Abstract
The health benefits of cranberries have long been recognized. However, the mechanisms behind its function are poorly understood. We have investigated the iron-binding properties of quercetin, the major phenolic phytochemical present in cranberries, and other selected phenolic compounds (chrysin, 3-hydroxyflavone, 3′,4′-dihydroxy flavone, rutin, and flavone) in aqueous media using UV/vis, NMR and EPR spectroscopies and ESI-Mass spectrometry. Strong iron-binding properties have been confirmed for the compounds containing the “iron-binding motifs” identified in their structures. The apparent binding constants are estimated to be in the range of 106 M−1 to 1012 M−2 in phosphate buffer at pH 7.2. Surprisingly, quercetin binds Fe2+ even stronger than the well known Fe2+-chelator ferrozine at pH 7.2. This may be the first example of an oxygen-based ligand displaying stronger Fe2+-binding affinity than a strong nitrogen-based Fe2+-chelator. The strong Fe-binding properties of these phenolics argue that they may be effective in modulating cellular iron homeostasis under physiological conditions. Quercetin can completely suppress Fenton chemistry both at micromolar levels and in the presence of major cellular iron chelators like ATP or citrate. However, the radical scavenging activity of quercetin provides only partial protection against Fenton chemistry-mediated damage while Fe chelation by quercetin can completely inhibit Fenton chemistry, indicating that the chelation may be key to its antioxidant activity. These results demonstrate that quercetin and other phenolic compounds can effectively modulate iron biochemistry under physiologically relevant conditions, providing insight into the mechanism of action of bio-active phenolics.
Introduction
Cranberries have long been used as food and beverages, as well as for medicinal purposes. Health benefits of the North American cranberry (Vaccinium macrocarpon, Ait. Ericaceae) have been recognized since the early 1900s.1 The effectiveness of cranberries in the prevention and treatment of urinary tract infections (UTIs) has been confirmed by randomized, double-blind placebo-controlled clinical trials.2 Recent studies have revealed that cranberries and cranberry products are also beneficial in the prevention and treatment of stomach ulcers, gum diseases and dental infections. More recently, cranberries have demonstrated anti-oxidative and antiviral effects, which are linked to potential protection against aging, stroke, cardiovascular diseases, neurological disorders and certain cancers.3-5 Many of these biological effects have been linked to the presence of various phenolic compounds in cranberries called flavonoids.6,7 However, the molecular mechanism of action is poorly understood. The antioxidant activities of flavonoids have been ascribed to their ability to act as free radical scavengers but their metal-binding properties have been implicated as well.8,9
The major flavonoids found in cranberries are anthocyanins, flavonol glycosides and polymeric proanthocyanidins. Other potentially healthful components include triterpenoids, catechins, β-hydroxybutyric acid, citric, malic, glucuronic, quinic and benzoic acids, substituted cinnamic acids, ellagic acid, and ascorbic acid.10,11 Cranberries have been shown to contain the highest total phenolic content among many common fruit species,12 with a diverse composition of small phenolic acids, flavonols, anthocyanins, and flavan-3-ol oligomers (proanthocyanidins).13 For example, the concentration of quercetin (a flavonol) in fresh cranberry fruit is as high as ca 11−25 mg per 100 g.14
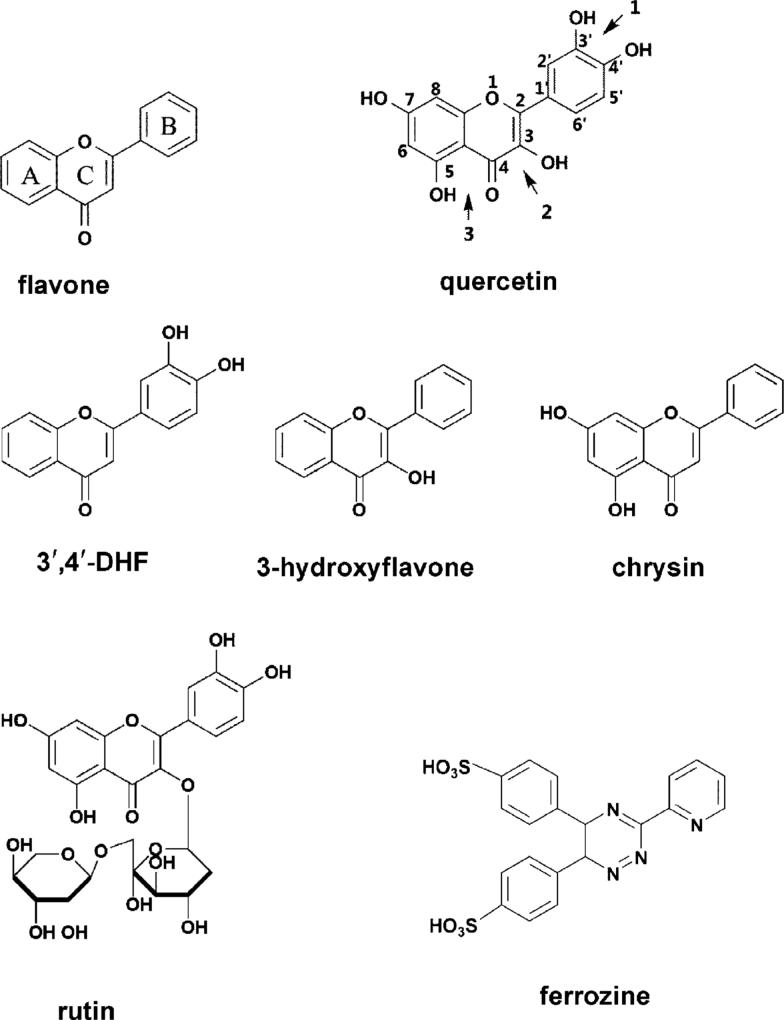
The basic structure of flavonoids (Chart 1) consists of a 3-ring system including two aromatic rings (A and B) linked through three carbons that usually form an oxygenated heterocycle (C ring). Most flavonoids contain one or more ortho hydroxyl phenolic group, or one phenolic with a nearby carboxylate or carbonyl group in a cis conformation. Proanthocyanidins, epicate-chins, flavonols and anthocyanins contain multiples of such structures. The ortho-hydroxyphenol functionality and “di-tyrosine iron-binding motif” (Fig. S1†) are found in human and bacterial iron transport proteins and siderophores.15-20 The critical role of phenolic groups in iron chelation has been further supported by our recent studies on developing synthetic prochelators.21 Thus, these phenolic compounds with an “iron-binding motif” are predicted to be strong iron-chelating agents which may modulate the bioactivity and bioavailability of iron in the body.
Chart 1.
Structures and atom numbering of the flavonoids and ferrozine.
Iron is an essential element for almost all organisms22-24 and is also considered the primary limiting nutrient for bacteria during infection. Thus, it is a decisive factor in bacterial virulence.25-27 However, free iron (either as Fe3+ or Fe2+) is toxic even at concentrations below 10−18 M,28,29 mainly because it catalyzes the Fenton reaction (eqn (1)) to produce hydroxyl radicals which are damaging to cells.30,31 Cellular reductants such as ascorbate (AscH−) and NADH can recycle Fe3+(Cu2+) back to Fe2+(Cu+), making the Fenton reaction catalytic when excess H2O2 is available. It is not surprising that variations in cellular iron status have a major influence on human health. Thus phenolic compounds with an “iron-binding motif” could influence iron homeostasis which may explain several health benefits of cranberries. The sequestration and subsequent depletion of free iron may attenuate iron-promoted radical production and inhibit bacteria and iron acquisition by tumors, thus providing an explanation for the cranberry’s bio-effects in the prevention and treatment of bacterial infections, antioxidant effects, and its antitumor activity.
| (1) |
| (2) |
Iron binding by quercetin and related compounds has been studied by a number of groups.32-42 Afanas’ef and co-workers34 studied Fe2+complexes with rutin in aqueous solution by UV-vis spectroscopy and found that the iron : rutin complex has a stoichiometry of 1 : 2 and that chelation may inhibit ferrous ion-dependent lipid peroxidation. Bodinini et al.35 examined the electrochemistry of quercetin and its Fe2+ complexes in dimethylsulfoxide and proposed chelation through the catechol group (i.e., site 1). A similar binding mode was proposed by Erdogan et al. based on a potentiometric study.32 In contrast, a recent density functional theory (DFT) calculation36 suggests that the oxygen atoms belonging to the 3-OH and 4-oxo group (i.e., site 2), and to the 5-OH and 4-oxo group (i.e., site 3) are the preferred ones for Fe2+ binding. The formation of 1 : 1 and 2 : 1 Fe3+–quercetin complexes have been suggested by Escandar and Sala37 from a potentiometric titration study in which Fe3+ coordination probably occurs with the catechol group for the first Fe3+ and carbonyl oxygen and either the C-5 OH group (i.e., site 3) or the C-3 OH group (i.e., site 2) for the second Fe3+. A recent potentiometric titration study38 with flavones processing only one of the metal binding sites has also suggested that the flavone with a catechol group displays the highest affinity for Fe3+. However, no direct evidence has been available to support the proposed binding modes.
On the basis of electrospray ionisation mass spectrometry (ESI-Mass) investigations in methanol–water (1 : 1, with 0.1% acetic acid), Fernandez et al.39 have reported that quercetin is able to reduce Fe3+ to Fe2+. With the iron source provided as FeCl3, they observed 1 : 1 Fe2+ complexes with oxidized quercetin, 1 : 2 Fe2+ complexes with quercetin and oxidized quercetin, and a 1 : 2 Fe3+ complex with quercetin. Reduction of Fe3+ to Fe2+ has also been observed in 50 mM acetate buffer at pH 5.5 by Mira and co-workers in a more detailed study using UV/Vis and ESI-mass spectrometry.40 They suggested that the chelation site is probably through the 5-hydroxyl and the 4-oxo groups (i.e., site 3) in contrast to the catechol-chelation model proposed by Bodinini et al.35 and Erdogan et al.32 More recently, the kinetics of Fe2+ and Fe3+ binding to quercetin has been investigated.41 Quercetin binds both Fe2+ and Fe3+ quickly (over ca. 1 min) at pH 5 and even more quickly at pH 7.4. In the presence of oxygen, quercetin may be oxidized by dioxygen to produce 2-(hydroxybenzoyl)-2-hydroxybenzofuran-3(2H)-ones, as demonstrated by Jungbluth et al.42 However, the iron complexes of quercetin are relatively inert in aqueous solution at pH 5 and 7.4 and react with dioxygen even more slowly than free quercetin.41
To clarify the iron chemistry of quercetin and to further elucidate the role of plant phenolics in modulating iron homeostasis, we have carried out a detailed investigation of iron binding to a series of phenolic compounds including quercetin using a variety of physical methods. The diamagnetic metal ions Zn2+ and Ga3+ were used as probes for the binding of Fe2+ and Fe3+, respectively. The effects of these phenolic compounds in attenuating the Fenton reaction also were studied under physiologically relevant conditions.
Experimental
Materials
The flavonoids quercetin dihydrate, chrysin and rutin were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA), and 3′,4′-dihydroxyl flavone was purchased from Lancaster (Windham, NH, USA). Tris(hydroxymethyl)aminomethane (Tris), potassium phosphate monobasic and dibasic, ethylenediaminetetraacetic acid (EDTA), ferrous ammonium sulfate, ferric chloride, zinc acetate, gallium chloride, dimethyl sulfoxide (DMSO), ferrozine monosodium salt, hydrogen peroxide (30%) and 2-deoxyribose were obtained from Sigma-Aldrich Corp. Flavonol and ferrous chloride were obtained from TCI America (Portland, OR, USA) and NMR solvents from Cambridge Isotopes Laboratory (Cambridge, MA, USA). The stock solutions of flavonoids were prepared in methanol or ethanol. Iron salts were dissolved in 0.1 M HCl, other chemicals were dissolved in deionized water or buffer and 2-thiobarbituric acid (TBA) in 50 mM NaOH.
UV/Vis spectroscopic studies
UV/Vis spectra were recorded on a Perkin Elmer Lambda 25 spectrometer at 25 °C. The kinetics of the formation of the complexes between flavonoids and metal ions was measured by monitoring the changes in UV/Vis spectrum. Typical titration experiments were performed by sequential additions of 1−2 μL of metal ion solution (1 mM stock solution, freshly made in 0.1 M HCl) to the same 1 mL flavonoid solution in a quartz cuvette (10 μM, from 1 mM stock solution in MeOH). The mixture was equilibrated at 25 °C until no further spectroscopic change was observed (ca. 2 to 10 min). All titrations were performed in 20 mM KBP buffer, pH 7.2 unless otherwise noted.
Apparent binding constants
Three different approaches were applied to estimate the apparent binding constants between the flavonoids and iron under physiologically relevant conditions.
Approach 1
Clear isosbestic points were observed in the spectroscopic titration spectra of Fe2+ with many of the flavonoids studied, indicating a complex formation process involving only two chromophores in equilibrium with one another. Estimation of the apparent binding constants for the formation of the flavonoid–Fe2+ complexes can be made as follows. For the ligand–substrate reaction with the formation of 1 : 1 complex LS:
| (3) |
where L = flavonoid, S = Fe2+, and LS = flavonoid–Fe2+ complex, the equilibrium constant (conditional binding constant) is given by:
| (4) |
The subscript e designates equilibrium concentrations. The ratios of the equilibrium LS complex concentration, [LS]e, and the initial L concentration, [L]o, can be derived from the absorbance of the solutions at a chosen wavelength both at equilibrium and far from equilibrium (see ES1†), namely
| (5) |
where Fc is the fraction of L complexed.
For the formation of a 1 : 2 complex SL2, the following equations apply (see ES1†)
| (6) |
| (7) |
| (8) |
Approach 2
This was performed similarly to that described by Gibbs for the determination of the binding constant between ferrozine and Fe2+.43 After determining the stoichiometry of the metal–ligand complex, the extinction coefficient of the complex was found for a chosen wavelength. Since the ligand has a minor absorbance in the region where the complex absorbs, it was always subtracted from the complex absorption after correcting for the fraction of free ligand present. The extinction coefficients were calculated using a linear regression calculation in Excel (Beer–Lambert law). With the extinction coefficient for the complex known, along with the concentrations of the reagents and molar ratios, a chemical equilibrium equation was set up to find the concentrations of the reagents and complex at equilibrium. With this information Keq was determined for each metal–ligand pairing.
Approach 3
A third approach was carried out by using competitive binding of Fe2+ between flavonoid and ferrozine, a specific Fe2+ chelator.43,44 The results are qualitatively in agreement with binding constants obtained by the first two approaches.
NMR spectroscopic studies
1H NMR spectra were recorded on a Bruker AC 300 spectrometer as described previously.21 Quercetin and other flavonoid stock solutions (300 mM) were prepared in d6-DMSO solution. Freshly prepared stock solutions of Fe2+ (50 mM ferrous ammonium sulfate in D2O/HCl), Fe3+ (50 mM FeCl3 in D2O/HCl), Zn2+ (300 mM ZnAc in D2O) and Ga3+ (600 mM GaCl3 in D2O/HCl) were used for the NMR titrations. Pure organic solvents d4-MeOH, d6-acetone, d6-DMSO and their mixtures with D2O or with 10−50 mM D2O/KBP buffer were used for the initial NMR study. However, the initial studies were hampered by the poor solubility of either the metal ions or the complexes in these solvent systems. Finally, a mixed solvent system containing d6-DMSO/D2O (v/v : 50/50), 50 mM Tris-HCl, pH* 7.20 was found to be a suitable media for the NMR study, in which a solubility up to 5 mM is achievable for most of the complexes.
The pH values of the solutions were determined using a Corning pH meter equipped with a Sigma-Aldrich micro combination electrode calibrated with Aldrich buffer solutions at pH 4, 7, and 10. The pH meter readings for D2O solutions are recorded as pH* values, i.e., uncorrected for the effect of deuterium.
EPR spectroscopic studies
EPR experiments were carried out on a Bruker EleXsys E560 spectrometer fitted with SHQ cavity and a liquid nitrogen Dewar insert. Samples were placed in 3 mm i.d./4 mm o.d. quartz tubes. Spectra were measured with the following parameters: Power: 0.271 mW (29 dB); modulation amplitude: 10 G; frequency: 9.157 GHz; time constant: 163.84 ms; scan time: 167.77 s; scan range: 2000 G; no. of points: 8192; central field: 1500 G; conversion time: 20.48 ms; receiver gain: 60 dB; no. of scans: 1; temperature: 77 K (liquid nitrogen temperature); reaction time: ∼3 min unless otherwise stated. Quercetin stock solution (10 mM) was prepared in 50 : 50 methanol : H2O solution. Stock solutions of Fe2+ (0.042 M, from FeSO4), Fe3+ (0.042 M from FeCl3 in a 1000 ppm atomic absorption standard, Ricca Chemical Co., Arlington, TX, USA) and EDTA (0.042 M, disodium salt dihydrate) were prepared in H2O/HCl, pH 2.0.
Electrospray ionisation mass spectrometry (ESI-mass) studies
ESI mass spectrometry experiments were carried out at the University of Massachusetts Amherst Mass Spectrometry Facility on an Esquire-LC ion trap instrument (Bruker Daltonics, Inc., Billerica MA) equipped with an electrospray ionization source. Both positive and negative ion modes of detection were employed. Sample solutions (∼10 μM, in 1 : 1 methanol:water) were continually infused via a syringe pump at a flow rate of 2 μL min−1. Default parameters were used in most cases (smart mode optimized for 600 m/z), except that capillary exit and skimmer 1 voltages were reduced to insure metal complexes remained intact upon transfer through the ion optics. Averages of 10 scans were analyzed and all data were processed using the Bruker Data Analysis software.
Typically, samples were prepared by addition of iron solutions (in 0.1 M HCl) of ferrous ammonium sulfate or ferric chloride into a 10 μM flavonoid solution in methanol/water (1 : 1, v/v) with a molar ratio 1 : 1. ESI mass spectra were acquired ca. 3−10 min after the samples were prepared. The pH values of the leftover solutions were measured and found in the range of 3.7 to 5.0. In order to study the species formed in buffered solutions, 1 mM potassium phosphate buffer (KPB) (pH 7.2) or ammonium acetate (pH 8.0) was introduced to the samples containing iron and quercetin. However, after several trials, no meaningful spectrum could be obtained for the samples with the buffer either by positive or negative mode.
Assay of 2-deoxyribose degradation
Formation of hydroxyl radicals was quantified using a 2-deoxyribose oxidative degradation assay as conducted by Lopes et al. with modification.45 Typical reactions started with the addition of Fe2+ and chelator (EDTA, citrate, ATP, or flavonoids) to 10 mM KPB at pH 7.2 prepared with doubly deionised H2O. This solution was allowed to incubate for approximately 45 min to ensure complexation was complete. After the preliminary incubation, a flavonoid was added if it had not been added already. If the flavonoid was added at this time, the solution was allowed to incubate for an additional 30 min. 10 mM 2-Deoxyribose and 100 μM ascorbic acid (final concentrations) were then added to complete the reaction mixture (total volume 500 μL). The reaction was initiated by the addition of 25 μL of 4 mM H2O2 and allowed to run for 10 min and then stopped by the addition of 500 μL of 10% (w/v) trichloroacetic acid (TCA) followed by 0.5 mL 1% 2-thiobarbituric acid (TBA, w/v, in 40 mM NaOH). After heating at 80 °C for 15 min, the absorbance was measured at 532 nm.
Results and discussion
UV/Vis spectroscopic studies of quercetin with Fe2+ and Fe3+
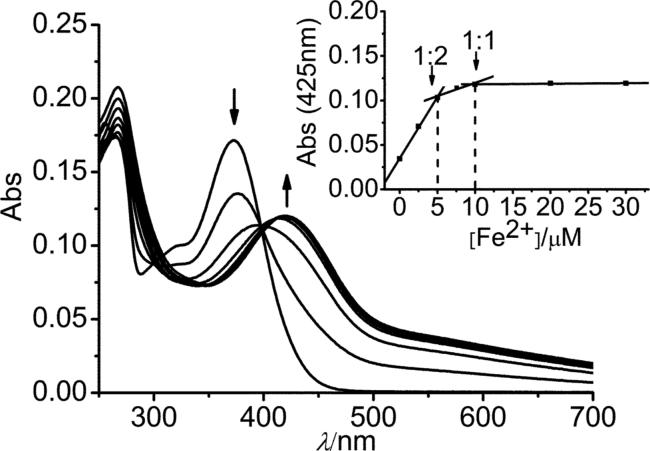
UV/Vis titration of quercetin with Fe2+ was carried out in 20 mM KPB at pH 7.2. With the addition of Fe2+, the free quercetin absorption (λmax = 372 nm, assigned to the π to π* transitions of the B-ring46) decreased rapidly and a new absorption band (λmax = 425 nm) from the Fe2+−complexes appeared and increased in intensity (Fig. 1). The presence of a clear isosbestic point (398 nm) suggests clean formation of the quercetin–Fe2+ complex. Beyond 0.50 mol eq. of Fe2+, the spectrum underwent further changes including a small decrease in the quercetin band at 372 nm accompanied by a minor increase in intensity of the new complex band red shifted at ca. 425 nm. The isosbestic point at 398 nm shifted to 410 nm.
Fig. 1.
Titration of quercetin by Fe2+ in 20 mM KPB buffer, pH 7.2. Top to bottom: 10 μM quercetin in the presence of 0, 2.5, 5.0, 7.5, 10, 20 and 30 μM Fe2+. Inset, titration curve: absorbance of the quercetin–Fe2+ complex at 425 nm versus Fe2+ concentrations, with 10 μM quercetin initially.
Fig. 1 inset displays a plot of the quercetin–Fe2+ complex absorbance at 425 nm versus Fe2+ concentration. The titration curve reveals the formation of 1 : 2 and 1 : 1 Fe2+ : quercetin complexes, a result comfirmed by ESI-mass spectrometry measurements (vide infra). A kinetics study of a 1 : 1 Fe2+ : quercetin mixture (10 μM each) monitored by UV/Vis spectroscopy (Fig. S2†) revealed a fast reaction which is completed within 1 min, in agreement with Hajji et al.41 Thereaction of a 1 : 2 Fe2+ : quercetin mixture is also completed in ∼1 min (Fig. S3†), producing a new UV/Vis band centred ca. 398 nm.
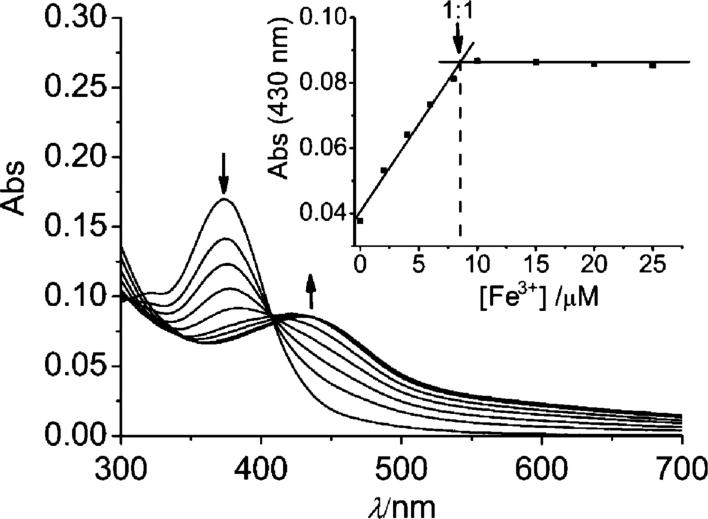
The binding between quercetin and Fe3+ was studied similarly. The addition of Fe3+ to quercetin produced a new band at 415 nm of lower intensity than the 425 nm band of Fe2+–quercetin. The spectra during the later stage of the titration showed some characteristics similar to the Fe2+–quercetin complex (e.g., the Fe-quercetin band shifted to ∼425 nm), suggesting that part of the Fe3+ was probably reduced to Fe2+, a phenomenon known for hydroxybenzene compounds in aqueous media.47 The partial reduction of Fe3+ to Fe2+ by quercetin was confirmed by ESI-mass and EPR measurements (vide infra), in accord with recent findings by others.39,40 The titration (Fig. 2 inset) indicates that a 1 : 1 complex is formed, probably a mixture of 1 : 1 Fe3+ and Fe2+ complexes. A similar spectrum to that shown in Fig. 2 was observed by mixing quercetin with equimolar Fe3+ at pH 5.5.40 A kinetics study of a 1 : 1 Fe3+: quercetin mixture (10 μM each) showed that reaction was completed in ca. 5 min (t1/2 ∼ 30 s). The reaction is much slower than that with Fe2+, perhaps due to the stronger competition for Fe3+ from the phosphate ions in the buffer as discussed by Hajji et al. recently.41
Fig. 2.
Titration of quercetin with Fe3+ in 20 mM KPB buffer, pH 7.2. From top to bottom: 10 μM quercetin in the presence of 0, 2, 4, 6, 8, 10, 15, 20, 25 and 30 μM Fe3+. Inset, titration curve: absorbance at 430 nm versus Fe3+ concentrations, with 10 μM quercetin initially.
Quercetin itself exhibits two major π–π* bands (Fig. 1),46,48 the absorption band at 372 nm corresponding to the B ring portion (cinnamoyl system, band I), and that at 268 nm to the A ring portion (benzoyl system, band II).48 Upon binding to Fe2+/Fe3+, the absorption of band I diminished while a new band appeared at ∼425 nm. The new band has an extinction coefficient of approximately 12 000 M−1 cm−1 and 8600 M−1 cm−1 for the 1 : 1 complexes with Fe2+ and Fe3+, respectively; although the latter value more appropriately represents a mixture. This band may be assigned to a shift in the π–π* quercetin absorption band at 372 nm upon metal binding. Charge transfers (π → dπ) in catechol–Fe3+ complexes produce broader bands with extinction coefficients of 1000–3300 M−1 cm−1.49-51 Similar spectral changes with quercetin have been observed for some other transition metal ions such as Zn2+,56 Ti4+ (unpublished data), V4+,52 Cu2+,40 and the main group metal ions such as Pb2+,53 Al3+,54 and Ga3+ (Fig. S4†). It is unlikely that the coordination with all of these metal ions produce electronic transitions that generate a charge transfer band at a similar wavelength nor are such transitions possible for d10 transition metal ions or the main group metal ions, a conclusion contrary to a recent DFT calculation suggesting a ligand-to-metal charge transfer transition.36 In metal–quercetin complexes the red shift of the π–π* band I may be due to a further delocalization of the π electrons on the ligand induced by the positively charged metal centers.
ESI-Mass studies on the complex formation between quercetin with Fe2+ and Fe3+
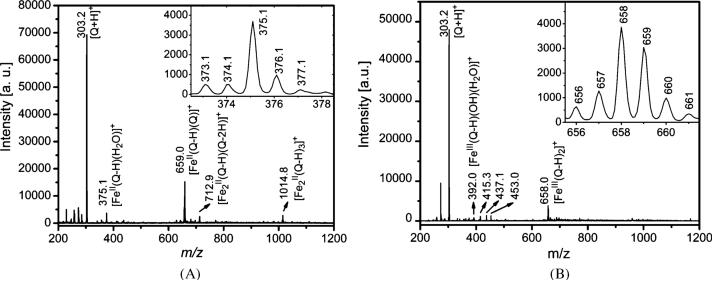
To determine more accurately the stoichiometry and iron oxidation states of the complexes formed between quercetin and iron, electrospray ionization mass spectroscopy (ESI-Mass) experiments were carried out.55 ESI-Mass studies on quercetin with Fe3+ have been reported39,40 recently but no report has been available on Fe2+.
The ESI-Mass spectra of the sample prepared by addition of 1 mol equiv. of Fe2+ to quercetin solution is shown in Fig. 3A. In addition to the free ligand quercetin (m/z = 303.2, [Q + H]+), four Fe2+-complexes containing quercetin were readily detected at m/z = 375.1, 659.0, 712.9 and 1014.8, respectively, all displaying the expected isotopic pattern of iron. The species with m/z 375.1 is assigned to a 1 : 1 complex with a bound water [FeII(Q – H)(H2O)]+. The isotopic pattern (inset) is consistent with an Fe2+ species and about 5% of Fe3+. The species with m/z = 659.0 is assigned to a 1 : 2 complex [FeII(Q – H)(Q)]+ and that with m/z = 712.9 to a 2 : 2 complex [(FeII)2 (Q – H)(Q – 2H)]+ which may be a result of the loss of quercetin from a 2 : 3 complex [(FeII)2(Q – H)3]+ (m/z = 1014.8).
Fig. 3.
Electrospray mass spectra of solutions of Fe2+/Fe3+quercetin and (10 μM each, 1 : 1) in methanol/water (1 : 1, v/v). (A), with Fe2+, the inset is the isotopic pattern of the peak at m/z = 375.1; (B), with Fe3+, the inset is the isotopic pattern of the peak at m/z = 658.
Similar ESI-Mass experiments were performed with Fe3+. The major iron complex at m/z = 658.0 can be assigned to a 1 : 2 complex [FeIII(Q – H)2]+ (Fig. 3B). However, the isotopic pattern (shown in inset) suggests a mixture of Fe2+ (e.g., the peak at m/z = 659) and Fe3+ (e.g. the peak at m/z = 658) species, with ca. 35% being Fe2+. This finding implies a reduction of Fe3+to Fe2+ with quercetin under the ESI-Mass conditions. A closer inspection of the small peak at m/z = 392.0 suggests a 1 : 1 complex, [FeIII(Q – H)(OH)(H2O)]+, fitting well the iron isotopic pattern. However the small peaks at m/z = 437.1 and 453.0 cannot be iron complexes because they do not fit the isotopic pattern of iron. No further analysis was made for these species.
The 1 : 2 complex [FeIII(Q – H)2]+ (m/z = 658) and its reduced product [FeII(Q − H)(Q)]+ (m/z = 659) were also observed by Fernandez et al.39 and Mari et al.40 by reacting quercetin with FeCl3 in methanol/water 1 : 1 with 0.1% acetic acid. However, the 1 : 1 complex (m/z = 392.0) was not identified in those studies.40 Instead, Fernandez et al.39 observed additional 1 : 1 Fe2+ adduct (37.9%) and a 1 : 2 adduct (19.8%) with oxidized quercetin as the major species in ESI-Mass spectra after reaction with FeCl3. A closer inspection of our data suggests that oxidized quercetin species are also present in our spectrum but only in small amounts (< 4%).
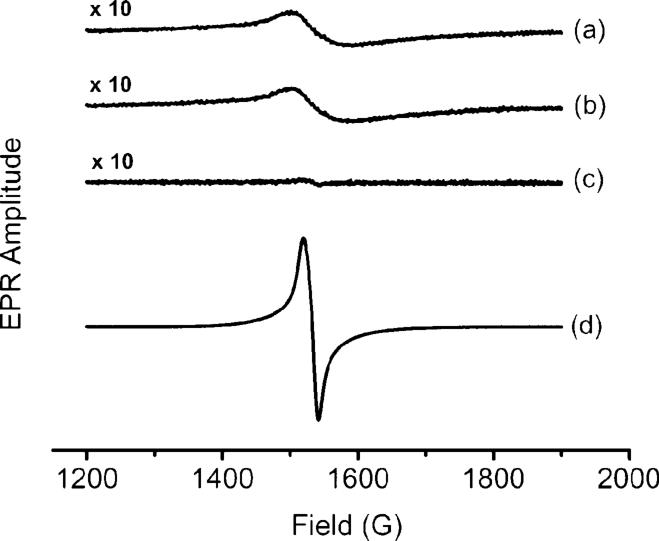
EPR studies
EPR studies were carried out in 1 : 1 MeOH/H2O to further probe the oxidation state of iron in quercetin complexes. When Fe3+ was added to quercetin in a 1 : 1 ratio either aerobically or anaerobically, a broad g′ = 4.3 signal (peak-to-peak line width = 77 G) from mononuclear high spin (S = 5/2) Fe3+ in a site of rhombic symmetry was observed (cf. Fig. 4, spectra a and b). The double integral of either spectrum a or b was 30% of that of control Fe3+–EDTA (peak-to-peak line width = 23 G) (spectrum d), a result suggesting that ∼ 70% of the added Fe3+ had been reduced to Fe2+ by quercetin. Addition of excess ferrozine caused immediate formation of an intense purple color from the Fe2+(ferrozine)3 complex, confirming the EPR result. It is noteworthy that the presence of O2 does not cause the Fe2+ to re-oxidize, indicating that quercetin stabilizes the +2 oxidation state relative to the +3 oxidation state of iron (cf. spectra a and b). As expected for a high spin (S = 2) complex, Fe2+–quercetin showed no EPR signal (spectrum c).
Fig. 4.
EPR spectra of quercetin, Fe2+/Fe3+ and EDTA in MeOH/H2O (v/v : 50/50) under anaerobic conditions except for (a). (a) quercetin (0.6 mM) + Fe3+ (0.3 mM), in air; (b) quercetin (0.6 mM) + Fe3+ (0.3 mM); (c) quercetin (0.4 mM) + Fe2+ (0.3 mM); (d) EDTA (0.6 mM) + Fe3+ (0.3 mM). Spectra (a), (b) and (c) have been amplified 10-fold.
The binding sites of Fe2+ on quercetin and other flavonoids
Crystallization of the Fe–quercetin complexes was attempted in various aqueous, organic and mixed solvents (producing a similar UV/Vis spectrum as for the Fe–quercetin complex in KPB). However, none of the attempts were successful, nor has any metal–quercetin complex structure been reported in the literature. Therefore alternative strategies were applied to elucidate the logical Fe-binding site on quercetin.
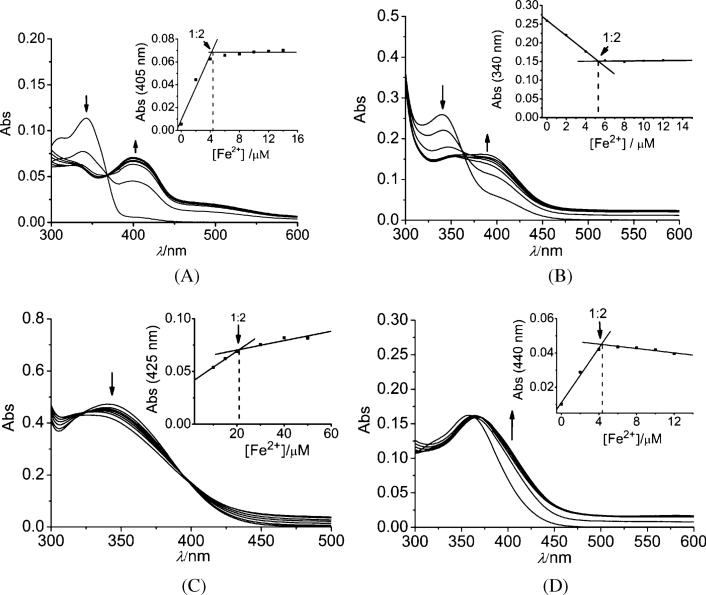
First, the Fe2+-binding properties of flavone (with the 3-ring structure but no “iron-binding motif) and three model flavonoids (Chart 1), 3-hydroxyflavone (3-HF, flavonol), 5,7-dihyroxyflavone (chrysin), and 3′,4′-dihydroxy flavone (3′,4′-DHF), one of each containing only one of the potential iron-binding motif at the equivalent positions of quercetin, were studied and their binding constants with Fe2+ measured in the KPB medium. In addition, rutin, a quercetin derivative with one of the potential binding sites (the C-ring site) blocked by a glycoside moiety, was also studied to probe the influence of glycosidation on the iron-binding property. These spectroscopic changes and binding constants were then used to suggest the preferred binding site of Fe2+ on quercetin.
The titration of Fe2+ or Fe3+ into flavone lacking an “iron-binding motif” produced no spectral changes in the UV/Vis region as expected (data not shown). As shown in Fig. 5, the other three model flavonoids with an “iron-binding motif” produced spectral changes upon addition of Fe2+. The titrations suggest the formation of 1 : 2 Fe2+–flavonoid complexes. These data demonstrate that each of the 3 predicted “iron-binding motifs” is capable of iron-binding under physiological relevant conditions. Among these, 3′,4′-dihydroxy flavone (3′,4′-DHF) and 3-hydroxyflavone (3-HF, flavonol) produced similar spectral changes as those with quercetin, while 5,7-dihyroxyflavone (chrysin) gave relatively small changes. The Fe2+ binding constants are all similar but follow the order 3-HF > chrysin ∼ 3′,4′-DHF for the 2 : 1 complexes (Table 1). Together with the spectroscopic data, this result suggests that the 3-hydroxyl and carbonyl site (C ring, site 2) is the most preferred Fe2+-binding site followed by the 5-hydroxyl and carbonyl site (A and C rings, site 3) and the 3′,4′-dihydroxy site (B ring, site 1). This order of binding constants for Fe2+ is different from that reported for the 1 : 1 complexes with Fe3+ which follows the order 3′,4′-DHF > 3-HF > chrysin, based on a potentiometric titration study.38
Fig. 5.
Titration of 3-hydroxyflavone (A), 3′,4′-dihydroxyflavone (B), chrysin (C) and rutin (D) with Fe2+ in 20 mM KPB, pH 7.2. Insets: titration curves. (A), from top to bottom: 10 μM 3-hydroxyflavone in the presence of 0, 2, 4, 6, 8, 10, 12 and 14 μM Fe2+, respectively. (B), from top to bottom: 10 μM 3′4′-dihydroxyflavone in the presence of 0, 2, 4, 6, 8, 10 and 12 μM Fe2+, respectively. (C), from top to bottom: 40 μM chrysin in the presence of 0, 10, 15, 20, 30, 40 and 50 μM Fe2+, respectively. (D), from bottom to top: 10 μM rutin in the presence of 0, 2, 4, 6, 8, 10, 12 and 14 μM Fe2+, respectively.
Table 1.
Estimated conditional binding constants with Fe2+ in 20 mM KPB, pH 7.2 at 25 °C
| Fe2+/flavonoid | Kapp by approach 1 | Kapp by approach 2 |
|---|---|---|
| Quercetin (1 : 1 complex) | 2 × 106/M | 7 × 106/M |
| Quercetin (1 : 2 complex) | 5 × 1010/M2 | Not determined |
| 3-Hydroxyflavone (1 : 2 complex) | 2 × 1011/M2 | 2 × 1011/M2 |
| Chrysin (1 : 2 complex) | 8 × 1010/M2 | Not determined |
| 3′,4′-Dihydroxyflavone (1 : 2 complex) | 3 × 1010/M2 | 2 × 1011/M2 |
| Rutin (1 : 2 complex) | 4 × 1011/M2 | 1 × 1012/M2 |
Titration of Fe2+ into rutin induces a small red shift of the band at ∼357 nm, with an isosbestic point at 368 nm and a stoichiometry consistent with a 1 : 2 Fe2+–rutin complex (Fig. 5D). Rutin has only two possible Fe-binding sites (sites 1 and 3) because site 2 is blocked by the rutinose. The spectral changes upon Fe2+-binding are very different from those of quercetin, 3′,4′-DHF or 3-HF, but closer to that of chyrsin, indicating a similar binding site as in chyrsin, i.e., site 3 of the A and C rings.
NMR studies of the metal-binding sites
NMR studies were carried out to confirm the iron-binding sites on quercetin and the other flavonoids. Quercetin displayed a better resolved 1H NMR spectrum in D2O and mixed solvents than previously reported.56 Upon addition of 0.25 mol eq. of Fe2+ (freshly prepared in 0.1 M D2O/HCl) into quercetin solution (DMSO-d6/D2O, 1 : 1, v/v, 20 mM KPB or 50 mM Tris-Cl, pH* 7.2), the 1H NMR peaks for quercetin broadened beyond detection, suggesting that a high-spin Fe2+ binds to quercetin, low-spin Fe2+ being diamagnetic. Oxidation does not occur as indicated by the aforementioned UV/Vis, ESI-Mass studies, and EPR measurements, indicating that the broadening is not due to Fe3+.
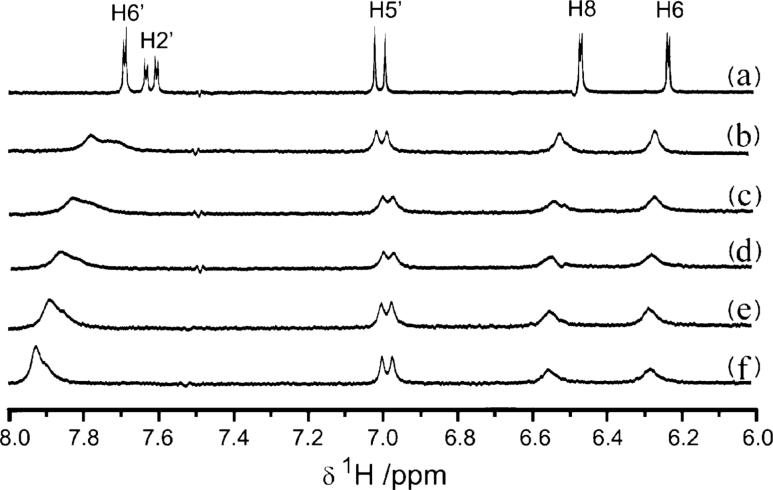
Diamagnetic metal ions Zn2+ and Ga3+ were employed to probe the binding site on quercetin for Fe2+ and Fe3+, respectively. UV/Vis studies demonstrated that Zn2+-binding to quercetin induces similar spectral changes to those of Fe2+-binding,56 while Ga3+ induces similar changes to Fe3+-binding (Fig. S4†). 1H NMR spectra of the titration of Zn2+ into a quercetin solution are shown in Fig. 6. Zn2+ perturbed the proton resonances on both the B-ring and the A-ring, i.e. the H2′ (broadening) and the H6′ (shifting downfield), H6 and H8 (broadening and shifting downfield). However, little change was observed in the H5′ doublet of the B-ring. Thus Zn2+ likely binds at site 2 on the C-ring, in accord with the UV/Vis data. In this binding mode, the 2′, 6′, 6 and 8 carbons are all connected to a Zn-bonded oxygen atom by 4 conjugated bonds, while the 5′ carbon is located one bond away. Little further change in the NMR spectrum was observed as the Zn/quercetin ratio progressed beyond 1, consistent with the formation of a specific 1 : 1 complex as for Fe2+.
Fig. 6.
1H NMR spectra of quercetin (5 mM) and the titration with Zn2+ in d6-DMSO/D2O (v/v : 50/50), 50 mM Tris-HCl, pH* 7.20. (a) quercetin only; (b) to (f), with the addition of 0.25, 0.50, 0.75, 1.0 and 2.0 mol eq. of Zn2+, respectively.
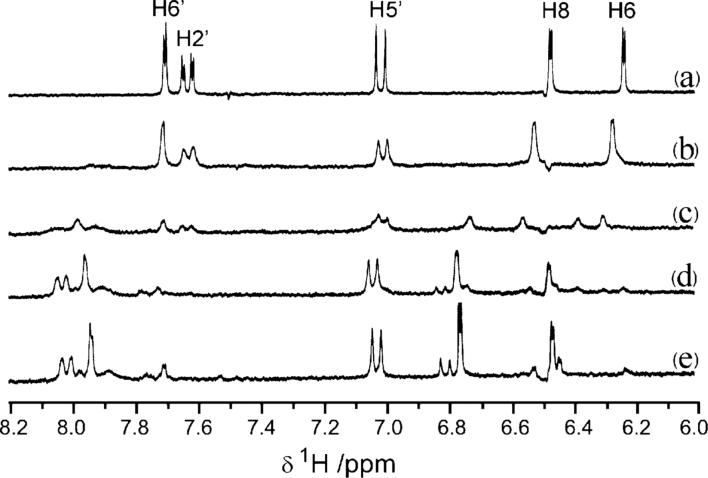
1H NMR spectra of the titration of Ga3+ into quercetin solution are shown in Fig. 7. The addition of Ga3+ perturbed the H6 and the H8 resonances on the A-ring (shifting downfield by 0.2–0.4 ppm) and the H2′ and H6′ resonances on the B-ring (shifting downfield by 0.2–0.4 ppm) but not the H5′ resonance. For the same reasons as discussed for Zn2+-binding, we conclude that Ga3+ binds to the site 2 (C-ring) of quercetin, a conclusion further supported by comparison of Ga3+-induced specific NMR changes with the other model flavonoids (unpublished data).
Fig. 7.
1H-NMR spectra of quercetin (5 mM) and the titration with Ga3+ in d6-DMSO/D2O (v/v : 50/50), 50 mM Tris-HCl, pH* 7.20. (a) quercetin only; (b) to (e), with the addition of 0.25, 0.50, 1.0 and 2.0 mol eq. of Ga3+, respectively.
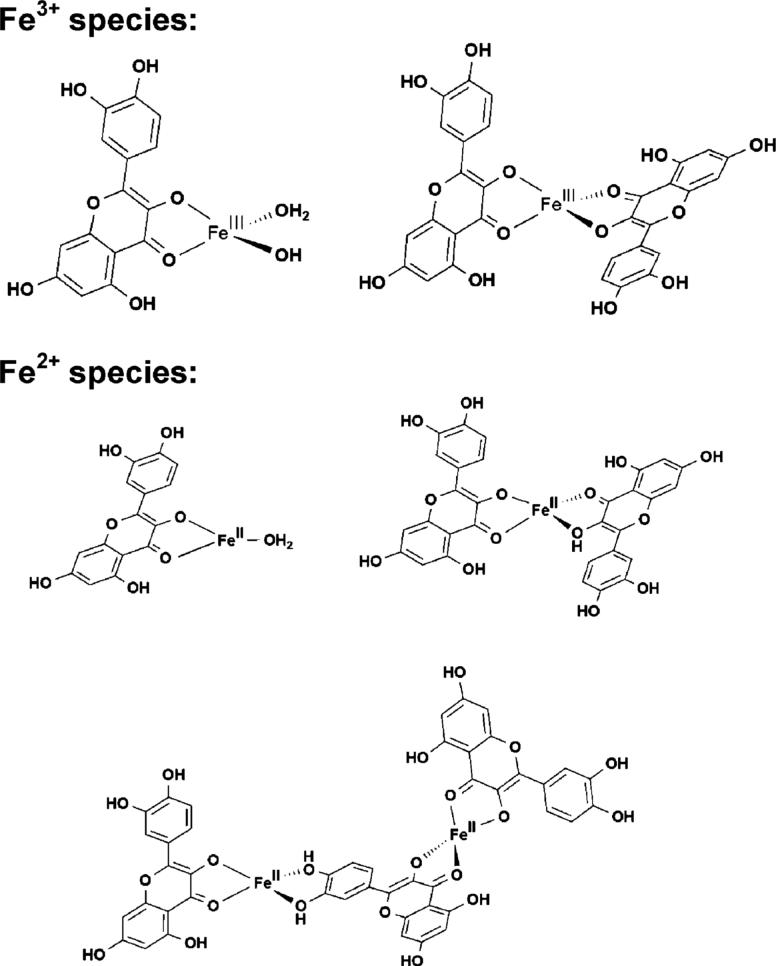
Taken together, we propose the following structures for the complexes formed under ESI-Mass conditions between quercetin with Fe2+ and Fe3+, respectively (Chart 2). In the 1 : 1 complex [FeII(Q–H)(H2O)]+, Fe2+ may be 3-coordinated by oxygen donors, two from the quercetin and one from a water molecule. Though 3-coordination for Fe2+ is not common, 3-coordinate planar Fe2+ complexes have been demonstrated recently.57 In the 1 : 2 complex [FeII(Q – H)(Q)]+, Fe2+ may be coordinated by 4 oxygen donors, two from each of the quercetin molecules. In the 2 : 3 complex [(FeII)2(Q – H)3]+ (m/z = 1014.8), both of the Fe2+ may be 4-coordinated by oxygen from the two quercetin molecules, while one quercetin may serve as a bridge with two Fe2+ coordinated. Ternary complexes with phosphate may be formed in phosphate buffer, though our attempts to detect such species by ESI-Mass were unsuccessful.
Chart 2.
Proposed structures for the complexes formed between quercetin and Fe3+ or Fe2+ under ESI-Mass conditions.
The binding affinities of Fe2+/Fe3+ with quercetin and other flavonoids
Simple isosbestic points were observed in the spectroscopic titration spectra of Fe2+ with many of the flavonoids studied, indicating a single step complexation process. Estimation of the conditional binding constants for the formation of the various flavonoid–Fe2+ complexes were obtained using the equations described in the Experimental section. The constants from the two approaches are comparable with the second approach giving values 4 to 6 times higher in a few cases (Table 1). The magnitude of the constants are in accord with the results from the competition experiments with ferrozine and EDTA (vide infra). Recently, Hajji et al.41 reported a binding constant of 4 × 105 M−1 for 1 : 1 Fe2+–quercetin complex at pH 7.4 and 37 °C, a value within a factor of 4 of that measured here by approach 1. The conditional binding constant we obtained by approach 1 for the 1 : 1 complex between Fe2+ and quercetin is also in good agreement with that reported recently by Erdogan et al.32
UV/Vis spectroscopic analysis of quercetin's ability to compete with ferrozine, EDTA and other cellular iron-chelators
Since strong binding between Fe2+ with the oxygen-based ligands of flavonoids is unexpected, competition experiments were performed with ferrozine, a well known strong Fe2+-chelator with a known binding constant (K = 3.65 × 1015 M−3).43 Fe2+ was added firstly to ferrozine, forming the (ferrozine)3–Fe2+ complex, then 1 mol eq. of quercetin was added and the kinetics was followed (Fig. 8). Upon the addition of quercetin, the free quercetin absorption at 372 nm decreased in intensity (lines c–e in Fig. 8), and this was accompanied by the growth of a new peak at ca. 425 nm, indicating the formation of the Fe2+–quercetin complex, while the absorption in the region of 560 nm where the Fe2–ferrozine complex absorbs rises slightly (lines c–e in Fig. 8) in the first hour, perhaps from mixed ligand complex formation, as found for iron–ferrozine–amino acid complexes.58 After 1 h, the spectral changes corresponding to the formation of Fe2+–quercetin continued (lines e–h in Fig. 8) but the absorption at 562 nm decreased, suggesting the dissociation of Fe2+–ferrozine and transfer of Fe2+ to quercetin. Beyond 6 h the absorptions of both quercetin–Fe2+ and ferrozine–Fe2+ decreased in intensity at 425 nm and 562 nm, respectively (data not shown), implying the degradation of the complexes. Because equilibrium was not achieved, calculations of a binding constant for quercetin–Fe2+ based on the competition data is not possible. However, the data demonstrate that quercetin binds Fe2+ more strongly than ferrozine itself under these conditions.
Fig. 8.
Competition between ferrozine and quercetin for Fe2+ in 20 mM KPB, pH 7.2. Addition of quercetin into (ferrozine)3–Fe2+: (a) 30 μM ferrozine only; (b) addition of 10 μM Fe2+. Then addition of 10 μM quercetin (c) and the kinetics of the competition was monitored in 5 min (d), 1 h (e), 2 h (f), 3 h (g), 4 h (h) and 5 h (i).
Competition with EDTA was also performed in 20 mM KPB buffer, pH 7.2. However, the addition of 1 eq. quercetin to Fe2+–EDTA solution produced only the free quercetin band (Fig. S5†), and the addition of 1 eq. EDTA to Fe2+–quercetin solution shifted the Fe2+–quercetin band to that of the free quercetin position (Fig. S6†), indicating that EDTA is a much stronger chelator of iron than quercetin, a result borne out by EPR experiment (data not shown).
Similar competition experiments were also performed with the well known cellular iron chelators ATP and citrate. As illustrated in Fig. S5 and S6†, no matter the order of the addition of the chelators, the resulting UV/Vis spectra of the ternary systems were located between those of the free quercetin and the quercetin–Fe2+ complex, indicating partial formation of the quercetin–Fe2+ complex under these conditions. Thus quercetin is capable of competition with ATP and citrate for Fe2+ under physiologically relevant conditions.
Attenuation of the Fenton reaction by flavonoids under physiologically relevant conditions
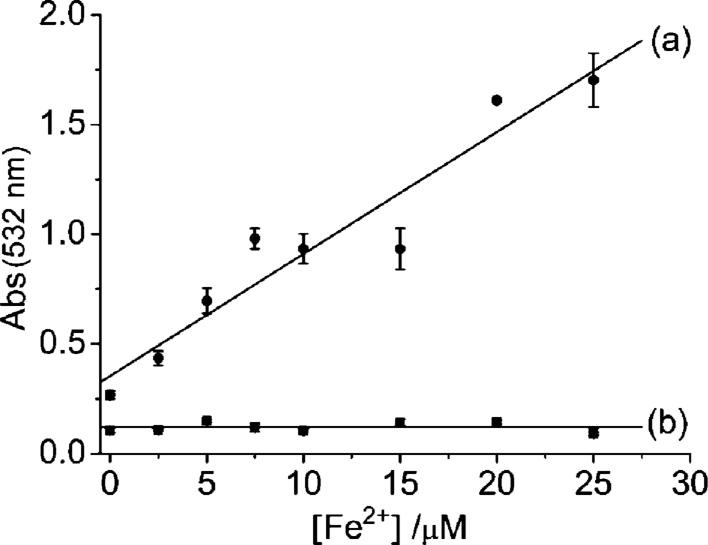
The ability of quercetin to attenuate the Fe-promoted Fenton reaction was evaluated by the 2-deoxyribose degradation assay. The Fenton reaction was generated according to eqn(1) by incubating ferrous ammonium sulfate or FeCl2 with H2O2 in the presence of 100 μM ascorbate and 10 mM 2-deoxyribose in 20 mM KPB, pH 7.2. The 2-deoxyribose degradation product, malonaldehyde (MDA) was quantified by its condensation with thiobarbituric acid (TBA) to form a chromophore with characteristic absorption at 532 nm.45 We observed 2-deoxyribose degradation in the presence of H2O2 and Fe2+ and the degree of degradation increased with an increasing Fe2+ concentration (Fig. 9) as per the Fenton reaction. In contrast, the presence of quercetin, 2-deoxyribose degradation was greatly inhibited, even at high Fe2+ concentrations (Fig. 9), indicating that quercetin is very effective at minimizing the Fenton chemistry.
Fig. 9.
Absorbance of malonaldehyde–TBA complex at 532 nm at various Fe2+ concentrations in the absence (a) or presence (b) of 10 μM quercetin.
To investigate whether quercetin itself is degraded under “catalytic” Fenton reaction conditions, the UV/Vis spectrum of Fe2+–quercetin was monitored in the presence of excess H2O2 and ascorbate in 20 mM KPB, pH 7.2. Little change in the quercetin–Fe2+ complex over 10 min was observed (Fig. S7).† These data suggest that no Fenton reaction occurred in the presence of quercetin, in agreement with a recent report59 that quercetin completely suppresses the voltammetric catalytic wave of the iron–ATP/H2O2 system.
To determine whether Fenton chemistry occurs during oxidation of 1 : 1 Fe2+–quercetin by H2O2, two mol equiv. of H2O2 were introduced and the sample was analyzed by ESI-Mass after 5−10 min (Fig. S8).† The data in Fig. S8† show no evidence of quercetin degradation, only the conversion of Fe2+–quercetin complexes to Fe3+–quercetin complexes, confirming the lack of significant Fenton chemistry.
Attenuation of Fe-promoted Fenton reaction by quercetin in the presence of ATP or citrate
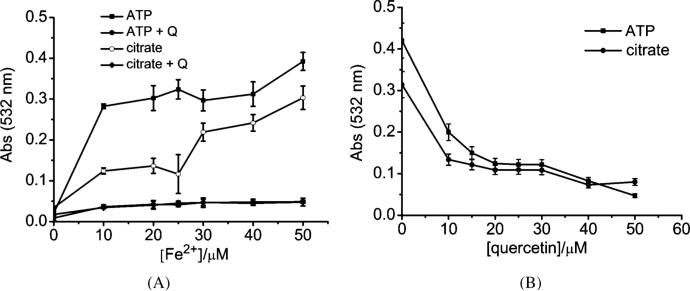
The ability of quercetin to attenuate the Fe-promoted Fenton reaction in the presence of ATP or citrate was also investigated since ATP and citrate are major iron-chelators in the cellular labile iron pool. In Fig. 10A, samples with increasing concentration of Fe2+ were incubated with 25 μM ATP or citrate for 1 h, then 25 μM of quercetin was added to the mixtures (3 repeats) and then allowed to incubate for an additional 30 min. In Fig. 10B, 25 μM Fe2+ was incubated with 25 μM ATP or citrate for 1 h, then quercetin was added to the mixtures at increasing concentration and then allowed to incubate for an additional 30 min. After the incubation, the 2-deoxyribose degradation assays were carried out. Significant degradation of 2-deoxyribose occurred in the presence of ATP or citrate (Fig. 10), but quercetin demonstrated its ability to effectively minimize the Fenton chemistry in the presence of ATP or citrate (Fig. 10A), and the effectiveness is increased with quercetin concentration (i.e., the dose) in the system (Fig. 10B).
Fig. 10.
Effect of quercetin (Q) on Fe-promoted 2-deoxyribose degradation in the presence of ATP or citrate in 20 mM KPB at pH 7.4, 100 μM ascorbate and 200 μM H2O2. (A), with increasing concentration of Fe2+, [ATP] = 25 μM, [citrate] = 25 μM and [quercetin] = 25 μM; (B), with increasing concentration of quercetin, [ATP] = 25 μM, [citrate] = 25 μM and [Fe2+] = 25 μM.
Mechanisms of quercetin attenuation of the Fenton reaction
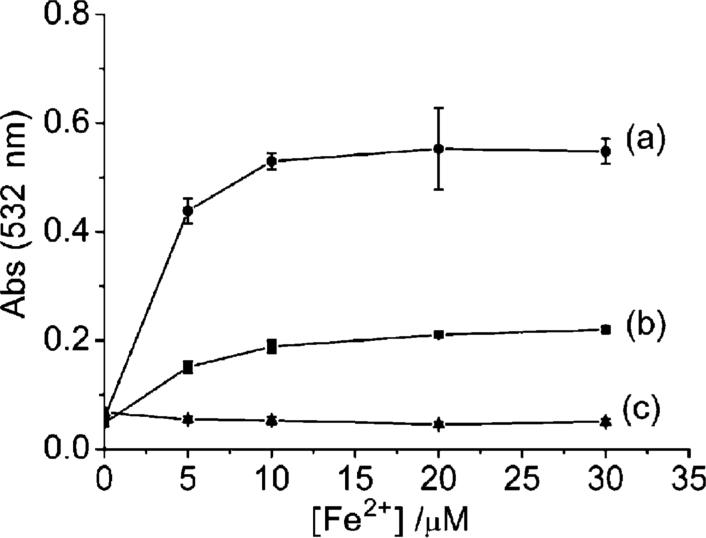
In order to determine whether quercetin functions primarily as a scavenger of hydroxyl radicals or as a chelator of Fe2+ in attenuating the Fenton reaction, additional experiments were carried out. Since EDTA can remove Fe2+ from quercetin and the Fe2+–EDTA itself is capable of generating hydroxyl radicals via the Fenton reaction,60 EDTA was used to distinguish between the scavenging and chelation activities of quercetin. In the absence of quercetin, hydroxyl radicals were readily produced in the Fe–EDTA system (Fig. 11, curve a). When quercetin was added, however, 2-deoxyribose degradation was 65% lower (curve b) but higher than that of the Fe–quercetin control (curve c). Thus quercetin is capable of scavenging ca. 65% of the hydroxyl radical produced in the system; whereas, when EDTA is absent, the chelation activity of quercetin is fully functional and 100% protection is achieved. These findings further support the crucial role of Fe-chelation in quercetin's anti-Fenton activity. A possible mechanism for iron interaction with quercetin is to shift the redox potential of Fe2+/Fe3+ upon coordination thus shutting down Fenton chemistry. Quercetin reduces Fe3+ to Fe2+ upon binding and binds Fe2+more tightly than Fe3+ in phosphate buffer. In contrast, other Fe-chelators such as EDTA or NTA (nitrilotriacetate) bind Fe3+ more tightly than Fe2+ and favor Fe2+ oxidation and the Fenton reaction.
Fig. 11.
Effect of EDTA on the ability of quercetin to prevent 2-deoxyribose degradation in 20 mM KPB at pH 7.2. (a) 25 μM EDTA with Fe2+; (b) 25 μM EDTA with Fe2+ in the presence of 10 μM quercetin; (c) Fe2+ in the presence of 10 μM quercetin. [H2O2] = 200 μM.
Biological relevance and conclusions
Recent studies on bioavailability, pharmacokinetics and metabolism of flavonoids in humans61,62 have revealed large differences from one flavonoid to another. The plasma concentrations of total metabolites generally range from 0 to 4 μM with an intake of 50 mg aglycone equivalents, and the relative urinary excretion ranged from 0.3% to 43%. The most well absorbed flavonoids by humans are isoflavones, gallic acid, catechins, flavanones and quercetin glucosides, each with different kinetics. Flavonols including quercetin are rapidly absorbed and reach maximum plasma concentration within a few hours. The elimination of flavonols (such as quercetin metabolites) is quite slow, with reported elimination half-lives ranging from 11 to 28 h. Moreover, the flavonoids (and their metabolites) from the circulation are further delivered into various organs and tissues such as liver, skin, and brain.63
A recent study with rats64 at higher doses of flavonoids in the diet (corresponding to a daily ingestion of 45–47 mg of quercetin or catechin equivalents for 21 days) revealed a much higher concentration of flavonoid metabolites in plasma (61.2 ± 4.5 μM for quercetin and 3′-O-methylquercetin; 51.2 ± 4.4 μM for catechin and 3′(or 4′)-O-methylcatechin) and in the liver (16.3 ± 3.0 μmol kg−1 tissue for quercetin and 15.9 ± 3.0 μmol kg−1 tissue for catechin). These high concentrations of polyphenols (and their metabolites) generally exceed the 10 μM quercetin used in the in vitro experiments reported here.
The strong iron-binding properties of the predicted “iron-binding motif” in phenolic compounds present in cranberries and other plants have been confirmed in this study. All the three predicted “iron-binding motifs” on quercetin, are capable of binding iron strongly under physiologically relevant conditions. The conditional binding constants with Fe2+ have been determined to be ca 106−107 M−1 (for 1 : 1 complexes) and ca 1010−1012 M−2 (for 1 : 2 complexes) at pH 7.2 in KPB buffer, with quercetin binding Fe2+ stronger than the well known Fe2+-chelator ferrozine. Quercetin can also bind Fe3+,Ga3+ and Zn2+. The most preferred site on quercetin for Fe2+ and Fe3+-binding appears to be site 2 on the C-ring. The strong Fe-binding properties suggest that quercetin may be effective in modulating cellular iron homeostasis under physiological conditions. Interestingly, quercetin can completely suppress Fe-promoted Fenton chemistry at micromolar levels even in the presence of the major cellular iron chelators ATP or citrate. Furthermore, our data indicate that the radical scavenging activity of quercetin provides only partial protection against Fenton chemistry-mediated damage while Fe-chelation by quercetin can completely inhibit the Fenton chemistry. Thus the iron-chelation activity of quercetin may be key to its antioxidant activity. Phenolics containing iron-binding motifs have also been identified in other bio-active plants such as grapes, tea and traditional Chinese medicine plants. Hence, the regulation of iron homeostasis and the inhibition of Fenton chemistry may also be important in their bio-effects as found here for quercetin.
Supplementary Material
Acknowledgements
We thank Dr Stephen J. Eyles at the UMass Amherst mass spectrometry facility for help with the ESI-Mass work as well as Dr T. Su and Dr C. Neto (UMass Dartmouth, MA, USA) for helpful discussions. Financial support from National Institutes of Health and University of Massachusetts Dartmouth are greatly acknowledged. This publication or project was made possible by grant 1 R21 AT002743 (M.G.) from the National Center for Complementary and Alternative Medicine (NCCAM) and grant R01 GM20194 (N.D.C.) from the National Institute of General Medical Science. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCCAM, or the National Institutes of Health. We also thank Mr Mark Sullivan (Bishop Stang High School, Dartmouth, MA) for participating in part of this work in the summer of 2005.
Footnotes
Electronic supplementary information (ESI) available: Apparent binding constant equations and Fig. S1–S8.
References
- 1.Blatherwick NR. Arch. Intern. Med. 1914;14:409–450. [Google Scholar]
- 2.Avorn J, Monane M, Gurwitz JH, Glynn RJ, Choodnovskil I, Lipsitz LA. J. Am. Med. Assoc. 1994;271:751–754. doi: 10.1001/jama.1994.03510340041031. [DOI] [PubMed] [Google Scholar]
- 3.Neto CC. J. Nutr. 2007;137:186S–193S. doi: 10.1093/jn/137.1.186S. [DOI] [PubMed] [Google Scholar]
- 4.Neto CC, Krueger CG, Lamoureaux TL, Kondo M, Vaisberg AJ, Hurta RAR, Curtis S, Matchett MD, Yeung H, Sweeney MI, Reed JD. J. Sci. Food Agric. 2006;86:18–25. [Google Scholar]
- 5.Weiss EI, Houri-Haddad Y, Greenbaum E, Hochman N, Ofek I, Zakay-Rones Z. Antiviral Res. 2005;66:9–12. doi: 10.1016/j.antiviral.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Vinson JA, Su X, Zubik L, Bose P. J. Agric. Food Chem. 2001;49:5315–5321. doi: 10.1021/jf0009293. [DOI] [PubMed] [Google Scholar]
- 7.Pietta P. J. Nat. Prod. 2000;63:1035–1042. doi: 10.1021/np9904509. [DOI] [PubMed] [Google Scholar]
- 8.Morel I, Lescoat G, Cogrel P, Sergent O, Pasdeloup N, Brissot P, Cillard P, Cillard J. Biochem. Pharmacol. 1993;45:13–19. doi: 10.1016/0006-2952(93)90371-3. [DOI] [PubMed] [Google Scholar]
- 9.T Nest GL, Caille O, Woudstra M, Roche S, Burlat B, Belle V, Guigliarelli B, Lexa D. Inorg. Chim. Acta. 2004;357:2027–2037. [Google Scholar]
- 10.Siciliano AA. Herbalgram. 1996;38:51–54. [Google Scholar]
- 11.Neto CC, Sweeney-Nixon MI, Lamoureaux TL, Solomon F, Kondo M, MacKinnon SL. In: Symposium Series: Phenolics in Foods and Natural Health Products. Chi-Tang Ho., editor. ACS Books; 2005. [Google Scholar]
- 12.Sun J, Chu Y, Wu X, Liu RH. J. Agric. Food Chem. 2002;50:7449–7454. doi: 10.1021/jf0207530. [DOI] [PubMed] [Google Scholar]
- 13.Vvedenskaya IO, Rosen RT, Guido JE, Russell DJ, Mills KA, Vorsa N. J. Agric. Food Chem. 2004;52:188–195. doi: 10.1021/jf034970s. [DOI] [PubMed] [Google Scholar]
- 14.Bilyk A, Sapers GM. J. Agric. Food Chem. 1986;34:585–588. [Google Scholar]
- 15.Guo M, Harvey I, Yang W, Coghill L, Campopiano D, Parkinson JA, MacGillivray R, Harris W, Sadler PJ. J. Biol. Chem. 2003;278:2490–2502. doi: 10.1074/jbc.M208776200. [DOI] [PubMed] [Google Scholar]
- 16.Baker HM, Anderson BF, Baker EN. Proc. Natl. Acad. Sci. USA. 2003;100:3579–3583. doi: 10.1073/pnas.0637295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexeev D, Zhu H, Guo M, Zhong W, Hunter D, Yang W, Campopiano D, Sadler PJ. Nat. Struct. Biol. 2003;10:297–302. doi: 10.1038/nsb903. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, Alexeev D, Hunter D, Campopiano D, Sadler PJ. Biochem. J. 2003;376:35–41. doi: 10.1042/BJ20031283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo M, Harvey I, Campopiano D, Sadler PJ. Angew. Chem., Int. Ed. 2006;45:2758–2761. doi: 10.1002/anie.200600260. [DOI] [PubMed] [Google Scholar]
- 20.Raymond KN, Dertz EA, Kim SS. Proc. Natl. Acad. Sci. USA. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Y, Guo M. Angew. Chem., Int. Ed. 2007;46:4722–4725. doi: 10.1002/anie.200604859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Einstein RS, Blemings KP. J. Nutr. 1998;128:2295–2298. doi: 10.1093/jn/128.12.2295. [DOI] [PubMed] [Google Scholar]
- 23.Ratledge C, Dover LG. Annu. Rev. Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 24.Stephens PJ, Jollie DR, Warshel A. Chem. Rev. 1996;96:2491–2513. doi: 10.1021/cr950045w. [DOI] [PubMed] [Google Scholar]
- 25.Braun V. Int. J. Med. Microbiol. 2001;291:67–79. doi: 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths E. Biol. Met. 1991;4:7–13. doi: 10.1007/BF01135551. [DOI] [PubMed] [Google Scholar]
- 27.Andrews SC, Robinson AK, Rodriguez-Quinones FEMS Microbiol. Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 28.Martin RB, Savory J, Brown S, Bertholf RL, Wills WR. Clin. Chem. (Washington, DC, U. S.) 1987;33:405–407. [PubMed] [Google Scholar]
- 29.Braun V, Killmann H. Trends Biochem. Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 30.Fenton HJH. J. Chem. Soc., Trans. 1894;65:899–910. [Google Scholar]
- 31.Ames BN, Shigenaga MK, Hagen TM. Proc. Natl. Acad. Sci. USA. 1993;90:7915–22. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erdogan G, Karadag R, Dolen E. Rev. Anal. Chem. 2005;24:247–261. [Google Scholar]
- 33.Khokhar S, Apenten SRKO. Food Chem. 2003;81:133–140. [Google Scholar]
- 34.Afanas'ef IB, Dorozhko AI, Brodskii AV, Kostiuk VA, Potapovitch AI. Biochem. Pharmacol. 1989;38:1763–1769. doi: 10.1016/0006-2952(89)90410-3. [DOI] [PubMed] [Google Scholar]
- 35.Bodini ME, Copia G, Tapia R, Leighton F, Herrera L. Polyhedron. 1999;18:2233–2239. [Google Scholar]
- 36.Leopoldini M, Russo N, Chiodo S, Toscano M. J. Agric. Food Chem. 2006;54:6343–6351. doi: 10.1021/jf060986h. [DOI] [PubMed] [Google Scholar]
- 37.Escandar GM, Sala LF. Can. J. Chem. 1991;69:1994–2001. [Google Scholar]
- 38.Engelmann MD, Hutcheson R, Cheng IF. J. Agric. Food Chem. 2005;53:2953–2960. doi: 10.1021/jf048298q. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez MT, Mira ML, Florencio MH, Jennings KR. J. Inorg. Biochem. 2002;92:105–111. doi: 10.1016/s0162-0134(02)00511-1. [DOI] [PubMed] [Google Scholar]
- 40.Mira L, Fernandez MT, Santos M, Rocha R, Florencio MH, Jennings KR. Free Radical Res. 2002;36:1199–1208. doi: 10.1080/1071576021000016463. [DOI] [PubMed] [Google Scholar]
- 41.Hajji H, Nkhili E, Tomao V, Dangles O. Free Radical Res. 2006;40:303–320. doi: 10.1080/10715760500484351. [DOI] [PubMed] [Google Scholar]
- 42.Jungbluth G, Ruhling I, Ternes W. J. Chem. Soc., Perkin Trans. 2000;2:1946–1952. [Google Scholar]
- 43.Gibbs CR. Anal. Chem. 1976;48:1197–1201. [Google Scholar]
- 44.Berlett BS, Levine RL, Chock PB, Chevion M, Stadtman ER. Proc. Natl. Acad. Sci., USA. 2001;98:451–456. doi: 10.1073/pnas.021552298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopes GKB, Schulman HM, Hermes-Lima M. Biochim. Biophys. Acta. 1999;1472:142–152. doi: 10.1016/s0304-4165(99)00117-8. [DOI] [PubMed] [Google Scholar]
- 46.Cook NC, Samman S. J. Nutr. Biochem. 1996;7:66–76. [Google Scholar]
- 47.Hider RC, Mohd-Nor AR, Silver J, Morrison IEG, Rees LVC. J. Chem. Soc., Dalton Trans. 1981:609. [Google Scholar]
- 48.Souza RFV, Giovani WF. Spectrochim. Acta, Part A. 2005;61:1985–1990. doi: 10.1016/j.saa.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 49.Salama S, Stong JD, Neilands JB, Spiro TG. Biochemistry. 1978;17:3781–3785. doi: 10.1021/bi00611a017. [DOI] [PubMed] [Google Scholar]
- 50.Abergel RJ, Warner JA, Shuh DK, Raymond KN. J. Am. Chem. Soc. 2006;128:8920–8931. doi: 10.1021/ja062046j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawabata T, Schepkin V, Haramki N, Phadke RS, Packer L. Biochem. Pharmacol. 1996;51:1569–1577. doi: 10.1016/0006-2952(96)00101-3. [DOI] [PubMed] [Google Scholar]
- 52.Ferrer EG, Salinas MV, Correa MJ, Naso L, Barrio DA, Etcheverry SB, Lezama L, Rojo T, Williams PAM. JBIC, J. Biol. Inorg. Chem. 2006;11:791–801. doi: 10.1007/s00775-006-0122-9. [DOI] [PubMed] [Google Scholar]
- 53.Cornard JP, Dangleterre L, Lapouge C. J. Phys. Chem. A. 2005;109:10044–10051. doi: 10.1021/jp053506i. [DOI] [PubMed] [Google Scholar]
- 54.Cornard JP, Merlin JC. J. Inorg. Biochem. 2002;92:19–27. doi: 10.1016/s0162-0134(02)00469-5. [DOI] [PubMed] [Google Scholar]
- 55.Marco VB, Bombi GG. Mass Spectrom. Rev. 2006;25:347–379. doi: 10.1002/mas.20070. [DOI] [PubMed] [Google Scholar]
- 56.Nest GL, Woudstra OCM, Guerlesquin SRF, Lexa D. Inorg. Chim. Acta. 2004;357:775–784. [Google Scholar]
- 57.Andres H, Bominaar EM, Smith JM, Eckert NA, Holland PL, Münck E. J. Am. Chem. Soc. 2002;124:3012–3025. doi: 10.1021/ja012327l. [DOI] [PubMed] [Google Scholar]
- 58.Berlett BS, Levine RL, Chock PB, Chevion M, Stadtman ER. Proc. Natl. Acad. Sci., USA. 2001;98:451–456. doi: 10.1073/pnas.021552298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng IF, Breen K. Biometals. 2000;13:77–83. doi: 10.1023/a:1009229429250. [DOI] [PubMed] [Google Scholar]
- 60.Matsufuji H, Shibamoto T. J. Agric. Food Chem. 2004;52:3136–3140. doi: 10.1021/jf035337+. [DOI] [PubMed] [Google Scholar]
- 61.Manach C, Williamson G, Morand C, Scalbert A, Rémésy C C. Am. J. Clin. Nutr. 2005;81:230S–242S. doi: 10.1093/ajcn/81.1.230S. [DOI] [PubMed] [Google Scholar]
- 62.Williamson G, Manach C. Am. J. Clin. Nutr. 2005;81:243S–255S. doi: 10.1093/ajcn/81.1.243S. [DOI] [PubMed] [Google Scholar]
- 63.Spencer JPE, Mohsen MMA, Rice-Evans C. Arch. Biochem. Biophys. 2004;423:148–161. doi: 10.1016/j.abb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 64.Silberberg M, Morand C, Manach C, Scalbert A, Remesy AC. Life Sci. 2005;77:3156–3167. doi: 10.1016/j.lfs.2005.03.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.