Abstract
Mammalian defensins are cationic antimicrobial peptides that play a central role in host innate immunity and as regulators of acquired immunity. In animals, three structural defensin subfamilies, designated as α, β, and θ, have been characterized, each possessing a distinctive tridisulfide motif. Mature α- and β-defensins are produced by simple proteolytic processing of their prepropeptide precursors. In contrast, the macrocyclic θ-defensins are formed by the head-to-tail splicing of nonapeptides excised from a pair of prepropeptide precursors. Thus, elucidation of the θ-defensin biosynthetic pathway provides an opportunity to identify novel factors involved in this unique process. We incorporated the θ-defensin precursor, proRTD1a, into a bait construct for a yeast two-hybrid screen that identified rhesus macaque stromal cell-derived factor 2-like protein 1 (SDF2L1), as an interactor. SDF2L1 is a component of the endoplasmic reticulum (ER) chaperone complex, which we found to also interact with α- and β-defensins. However, analysis of the SDF2L1 domain requirements for binding of representative α-, β-, and θ-defensins revealed that α- and β-defensins bind SDF2L1 similarly, but differently from the interactions that mediate binding of SDF2L1 to pro-θ-defensins. Thus, SDF2L1 is a factor involved in processing and/or sorting of all three defensin subfamilies.
Mammalian defensins are tridisulfide-containing antimicrobial peptides that contribute to innate immunity in all species studied to date. Defensins are comprised of three structural subfamilies: the α-, β-, and θ-defensins (1). α- and β-Defensins are peptides of about 29–45-amino acid residues with similar three-dimensional structures. Despite their similar tertiary conformations, the disulfide motifs of α- and β-defensins differ. Expression of human α-defensins is tissue-specific. Four myeloid α-defensins (HNP1–4) are expressed predominantly by neutrophils and monocytes wherein they are packaged in granules, while two enteric α-defensins (HD-5 and HD-6) are expressed at high levels in Paneth cells of the small intestine. Myeloid α-defensins constitute about 5% of the protein mass of human neutrophils. HNPs are discharged into the phagosome during phagocytic ingestion of microbial particles. HD-5 and HD-6 are produced and stored as propeptides in Paneth cell granules and are processed extracellularly by intestinal trypsin (2). β-Defensins are produced primarily by various epithelia (e.g. skin, urogenital tract, airway) and are secreted by the producing cells in their mature forms. In contrast to pro-α-defensins, which contain a conserved prosegment of ∼40 amino acids, the prosegments in β-defensins vary in length and sequence. θ-Defensins are found only in Old World monkeys and orangutans and are the only known circular peptides in animals. These 18-residue macrocyclic peptides are formed by ligation of two nonamer sequences excised from two precursor polypeptides, which are truncated versions of ancestral α-defensins. Like myeloid α-defensins, θ-defensins are stored primarily in neutrophil and monocyte granules (3).
Numerous laboratories have demonstrated that the antimicrobial properties of defensins derive from their ability to bind and disrupt target cell membranes (4), and studies have shown defensins to be active against Gram-positive and -negative bacteria (5), viruses (6–9), fungi (10, 11), and parasites such as Giardia lamblia (12). Defensins also play a regulatory role in acquired immunity as they are known to chemoattract T lymphocytes, monocytes, and immature dendritic cells (13, 14), act as adjuvants, stimulate B cell responses, and up-regulate proliferation and cytokine production by spleen cells and T helper cells (15, 16).
Defensins are produced as pre-propeptides and undergo post-translational processing to form the mature peptides. While much has been learned about regulation of defensin expression, little is known about the factors involved in their biosynthesis. Valore and Ganz (17) investigated the processing of defensins in cultured cells and demonstrated that maturation of HNPs occurs through two proteolytic steps that lead to formation of mature α-defensins, but the proteases involved have yet to be identified. Moreover, there are virtually no published data regarding endoplasmic reticulum (ER)2 factors that are responsible for the folding, processing, and sorting steps necessary for defensin maturation and secretion or trafficking to the proper subcellular compartment. It is likely that several chaperones, proteases, and protein-disulfide isomerase (PDI) family proteins are involved. Consistent with this possibility, Gruber et al. (18) recently demonstrated the role of a PDI in biosynthesis of cyclotides, small ∼30-residue macrocyclic peptides produced by plants.
The primary structures of α- and θ-defensin precursors are closely related. We therefore undertook studies to identify proteins that interact with representative propeptides of each defensin subfamily with the goal of determining common and unique processes that regulate biosynthesis of α- and θ-defensins. We used two-hybrid analysis to first identify interactors of the θ-defensin precursor, proRTD1a. As described, we identified SDF2L1, a component of the ER-chaperone complex as an interactor, and showed that it also specifically interacts with α- and β-defensins. This suggests that SDF2L1 is involved in the maturation/trafficking of defensins at a step common to all three subfamilies of mammalian defensins.
EXPERIMENTAL PROCEDURES
The plasmids and strains used in this study are listed in Table 1. The primers used for polymerase chain reactions (PCR), and the details for plasmid construction are described in supplemental information.
TABLE 1.
Plasmids and yeast strains used in this study
| Plasmid (description) | Comments |
| PEC2036 (AD-vector) | pGADT7-Rec from Clontech TRP1 Ampr |
| PEC2037 (DBD-vector) | pGBKT7 from Clontech LEU2 Kanr |
| PEC2038 (DBD-proRTD1a) | proRTD1a cloned in the NcoI/BamHI sites of pGBKT7 |
| PEC2069 (AD-SDF2L1) | SDF2L1/pGADT7-Rec; plasmid expressing interactor of proRTD1a, isolated from yeast |
| PEC2073 (DBD-proseg) | The prosegment of RTD1a cloned in the EcoRI/BamHI sites of pGBKT7 |
| PEC2082 (DBD-proHNP3) | PEC2080 (proHNP3/pCR2.1) was digested with Acc65I, filled-in with Klenow and then digested with EcoRI. The fragment containing proHNP3 was then ligated into EcoRI/SmaI-digested pGBKT7 |
| PEC2084 (AD-ΔN28SDF2L1) | ΔN28SDF2L1 cloned into EcoRI and XhoI sites of pGADT7-Rec |
| PEC2105 (AD-ΔN28SDF2L1ΔMIR1) | SDF2L1ΔMIR1 (N-terminal deletion, ΔN87) cloned into EcoRI and XhoI sites of pGADT7-Rec |
| PEC2106 (AD-SDF2L1ΔMIR3) | SDF2L1ΔMIR3 (C-terminal deletion, ΔC151) cloned into EcoRI and XhoI sites of pGADT7-Rec |
| PEC2113 (DBD-proHBD1) | proHBD1 cloned as EcoRI/BamHI fragment into pGBKT7 |
| PEC2114 (DBD-proHD5) | proHD5 cloned as EcoRI/BamHI fragment into pGBKT7 |
| PEC2122 (DBD-ΔN28SDF2L1ΔMIR2) | ΔN28SDF2L1ΔMIR2 (internal deletion of residues 95-149) cloned into the EcoR1/XhoI sites of pGADT7-Rec |
| PEC2123 (DBD-proHNP3Δ35-49) | proHNP3Δ35-49 fragment cloned into EcoRI/BamHI sites of pGBKT7 |
| PEC2124 (DBD-proRTD1aΔRLL) | proRTD1aΔRLL cloned into NcoI and BamHI sites of pGBKT7 |
| PEC2125 (DBD-RTD1a-9mer) | RTD1a was cloned into the EcoRI and XhoI sites of pGBKT7 |
| PEC2126 (DBD-HNP3) | HNP3 fragment was cloned into EcoRI and XhoI sites of pGBKT7 |
| PEC2127 (AD-MIR3) | MIR3 fragment (residues 151-221 of SDF2L1) was cloned into EcoRI and XhoI sites of pGADT7-Rec |
| PEC2003 (GST) | NcoI site in PEC615 (UAF30/pCBGST1) was filled in with Klenow and re-ligated to render the UAF30 sequence out of frame, for expression of GST TRP1 Ampr |
| PEC2007 (GST-proRTD1a) | proRTD1a cloned in the NcoI/Acc65I sites of pCBGST1 for expression of GST-proRTD1a |
| PEC2075 (GST-ΔN28SDF2L1) | ΔN28SDF2L1 cloned into the NcoI/Acc65I sites of pCBGST1 for expression of GST-ΔN28SDF2L1 |
| PEC2121 (GST-proHNP3) | proHNP3 cloned into the NcoI/Acc65I sites of pCBGST1 for expression of GST-proHNP3 |
| Yeast strains | Phenotype |
| AH109 | MATa trp1-901 leu2-3, 112 ura3-52 his3-Δ200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ MEL1 |
| JD47-13C | MATa his3-Δ200 trp1-Δ63 lys2-801 ura3-52 leu2-3, 112 |
Yeast Two-hybrid Screening—The cDNA encoding the θ-defensin propeptide proRTD1a was cloned into the pGBKT7 vector inframe with the GAL4-DNA binding domain (DBD). Total RNA prepared from rhesus macaque bone marrow (obtained from the Oklahoma National Primate Research Center) was reverse-transcribed and first strand cDNA was amplified using long distance PCR and purified using spin columns. The amplified cDNA and linearized pGADT7-Rec plasmid were then transformed into yeast AH109 cells containing proRTD1a/pGBKT7 (DBD-proRTD1a) plasmid as recommended by the manufacturer (Matchmaker 3; Clontech). Homologous recombination between the amplified PCR product and pGADT7-Rec produced circular plasmids with the amplified cDNAs cloned downstream of the GAL4 activation domain (AD). Transformants were plated on selective synthetic medium containing dextrose (SDex) and lacking histidine, adenine, tryptophan, and leucine (quadruple drop out; SDex/QDO), disclosing positive interactors. An aliquot of the transformant mixture was also plated on SDex lacking tryptophan and leucine (SDex-TL) to calculate the number of colonies screened, estimated to be ∼2 × 106. Colonies from SDex/QDO plates were replated on SDex/QDO plates containing 20 mg/l 5-bromo-4-chloro-in-doyl-α-d-galactopyranoside (X-α-gal; SDex/QDO+X-α-gal plate) and the pGADT7-Rec plasmid was isolated from blue colonies. The pGADT7-Rec plasmid thus isolated was transformed into Escherichia coli and selected on LB-ampicillin plates. The size of inserts in pGADT7-Rec transformants was determined by agarose electrophoresis of PCR products. Plasmids with inserts of >300 bp were retransformed into yeast AH109 containing proRTD1a/pGBKT7 or control pGBKT7 vector. Inserts that showed two-hybrid interactions in the presence of DBD-proRTD1a but not in the presence of vector alone (putative interactors) were then sequenced.
Spot Test Analysis—Triplicate transformant colonies from SDex-TL plates were grown in SDex-TL medium to saturation, and 3 μl of each culture were spotted on SDex/QDO+X-α-gal and control SDex-TL plates. All DBD-fusion and AD-fusion constructs were tested to confirm that they did not activate the reporters by themselves. AH109 cells transformed with plasmids for AD-SV40 large T/DBD-p53 and AD-SV40 large T/DBD-laminin A were used as two-hybrid positive and negative controls, respectively, on all plates (denoted by “+” and “-” in figures). Plates were incubated for 3–5 days at 30 °C.
In Vitro Transcription Translation of [35S]HA-SDF2L1—The pGADT7-Rec plasmid, which contains the T7 promoter and an HA-epitope tag downstream of the GAL4-AD, was used for biosynthesis of HA-tagged proteins in vitro. [35S]HA-SDF2L1 was produced by in vitro transcription/translation using the TnT T7-coupled rabbit reticulocyte lysate system (Promega) and the AD-SDF2L1 plasmid (see Table 1) per the supplier's protocol. Reactions were carried out in triplicate (90 min, 30 °C) in the presence of [35S]Met (>1000 Ci/mmol). Reaction mixtures were pooled and desalted on Zeba desalt spin columns (Pierce) equilibrated in HEN buffer (20 mm HEPES pH 7.2, 2 mm EDTA, and 100 mm NaCl), and stored at –20 °C.
Glutathione S-Transferase (GST) Pull-down Assays—pCBGST1 plasmids (19) expressing GST or GST fusions were transformed into yeast JD47–13C (20) (Table 1). Cells were grown in SDex-Trp medium containing 0.1 mm CuSO4, harvested by centrifugation, washed with water, and resuspended in 500 μl of buffer A (50 mm HEPES pH 7.5, 150 mm NaCl, 5 mm EDTA, and 1% Triton X-100; plus EDTA-free protease inhibitors (Roche Applied Science)) per 50 ml of cell culture. Cells were lysed by vortexing with glass beads, and the supernatant was collected by centrifugation. The pellet was re-extracted with buffer A containing protease inhibitors, and this extract was combined with the initial supernatant and clarified by centrifugation. Affinity columns were prepared in duplicate by adding glutathione beads to yeast extracts containing GST fusion proteins and incubating the mixture at 4 °C for 3 h, after which beads were washed with buffer A.
Pull-down assays were performed using affinity columns prepared from yeast lysates expressing GST alone, GST-proRTD1a, or GST-proHNP3. One set of columns was loaded with 85 μl of HEN buffer plus 15 μl of [35S]HA-SDF2L1 and incubated at 4 °C for 3 h. Beads were then washed twice with HEN buffer and boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Samples were then electrophoresed on a 15% acrylamide SDS-Tricine gel and analyzed by autoradiography. In parallel, a second set of affinity columns was similarly processed for electrophoresis, but bound proteins were analyzed by Coomassie Blue staining of SDS-Tricine gels to assess the quantity of GST fusion protein bound to each affinity column.
Interaction Studies using Human Myeloid α-Defensins (HNPs) Produced in HL-60 Cells—HL-60 cells, grown in Iscove's modified Dulbecco medium with 20% fetal bovine serum and antibiotics (penicillin and streptomycin), were harvested and duplicate 2 × 106 cell-samples were suspended in 1 ml of labeling medium (Cys- and Met-free DMEM, 10% dialyzed fetal bovine serum, 4 mm glutamine, antibiotics, and 30 mg/liter Met). After 1 h of incubation at 37 °C, 80 μCi/ml of [35S]Cys (1075 Ci/mmol) was added for 6 h. Cells were harvested by centrifugation, and extracts were prepared by addition of 1 ml of cell lysis buffer (50 mm Tris-HCl pH 7.5, 150 mm NaCl, 0.5% sodium deoxycholate, 0.5% Igepal CA630) containing complete protease inhibitor mix (Roche Applied Science) and extracted for 0.5–1 h. Clarified extracts were added to either GST or GST-ΔN28SDF2L1 (Table 1) columns (prepared as above) and mixed at 4 °C for 3 h. As controls, HNPs from HL-60 extracts were immunoprecipitated with either α-HNP IgG or pre-immune IgG and protein A-Sepharose. The columns were washed twice with cell lysis buffer, and bound proteins were desorbed and analyzed on SDS-tricine gels by autoradiography as described above.
Immunoprecipitation and Western Blot Analysis of DBD Fusion Proteins—Extracts of AH109 cells transformed with DBD-vector (pGBKT7), DBD-proRTD1a, or DBD-proseg plasmids (Table 1) were prepared in buffer A containing EDTA-free protease inhibitors by lysis with glass beads. Extracts containing ∼0.7 mg of protein were immunoprecipitated with α-DBD antibody (Santa Cruz Biotechnology.) and Protein A/G beads (Pierce). Beads were washed twice with buffer A and bound proteins were released by boiling in SDS-PAGE sample buffer. Samples were resolved on 15% acrylamide SDS-Tricine gel, transferred to nitrocellulose membrane, probed with α-Myc antibody (Covance), and the chemiluminescence signal was detected with x-ray film.
Bioinformatics Analyses—The ExPASy proteomics server of the Swiss Institute of Bioinformatics was used for BLAST searches of the SWISSPROT data base, for alignment of protein sequences and for homology modeling using SWISS-MODEL. The Ensembl data base was used for BLASTN search of the Macaca mulatta genome data base (21).
RESULTS
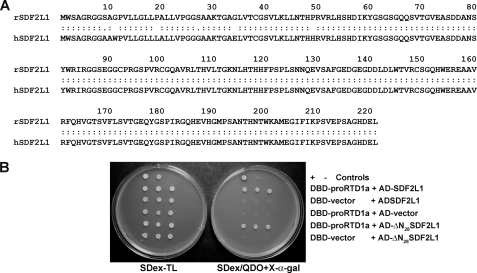
Rhesus SDF2L1 Interacts with proRTD1a—θ-Defensins are synthesized by immature myeloid cells in the bone marrow (3). To identify proteins involved in θ-defensin biosynthesis, we employed two-hybrid analyses wherein the θ-defensin precursor, proRTD1a (fused with DBD) was used as the bait, and rhesus bone marrow cDNA cloned into pGADT7 provided a library of potential interactors that were screened in the AH109 yeast strain. After eliminating false positives, pGADT7-Rec plasmids bearing interactors were sequenced and identified by BLAST analysis. Among the dozen interactors identified was a 221-amino acid residue protein that was 97.3% identical to human SDF2L1 (Swiss-Prot Accession Number: Q9HCN8; Fig. 1A). A BLASTN search of the M. mulatta genomic data base identified the cloned cDNA as rhesus macaque SDF2L1, a three-exon gene located on chromosome 10. SDF2L1 is an ER-resident protein expressed in a wide variety of tissues. Given its subcellular address and previously characterized interactions with chaperone proteins (22–24), we conducted experiments to characterize the interaction of SDF2L1 with defensin precursors in vitro and in vivo.
FIGURE 1.
Primary structure of SDF2L1 and its interaction with proRTD1a. A, amino acid sequence alignment of rhesus SDF2L1 (rSDF2L1) with human SDF2L1 (hSDF2L1) using the LALIGN program. B, spot tests demonstrating two-hybrid interactions using the listed plasmids transformed in AH109 cells. Transformants were plated in triplicate.
Interaction of SDF2L1 with proRTD1a was confirmed by retransforming AH109 cells with DBD-proRTD1a and SDF2L1/pGADT7-Rec (AD-SDF2L1) plasmids. Colonies from SDex-TL plates were replated and grew as blue colonies on SDex/QDO+X-α-gal plates. In contrast AH109 cells with pGBKT7 vector (DBD-vector) and AD-SDF2L1 or DBD-proRTD1a and pGADT7-Rec (AD-vector) plasmids were incapable of growth on SDex/QDO+X-α-gal plates (Fig. 1B).
The plasmid bearing the SDF2L1 sequence encodes the full length protein. To determine whether the N-terminal signal sequence was required for interaction with proRTD1a (which would cast doubt on the physiologic relevance of the interaction) we produced pGADT7-Rec encoding a truncated form of SDF2L1 lacking the N-terminal signal peptide (AD-ΔN28SDF2L1). As shown in Fig. 1B, cells bearing the AD-ΔN28SDF2L1 and DBD-proRTD1a plasmids grew as blue colonies on SDex/QDO+X-α-gal plates, whereas negative controls did not. These data demonstrate that the interaction of proRTD1a with SDF2L1 is not mediated through (nonspecific) interactions with the SDF2L1 signal peptide.
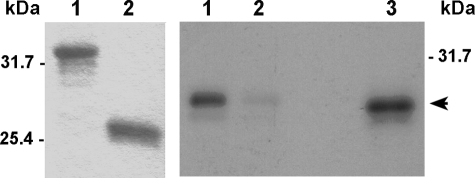
To further analyze the interaction of proRTD1a with SDF2L1 we conducted GST pull-down experiments. GST and GST-proRTD1a columns were prepared from yeast lysates as described under “Experimental Procedures” (Fig. 2, left panel). Columns were incubated with [35S]HA-SDF2L1 and washed with HEN-buffer. Bound proteins were eluted and analyzed by SDS-PAGE and autoradiography. As shown in Fig. 2 (right panel), [35S]HA-SDF2L1 was specifically retained on the GST-proRTD1a column, but not on GST columns, demonstrating an in vitro interaction between proRTD1a and SDF2L1.
FIGURE 2.
In vitro interaction of SDF2L1 with proRTD1a. Left panel, equivalent amounts of GST-proRTD1a (lane 1) or GST (lane 2) were precipitated from yeast extracts with glutathione-Sepharose beads, electrophoresed on SDS-Tricine gels and stained with Coomassie Blue. Right panel, GST-proRTD1a (lane 1) and GST (lane 2) columns were incubated with [35S]HA-SDF2L1, and the bound proteins were analyzed on 15%-acrylamide SDS-Tricine gels and autoradiography. Lane 3 was loaded with 5% of input [35S]HA-SDF2L1 (arrowhead).
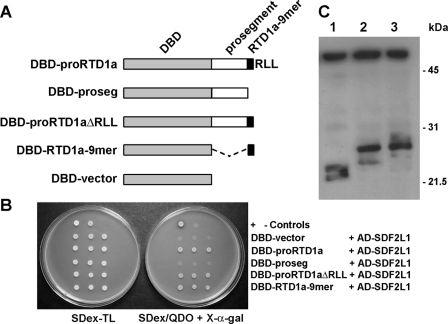
A Nonamer Sequence in RTD1a Is Sufficient for SDF2L1 Interaction—To delineate the proRTD1a sequence that interacts with SDF2L1, we expressed selected truncated forms of proRTD1a fused to the DBD, including the prosegment sequence (DBD-proseg), the propeptide lacking the C-terminal RLL-tripeptide (DBD-proRTD1aΔRLL), and a fusion of DBD to the RTD1a nonapeptide that constitutes half of the mature cyclic θ-defensin structure (DBD-RTD1a-9mer; Fig. 3A). Two hybrid analysis demonstrated an interaction between AD-SDF2L1 and DBD-proRTD1aΔRLL as well as DBD-RTD1a-9mer. However, neither the DBD vector nor DBD-proseg showed two-hybrid interaction with AD-SDF2L1 (Fig. 3B). The lack of interaction was not due to instability of the mutant protein as evidenced by the presence of comparable amounts of immunoprecipitable DBD, DBD-proRTD1a, and DBD-proseg in yeast lysates (Fig. 3C). These data demonstrate that the θ-defensin prosegment is not required for interaction with SDF2L1, and that the RTD1a nonapeptide is sufficient for this interaction.
FIGURE 3.
The RTD1a nonamer sequence is sufficient for interaction with SDF2L1. A, schematic representation of the DBD-fusion constructs of proRTD1a and its mutants, described in Table 1. B, spot tests were performed using three colonies of AH109 cells transformed with plasmids listed. C, immunoprecipitation was performed by incubating extracts of AH109 cells containing DBD-vector (lane 1), DBD-proRTD1a (lane 2), and DBD-proseg (lane 3) with anti-DBD antibodies and protein A-Sepharose. The immunoblot was probed using anti-Myc antibodies. The upper band present in all three lanes is antibody heavy chain.
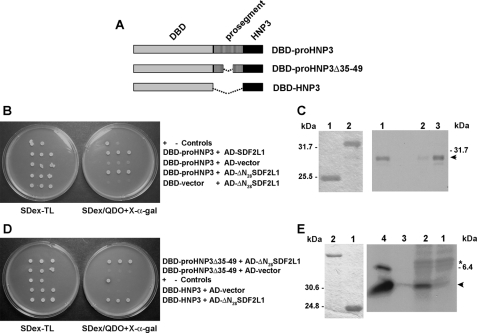
Interactions of Human Defensins with SDF2L1—Because human myeloid α-defensins (HNPs) have significant homology to the (pro-θ-defensin) proRTD1a (3), we tested for the ability of proHNPs to interact with SDF2L1. For these studies we produced a DBD-proHNP3 fusion and observed a positive interaction that was judged to be specific since DBD-proHNP3 did not activate the reporters in the presence of pGADT7-Rec vector alone. As with proRTD1a, deletion of the SDF2L1 signal sequence did not abolish this interaction, indicating that the interaction was not mediated through nonspecific hydrophobic interactions with the signal peptide (Fig. 4B). Pull-down experiments were performed using GST or GST-proHNP3 columns incubated with [35S]HA-SDF2L1. Analysis of the bound proteins demonstrated that [35S]HA-SDF2L1 was specifically retained on the GST-proHNP3 column (Fig. 4C). Thus, like proRTD1a, proHNP3 interacts with SDF2L1 in yeast and in vitro.
FIGURE 4.
SDF2L1 interacts with proHNP3 and the mature HNP3 sequence is sufficient for this interaction. A, schematic representations of the DBD-fusion constructs used in this figure. B, spot tests were performed using three colonies of AH109 cells transformed with plasmids DBD-proHNP3 and AD-SDF2L1, DBD-proHNP3, and AD-ΔN28SDF2L1, and the controls: DBD-proHNP3 and AD-vector, and DBD-vector and AD-ΔN28SDF2L1 as indicated in the figure. C, In vitro interaction between proHNP3 and SDF2L1. Left panel, SDS-tricine PAGE of GST (lane 1) and GST-proHNP3 (lane 2) purified from yeast extracts with glutathione-Sepharose beads and then Coomassie Blue-stained. Right panel, GST (lane 2) and GST-proHNP3 columns (lane 3) were incubated with [35S]HA-SDF2L1. Bound proteins were analyzed by autoradiography. Lane 1, input [35S]HA-SDF2L1 (arrowhead). D, spot tests of AH109 cells transformed with plasmids listed (Table 1). E, in vitro interaction between HNPs and SDF2L1; left panel, Coomassie Blue-stained SDS-Tricine PAGE of GST (lane 1) and GST-ΔN28SDF2L1 (lane 2) prepared as in C above. Right panel, GST (lane 1) and GST-ΔN28SDF2L1 (lane 2) columns incubated with metabolically labeled HL60 extracts. Bound proteins were analyzed by autoradiography. Control immunoprecipitations with labeled HL60 extracts and protein A-Sepharose beads and either rabbit preimmune IgG (lane 3) or α-HNP IgG (lane 4). The arrowhead indicates 35S-labeled HNP, and the asterisk indicates the precursor, proHNP.
As noted above, processing of (α-defensin) proHNP3 gives rise to a linear peptide, whereas proRTD1a is converted via excision and splicing reactions to a cyclic octadecapeptide. Previously, Liu and Ganz (25) identified a 12-amino acid residue motif in the HNP prosegment (amino acid residues 40–51) that was required for correct processing and sorting of mature HNP peptides. We hypothesized that this sequence motif might also interact with SDF2L1. To test this, we produced a two-hybrid construct in which residues 35–49 were deleted (DBD-proHNP3Δ35–49). When tested in the two hybrid assay, we observed an interaction between proHNP3Δ35–49 and SDF2L1 and also between mature HNP3 sequence (DBD-HNP3) and SDF2L1 (Fig. 4D). These results indicate that the mature HNP sequence is sufficient for interaction with SDF2L1 and that the prosegment is not required.
The human promyelocytic cell line HL60 is known to synthesize HNPs (17, 26). To further characterize the interaction of HNPs with SDF2L1, GST-ΔN28SDF2L1 and GST columns were prepared (Fig. 4E, left panel) and incubated with extracts of [35S]Cys-labeled HL60 cells. Bound proteins were desorbed and characterized by SDS-PAGE. As shown in Fig. 4E (right panel), a radiolabeled peptide that comigrated with mature HNP bound to immobilized GST-ΔN28SDF2L1, but not to GST beads. These results demonstrate the interaction of SDF2L1 with endogenous HNPs expressed in human leukemia cells, confirming the interactions detected in the two-hybrid assays.
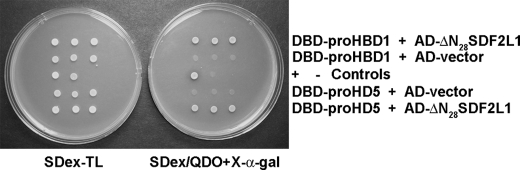
Because SDF2L1 interacted with precursors and mature sequences of structurally diverse (α- and θ-) myeloid defensins, we sought to determine if other defensins interact with SDF2L1. We fused DBD to the prodefensin sequences of human enteric α-defensin HD5 (2) and human β-defensin HBD-1 (27). In two hybrid analysis, AH109 cells expressing either DBD-proHBD1 or DBD-proHD5 in the presence of AD-ΔN28SDF2L1 plasmid grew as blue colonies on SDex/QDO+X-α-gal plates. No growth was observed for cells expressing either DBD-proHBD1 or DBD-proHD5 in the presence of the control pGBKT7 plasmid (Fig. 5), indicating that the interaction was specific. These data demonstrate that SDF2L1 interacts with precursors of α-, β-, and θ-defensins.
FIGURE 5.
SDF2L1 interacts with precursors of enteric α-defensins and human β-defensin. Expression plasmids containing DBD-proHBD1 and DBD-proHD5 were transformed into AH109 cells with AD-ΔN28SDF2L1 plasmid or the control AD-vector. Transformants were then spotted on SDex-TL or SDex/QDO+X-α-gal plates.
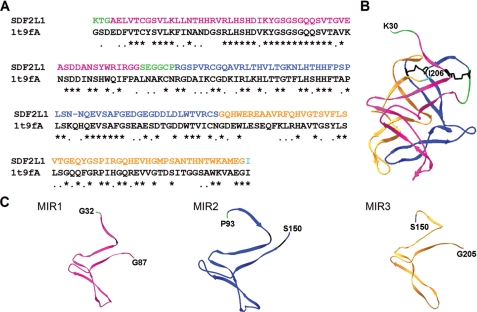
Interactions of Pro-defensins with SDF2L1 Mutants—SDF2L1 has three ∼50 residue MIR (Mannosyltransferase, IP3 receptor, Ryanodine receptor) domains (28), which correspond to residues 35–87 (MIR1), 95–150 (MIR2), and 151–205 (MIR3). To date no function has been attributed to any of the MIRs. We constructed a three-dimensional model of SDF2L1 using SWISS-MODEL (29, 30), building on the crystal structure of a Caenorhabditis elegans protein of unknown function which is 49% identical to SDF2L1 sequence between residues 30–206 (Fig. 6A). The SDF2L1 model has a compact structure composed of three MIR domains each made up of extended β-sheets and turns. Despite the low sequence homology (<30%) between the three MIR domains, these domains are structurally similar (Fig. 6, B and C).
FIGURE 6.
Homology model of SDF2L1 and the structure of its MIR domains. SWISS-MODEL was used to construct a three-dimensional structure of SDF2L1 that includes residues 30–206. The MIR1, MIR2, and MIR3 domains are represented in magenta, blue, and orange, respectively. The N-terminal sequence and the sequence between MIR1 and -2 are green, and the C-terminal residue is shown in cyan. The N- and C-terminal residues are indicated in the structures. A, alignment of rhesus SDF2L1 residues 30–206 with the chain A of 1t9f.PDB sequence (1t9fA). B, predicted structure of SDF2L1 displayed using the SWISS-PDBviewer. The Cys residues and the putative disulfide linkages in this structure are shown in black. C, structures of individual MIR1, -2, and -3 domains; an extra N-terminal residue was also included to indicate the sequence orientation of each domains.
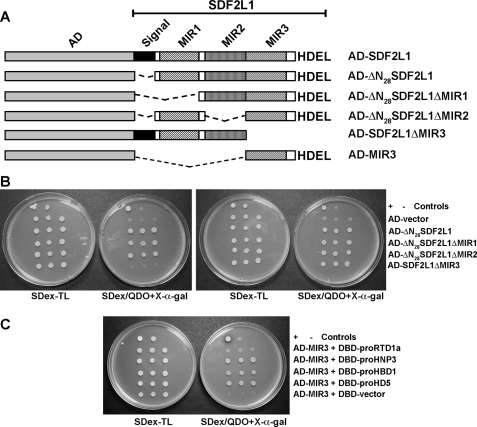
To identify the MIR domains of SDF2L1 involved in interactions with pro-defensins, we generated AD fusions of SDF2L1 in which the three MIR domains were individually deleted (Fig. 7A) and tested these constructs for their interaction with proRTD1a and proHNP3 in the two hybrid system. Deletions of the MIR1 and MIR2 domains did not affect the interactions of SDF2L1 with either proRTD1a or proHNP3, indicating that neither domain is essential for interaction with the defensin precursors. However, deletion of the MIR3 domain (AD-SDF2L1ΔMIR3) produced an SDF2L1 variant that interacted specifically with proHNP3 but not with proRTD1a (Fig. 7B). Moreover, when the MIR-deletion mutants were tested for their interactions with DBD fusions of proHD5 and proHBD1, the result was identical to that obtained with proHNP3 (data not shown). Thus SDF2L1 appears to interact differently with the θ-defensin precursor than it does with α- and β-prodefensins.
FIGURE 7.
Interactions of prodefensins with SDF2L1 mutants. A, schematic representation of SDF2L1 MIR domain deletion mutants. B, spot tests of AH109 cells transformed with the DBD-proRTD1a (left panel) or DBD-proHNP3 (right panel) plasmids with control AD-ΔN28SDF2L1 and corresponding MIR deletion mutant plasmids. C, spot tests of AH109 cells transformed with AD-MIR3 plasmid and DBD-prodefensin fusion constructs and controls as listed.
We next tested whether the MIR3 domain alone is sufficient to interact with pro-defensins. The SDF2L1 MIR3 was fused to AD (AD-MIR3, Fig. 7A) and tested for interaction with several prodefensins in the two hybrid assay. As shown in Fig. 7C, the MIR3 domain was sufficient for interaction with proHNP3, proHD5, and proHBD1; however, the MIR3 alone did not interact with proRTD1a. This result provides further evidence that SDF2L1 interacts with the θ-defensin precursor in a manner distinct from its interaction with α- and β-prodefensins.
DISCUSSION
In this study we used the two hybrid system and identified rhesus SDF2L1 as a novel interactor with α-, β-, and θ-prodefensins. SDF2L1 is an ER-resident protein found in diverse cells and tissues (22). It has an N-terminal 28 amino acid residue hydrophobic signal sequence and a C-terminal HDEL sequence for ER-retention (22, 23). In mouse lymphoma cells Meunier et al. (24) identified SDF2L1 as a component of an ER-chaperone complex. Consistent with these results Bies et al. (23) showed that SDF2L1 interacts with both the chaperone BiP and co-chaperone Erj3.
Our studies reveal that the prosegment of defensins is not required for interaction with SDF2L1. Indeed, only those elements expressed in mature α-, β-, and θ-defensins are required for interaction with SDF2L1. The in vitro interaction of HNPs (myeloid α-defensins) expressed in HL60 cells with GST-ΔN28SDF2L1 provides additional validation of the two-hybrid analyses. Interestingly, the precursors of human enteric α-defensin, proHD5, and the β-defensin, proHBD1, whose sequence is quite different from proRTD1a and proHNPs, also interacted with SDF2L1. It is also surprising that the proRTD1a nonamer sequence, which is homo/heterodimerized to form the mature cyclic θ-defensin, is sufficient to mediate this interaction. To our knowledge SDF2L1 is the first ER-resident protein reported to interact with defensins.
The mobilization of defensins in the cell varies depending among defensin subtype and site of expression: the myeloid α-defensins and θ-defensins are primarily stored in intracellular granules in neutrophils, human enteric α-defensins are stored in their unprocessed forms in Paneth cell granules, and epithelial β-defensins are secreted by the producing-cell. Therefore it is possible that the respective α-, β-, and θ-defensin prosegments are involved in interaction with defensin-specific and cell type-specific processing/sorting factors. The 12-residue motif previously identified in the HNP1 prosegment, which was required for correct processing and sorting of HNPs in vivo (25) was not required for interaction with SDF2L1 in the current study.
The SDF2L1 conformational model (Fig. 6), constructed by homology, has a compact structure with three structurally similar MIR domains that are independently folded. The role of the MIR domains is unclear but they have been identified in proteins with diverse functions; proteins with one to five MIR domains have been identified. Here we showed that the interaction of α- and β-defensins with SDF2L1 was not disrupted by deletion of any one MIR domain. Further, the MIR3 domain alone was sufficient for interaction with α-or β-prodefensins. Because the sequence conservation of the three MIR domains is only ∼30%, it is likely that the binding of diverse defensins depends on a structural rather than a sequential SDF2L1 motif. However, in the case of the θ-defensin precursor, proRTD1a, the interaction of the peptide did not require the MIR1 or MIR2 domains, but instead required the presence of MIR3. However, unlike the results obtained with the α- and β-prodefensins, the MIR3 domain alone was not sufficient for interaction with proRTD1a. These results indicate that the interaction of SDF2L1 with α- and β-prodefensins is different from its interaction with proRTD1a.
We speculate that the different structural requirements for SDF2L1 binding to different defensin subfamilies correlate with novel structural features of the peptides. The finding that the α- and β-defensin propeptides interacted similarly with SDF2L1 (Fig. 7) is consistent with the fact that mature α- and β-defensins have very similar three-dimensional structures. In contrast, there is a unique requirement for the MIR3 domain in the SDF2L1 interaction with proRTD1a, the θ-defensin precursor that contains only nine residues of mature peptide sequence. It is possible that the mature sequences of α- and β-defensins either by themselves or cooperatively with the prosegment sequence bind one or more MIR domains. This has been shown previously in studies on the inositol 1,4,5-trisphosphate (IP3) receptor, IP3R1 (31). In that case, multiple MIR domains were required for ligand binding as the IP3-binding core domain, encompassing residues 226–578, includes three MIR domains (28, 31). Indeed, the fact that no one MIR domain was required for interaction with α-or β-defensins suggests that these propeptides bind two or more domains in SDF2L1. As mentioned earlier, processing of the linear α- and β-defensins involves the proteolytic cleavage to form the mature peptide, whereas, biosynthesis of θ-defensins requires excision of the nonamer peptides from two precursors and then the head-to-tail ligation of these peptides to form mature cyclic θ-defensin. Interaction of SDF2L1 with the 9-mer sequence of proRTD1a rather than the full length α- or β-defensin sequence may lead to formation of a different processing complex unique to the cyclization reaction required for biosynthesis of θ-defensins. Additional experiments are required to delineate the critical role of MIR3 in binding of the θ-defensin precursor.
As noted, SDF2L1 is known to be associated with chaperone complexes in the ER. The ER-chaperone complex was identified by cross-linking studies and consisted of SDF2L1, chaperones BiP, GRP170, GRP194, the co-chaperone Erdj3, the peptidyl prolyl isomerase cyclophilin B, the protein-disulfide isomerases PDI, Erp72, CaBP1, and the glucosyl transferase UDP-GT (24). Folding of nascent polypeptides in the ER is a complex process mediated by both chaperones and protein-disulfide family proteins and multiple chaperones and PDI-family proteins associate with nascent polypeptides in the ER during their folding (32–34). Taking into consideration the association of SDF2L1 with both the ER-chaperone complex and all types of defensins tested, it is possible that SDF2L1 is involved in recognition of not only defensins but also other nascent polypeptides in the ER which may then be transferred to other factors such as chaperones or protein disulfide isomerases for correct folding. It is also possible that SDF2L1 functions downstream of the chaperones/disulfide isomerases to direct the localization of folded substrates by interacting with other factors.
Supplementary Material
Acknowledgments
We thank Kevin Roberts for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants AI058129, AI022931, and DE015517. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) FJ607063.
The on-line version of this article (available at http://www.jbc.org) contains supplemental information.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PDI, protein-disulfide isomerase; GST, glutathione S-transferase; SDF, stromal cell-derived factor; HA, hemagglutinin; AD, activation domain; DBD, DNA binding domain; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; MIR, mannosyl-transferase, IP3 receptor, ryanodine receptor; IP3, inositol 1,4,5-trisphosphate.
References
- 1.Selsted, M. E., and Ouellette, A. J. (2005) Nat. Immunol. 6 551–557 [DOI] [PubMed] [Google Scholar]
- 2.Ghosh, D., Porter, E., Shen, B., Lee, S. K., Wilk, D., Drazba, J., Yadav, S. P., Crabb, J. W., Ganz, T., and Bevins, C. L. (2002) Nat. Immunol. 3 583–590 [DOI] [PubMed] [Google Scholar]
- 3.Tang, Y. Q., Yuan, J., Osapay, G., Osapay, K., Tran, D., Miller, C. J., Ouellette, A. J., and Selsted, M. E. (1999) Science 286 498–502 [DOI] [PubMed] [Google Scholar]
- 4.Wimley, W. C., Selsted, M. E., and White, S. H. (1994) Protein Sci. 3 1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ericksen, B., Wu, Z., Lu, W., and Lehrer, R. I. (2005) Antimicrob. Agents Chemother. 49 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daher, K. A., Selsted, M. E., and Lehrer, R. I. (1986) J. Virol. 60 1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastian, A., and Schafer, H. (2001) Regul Pept. 101 157–161 [DOI] [PubMed] [Google Scholar]
- 8.Yasin, B., Wang, W., Pang, M., Cheshenko, N., Hong, T., Waring, A. J., Herold, B. C., Wagar, E. A., and Lehrer, R. I. (2004) J. Virol. 78 5147–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinones-Mateu, M. E., Lederman, M. M., Feng, Z., Chakraborty, B., Weber, J., Rangel, H. R., Marotta, M. L., Mirza, M., Jiang, B., Kiser, P., Medvik, K., Sieg, S. F., and Weinberg, A. (2003) Aids 17 F39–48 [DOI] [PubMed] [Google Scholar]
- 10.Mambula, S. S., Simons, E. R., Hastey, R., Selsted, M. E., and Levitz, S. M. (2000) Infect. Immun. 68 6257–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman, S. L., Gootee, L., Gabay, J. E., and Selsted, M. E. (2000) Infect. Immun. 68 5668–5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aley, S. B., Zimmerman, M., Hetsko, M., Selsted, M. E., and Gillin, F. D. (1994) Infect. Immun. 62 5397–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chertov, O., Michiel, D. F., Xu, L., Wang, J. M., Tani, K., Murphy, W. J., Longo, D. L., Taub, D. D., and Oppenheim, J. J. (1996) J. Biol. Chem. 271 2935–2940 [DOI] [PubMed] [Google Scholar]
- 14.Yang, D., Chertov, O., Bykovskaia, S. N., Chen, Q., Buffo, M. J., Shogan, J., Anderson, M., Schroder, J. M., Wang, J. M., Howard, O. M., and Oppenheim, J. J. (1999) Science 286 525–528 [DOI] [PubMed] [Google Scholar]
- 15.Tani, K., Murphy, W. J., Chertov, O., Salcedo, R., Koh, C. Y., Utsunomiya, I., Funakoshi, S., Asai, O., Herrmann, S. H., Wang, J. M., Kwak, L. W., and Oppenheim, J. J. (2000) Int. Immunol. 12 691–700 [DOI] [PubMed] [Google Scholar]
- 16.Lillard, J. W., Jr., Boyaka, P. N., Chertov, O., Oppenheim, J. J., and McGhee, J. R. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valore, E. V., and Ganz, T. (1992) Blood 79 1538–1544 [PubMed] [Google Scholar]
- 18.Gruber, C. W., Cemazar, M., Clark, R. J., Horibe, T., Renda, R. F., Anderson, M. A., and Craik, D. J. (2007) J. Biol. Chem. 282 20435–20446 [DOI] [PubMed] [Google Scholar]
- 19.Baniahmad, C., Baniahmad, A., and O'Malley, B. W. (1994) BioTechniques 16 194–196 [PubMed] [Google Scholar]
- 20.Madura, K., Dohmen, R. J., and Varshavsky, A. (1993) J. Biol. Chem. 268 12046–12054 [PubMed] [Google Scholar]
- 21.Flicek, P., Aken, B. L., Beal, K., Ballester, B., Caccamo, M., Chen, Y., Clarke, L., Coates, G., Cunningham, F., Cutts, T., Down, T., Dyer, S. C., Eyre, T., Fitzgerald, S., Fernandez-Banet, J., Graf, S., Haider, S., Hammond, M., Holland, R., Howe, K. L., Howe, K., Johnson, N., Jenkinson, A., Kahari, A., Keefe, D., Kokocinski, F., Kulesha, E., Lawson, D., Longden, I., Megy, K., Meidl, P., Overduin, B., Parker, A., Pritchard, B., Prlic, A., Rice, S., Rios, D., Schuster, M., Sealy, I., Slater, G., Smedley, D., Spudich, G., Trevanion, S., Vilella, A. J., Vogel, J., White, S., Wood, M., Birney, E., Cox, T., Curwen, V., Durbin, R., Fernandez-Suarez, X. M., Herrero, J., Hubbard, T. J., Kasprzyk, A., Proctor, G., Smith, J., Ureta-Vidal, A., and Searle, S. (2008) Nucleic Acids Res. 36 D707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukuda, S., Sumii, M., Masuda, Y., Takahashi, M., Koike, N., Teishima, J., Yasumoto, H., Itamoto, T., Asahara, T., Dohi, K., and Kamiya, K. (2001) Biochem. Biophys. Res. Commun. 280 407–414 [DOI] [PubMed] [Google Scholar]
- 23.Bies, C., Blum, R., Dudek, J., Nastainczyk, W., Oberhauser, S., Jung, M., and Zimmermann, R. (2004) Biol. Chem. 385 389–395 [DOI] [PubMed] [Google Scholar]
- 24.Meunier, L., Usherwood, Y. K., Chung, K. T., and Hendershot, L. M. (2002) Mol. Biol. Cell 13 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, L., and Ganz, T. (1995) Blood 85 1095–1103 [PubMed] [Google Scholar]
- 26.Herwig, S., Su, Q., Zhang, W., Ma, Y., and Tempst, P. (1996) Blood 87 350–364 [PubMed] [Google Scholar]
- 27.Hiratsuka, T., Nakazato, M., Ihi, T., Minematsu, T., Chino, N., Nakanishi, T., Shimizu, A., Kangawa, K., and Matsukura, S. (2000) Nephron 85 34–40 [DOI] [PubMed] [Google Scholar]
- 28.Ponting, C. P. (2000) Trends Biochem. Sci. 25 48–50 [DOI] [PubMed] [Google Scholar]
- 29.Guex, N., and Peitsch, M. C. (1997) Electrophoresis 18 2714–2723 [DOI] [PubMed] [Google Scholar]
- 30.Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003) Nucleic Acids Res. 31 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshikawa, F., Morita, M., Monkawa, T., Michikawa, T., Furuichi, T., and Mikoshiba, K. (1996) J. Biol. Chem. 271 18277–18284 [DOI] [PubMed] [Google Scholar]
- 32.Kuznetsov, G., Chen, L. B., and Nigam, S. K. (1994) J. Biol. Chem. 269 22990–22995 [PubMed] [Google Scholar]
- 33.Linnik, K. M., and Herscovitz, H. (1998) J. Biol. Chem. 273 21368–21373 [DOI] [PubMed] [Google Scholar]
- 34.Di Jeso, B., Park, Y. N., Ulianich, L., Treglia, A. S., Urbanas, M. L., High, S., and Arvan, P. (2005) Mol. Cell. Biol. 25 9793–9805 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.