize into reduced monomers and fully native dimers, the relative fractions of which are under the control of CCS and metal accessibility. The existence of such a biased distribution of SOD1 species in vivo lends further support to the idea that the disease-provoking properties emerge from immature apo protein (Assfalg et al., 2003; Baglioni et al., 2006; Furukawa et al., 2008; Hough et al., 2004; Jonsson et al., 2006; Khare and Dokholyan, 2006; Lindberg et al., 2005; 2002b; Lynch et al., 2004; Rakhit et al., 2004; Sherman and Goldberg, 2001; Silveira et al., 2005; Tiwari and Hayward, 2003).
DISEASE FACTORS IMPLICATED BY ALS-ASSOCIATED MUTATIONS 1: DECREASED SOD1 STABILITY
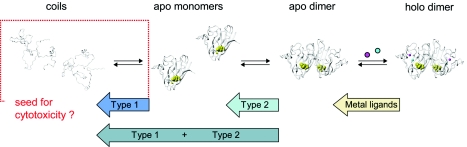
As the vast majority of ALS-associated SOD1 mutations have full penetrance, the disease-provoking properties reside by necessity in each of their structural characteristics: if you have the mutation you inevitably get the disease. In this perspective, the more than 120 ALS-associated mutations provide a unique research material for mapping out the underlying, disease-triggering factors at a molecular level. Of immediate notice is that the ALS-associated SOD1 mutations investigated so far have their main impact on the unfolding kinetics (Lindberg et al., 2005). The corresponding effect on the refolding rate constant is overall marginal. This shows that the energetic impact of these mutations is predominantly after the transition state, i.e., they perturb late-forming structure on the native side of the monomer barrier. It is further apparent that the way the ALS mutations affect the late-folding features of SOD1 is not the same but varies. The mutations can be categorised as follows. Type-1 mutations that affect selectively the structure of the monomer, type-2 mutations that weaken exclusively the dimer interface, and type 1+3 mutations that perturb both the monomer and dimer formation (Fig. 5). In addition, there is a class of mutations that destabilizes the holo species by interfering with the metal coordination (Crow et al., 1997; Roberts et al., 2007; Shaw and Valentine, 2007) and, finally, a group of mutations that decreases the protein’s repulsive charge with only very small or negligible effects on protein stability (Sandelin et al., 2007). The disease-provoking property of the ALS mutations can thus not be linked to a defined part of the structure but is more likely to act on the conformational equilibrium (Lindberg et al., 2005). Consistently, the mutations that do not affect repulsive charge display a unifying denominator in the form of a shift of the folding equilibrium towards denatured and partly folded apo monomers. In other words, the destabilizing ALS mutations seem to accentuate the already questionable trait of the wild-type protein: the predisposition to populate incompletely folded apo species.
Figure 5. The ALS-associated SOD1 mutations describe a common shift of the folding equilibrium towards denatured and partly unfolded apo monomers, by either destabilize the monomer structure (type 1), weaken the dimer interface (type 2), or obstruct the formation of the holo dimer through replacement the metal ligands.
This common pattern implicates that the seed for cytotoxicity is to be found among the immature apo monomers. For simplicity, the scheme only outlines the species encountered under oxidizing conditionsin vitro.
The detailed quantitative description of the SOD1 mutations is unique for a protein associated with neurodegenerative disease and provides an important reference for investigations of putative couplings to the clinical manifestation of ALS.
Accordingly, analysis of ALS-mutations selected on the basis of reliable clinical statistics has indicated a correlation between increased population of denatured apo monomers and decreased survival time after first diagnosis (Lindberg et al., 2005). The implication of this observation, however, is yet uncertain and recent extensions of the data set seem to reveal several outliers. A more robust interpretation is possibly that severely destabilizing mutations are overrepresented among patients with exclusively short survival times (<5 years), and mutations with small or marginal impact on protein stability are overrepresented among long (>5 years) and variable survival times. It is also notable that similar correlations between loss of protein stability and clinical impact have been implicated for X-linked hydrocephalus (Randles et al., 2006) and the cancer suppressor p53 (Joerger and Fersht, 2008). In contrast to ALS, the latter diseases arise from loss of protein function. Nevertheless, it is conceivable that mutational decrease of protein stability below a certain threshold is coupled to the same basic problem: structural integrity is lost and the cell is burdened with increased load of denatured protein. If the function of the affected protein is critical, this leads to acute viability problems. If not, gain-of-function phenomena could arise as a secondary complication on longer time scales.
DISEASE FACTORS IMPLICATED BY ALS-ASSOCIATED MUTATIONS 2: DECREASED REPULSIVE CHARGE
A distinct group of outliers in the putative relation between SOD1 stability and disease progression is the mutations altering the proteins net charge. In the extreme cases, these mutations can lead to ALS without affecting the protein stability (Shaw and Valentine, 2007). Some representative example are the mutations D76V and D101N that reduce the monomer net charge from −6 to −5 and still maintain wild-type-like chevron plots (Roberth Byström and M.O., unpublished). Yet, these mutations are observed to be fully penetrating (Andersen et al., 2003), indicating that even minor charge alterations can trigger neural damage. Consistently, bioinformatic analysis of the extensive set of ALS-associated SOD1 mutations shows that these are site selective by targeting preferentially side chains with negative charge (Sandelin et al., 2007). The accompanying charge alteration is on average +0.30 per ALS mutation, which is an order of magnitude larger than what is expected for random mutations (Sandelin et al., 2007). The very existence of this signal at genetic level suggests that loss of negative charge is disease factor in its own right. If it was only modulating and required a second, dominant molecular factor to cause ALS it would not be penetrant. Moreover, the charge signal is correlative since ALS can be triggered in more than one ways: by decreased SOD1 stability, reduced negative charge, or a combination of both. As the sites of the charge mutations appear to be scattered rather randomly throughout the SOD1 structure their effect is unlikely to be structurally specific. More likely, the impact is global and coupled to a weakening of the Coulombic repulsion between the individual SOD1 molecules and their overall negatively charged cellular environment (Sandelin et al., 2007). In the few ALS mutations where the SOD1 mutations produce an increased repulsive charge the sidechain substitutions tend also to be structurally obstructive and to occur in strictly conserved positions. The opposed effect of increased repulsive charge is here not strong enough to overcome the accompanying penalties in protein stability. Interestingly, the only other set of human mutations that seems to show a corresponding charge effect are those associated with the systemic amyloid disease FAP (Sandelin et al., 2007). Similar to SOD1, the disease-provoking protein is here a soluble tetramer of four identical immunoglobuline-like subunits (Sekijima et al., 2005). Taken together, the combined dependence on protein stability and net charge suggests that the causative event in ALS involves protein aggregation. Alternatively, a similar pattern could arise from a disease mechanism triggered by erroneous or disproportionate interactions between non-native SOD1 and other negatively charged biomolecules, for example other proteins (Zhang et al., 2007), lipids (Aisenbrey et al., 2008), genetic material or chaperones (Okado-Matsumoto and Fridovich, 2002). The latter scenario needs not to exclude that SOD1 aggregation occurs in parallel, or even augments, the disease progression. Such parallel accumulation of protein aggregates could, for example, result from exhaustive overload of the cellular housekeeping system by denatured monomers. The “key” disease-provoking event would here disperse to systems level, involving a complex network of modulating factors (Balch et al., 2008).
STICKY SEQUENCE REGIONS: WHICH PART OF SOD1 FORMS THE PATHOLOGIC INTERACTION?
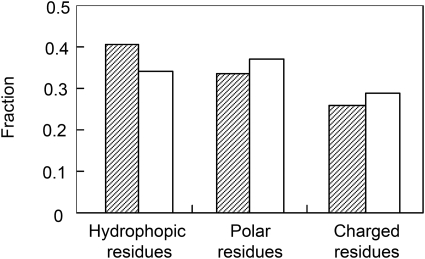
One of the first features that strike the experimentalist working with recombinant SOD1 is that the protein is remarkably soluble. Highly destabilized mutants like, e.g., G93A (Lindberg et al., 2005) can easily be purified and studied at nuclear magnetic resonance (NMR) concentrations in physiological buffer despite being fractionally unfolded. Under corresponding conditions, most other globular proteins of similar size would precipitate. An explanation for the high solubility of SOD1 can be found in the protein’s sequence composition that shows a distinct bias towards hydrophilic residues (Fig. 6). Accordingly, SOD1 lacks many of the (hydrophobic) sequence signatures that have been associated with aggregation in other proteins (Thirumalai et al., 2003) and model peptides (Rousseau et al., 2006a), and the aggregation propensity as calculated by TANGO (Fernandez-Escamilla et al., 2004) is essentially zero. It is tempting to speculate that this resistance to aggregation is an adaptation to the high in vivo concentration of SOD1 and the intrinsically low stability of the apo state. However, aggregation of SOD1 can readily be induced at pH values below 5 where the net repulsive charge is reduced (DiDonato et al., 2003; Stathopulos et al., 2003). Although it is uncertain whether such low pH is a parameter for aggregation in vivo, the observations point at an underlying propensity of SOD1 to aggregate upon loss of negative charges. The behavior is also in line with the charge selectivity of the ALS-associated SOD1 mutations. Aggregation of SOD1 can also be provoked in a more generic manner by subjecting the protein to organic solvents (Stathopulos et al., 2003), the effect of which is essentially to promote backbone hydrogen-bond formation by replacing competing water molecules (Thirumalai et al., 2003). An interesting example of a sequence-specific mode of SOD1 aggregation has been observed in the crystal lattice of the ALS-associated mutant S134N (Elam et al., 2003). In this case, the intermolecular interface is comprised of a short segment of loop VII that binds to an edge strand in the neighboring molecule (Elam et al., 2003). For the binding to occur, loop VII must first unfold locally. Yet another mode of SOD1 aggregation is by intermolecular disulphide bonding. Normally such oxidative crosslinking is not expected to take place in the reducing environment of the cytosol, but under conditions of cellular stress or in specific organelles the situation could well be different. For example, in neurons that were cultured aerobically, neurotoxicy was only observed if the overexpressed SOD1 contained C111 (Cozzolino et al., 2008), or both C6 and C111 (Niwa et al., 2007). The results are supported by observations that the free cysteines C6 and C111 are generally required for oligomerization of SOD1 into β-rich aggregates under oxidizing conditions in vitro (Banci et al., 2008). Of particular interest is here that metallation of SOD1 is observed to greatly suppress the disulphide-promoted aggregation (Banci et al., 2007), consistent with the view that the apo protein is the cytotoxic precursor. However, there are also contradictory reports showing that the common denominator of ALS mutations in transgenic mice is reduced material (Zetterstrom et al., 2007) and that disulphide-reduced apo is the most facile precursor for amyloid-like aggregation in vitro (Furukawa et al., 2008). The occurrence of disulphide-linked aggregates seems thus to be highly conditional and to depend in vivo on the redox stress of the cells under investigation. Nevertheless, mutant dependent formation of disulphide bonds is interesting since it could report on structural fluctuations of SOD1 structure, i.e., the disulphide crosslinks could serve to trap high-energy clusters of SOD1 intermediates that would otherwise escape detection.
Figure 6. The residue composition in SOD1 (open bars) contains higher fractions of polar and charged residues that the average protein in the human proteome (filled bars), providing a simple explanation for the relatively high solubility of its denatured state.
THE LINK BETWEEN PROTEIN FOLDING AND NATIVE-STATE INTEGRITY
The free-energy barrier separating the denatured and native states is not only contributing to make folding slow, but also plays an important role in safeguarding the structural integrity of the native state (Lindberg et al., 2002a). If the barrier gradient towards the native basin is too shallow, this will result in excessive sampling of locally unfolded conformations, i.e., the structure of the folded protein becomes “floppy.” Such locally unfolded, high energy, species have been implicated as starting material for aggregation of SOD1 (Nordlund and Oliveberg, 2006), lysozyme (Canet et al., 2002), and β2-microglobuline (Jahn and Radford, 2008;McParland et al., 2002; Platt et al., 2005). Notably, the detailed shape of the folding barrier, and thus the dynamic properties of the native structure, are not primarily determined by native-state stability [Eqs. 1, 2], but are rather controlled by the features of the folding-energy landscape (Bryngelson et al., 1995; Oliveberg and Wolynes, 2005). Studies of protein folding have shown that natural proteins generally fold in highly concerted two-state processes where most of the native contacts appear to be fractionally formed in the transition-state ensemble (‡) at the top of the folding barrier (Oliveberg and Wolynes, 2005). From the perspective of an individual protein molecule this implicates that the folding progression is broad and that the molecule is likely to sample different regions of the energy landscape in each consecutive jump along the folding barrier (Plotkin and Onuchic, 2000). The probability forming or losing the different native contacts is relatively unbiased. Such broad progressions across the folding-energy landscapes contribute also to efficiently reduce the occupancy of specific intermediates since none of the partly structured states are significantly more (im)probable than the rest. Interestingly, this diffuse folding behavior seems not necessary for the acquisition of native structure but can be an evolutionary adaptation to reduce the population of partly folded states (Lindberg et al., 2002a). Accordingly, it has been possible to completely change by circular permutation the archetypically broad folding reaction of the ribosomal protein S6 into much more narrow trajectories that in some cases also reverse the order of folding events (Lindberg et al., 2002a; 2006). The accompanying effects on the refolding rate constant and the native structure are overall negligible, indicating that the narrow folding trajectories would be frequently observed in natural proteins unless they were associated with biological disadvantages. One such disadvantage could be that a narrow trajectory across the folding-energy landscape forces all molecules to undergo the same conformational events. At the extreme, all of the unfolding attempts that constantly occur through mild thermal fluctuations would begin in the same part of the structure leading to fraying or local unfolding of the native state. If this local unfolding is incompatible with biological function, decreases the resistance to proteases, or leads to the exposure of sticky interfaces, it is easy to imagine that problems are close at hand. From this perspective, the folding pathway could be as important as protein stability in controlling protein aggregation and disease.
STRUCTURAL HOTSPOTS INDICATED BY THE SOD1 FOLDING REACTION
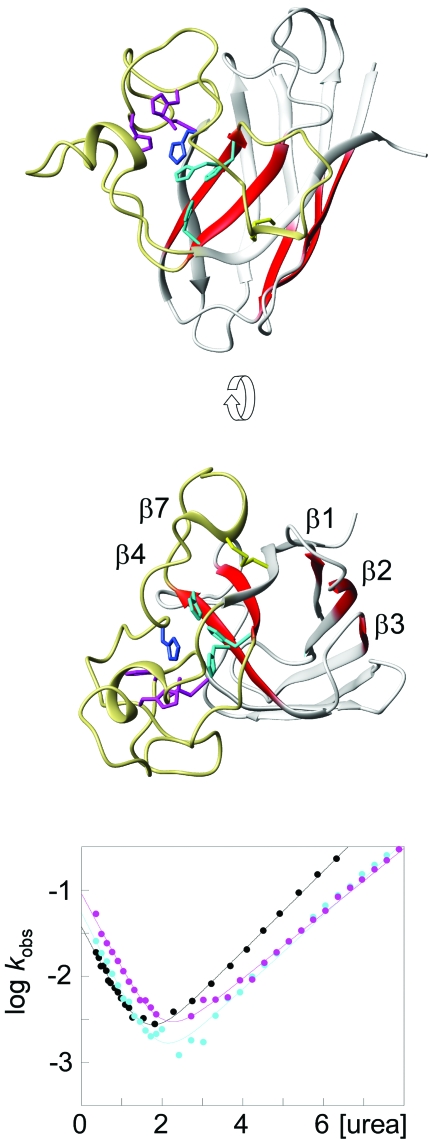
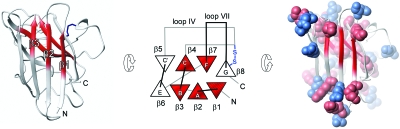
To see if there are any anomalies in the folding behavior of the disease-implicated apo SOD1 monomer, we characterized its folding transition state (‡) by ϕ-value analysis. The approach uses changes in the chevron data upon mutational truncation of individual sidechains to map out which of the native contacts that are present on top of the folding barrier (Fersht, 1999). From this snap-shot of the folding reaction it is then possible to infer in a qualitative manner which parts of the protein that unfold first, i.e., the parts that are most susceptible to unfold locally through native-state fluctuations. Vice versa, it sheds light on the structure of the species that form early in the folding reaction. The results show that the transition-state of the apo SOD1 monomer is mainly composed of contacts between β strands 1–3 and 4 and 7, spanning the center of the hydrophobic core. As the ϕ values of this nucleus are only fractional, the observed structure is not as consolidated as in the native state but more “expanded” (Fig. 7). For example, the hydrogen bonding between the involved strands appears only partially developed at macroscopic level and is likely to contain a diffuse pattern of bridging water molecules. This disperse nature of the transition-state structure is in good accordance with the nucleation-condensation behavior (Fersht, 1995) observed for the majority of other globular proteins (Jackson, 1998; Oliveberg and Wolynes, 2005). The interesting parts of the SOD1 nucleus, however, are the parts that are missing. Looking at the transition state from the unfolding side of the barrier, the SOD1 monomer has lost the capping strands on either sides of the central core. In the native state, these strands offer protection against aggregation by either being decorated with charged gatekeepers (β5 and β6) (Otzen et al., 2000; Richardson and Richardson, 2002), or by sealing up the continuous hydrogen bonding round the hinge of the SOD1 half barrel (β8) (Richardson and Richardson, 2002) (Fig. 8). Moreover, the protein has lost the Zn site, and the Cu site spanning strands 4 and 7 across the folding nucleus appears partially ruptured, consistent with complete disorder in the loops surrounding the catalytic site. Taken together, this means that the apo SOD1 monomer loses the majority of its aggregation-suppressing features early in the unfolding process, exposing the naked core to the solvent. The result is consistent with earlier reports from NMR showing that the loop regions of the apo SOD1 monomer, but not the edge strands, undergo considerable conformational exchange (Banci et al., 2003). Judged by the chevron data, however, the observed exchange would not indicate complete disorder in the folded ground state but rather fractional occupancy of an intermediate in which the loops, but not the edge strands, are unfolded. The compiled picture would thus be that the apo SOD1 monomer relatively frequently undergoes local unfolding of the loops, and more rarely samples a high-energy species in which also the β caps are gone (Fig. 7). Considering, the cellular concentrations of SOD1 is several μM, even very low occupancy of these partly unfolded species would match, or even exceed, the levels of the protein precursors in other neurodegenerative diseases. For comparison, the plasma concentration of Aβ peptides in patients with Alzheimer’s disease is only in the nM regime (Giedraitis et al., 2007). With respect to the ALS mechanism, both of the putative unfolding intermediates of the apo SOD1 monomer have some quite intriguing features. The more abundant species with disordered loops is predisposed to form the fibrillar arrays implicated in the crystal packing of the ALS-associated mutation S134N (Elam et al., 2003). Also, unfolding of the loops would increase the solvent accessibility of C111 implicated in erroneous oxidative crosslinking (Banci et al., 2008, 2007; Cozzolino et al., 2008; Niwa et al., 2007). It is apparent, however, that the exposure of C6, which has been put forward as the other partner in this crosslinking, needs further breaching of the SOD1 core since C6 forms part of the folding nucleus. The second SOD1 species lacking also the β caps appear to encompass the archetypical features of a sticky intermediates and shows also a striking resemblance with the amyloid precursor of β2 microglobulin (McParland et al., 2002). Besides these structural aspects of the putative SOD1 intermediates, it can be noted that the vast majority of the ALS-associated mutations enhance their occupancy by being localised outside the folding nucleus or at its interface (Nordlund and Oliveberg, 2006). The already questionable properties of the wild-protein seem to be made worse. Correspondingly, it is easy to envisage that metallation of the protein would efficiently contribute to reduce the extent of local unfolding by efficiently stabilizing the native ground state.
Figure 7. The folding nucleus of the oxidized apo SOD1 monomer is comprised by the strands β1, β2, β3, β4, and β7 forming diffuse contacts through the centre of the hydrophobic core (red).
The most frequently sampled unfolding intermediate of the SOD1 monomer has loops IV and VII locally unfolded (yellow). The second unfolding step is likely to involve unfolding of the protective β-strands β5, β6, and β8 on either side of the folding nucleus. The bottom panel shows the chevron plots for mutation of the Cu (cyan) and Zn (magenta) ligands to serines. The corresponding ϕ values are about 0.5 and 0, respectively.
Figure 8. The folding nucleus of the apo SOD1 monomer (red) and the protective β-strands β5, β6, and β8 (white).
β5 and β6 contain aggregation gatekeepers in the form of positive (red) and negative (blue) charges that prevents erroneous side-to-side contacts of the sheets, and β8 seals up the continuous hydrogen-bonds around the hinge of the SOD1 half barrel.
CONCLUSIONS
From a reductionist perspective, it is thus apparent that the molecular determinants for ALS fit nicely into a simplistic scheme for protein aggregation (Fig. 2). Although the vast number and broad structural distribution of the ALS-associated SOD1 mutations may at first be perplexing, they seem all to affect protein stability or repulsive charge. Decreased stability towards unfolding is either through mutations that sterically perturb the SOD1 structure (DiDonato et al., 2003; Lindberg et al., 2002b; Rodriguez et al., 2002) or loss of metal affinity (Banci et al., 2007; Shaw and Valentine, 2007), whereas the decrease of repulsive charge is preferentially through loss of negatively charged sidechains at the SOD1 surface. In the few cases where the ALS mutations produce an increased repulsive charge, the sidechain substitutions are accompanied by severe structural obstruction or occur in strictly conserved positions (Sandelin et al., 2007): the opposed effect of increased repulsive charge here is not sufficiently strong to compensate for the concomitant loss of protein stability. Taken together, these disease factors constitute the classical hallmarks for a toxicity mechanism based on aggregation, either in the form of self association (Chiti et al., 2003) or erroneous interaction with other negatively charged biomolecules. An important task is now to establish if there are any ALS mutations that deviate from this general pattern. For example, it is conceivable that ALS would be triggered by mutations that enhance the intrinsic stickiness of the SOD1 molecule (Otzen et al., 2000; Richardson and Richardson, 2002; Rousseau et al., 2006a; b; Thirumalai et al., 2003), increase the extent of misfolding by interfering with negative-design features (Otzen and Oliveberg, 1999; Richardson et al., 1992), or interfere negatively with binding to the SOD1-specific chaperone CCS (Lamb et al., 2001). It is also not clear which of the SOD1 features that actually promote SOD1 deposition in ALS. There seems to be many ways to aggregate proteins in vitro and all of these needs not to be disease relevant (Otzen and Oliveberg, 2004).
References
- Aisenbrey, C, Borowik, T, Bystrom, R, Bokvist, M, Lindstrom, F, Misiak, H, Sani, M A, and Grobner, G (2008). “How is protein aggregation in amyloidogenic diseases modulated by biological membranes?” Eur. Biophys. J. 37, 247–255. [DOI] [PubMed] [Google Scholar]
- Andersen, P M, et al. (2003). “Sixteen novel mutations in the Cu∕Zn superoxide dismutase gene in amyotrophic lateral sclerosis: a decade of discoveries, defects and disputes.” Amyotroph. Lateral Scler. Other Motor Neuron Disord. 4, 62–73. [DOI] [PubMed] [Google Scholar]
- Assfalg, M, Banci, L, Bertini, I, Turano, P, and Vasos, P R (2003). “Superoxide dismutase folding?unfolding pathway: role of the metal ions in modulating structural and dynamical features.” J. Mol. Biol. 330, 145–158. [DOI] [PubMed] [Google Scholar]
- Baglioni, S, Casamenti, F, Bucciantini, M, Luheshi, L M, Taddei, N, Chiti, F, Dobson, C M, and Stefani, M (2006). “Prefibrillar amyloid aggregates could be generic toxins in higher organisms.” J. Neurosci. 26, 8160–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch, W E, Morimoto, R I, Dillin, A, and Kelly, J W (2008). “Adapting proteostasis for disease intervention.” Science 319, 916–919. [DOI] [PubMed] [Google Scholar]
- Banci, L, Bertini, I, Boca, M, Girotto, S, Martinelli, M, Valentine, J S, and Vieru, M (2008). “SOD1 and amyotrophic lateral sclerosis: mutations and oligomerization.” PLoS ONE 3, e1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci, L, Bertini, I, Cantini, F, D’Amelio, N, and Gaggelli, E (2006). “Human SOD1 before harboring the catalytic metal: solution structure of copper-depleted, disulfide-reduced form.” J. Biol. Chem. 281, 2333–2337. [DOI] [PubMed] [Google Scholar]
- Banci, L, Bertini, I, Cramaro, F, Del Conte, R, and Viezzoli, M S (2003). “Solution structure of Apo Cu,Zn superoxide dismutase: role of metal ions in protein folding.” Biochemistry 42, 9543–9553. [DOI] [PubMed] [Google Scholar]
- Banci, L, Bertini, I, Durazo, A, Girotto, S, Gralla, E B, Martinelli, M, Valentine, J S, Vieru, M, and Whitelegge, J P (2007). “Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS.” Proc. Natl. Acad. Sci. U.S.A. 104, 11263–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M J, Choe, S, and Eisenberg, D (1994). “Domain swapping: entangling alliances between proteins.” Proc. Natl. Acad. Sci. U.S.A. 91, 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini, I, Piccioli, M, Viezzoli, M S, Chiu, C Y, and Mullenbach, G T (1994). “A spectroscopic characterization of a monomeric analog of copper, zinc superoxide dismutase.” Eur. Biophys. J. 23, 167–176. [DOI] [PubMed] [Google Scholar]
- Bruijn, L I, Miller, T M, and Cleveland, D W (2004). “Unraveling the mechanisms involved in motor neuron degeneration in ALS.” Annu. Rev. Neurosci. 27, 723–749. [DOI] [PubMed] [Google Scholar]
- Bryngelson, J D, Onuchic, J N, Socci, N D, and Wolynes, P G (1995). “Funnels, pathways, and the energy landscape of protein folding: a synthesis.” Proteins 10.1002/prot.340210302 21, 167–195. [DOI] [PubMed] [Google Scholar]
- Canet, D, Last, A M, Tito, P, Sunde, M, Spencer, A, Archer, D B, Redfield, C, Robinson, C V, and Dobson, C M (2002). “Local cooperativity in the unfolding of an amyloidogenic variant of human lysozyme.” Nat. Struct. Biol. 9, 308–315. [DOI] [PubMed] [Google Scholar]
- Chiti, F, Stefani, M, Taddei, N, Ramponi, G, and Dobson, C M (2003). “Rationalization of the effects of mutations on peptide and protein aggregation rates.” Nature (London) 10.1038/nature01891 424, 805–808. [DOI] [PubMed] [Google Scholar]
- Clarke, J, Cota, E, Fowler, S B, and Hamill, S J (1999). “Folding studies of immunoglobulin-like beta-sandwich proteins suggest that they share a common folding pathway.” Folding Des. 7, 1145–1153. [DOI] [PubMed] [Google Scholar]
- Cozzolino, M, Amori, I, Pesaresi, M G, Ferri, A, Nencini, M, and Carri, M T (2008). “Cysteine 111 affects aggregation and cytotoxicity of mutant Cu,Zn-superoxide dismutase associated with familial amyotrophic lateral sclerosis.” J. Biol. Chem. 283, 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, J P, Sampson, J B, Zhuang, Y, Thompson, J A, and Beckman, J S (1997). “Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite.” J. Neurochem. 69, 1936–1944. [DOI] [PubMed] [Google Scholar]
- Deng, H X, et al. (1993). “Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase.” Science 261, 1047–1051. [DOI] [PubMed] [Google Scholar]
- Dickson, D (2003). “Neurodegeneration: the molecular pathology of dementia and movement disorders.” ISN NEUROPATH PRESS.
- DiDonato, M, et al. (2003). “ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization.” J. Mol. Biol. 332, 601–615. [DOI] [PubMed] [Google Scholar]
- Dobson, C M (1999). “Protein misfolding, evolution and disease.” Trends Biochem. Sci. 10.1016/S0968-0004(99)01445-0 24, 329–332. [DOI] [PubMed] [Google Scholar]
- Dunker, A K, Brown, C J, Lawson, J D, Iakoucheva, L M, and Obradovic, Z (2002). “Intrinsic disorder and protein function.” Biochemistry 10.1021/bi012159+ 41, 6573–6582. [DOI] [PubMed] [Google Scholar]
- Elam, J S, et al. (2003). “Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS.” Nat. Struct. Biol. 10, 461–467. [DOI] [PubMed] [Google Scholar]
- Fernandez-Escamilla, A M, Rousseau, F, Schymkowitz, J, and Serrano, L (2004). “Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins.” Nat. Biotechnol. 22, 1302–1306. [DOI] [PubMed] [Google Scholar]
- Fersht, A R (1995). “Optimization of rates of protein folding: the nucleation-condensation mechanism and its implications.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.92.24.10869 92, 10869–10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht, A R (1999). Structure and Mechanism in Protein Science: a Guide to Enzyme Catalysis and Protein Folding, WH Freeman and Co., New York. [Google Scholar]
- Furukawa, Y, Kaneko, K, Yamanaka, K, O’Halloran, T V, and Nukina, N (2008). “Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in familial form of ALS.” J. Biol. Chem. 283, 24167–24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, Y, Torres, A S, and O’Halloran, T V (2004). “Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS.” EMBO J. 23, 2872–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedraitis, V, Sundelof, J, Irizarry, M C, Garevik, N, Hyman, B T, Wahlund, L O, Ingelsson, M, and Lannfelt, L (2007). “The normal equilibrium between CSF and plasma amyloid beta levels is disrupted in Alzheimer’s disease.” Neurosci. Lett. 427, 127–131. [DOI] [PubMed] [Google Scholar]
- Glover, J R, Kowal, A S, Schirmer, E C, Patino, M M, Liu, J J, and Lindquist, S (1997). “Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae.” Cell 89, 811–819. [DOI] [PubMed] [Google Scholar]
- Gurney, M E, et al. (1994). “Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation.” Science 264, 1772–1775. [DOI] [PubMed] [Google Scholar]
- Hornberg, A, Logan, D T, Marklund, S L, and Oliveberg, M (2007). “The coupling between disulphide status, metallation and dimer interface strength in Cu∕Zn superoxide dismutase.” J. Mol. Biol. 365, 333–342. [DOI] [PubMed] [Google Scholar]
- Hough, M A, et al. (2004). “Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0305143101 101, 5976–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S E (1998). “How do small single-domain proteins fold?” Folding Des. 10.1016/S1359-0278(98)00033-9 3, R81–R91. [DOI] [PubMed] [Google Scholar]
- Jahn, T R, and Radford, S E (2008). “Folding versus aggregation: polypeptide conformations on competing pathways.” Arch. Biochem. Biophys. 469, 100–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger, A C, and Fersht, A R (2008). “Structural biology of the tumor suppressor p53.” Annu. Rev. Biochem. 77, 557–582. [DOI] [PubMed] [Google Scholar]
- Jonsson, P A, Graffmo, K S, Andersen, P M, Brannstrom, T, Lindberg, M, Oliveberg, M, and Marklund, S L (2006). “Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models.” Brain 129, 451–464. [DOI] [PubMed] [Google Scholar]
- Khare, S D, and Dokholyan, N V (2006). “Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants.” Proc. Natl. Acad. Sci. U.S.A. 103, 3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, A L, Torres, A S, O’Halloran, T V, and Rosenzweig, A C (2001). “Heterodimeric structure of superoxide dismutase in complex with its metallochaperone.” Nat. Struct. Biol. 8, 751–755. [DOI] [PubMed] [Google Scholar]
- Levy, Y, Wolynes, P G, and Onuchic, J N (2004). “Protein topology determines binding mechanism.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.2534828100 101, 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, M, Tangrot, J, and Oliveberg, M (2002a). “Complete change of the protein folding transition state upon circular permutation.” Nat. Struct. Biol. 9, 818–822. [DOI] [PubMed] [Google Scholar]
- Lindberg, M J, Bystrom, R, Boknas, N, Andersen, P M, and Oliveberg, M (2005). “Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants.” Proc. Natl. Acad. Sci. U.S.A. 102, 9754–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, M J, Normark, J, Holmgren, A, and Oliveberg, M (2004). “Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers.” Proc. Natl. Acad. Sci. U.S.A. 101, 15893–15898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, M J, Tibell, L, and Oliveberg, M (2002b). “Common denominator of Cu∕Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis: decreased stability of the apo state.” Proc. Natl. Acad. Sci. U.S.A. 99, 16607–16612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, M O, Haglund, E, Hubner, I A, Shakhnovich, E I, and Oliveberg, M (2006). “Identification of the minimal protein-folding nucleus through loop-entropy perturbations.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0508863103 103, 4083–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y, Gotte, G, Libonati, M, and Eisenberg, D (2001). “A domain-swapped RNase A dimer with implications for amyloid formation.” Nat. Struct. Biol. 8, 211–214. [DOI] [PubMed] [Google Scholar]
- Lynch, S M, Boswell, S A, and Colon, W (2004). “Kinetic stability of Cu∕Zn superoxide dismutase is dependent on its metal ligands: implications for ALS.” Biochemistry 43, 16525–16531. [DOI] [PubMed] [Google Scholar]
- Lynch, S M, and Colon, W (2006). “Dominant role of copper in the kinetic stability of Cu∕Zn superoxide dismutase.” Biochem. Biophys. Res. Commun. 340, 457–461. [DOI] [PubMed] [Google Scholar]
- McParland, V J, Kalverda, A P, Homans, S W, and Radford, S E (2002). “Structural properties of an amyloid precursor of beta(2)-microglobulin.” Nat. Struct. Biol. 10.1038/nsb791 9, 326–331. [DOI] [PubMed] [Google Scholar]
- Michalik, A, and Van Broeckhoven, C (2003). “Pathogenesis of polyglutamine disorders: aggregation revisited.” Hum. Mol. Genet. 12, R173–R186. [DOI] [PubMed] [Google Scholar]
- Niwa, J, Yamada, S, Ishigaki, S, Sone, J, Takahashi, M, Katsuno, M, Tanaka, F, Doyu, M, and Sobue, G (2007). “Disulfide bond mediates aggregation, toxicity, and ubiquitylation of familial amyotrophic lateral sclerosis-linked mutant SOD1.” J. Biol. Chem. 282, 28087–28095. [DOI] [PubMed] [Google Scholar]
- Nordlund, A, and Oliveberg, M (2006). “Folding of Cu∕Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease.” Proc. Natl. Acad. Sci. U.S.A. 103, 10218–10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okado-Matsumoto, A, and Fridovich, I (2002). “Amyotrophic lateral sclerosis: a proposed mechanism.” Proc. Natl. Acad. Sci. U.S.A. 99, 9010–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveberg, M (1998). “Alternative explanations for multi-state kinetics in protein folding: transient aggregation and changeing transition-state ensambles.” Acc. Chem. Res. 10.1021/ar970089m 31, 765–772. [DOI] [Google Scholar]
- Oliveberg, M, and Wolynes, P G (2005). “The experimental survey of protein-folding energy landscapes.” Q. Rev. Biophys. 10.1017/S0033583506004185 38, 245–288. [DOI] [PubMed] [Google Scholar]
- Otzen, D E, Kristensen, O, and Oliveberg, M (2000). “Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: a structural clue to amyloid assembly.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.160086297 97, 9907–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen, D E, and Oliveberg, M (1999). “Salt-induced detour through compact regions of the protein folding landscape [in process citation],” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.96.21.11746 96, 11746–11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen, D E, and Oliveberg, M (2004). “Transient formation of nano-crystalline structures during fibrillation of an Abeta-like peptide.” Protein Sci. 13, 1417–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt, G W, McParland, V J, Kalverda, A P, Homans, S W, and Radford, S E (2005). “Dynamics in the unfolded state of beta2-microglobulin studied by NMR.” J. Mol. Biol. 346, 279–294. [DOI] [PubMed] [Google Scholar]
- Plotkin, S S, and Onuchic, J N (2000). “Investigation of routes and funnels in protein folding by free energy functional methods.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.97.12.6509 97, 6509–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, S Z, and Valentine, J S (2003). “The perplexing role of copper-zinc superoxide dismutase in amyotrophic lateral sclerosis (Lou Gehrig’s disease).” JBIC, J. Biol. Inorg. Chem. 8, 373–380. [DOI] [PubMed] [Google Scholar]
- Rakhit, R, Crow, J P, Lepock, J R, Kondejewski, L H, Cashman, N R, and Chakrabartty, A (2004). “Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis.” J. Biol. Chem. 279, 15499–15504. [DOI] [PubMed] [Google Scholar]
- Randles, L G, Lappalainen, I, Fowler, S B, Moore, B, Hamill, S J, and Clarke, J (2006). “Using model proteins to quantify the effects of pathogenic mutations in Ig-like proteins.” J. Biol. Chem. 281, 24216–24226. [DOI] [PubMed] [Google Scholar]
- Richardson, J S, and Richardson, D C (2002). “Natural beta -sheet proteins use negative design to avoid edge-to-edge aggregation.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.052706099 99, 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, J S, et al. (1992). “Looking at proteins: representations, folding, packing, and design.” Biophys. J. 63, 1185–1209. [PMC free article] [PubMed] [Google Scholar]
- Ripps, M E, Huntley, G W, Hof, P R, Morrison, J H, and Gordon, J W (1995). “Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis.” Proc. Natl. Acad. Sci. U.S.A. 92, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, B R, Tainer, J A, Getzoff, E D, Malencik, D A, Anderson, S R, Bomben, V C, Meyers, K R, Karplus, P A, and Beckman, J S (2007). “Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS.” J. Mol. Biol. 373, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, J A, Valentine, J S, Eggers, D K, Roe, J A, Tiwari, A, Brown, R H, Jr., and Hayward, L J (2002). “Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper∕zinc superoxide dismutase.” J. Biol. Chem. 277, 15932–15937. [DOI] [PubMed] [Google Scholar]
- Rousseau, F, Schymkowitz, J, and Serrano, L (2006a). “Protein aggregation and amyloidosis: confusion of the kinds?” Curr. Opin. Struct. Biol. 16, 118–126. [DOI] [PubMed] [Google Scholar]
- Rousseau, F, Serrano, L, and Schymkowitz, J W (2006b). “How evolutionary pressure against protein aggregation shaped chaperone specificity.” J. Mol. Biol. 355, 1037–1047. [DOI] [PubMed] [Google Scholar]
- Sandelin, E, Nordlund, A, Andersen, P M, Marklund, S S, and Oliveberg, M (2007). “Amyotrophic lateral sclerosis-associated copper∕zinc superoxide dismutase mutations preferentially reduce the repulsive charge of the proteins.” J. Biol. Chem. 282, 21230–21236. [DOI] [PubMed] [Google Scholar]
- Sekijima, Y, Wiseman, R L, Matteson, J, Hammarstrom, P, Miller, S R, Sawkar, A R, Balch, W E, and Kelly, J W (2005). “The biological and chemical basis for tissue-selective amyloid disease.” Cell 121, 73–85. [DOI] [PubMed] [Google Scholar]
- Shaw, B F, and Valentine, J S (2007). “How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein?” Trends Biochem. Sci. 32, 78–85. [DOI] [PubMed] [Google Scholar]
- Sherman, M Y, and Goldberg, A L (2001). “Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases.” Neuron 29, 15–32. [DOI] [PubMed] [Google Scholar]
- Silow, M, Tan, Y J, Fersht, A R, and Oliveberg, M (1999). “Formation of short-lived protein aggregates directly from the coil in two-state folding.” Biochemistry 38, 13006–13012. [DOI] [PubMed] [Google Scholar]
- Silveira, J R, Raymond, G J, Hughson, A G, Race, R E, Sim, V L, Hayes, S F, and Caughey, B (2005). “The most infectious prion protein particles.” Nature (London) 10.1038/nature03989 437, 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos, P B, Rumfeldt, J A, Scholz, G A, Irani, R A, Frey, H E, Hallewell, R A, Lepock, J R, and Meiering, E M (2003). “Cu∕Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.1237797100 100, 7021–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroppolo, M E, Malvezzi-Campeggi, F, Mei, G, Rosato, N, and Desideri, A (2000). “Role of the tertiary and quaternary structures in the stability of dimeric copper, zinc superoxide dismutases.” Arch. Biochem. Biophys. 377, 215–218. [DOI] [PubMed] [Google Scholar]
- Svensson, A K, Bilsel, O, Kondrashkina, E, Zitzewitz, J A, and Matthews, C R (2006). “Mapping the folding free energy surface for metal-free human Cu,Zn superoxide dismutase.” J. Mol. Biol. 364, 1084–1102. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D, Klimov, D K, and Dima, R I (2003). “Emerging ideas on the molecular basis of protein and peptide aggregation.” Curr. Opin. Struct. Biol. 10.1016/S0959-440X(03)00032-0 13, 146–159. [DOI] [PubMed] [Google Scholar]
- Tiwari, A, and Hayward, L J (2003). “Familial amyotrophic lateral sclerosis mutants of copper∕zinc superoxide dismutase are susceptible to disulfide reduction.” J. Biol. Chem. 278, 5984–5992. [DOI] [PubMed] [Google Scholar]
- Vande Velde, C, Miller, T M, Cashman, N R, and Cleveland, D W (2008). “Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria.” Proc. Natl. Acad. Sci. U.S.A. 105, 4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska, M, Lobov, S, Ohlsson, P I, and Ny, T (2003). “A redox-sensitive loop regulates plasminogen activator inhibitor type 2 (PAI-2) polymerization.” EMBO J. 22, 1753–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittung-Stafshede, P (2004). “Role of cofactors in folding of the blue-copper protein azurin.” Inorg. Chem. 43, 7926–7933. [DOI] [PubMed] [Google Scholar]
- Yang, S, Cho, S S, Levy, Y, Cheung, M S, Levine, H, Wolynes, P G, and Onuchic, J N (2004). “Domain swapping is a consequence of minimal frustration.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0403724101 101, 13786–13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterstrom, P, Stewart, H G, Bergemalm, D, Jonsson, P A, Graffmo, K S, Andersen, P M, Brannstrom, T, Oliveberg, M, and Marklund, S L (2007). “Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models.” Proc. Natl. Acad. Sci. U.S.A. 104, 14157–14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F, Strom, A L, Fukada, K, Lee, S, Hayward, L J, and Zhu, H (2007). “Interaction between familial amyotrophic lateral sclerosis (ALS)-linked SOD1 mutants and the dynein complex.” J. Biol. Chem. 282, 16691–16699. [DOI] [PubMed] [Google Scholar]