Abstract
The steroid and xenobiotic receptor (SXR) (also known as pregnane X receptor or PXR) is a nuclear hormone receptor activated by a diverse array of endogenous hormones, dietary steroids, pharmaceutical agents, and xenobiotic compounds. SXR has an enlarged, flexible, hydrophobic ligand binding domain (LBD) which is remarkably divergent across mammalian species and SXR exhibits considerable differences in its pharmacology among mammals. The broad response profile of SXR has led to the development of "the steroid and xenobiotic sensor hypothesis". SXR has been established as a xenobiotic sensor that coordinately regulates xenobiotic clearance in the liver and intestine via induction of genes involved in drug and xenobiotic metabolism. In the past few years, research has revealed new and mostly unsuspected roles for SXR in modulating inflammation, bone homeostasis, vitamin D metabolism, lipid homeostasis, energy homeostasis and cancer. The identification of SXR as a xenobiotic sensor has provided an important tool for studying new mechanisms through which diet, chemical exposure, and environment ultimately impact health and disease. The discovery and pharmacological development of new PXR modulators might represent an interesting and innovative therapeutic approach to combat various diseases.
SXR, a steroid and xenobiotic sensor
The mammalian xenobiotic response is mediated primarily through the activity of four families of cytochrome P450 (CYP) monooxygenases (CYP1, CYP2, CYP3, and CYP4) [Nelson et al., 1996]. CYP enzymes are the main enzymatic system for metabolism of lipophilic substrates of diverse structures and are important in the oxidative, peroxidative, and reductive metabolism of numerous endogenous compounds including steroids, bile acids, fatty acids, prostaglandins, leukotrienes, biogenic amines and retinoids [Nebert and Russell, 2002; Waxman, 1999]. Among four families of CYP enzymes, the CYP3A family is one of the most important since it is responsible for the metabolism of more than 50% of clinically used drugs and many xenobiotic chemicals [Guengerich, 1999]. The CYP2B family is responsible for another 25-30% of drug and xenobiotic metabolism [Xie and Evans, 2001]. The ability of organisms to induce P450 enzymes in response to elevated xenobiotic levels is crucial for their survival and normal homeostasis.
CYP3A genes are responsible for the initial metabolism of numerous xenobiotic chemicals, making them a first line of defense against toxic substances in the diet and environment. It is notable that a large number of CYP3A substrates are also capable of inducing or up-regulating expression of the mRNAs encoding the enzymes themselves [Denison and Whitlock, 1995; Guengerich, 1999; Wrighton et al., 2000]. Because the CYP3A family genes produce key enzymes for the metabolism of more than 50% of prescription drugs, there has been a strong interest in understanding the mechanistic basis of CYP3A gene regulation. It is well known that drugs capable of inducing expression of CYP3A genes are likely to lead to interactions with other drugs. This is a serious problem that is compounded by the extreme diversity of the compounds that are known CYP3A inducers.
In 1998, Blumberg et al. [Blumberg and Evans, 1998], Kliewer et al. [Kliewer et al., 1998], and Bertilsson et al. [Bertilsson et al., 1998] isolated cDNAs encoding a novel orphan nuclear receptor which was subsequently shown to play a central role in the transcriptional regulation of CYP3A4. It was named SXR (steroid and xenobiotic receptor), PXR (pregnane ‘X’ receptor), and PAR (pregnane activated receptor), respectively by these groups, and was designated as NR1I2 in the standard nomenclature [Nuclear Receptors Nomenclature Committee, 1999]. Despite the different names, this receptor has properties predicted for a steroid and xenobiotic sensor as shown by several lines of evidence. First, SXR is expressed predominantly in liver and intestine, both sites of steroid and xenobiotic metabolism. Second, SXR activation directly stimulates the transcription of CYP enzymes in response to the presence of its ligands and plays a central role in the transcriptional regulation of CYP3A4. Moreover, SXR not only regulates the expression of CYP enzymes such as CYP3A4, CYP2B6 and CYP2C8, but also conjugation enzymes (e.g., UDP-glucuronosyltransferase (UGT1A1) and sulfotransferase (SULT)) and ABC family transporters such as MDR1 and MRP2 [Dussault et al., 2001; Synold et al., 2001]. Finally, SXR is activated by a diverse array of pharmaceutical agents including taxol, rifampicin (RIF), SR12813, clotrimazole, phenobarbital , the herbal antidepressant St. John’s wort , and peptide mimetic HIV protease inhibitors such as ritonavir (reviewed in [Dussault and Forman, 2002; Kliewer et al., 2002]). These studies indicate that SXR functions as a xenobiotic sensor [Blumberg et al., 1998; Lehmann et al., 1998] that coordinately regulates drug clearance in the liver and intestine via induction of genes involved in drug and xenobiotic metabolism, including oxidation (phase I), conjugation (phase II), and transport (phase III) [Dussault and Forman, 2002; Kliewer et al., 2002]. Therefore, we will use the name SXR to refer to this endobiotic and xenobiotic sensor throughout this review, rather than the more commonly used but inaccurate PXR.
Gene knockout studies have confirmed a role for SXR in regulating the metabolism of endogenous steroids, dietary and xenobiotic compounds [Staudinger et al., 2001; Xie et al., 2000a]. At least two types of knockout animals have been developed. Xie et al. generated SXR knockout mice, which have a deletion of two exons including amino-acid residues 63–170 of the DNA-binding domain [Xie et al., 2000a]. The mice produced by Staudinger et al. have a deletion of the first coding exon, which includes the translation start site and the first zinc finger of the SXR DNA-binding domain (amino acids 1 to 63) [Staudinger et al., 2001]. In both models, complete gene disruption was confirmed by the absence of SXR expression in the liver and small intestine where it is predominantly expressed; although, there are subtle phenotypic differences between the two types of mice. Targeted disruption of SXR abolishes the induction of CYP3A genes in response to prototypic inducers such as dexamethasone or PCN. Notably, these mice are also susceptible to severe liver damage induced by toxic bile acid such as lithocholic acid (LCA). The loss of hepatoprotection to bile acids results from aberrant regulation of genes involved in the biosynthesis, transport, and metabolism of bile acids including cholesterol 7α-hydroxylase (CYP7A1) and the Na+-independent organic anion transporter 2 (OATP2). This indicates that SXR plays a critical role in both xenobiotic and bile acid detoxification.
SXR gene and protein structure
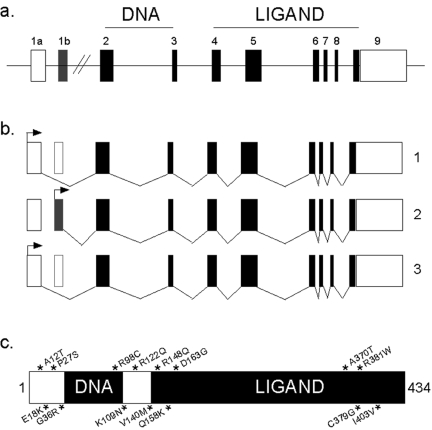
The SXR gene consists of nine exons and spans approximately 35 kb in chromosome 13q12-13.3 (Figure 1a). Three alternatively spliced transcripts that encode different isoforms of SXR have been described. Transcript variant 1 encodes two products (isoforms 1l and 1s) through the use of alternative translation initiation codons, which are in the same reading frame (Figure 1b). Transcript variant 2 encodes the longest isoform and initiates translation from the standard AUG codon present in its 5' terminal exon. Variant 3 contains an alternate 5' terminal exon, and uses a different acceptor splice site at exon 5 in comparison to transcript variant 2. It initiates translation from an in-frame, downstream non-AUG (CUG) codon, resulting in a shorter isoform 3 with a different N-terminus and is also missing an internal segment, compared to isoform 2 (Figure 1b). All three isoforms are present at varying levels in the liver and gut. Isoform 3 is largely non-responsive to ligands in transient transfection assays [Gardner-Stephen et al., 2004]. Additional transcript variants derived from alternative promoter usage, alternative splicing, and/or alternative polyadenylation may exist, but, they have not been completely characterized [Fukuen et al., 2002; Lamba et al., 2004].
Figure 1. Structure of SXR gene, protein and mRNA isoforms.
a) The genomic structure of SXR and its three splice variants. The protein coding regions are depicted as filled boxes and the untranslated 5’ and 3’ regions are shown as white boxes. The horizontal line represents introns. b) The structure of splice variants of SXR. Variant 1 originates from exon 1a and gives rise to proteins 1l and 1s through the use of the alternative initiation codons shown by arrows. Variant 2 originates from exon 1b and it makes the longest protein. Variant 3 represents an in-frame deletion of 111bp at the 5’ end of exon 5 (shown by white box). The arrows depict translation initiation codon. c) Single nucleotide polymorphisms (SNPs) in SXR. Fifteen known non-synonymous SNPs of SXR are represented by * and arranged according to their position on the wild-type (variant 1l) SXR protein. The DNA and ligand binding domains of the protein are shown in black boxes.
Although it shares the same overall arrangement of its functional domains with other nuclear receptors, in contrast to most other receptors SXR can bind structurally diverse ligands. SXR can be activated by a range of ligands that is almost as diverse as the substrates of CYP3A4, including many drugs. Yet, SXR can also display considerable discretion in ligand binding. The crystal structure of the SXR ligand binding domain (LBD) reveals its special features [Chrencik et al., 2005; Watkins et al., 2003a; Watkins et al., 2003b; Watkins et al., 2001]. SXR has three α-helical domains common to other nuclear receptors. Uniquely, SXR has five strands of β-sheet, whereas other nuclear receptors typically contain only two to three strands. SXR has a large insertion of approximately 60-residues between helices 1 and 3 that contains two additional β-strands. These extend the usual two- to three-stranded LBD β-sheet to five strands. This insert also adds a novel α-helix that folds along the underside of the SXR ligand-binding pocket. Together, these elements lead to an enlarged, flexible, hydrophobic LBD which is capable of fitting large and varied ligands and that can also change shape depending on the nature of its bound ligand. The crystal structures of SR12813- or hyperforin-bound SXR have also been characterized [Watkins et al., 2003b; Watkins et al., 2001]. These structures show that hyperforin makes more nonpolar and polar interactions than does SR12813, which helps to explain the latter’s high affinity binding to SXR. This study also found that SR12813 is able to interact with SXR in a variety of different orientations, suggesting that the ligands can make more than one set of hydrogen bonds in order to interact with the ligand-binding domain of SXR. This increases the number of ligands that can activate SXR. Interestingly, as a main target gene of SXR, CYP3A4 can also metabolize a wide range of endogenous compounds and xenobiotics. However, the crystal structures of un-liganded and substrate-bound CYP3A4 revealed a surprisingly small active site, with little conformational change associated with the binding of an inhibitor (metyapone) or substrate (progesterone) [Williams et al., 2004]. Future studies that investigate the potential mechanisms of molecular recognition by SXR and CYP3A4 may help us to understand the complex of xenobiotic metabolism regulated by SXR-CYP3A4 axis.
Chrencik et al. also described the 2.8 Å crystal structure of the ligand-binding domain of human SXR in complex with rifampicin, one of the largest known ligands for the receptor [Chrencik et al., 2005]. They found that the macrocyclic ring of rifampicin fits within the SXR ligand-binding pocket. In contrast, the drug’s 4-methyl-1-piperazinyl ring and several protein loops are disordered. Their observations suggest that the structural flexibility of SXR allows it to respond to large ligands by changing the effective size of its ligand-binding pocket. This highlights the key role that structural flexibility plays in SXR's promiscuous response to xenobiotics. Their structure also provides insights into the selective response to rifampicin exhibited by SXR from different species. Very recently, Xue et al. and colleagues presented a crystal structure for the SXR LBD in complex with T0901317 (T1317), which is also an agonist of another nuclear receptor, liver X receptor (LXR) [Xue et al., 2007a]. They found that despite differences in the size and shape of the ligand binding pockets, key interactions with this ligand are conserved between human SXR and human LXR. The same group subsequently reported the crystal structure of the human SXR ligand-binding domain (LBD) in complex with 17β-estradiol, a representative steroid ligand, at 2.65 Å resolution [Xue et al., 2007b]. They compared the SXR-estradiol complex with other nuclear receptors, including the estrogen receptor, in complexes with analogous ligands. The results showed that the placement of the steroid within the ligand-binding pocket is remarkably different for SXR compared with other members of the nuclear receptor superfamily. Their results provide detailed insights into the manner in which SXR responds to a wide range of endobiotic compounds. This suggests that determining the crystal structure of the SXR-ligand complex may be the most effective method to elucidate how SXR detects and protects the body from harmful chemicals.
The subcellular localization of SXR is currently controversial. Two groups have shown that un-liganded SXR stays predominantly in the cytoplasm and that it translocates to the nucleus after ligand addition. Based on microinjection studies and transient transfection assays, Kawana et al. reported that amino acids 66-92 are important for nuclear import of SXR [Kawana et al., 2003]. In agreement with these studies, Negishi and co-workers showed that in the absence of ligand, SXR resides in a complex with CCRP and heat-shock proteins (HSPs), remaining in the cytoplasm until ligand binding which triggers nuclear translocation [Squires et al., 2004]. They also showed that nuclear localization signal (NLS), xenobiotic response signal (XRS) and AF2 domain are all required for nuclear translocation of SXR. However, other studies showed that human SXR is exclusively nuclear, irrespective of ligand binding [Koyano et al., 2004; Saradhi et al., 2005]. The same results were obtained by overexpressing GFP-tagged proteins or by immunostaining transiently or stably-transfected HepG2 cells. Saradhi et al. also could not find any association between CCRP complex and human SXR [Saradhi et al., 2005]. Interestingly, they also reported that SXR was associated with condensed mitotic DNA during mitosis. We found, using a mouse monoclonal antibody directed against human SXR that it was localized to the nucleus in breast cancers in the absence of added ligands [Miki et al., 2006].
While it is difficult to reconcile these disparate results, it is notable that the earlier studies showing cytoplasmic localization employed the rodent receptor [Kawana et al., 2003; Squires et al., 2004], whereas those showing nuclear localization used the human receptor [Koyano et al., 2004; Miki et al., 2006; Saradhi et al., 2005]. Therefore, one possibility is that species differences may partly explain the different localization. Other possible explanations could be cell-type specificity and differences in the specificity of the antibodies employed. Further studies using other cell types and antibodies of well-demonstrated specificity (which is largely not the case with most of the widely used commercial antibodies) will be required to fully resolve the issue of sub-cellular localization.
SXR and species-specific xenobiotic metabolism
SXR can be activated by a wide diversity of natural steroids (e.g., pregnanes, estranes and androstanes), dietary compounds (e.g., hyperforin, tocotrienols, menaquinones), and xenobiotics (e.g., rifampicin, nifedipine, PCN). Interestingly, SXR exhibits considerable differences in its pharmacology among mammals [Blumberg et al., 1998; Jones et al., 2000; LeCluyse, 2001], which may explain species-specific differences in xenobiotic induction by CYP3A. Compared with most other nuclear receptors, SXR is remarkably divergent across mammalian species with the ligand binding domains sharing ~70-80% identity compared with the ~90% typically exhibited by orthologous nuclear receptors. Differences in amino acid sequences among the mammalian receptors are responsible for species-specific induction of CYP3A by drugs and xenobiotics [LeCluyse, 2001; Watkins et al., 2001]. One general observation is that there are significant differences in the xenobiotic response between humans and rodents and that these differences are completely explained by the pharmacology of SXR [Blumberg et al., 1998; Jones et al., 2000; Kliewer et al., 1998; LeCluyse, 2001; Lehmann et al., 1998; Xie et al., 2000a; Xie and Evans, 2001]. For example, the antibiotic rifampicin, the anti-diabetic drug troglitazone and the cholesterol-reducing drug SR12813 were found to be effective activators of both human and rabbit SXR, but had little activity on mouse or rat SXR [Jones et al., 2000]. In contrast, pregnane 16α-carbonitrile (PCN) was a more potent activator of rat and mouse SXR than of human or rabbit SXR [Jones et al., 2000; Masuyama et al., 2000; Takeshita et al., 2001]. Corresponding induction of the SXR-inducible CYP3A genes in humans, rats and rabbits was also observed in primary hepatocytes. The crystal structure of the SXR LBD suggested which amino acid differences in the LBD of SXR contributed to species differences in ligand activation of human vs. mouse receptor and the consequent induction of CYP3A. Conversion of four of the polar amino acids in the ligand binding pocket from the mouse sequence to the corresponding residues found in the human receptor resulted in a mutated mouse receptor that showed a humanized response profile to ligands [Watkins et al., 2001].
Since species differences exist in ligand specificity between human and mouse SXR, an important tool for the study of xenobiotic metabolism was the development of the “humanized” mouse [Xie et al., 2000a] that responds to human SXR activators such as rifampicin, but does not respond to the rodent activator PCN [Ma et al., 2007; Xie et al., 2000a]. Xie et al. first generated SXR-null mice containing the human SXR gene driven by an Albumin (Alb) promoter [Xie et al., 2000a]. This animal is deficient in the mouse SXR gene, while expressing a human SXR transgene specifically in the liver. Subsequently, Ma et al. generated a human SXR mouse model by bacterial artificial chromosome (BAC) transgenesis in SXR-null mice using a BAC clone containing the complete human SXR gene and 5'- and 3'-flanking sequences [Ma et al., 2007]. In this hSXR mouse model, SXR was expressed in both liver and intestine, and in many (but not all) of the same tissues where the endogenous gene and its CYP3A target are expressed (for example, lung is a notable exception). Treatment of both hSXR models with SXR ligands mimicked the human response since CYP3As were induced strongly by rifampicin, a human-specific SXR ligand, but not by PCN, a rodent-specific SXR ligand [Ma et al., 2007; Xie et al., 2000a]. These models demonstrate convincingly that SXR is the key regulator of CYP3A induction by xenobiotics. Moreover, the selective activation of target genes in response to species-specific activators was shown to reside in the ligand binding domain of the receptor, rather than in the DNA-binding domain or target DNA-binding elements [Ma et al., 2007; Xie et al., 2000a]. In addition to these two models, Xie et al. and Gong et al. also generated Alb- and Fatty Acid-Binding Protein (FABP)-activated hSXR (VP-hSXR) mice in which constitutively activated hSXR were expressed in the liver (Alb promoter driven) or in the liver and intestine (FABP promoter driven), respectively [Gong et al., 2006; Xie et al., 2000a] . These humanized mice are a powerful in vivo system to study the roles of human SXR in xenobiotic metabolism in an animal model.
Natural allelic variants of SXR
SXR displays a broad specificity for a variety of drugs and is a primary regulator of CYP3A4 induction. The levels of CYP3A enzymes show considerable sexual dimorphisms and variation in levels and function among individuals in the population [Gonzalez, 1992]. Variation in CYP3A expression may lead to important differences in drug metabolism, leading to clinically significant differences in drug toxicities and response. It may also influence the circulating levels of estrogens and the risk of breast cancer [Kuehl et al., 2001]. The molecular underpinnings of the variations in CYP3A4 expression are unknown at present, but it is likely that SXR plays a key role in this process. Approximately 90% of the inter-individual variability in hepatic CYP3A4 activity is genetically determined and several CYP3A4 variants have been reported [Ozdemir et al., 2000]. However, the reported allelic frequencies and the functional data demonstrate only a limited role of these variants in CYP3A4 expression and activity [Eiselt et al., 2001; Wandel et al., 2000]. Single nucleotide polymorphisms (SNPs) in SXR should be a major contributor to CYP3A4 expression and activity. Since SXR is a key xenobiotic ‘sensor’ that mediates the physiological response of multiple drug metabolism genes, identification of functional polymorphisms in SXR might explain the variable induction of CYP3A4 and other drug metabolizing enzymes in response to SXR ligands [Eichelbaum and Burk, 2001]. It has been reported that different strains of mice differ substantially in their sensitivity to estrogen treatment [Spearow et al., 1999]. It is reasonable to expect that some part of this differential sensitivity may result from differences in SXR; however, this remains to be demonstrated.
Several groups have investigated SXR allelic variants in different ethnic populations. In total, more than 70 SNPs have been identified so far including 15 in the coding region that are non-synonymous, creating new SXR proteins (Figure 1c) [Bosch et al., 2006; Hustert et al., 2001; King et al., 2007; Koyano et al., 2002; Lim et al., 2005; Zhang et al., 2001]. Four of the fifteen variants were located N-terminal to the DNA binding domain (A12T, E18K, P27S, and G36R) and have no significant effects on DNA-binding or transactivation compared with wild-type SXR. Three other variants were located in or near the DNA binding domain (R98C, K109N, and R122Q). The variant R98C failed to bind SXR response element or to transactivate CYP3A4 completely [Koyano et al., 2004]. The variant R122Q also shows significantly decreased affinity for DNA binding and attenuated transcriptional activity [Zhang et al., 2001]. The variant K109N has not yet been functionally described. The other eight variants are within the LBD of SXR (R148Q, Q158K, D163G, A370T, C379G, R381W and I403V) or close to the LBD (V140M). R148Q and Q158K are located in helix 1 of the ligand binding domain (LBD). In transient transfection assays R158Q showed a marked decrease in activity on the CYP3A4 promoter, whereas R148Q had no dramatic effect on transactivation ability of SXR [Koyano et al., 2004; Lim et al., 2005]. Amino acid residue 158 is well conserved among humans, rodent and rabbits, whereas AA 148 is not [Koyano et al., 2004]. The D163G variant exhibits lower basal activity and an eight-fold higher induction by rifampicin than wild-type SXR [Hustert et al., 2001]. In contrast, the variants A370T and V140M show 1.5-2 fold enhancement in the basal expression of a CYP3A4 promoter reporter gene, but lack any significant effect on transcriptional activation. Variants R381W and I403V have significantly reduced transactivation ability in comparison to wild-type receptor at 0.3 and 1 micromolar rifampicin, but had similar ability to that of wild-type at 10 and 20 micromolar rifampicin [Koyano et al., 2004]. Variant C379G has not been functionally described yet, but as this variant is in the LBD the authors speculated that this variant could have functional implications [Bosch et al., 2006].
In addition to SNPs in the protein coding region, two groups have found significant phenotypic association between polymorphisms in the promoter region (-566 and -1359, respectively) of SXR and CYP3A4 expression [King et al., 2007; Lamba et al., 2008]. A 6-bp deletion in the promoter region of NR1I2 at a putative hepatic nuclear factor binding site was suggested to have a possible influence on the promoter region and potentially inhibit SXR promoter activity and downregulate expression of SXR target genes [Lamba et al., 2008; Uno et al., 2003]. Another study reported strong phenotypic correlation between SNPs in the SXR promoter (at -25385 and -24381) with inflammatory bowel disease (IBD) [Dring et al., 2006].
These results are consistent with the possibility that SNPs in SXR can contribute to the inter-individual variability of CYP3A4 expression and drug response. However, a detailed study comparing the activity of all SXR SNPs toward ligand activation and the potential interactions between these SNPs and the ability of SXR to modulate the activity of CAR and other transcription factors remains to be performed. The relatively modest differences in the response of SXR harboring non-synonymous SNPs to ligands suggest that these SNPs are probably not the sole factors mediating variations in CYP3A expression. Thus, it is likely that other factors remaining to be identified are important for individual differences in drug metabolism. However, considering that SXR plays such a large role in the regulation of genes involved in drug metabolism and transport, bile acid detoxification and cholesterol metabolism, it remains likely that genetic variation contributing to altered SXR function will have important clinical implications. In addition, SXR polymorphisms could also influence individual predisposition to tumors caused by environmental carcinogens, including liver and lung cancer [Forrester et al., 1990; Paolini et al., 1999]. This area of research also remains largely unexplored.
SXR and natural products
Drug-drug interactions are a common problem in medical practice and activation of SXR represents the basis for several clinically important drug-drug interactions. Compared to drug-drug interaction, drug-nutrient interactions are less widely considered when prescribing medications. Modulation of SXR activity by many natural products has been reported. Below, we summarize the effects of a few widely consumed natural products on SXR signaling.
St. John’s wort
St. John’s wort (Hypericum perforatum) is a long-lived, wild-growing herb that has been used for centuries to treat a variety of ailments including bruises, dysentery, jaundice, diarrhea and a wide range of other complaints [Kumar et al., 2000]. In recent years, St. John’s wort has become increasingly used as an herbal alternative to antidepressant drugs for the treatment of mild to moderate clinical depression. It also is used to treat anxiety, seasonal affective disorder, and sleep disorders [Gaster and Holroyd, 2000; Linde et al., 1996]. St. John’s wort contains a dozen major components; however, hyperforin is believed to be the key compound responsible for the herb’s antidepressant effects [Bhattacharya et al., 1998; Laakmann et al., 1998]. There is accumulating evidence that St. John’s wort interacts with a variety of drugs. In 2000, two groups identified hyperforin as a natural ligand for SXR [Moore et al., 2000; Wentworth et al., 2000]. St. John’s wort is able to enhance the transcriptional activity of SXR comparably to rifampicin. It can also promote recruitment of the coactivator SRC-1 to SXR and displaces radiolabeled ligand bound to SXR, suggesting a direct interaction with the receptor. Of the two putative active constituents of SJW, hyperforin but not hypericin induces transcriptional activation and SRC-1 recruitment by SXR [Moore et al., 2000; Wentworth et al., 2000]. These data suggest that SXR activation by St. John’s wort mediates its adverse interactions with other drugs.
Vitamin E
Vitamin E is an essential nutrient with antioxidant activity. Vitamins E comprise a family of eight members, α-, β-, γ-, and δ-tocopherols and α-, β-, γ-, and δ-tocotrienols. All forms of vitamin E are initially metabolized by ω-oxidation, a reaction catalyzed by cytochrome P450 (CYP) enzymes. Tocopherols and tocotrienols differ in their side chain in that the tocopherols have an unsaturated phytol side chain, whereas tocotrienol side chains possess double bonds at the 3’, 7’ and 11’ positions [Kamat et al., 1997; Parker et al., 1993; Saito et al., 2003]. α-tocopherol is reported to be the most abundant form of Vitamin E in nature and has the highest biological activity as a vitamin in humans [Brigelius-Flohe and Traber, 1999]. Tocotrienols are minor plant constituents especially abundant in palm oil, cereal grains and rice bran that can provide a significant source of vitamin E activity [Sen et al., 2000]. Compared with tocopherols, the biology of tocotrienols has been poorly studied. Interestingly, all four tocotrienols were found to be able to specifically bind to and activate SXR, whereas tocopherols neither bind nor activate [Landes et al., 2003; Zhou et al., 2004]. Tocotrienols also selectively regulate the SXR target gene CYP3A4 in hepatic and intestinal cell lines, due to different expression levels of nuclear receptor corepressor NCoR in hepatic and intestinal cells [Zhou et al., 2004]. The ability of tocotrienols to regulate SXR target genes in a tissue-specific manner suggests that tocotrienols are selective SXR modulators and future development of compounds that selectively activate SXR target genes in certain tissues will be possible.
Sulforaphane
Sulforaphane (SFN) is one of the most biologically active phytochemicals in the human diet. SFN is present at high concentrations in some cruciferous vegetables, especially in broccoli and broccoli sprouts [Kushad et al., 1999; Zhang et al., 1992]. Epidemiologic and clinical studies have indicated that diets high in cruciferous vegetables protect against a number of cancers [Murillo and Mehta, 2001] and numerous studies in animal models and human cells support the putative chemopreventive effects of SFN as an active component in cruciferous vegetables [Chung et al., 2000; Conaway et al., 2002; Zhang et al., 1994]. The mechanisms of action of the putative chemopreventive effects of SFN appear to be multifactorial. SFN can induce apoptosis and cell cycle arrest in human cancer cells [Gamet-Payrastre et al., 2000], and is an inhibitor of histone deacetylases [Myzak et al., 2006]. SFN can also activate the Keap1/Nrf2 transcriptional factor complex that can bind to the antioxidant response element (ARE) and induce a series of detoxification enzymes. These include NAD(P)H:quinone oxidoreductase-1 (NQO1), certain GSTs and UDP-glucuronosyltransferases (UGTs), and other genes involved in antioxidant response [Conaway et al., 2002; Fahey and Talalay, 1999; Gao and Talalay, 2004; Talalay et al., 1995].
Interestingly, it has also been reported that SFN downregulated CYP3A4 transcription and enzymatic activity in cultured human hepatocytes, suggesting another mechanism that could also contribute to its anti-cancer effects [Maheo et al., 1997]. It was reported recently that SFN is a specific antagonist of SXR and inhibits SXR-mediated induction of drug clearance [Zhou et al., 2007]. SFN efficiently inhibits SXR-mediated transcription of the CYP3A4 gene in a concentration-dependent manner. SFN bound directly to SXR and inhibited SXR-coactivator interactions. SFN inhibited SXR-mediated CYP3A4 expression and CYP3A4-mediated midazolam (MDZ) clearance in human primary hepatocytes. Thus, SFN is the first naturally occurring antagonist identified for SXR. These findings point to a novel and complementary mechanism by which SFN exerts its putative chemoprotective effects - a reduction in CYP3A4-dependent reactive metabolite formation. These findings could also lead to potentially important new therapeutic and dietary approaches to reduce the frequency of adverse drug reactions that are secondary to SXR-mediated induction of drug clearance via CYP3A4, MDR1 and other genes regulated in part by SXR.
Other natural products
Many other natural products that have been shown to activate SXR include gugulipid, kava kava, paclitaxel, Coleus forskohlii, Hypoxis, Sutherlandia, qing hao, wu wei zi and gan cao. The impact of those products on SXR activities and drug metabolism has been discussed in detail in an excellent review to which the reader is referred [Staudinger et al., 2006].
SXR and the osteoprotective action of Vitamin K2
In addition to its high expression levels in the liver and intestine, SXR is also expressed at lower levels in the kidney and lung [Miki et al., 2005], bone [Tabb et al., 2003], and immune cells such as T cells, B cells, and mononuclear cells [Albermann et al., 2005; Owen et al., 2004; Siest et al., 2008]. It is not clear at present what role SXR is playing in other tissues. Interestingly, SXR is also expressed in osteosarcoma cell lines and SXR functions as a mediator of bone homeostasis in addition to its role as a xenobiotic sensor [Tabb et al., 2003]. Vitamin K2, a critical nutrient required for blood coagulation, plays an important role in bone formation. Vitamin K2 supplementation upregulates the expression of bone markers, increases bone density in vivo, and is used clinically in the management of osteoporosis. Tabb et al. showed that vitamin K2 can also act as a transcriptional regulator of gene expression in osteosarcoma cells [Tabb et al., 2003]. Vitamin K2 bound to and activated SXR and induced expression of the SXR target genes in osteosarcoma cells. Vitamin K2 treatment of osteosarcoma cells increased mRNA levels for the osteoblast markers bone alkaline phosphatase, osteoprotegerin, osteopontin and matrix Gla protein. The known SXR activators, rifampicin and hyperforin, induced this panel of bone markers to a similar extent, as did Vitamin K2. Vitamin K2 was able to induce bone markers in primary osteocytes isolated from wild-type murine calvaria, but not in cells isolated from SXR knockout animals.
To explore the SXR-mediated vitamin K2 signaling network, Ichikawa et al. identified several novel SXR target genes in osteoblastic cells using microarray and quantitative real-time PCR [Ichikawa et al., 2006]. They found that tsukushi, matrilin-2, and CD14 antigen are primary SXR target genes and that all three genes have bone-related functions. For example, collagen accumulation in osteoblastic cells was enhanced by vitamin K2 treatment and tsukushi, a small leucine-rich proteoglycan, contributes to this process, as demonstrated by gain- and loss-of-function analyses. Their results suggest a new function for vitamin K2 in bone formation as a transcriptional regulator of extracellular matrix-related genes involved in the collagen assembly. Very recently, Igarashi et al. showed that vitamin K2 induces osteoblast differentiation through SXR-mediated transcription control of Msx2, an osteoblastogenic transcription factor [Igarashi et al., 2007]. Upon activation by vitamin K2, SXR can be recruited together with a coactivator, p300, to the SXR response element in the Msx2 promoter. Knock-down of either SXR or Msx2 abolishes the effect of vitamin K2 on osteoblastic differentiation. Taken together, these lines of evidence show that the osteoprotective action of vitamin K2 is largely mediated through the activation of SXR, and that SXR plays a novel and unexpected role as a mediator of bone homeostasis. An important implication of this discovery is that a subset of SXR activators may function as effective therapeutic agents for the management of osteoporosis.
SXR and bile acid homeostasis
Bile acids are end products of hepatic cholesterol catabolism. They function as solubilizing detergents and play an important role in the digestion and absorption of lipids in the small intestine. Apart from the beneficial roles of bile acids, certain bile acids are toxic at high concentrations and secondary bile acids, especially lithocholic acid (LCA), are thought to participate in the pathogenesis of liver disease and colon cancer [Nagengast et al., 1995]. An intriguing link between SXR and bile acid homeostasis was uncovered by the discovery that the highly toxic LCA and its 3-keto metabolite can efficiently activate SXR [Staudinger et al., 2001; Xie et al., 2001]. Moreover, co-treatment of mice with LCA and the known activator, PCN, dramatically reduced the liver damage caused by LCA in wild-type mice as assessed by histology and serum levels of liver enzymes. No such hepatoprotection by PCN was detected in SXR null mice treated with LCA [Staudinger et al., 2001]. Mice expressing a constitutively active form of human SXR were also protected against LCA toxicity [Xie et al., 2001]. This suggests that SXR plays an important role in bile acid metabolism and protects against LCA toxicity in mouse liver. This notion was supported by the phenotype of SXR–FXR double knockout mice. Mice lacking both FXR and SXR exhibit more severe disturbances in cholesterol, lipid and bile acid metabolism than mice lacking only one of the two nuclear receptors [Guo et al., 2003]. Transgenic mice expressing CYP3A4-LacZ show significant induction of CYP3A4 and increases levels of 6-β hydroxylated bile acids when they undergo bile duct ligation, confirming the physiological relevance of induction of CYP3A4 in cholestatic conditions [Stedman et al., 2004].
Bile acid homeostasis through SXR is maintained by both feed-forward and feed-back mechanisms. Activation of SXR downregulated expression of CYP7A1, the first and rate limiting step in the metabolism of cholesterol to bile acids [Staudinger et al., 2001; Xie et al., 2001]. Inhibition of CYP7A1 is not mediated through SHP as in the case for FXR. Instead, it results from the interaction of SXR with PGC1α [Bhalla et al., 2004; Li and Chiang, 2005]. Bile acid induced activation of SXR also upregulates the expression of genes involved in bile acid metabolism and transport, such as MRP2, OATP2 and CYP3A [Frank et al., 2005; Guo et al., 2003; Kast et al., 2002; Kullak-Ublick, 2003]. MRP2 and OATP2 transport bile acids across canalicular and sinusoidal membranes, respectively, and CYP3A enzymes hydroxylate bile acids including LCA. Hydroxylation of LCA by CYP3A produces more polar 6α- and 6β-hydroxyl derivatives, which are more soluble and more easily renally secreted. Thus, activation of SXR maintains bile acid homeostasis both by repressing synthesis and by increasing metabolism and excretion by inducing CYP3A, OATP2 and MRP2 expression.
SXR, cholesterol metabolism and lipid homeostasis
The link between SXR and cholesterol homeostasis was uncovered when it was found that not only secondary bile acids, but also bile acid intermediate sterol compounds such as 5-cholestanoic acid-3,7,12-triols, 7α-hydroxy-4-cholesten-3-one and 4-cholesten-3-one can activate SXR. Indeed, bile acid precursors are endogenous ligands for SXR [Dussault et al., 2003]. Activation of SXR by these sterol compounds induces CYP3A expression and provides an alternative pathway for sterol clearance in CYP27A1 null mice [Dussault et al., 2003; Goodwin et al., 2003]. CYP27A1 catalyzes the cleavage of cholesterol side chains in classic bile acid biosynthetic pathway and the hydroxylation of cholesterol to 27-hydroxycholesterol (27-HOC) and 3β-hydroxy-5-cholestenoic acid in most tissues [Chiang, 1998; Russell, 2003]. Interestingly, the sterol intermediates of bile acid synthesis are more potent activators of mouse SXR than human SXR, which may explain the development of cerebrotendinous xanthomatosis (a neurodegenerative disease caused by deposition of cholesterol in various tissues) in humans with CYP27A1 deficiency, but not in corresponding knockout mice. The role of SXR in cholesterol clearance and homeostasis was further underlined by the finding that SXR null mice developed acute hepatorenal failure when they were fed a diet containing high cholesterol and cholic acid levels. This suggested that SXR plays an important role in the detoxification of cholesterol metabolites [Sonoda et al., 2005]. In agreement with this finding, it was recently shown that SXR can directly mediate CYP27A1 expression in cultured intestinal cells [Li et al., 2007]. The activity of CYP27A1 was confirmed by increased levels of 27-HOC in rifampicin treated cells. Moreover, the authors showed that SXR can also induce ABCA1 and ABCG1 expression and promote cholesterol efflux in the same cells.
In addition to playing important roles in cholesterol detoxification, SXR can also modulate SREBP-dependent and SREBP–independent lipogenic pathways in vitro and in vivo. SXR can mediate a SREBP-independent lipogenic pathway by activating the free fatty acid (FFA) uptake transporter CD36, PPARγ, and several accessory lipogenic enzymes, such as stearoyl CoA desaturase-1 (SCD-1) and long-chain free fatty acid elongase (FAE) [Zhou et al., 2006c]. SXR activation is also associated with induction of Insig-1, a protein with anti-lipogenic properties, and with reduced protein levels of the active form or SREBP-1 [Roth et al., 2008]. A functional SXR binding site was found in the Insig-1 promoter and it was suggested that Insig-1 induction by SXR could lead to decreased levels of active SREBP-1 and reduced triglyceride synthesis. These two studies showed opposite effects of SXR activation on lipid homeostasis, suggesting that further research will be required to sort out the details. Yet the overall findings are consistent with a role for SXR in mediating lipid homeostasis at multiple levels.
Although these studies indicate that SXR plays important roles in cholesterol detoxification and lipid homeostasis, it is not clear what effects, if any, long-term SXR activation has on cholesterol levels in human or in animal models. Interestingly, studies have shown that SXR activation can affect serum HDL-C and ApoA-I levels [Bachmann et al., 2004; Masson et al., 2005]. Induction of CYP3A by some SXR ligands was positively correlated with induction of ApoA-I mRNA and plasma HDL and ApoA-I levels in mice [Bachmann et al., 2004]. However, their interpretation that SXR is involved is complicated by the finding that the human specific ligand, rifampicin, which lacks the ability to activate the rodent receptor, gave positive results. This suggests that a non-SXR dependent mechanism may be at work.
Another study showed that the inhibitory effects of bile acids on HDL and ApoA-I levels were much more pronounced in SXR null mice, whereas these effects were blocked in transgenic humanized mice [Masson et al., 2005]. However, many clinically-relevant SXR ligands have been shown to increase cholesterol levels. For example, treatment with the potent and specific SXR ligand rifampicin has hyperlipidemia as a side effect [Khogali et al., 1974]. Patients treated with rifampicin for 6 days had an elevated ratio of lathosterol to cholesterol, indicating increased cholesterol synthesis [Lutjohann et al., 2004]. In addition, treatment of HIV patients with ritonavir, one of the first commercially available HIV protease inhibitors and a potent SXR activator, can cause hyperlipidemia and may be associated with increased risk of cardiovascular disease [Barbaro, 2006; Carr et al., 1998a; Carr et al., 1998b; Shafran et al., 2005]. Cafestol, presented in unfiltered brewed coffee and the most potent cholesterol-elevating compound known in the human diet was recently found to be an agonist of both SXR and farnesoid X receptor (FXR) [Ricketts et al., 2007]. Cafestol can induce intestinal CYP27A1 and ABCA1 expression and promotes cholesterol efflux to the liver via SXR activation, which is consistent with SXR effects in intestinal cells [Li et al., 2007].
Several clinical studies have shown that a mixed green vegetable and fruit beverage containing broccoli can lower cholesterol levels in hypercholesterolemic patients [Suido et al., 2002; Takai et al., 2003] and a pilot study indicated that intake of broccoli sprouts for 1 week can significantly reduce total cholesterol and LDL cholesterol levels in 12 healthy subjects [Murashima et al., 2004]. SFN is present at high concentrations in broccoli and broccoli sprouts and is a naturally occurring antagonist for human SXR [Zhou et al., 2007]. Therefore, it would be of great interest to investigate whether SFN is the active ingredient in broccoli that lowers cholesterol levels and whether it does so by inhibiting SXR activity.
Taken together, these studies suggest that SXR plays important roles in cholesterol metabolism and lipid homeostasis. However, the precise mechanisms through which SXR modulates lipid metabolism and cholesterol levels in vivo remain unclear. The discovery and pharmacological development of new selective SXR modulators could represent an interesting and innovative therapeutic approach to combat hyperlipidemia and atherosclerosis.
Interplay between SXR and other nuclear receptors
In addition to directly regulating expression of its own target genes, SXR can interact with other nuclear receptors to exert more complex effects on gene regulation. Crosstalk between SXR and several other nuclear receptors has been described, including constitutive androstane receptor (CAR), farnesoid X receptor (FXR), vitamin D receptor (VDR), small heterodimer partner (SHP), hepatocyte nuclear factor 4α (HNF4α), liver X receptor (LXR) and PPARγ. Here we discuss the crosstalk between SXR and CAR, FXR and VDR, all of which appear to be important for xenobiotic, bile acid, or vitamin D metabolism and disposition.
SXR and CAR
The orphan receptor CAR (NR1I3) was initially isolated and shown to activate a DR-5 type of retinoid acid response element (βRARE) in a ligand-independent manner, suggesting that CAR activates the response element in the absence of retinoic acid [Baes et al., 1994]. Unlike SXR, CAR shows relatively high basal activity to transactivate genes without ligand. The role of CAR as a xenobiotic receptor was first identified by the ability of selective androstane metabolites to inhibit its constitutive activity [Forman et al., 1998] and by phenobarbital to potentiate its activity [Wei et al., 2000]. Its role in positive xenobiotic regulation was suggested when CAR was shown to activate the PBREM found in promoters of phenobarbital-inducible CYP2B genes [Honkakoski et al., 1998]. It was found later that CAR has a broad role in xenobiotic metabolism by regulating additional phase I and phase II enzymes, as well as drug transporters [Xie and Evans, 2001].
Interestingly, SXR and CAR share a variety of ligands and activate an overlapping set of genes. For example, phenobarbital can also bind to and activate SXR. CAR can regulate CYP3A genes through SXR response elements and SXR can regulate CYP2B genes via adaptive recognition of the phenobarbital response element (PBRE), as revealed by receptor-DNA binding analysis and transfection assays [Xie et al., 2000b]. The cross-regulation of these two receptors was further evaluated in vivo. Constitutively active SXR (VP-SXR) transgenic mice showed sustained induction of CYP3A and CYP2B [Xie et al., 2000a; Xie et al., 2000b]. However, constitutively active CAR (VP-CAR) transgenic mice showed only CYP2B induction; CYP3A was unchanged [Saini et al., 2004]. It was subsequently shown that CAR only exhibits very weak binding and functional activation of the CYP3A4 promoter, instead showing a pronounced selectivity for CYP2B6 over CYP3A4 [Faucette et al., 2006]. These findings suggest that although SXR and CAR cross-regulate each other’s expression, that this cross-regulation of target genes is asymmetric.
SXR and FXR
Farnesoid X receptor (FXR) was originally identified as a receptor activated by farnesol [Forman et al., 1995]. Subsequent studies have convincingly shown that FXR functions as a bile acid receptor [Wang et al., 1999]. Bile acid activated FXR represses the expression of CYP7A1 and CYP2B, enzymes involved in bile acid synthesis, through an indirect mechanism. Bile acids activate FXR and induce the expression of small heterodimer partner (SHP), which binds to and inhibits the activity of liver receptor homolog-1 (LRH-1), which normally activates CYP7A1. In addition to inhibiting the activity of LRH-1, SHP can also interact with SXR and inhibit its transcriptional activity [Ourlin et al., 2003]. SXR and FXR not only share bile acid ligands, but they also regulate some common target genes such as SULT2A1 and MRP2 [Echchgadda et al., 2004; Kast et al., 2002]. SXR can also directly regulate SHP expression [Frank et al., 2005] and FXR regulates CYP3A4 transcription through two functional FXR response elements in the promoter of CYP3A4 [Goodwin et al., 1999]. Interestingly, SXR can be also transcriptionally regulated by FXR [Jung et al., 2006]. Feeding mice with cholic acid or the synthetic FXR agonist GW4064 resulted in strong induction of SXR expression. This effect was lost in FXR knockout mice [Jung et al., 2006]. Four FXR binding elements were identified in the SXR promoter; two of these were able to bind FXR protein and trigger a strong response to ligand treatment. The crosstalk between SXR and FXR leads to an efficient protection of the liver against bile acid induced toxicity. Bile acids activate FXR, which downregulates synthesis of bile acids and also leads to the transcriptional activation of SXR, thereby promoting bile acid metabolism.
SXR and VDR
Vitamin D exerts important biological functions in the maintenance of calcium homeostasis and in the development and maintenance of bones. Its active metabolite, 1,25-dihydroxyvitamin D3 (1,25(OH)2D3), elicits most of its effects through activation of the vitamin D receptor (VDR) [Jones et al., 1998; Omdahl and May, 2004]. The balance between bioactivation and degradation of 1,25(OH)2D3 is critical for ensuring appropriate biological effects of Vitamin D.
CYP24-mediated 24-hydroxylation is a critical step in the catabolism of 1,25(OH)2D3 and appears to be responsible for controlling intra-renal and systemic 1,25(OH)2D3 levels. CYP24 is directly regulated by VDR and it is expressed mainly in the kidney where VDR is also abundant. Although there is also a relatively high level of VDR expression in the small intestine, constitutive CYP24 expression in this tissue is very low or undetectable, in contrast to that in the kidney [Xu et al., 2006].
It is well recognized that long-term therapy with some antiepileptic drugs, including phenobarbital, phenytoin and carbamazepine, and the antimicrobial agent rifampicin (RIF), can cause a metabolic bone disease – osteomalacia [Andress et al., 2002; Burt et al., 1976; Karaaslan et al., 2000; Pack and Morrell, 2004; Shah et al., 1981]. Interestingly, many of the drugs that cause osteomalacia are able to activate SXR, suggesting a possible connection. To reveal the mechanism of drug-induced osteomalacia, several groups recently investigated the impact of SXR activation on CYP24 gene expression in vitro and in vivo.
Pascussi et al. first suggested that activation of SXR can enhance the expression of the VDR target gene, CYP24, which would increase the catabolism of 1,25(OH)2D3; thereby, leading to drug-induced osteomalacia [Pascussi et al., 2005]. They conducted a series of in vitro and in vivo studies which were interpreted as providing evidence that a drug, which can activate SXR is likely to enhance CYP24 expression and the catabolism of 25(OH)D3, leading to vitamin D deficiency. However, this model is of questionable significance with regard to the physiological functions of CYP24 in vivo, because CYP24 is found primarily in the kidney, where SXR is expressed at very low levels. In addition, CYP24 is expressed at very low levels in liver and intestine where SXR is abundant [Xu et al., 2006]. This suggests that enhanced CYP24 expression by SXR is unlikely to play an important role in the development of osteomalacia following long-term treatment with SXR activators.
In accord with this prediction, it was subsequently reported that CYP3A4, and not CYP24, played the dominant role in 23- and 24-hydroxylation of 1,25(OH)2D3 under constitutive and induced conditions in human small intestine and liver [Xu et al., 2006]. Heterologously-expressed CYP3A4 catalyzed the 23- and 24-hydroxylation of 1,25(OH)2D3; moreover, CYP3A4 exhibited opposite product stereochemical preference compared with that of CYP24A. Although the metabolic clearance of Vitamin D3 by CYP3A4 was less than that catalyzed by CYP24, comparison of metabolite profiles and experiments using CYP3A-specific inhibitors indicated that CYP3A4 was the dominant source of 1,25(OH)2D3 23- and 24-hydroxylase activity in both human small intestine and liver [Xu et al., 2006].
It was later shown that activation of SXR did not induce CYP24 expression in vitro or in vivo, nor did it transactivate the CYP24 promoter [Zhou et al., 2006a]. Instead, SXR was shown to be able to crosstalk with VDR to inhibit VDR-mediated CYP24 promoter activity. This provided a mechanism to explain the low levels of CYP24 expression in tissues containing high levels of SXR. 1,25(OH)2D3-induced CYP24 expression was enhanced in SXR knockout mice, and treatment of humans with the SXR agonist rifampicin had no effect on intestinal CYP24 expression, despite marked CYP3A4 induction [Zhou et al., 2006a]. Furthermore, feeding WT mice with diet containing the mouse SXR ligand, PCN, for two weeks strongly induced SXR target genes CYP3A11, GSTA1, and MDR1a expression, but failed to induce CYP24 expression in both liver and intestine (Zhou, C., unpublished observation). Interestingly, phenobarbital-activated SXR can also inhibit expression of CYP2D25 at the transcriptional level [Hosseinpour et al., 2007]. CYP2D25 is an important 25-hydroxylase involved in 1,25(OH)2D3 biosynthesis. Downregulation of CYP2D25 provides another potential new mechanism for drug-induced osteomalacia.
Taken together with the findings that CYP3A4, and not CYP24, dominated the hydroxylation of 1,25(OH)2D3 in human liver and intestine, these results indicate that SXR plays multiple roles in mediating vitamin D catabolism and drug-induced osteomalacia. It upregulates CYP3A4 expression, while repressing CYP24 and CYP2D25 expression in the liver and intestine.
SXR, NF-κB and inflammatory bowel disease (IBD)
SXR and NF-κB
It has long been known that inflammation and infection reduce hepatointestinal drug metabolism capacity [Aitken et al., 2006] and that exposure to xenobiotic chemicals can impair immune function. For example, expression of hepatic CYP genes can be profoundly decreased by various infectious and inflammatory stimuli, with concomitant clinical and toxicological consequences [Morgan, 1997]. Meanwhile, drug metabolism-inducing xenobiotics/drugs, such as the antibiotic rifampicin and the anticonvulsant phenytoin, have immunosuppressive side-effects [Badawy et al., 1991; Paunescu, 1970; Scheinfeld, 2003; Sorrell and Forbes, 1975; Sorrell et al., 1971]. However, the molecular mechanisms underlying both of these phenomena have remained largely unknown until recently. Two groups recently revealed a crosstalk between SXR and NF-κB, providing a potential molecular mechanism that links xenobiotic metabolism and inflammation [Gu et al., 2006; Xie and Tian, 2006; Zhou et al., 2006b].
Zhou et al. reported that activation of SXR by RIF and other agonists antagonized the activity of NF-κB, in vitro and in vivo. SXR inhibited NF-κB-mediated reporter activity and the expression of NF-κB target genes. Mice deficient in SXR showed increased expression of NF-κB target genes in multiple tissues and also had marked intestinal inflammation [Zhou et al., 2006b]. Activation of NF-κB also inhibited SXR activity and the expression of SXR target genes. Inhibition of NF-κB also enhanced the activity of SXR and the expression of its target genes [Zhou et al., 2006b]. Gu et al. also reported that NF-κB activation by lipopolysaccharide (LPS) and tumor necrosis factor-α (TNFα) plays a pivotal role in the suppression of CYP3A4 through interactions of NF-κB with the SXR-RXR complex [Gu et al., 2006]. Inhibition of NF-κB by NF-κB-specific suppressor SRIκBα reversed the suppressive effects of LPS and TNFα. Furthermore, they also performed electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation assays (ChIP) and showed that NF-κB p65 disrupted binding of the SXR-RXR complex to its binding motif. Thus, the negative crosstalk between SXR and NF-κB not only reveals the possible mechanism underlying the immunosuppressive effects of RIF, but also explains the well recognized decreased expression of hepatic CYP genes during inflammation or infection.
Although transrepression by nuclear receptors and crosstalk between nuclear receptors and other signaling pathways have been extensively studied, the molecular mechanisms are still far from being completely understood [De Bosscher et al., 2003]. For instance, many plausible models have been proposed for the crosstalk between NF-κB and GR, and each of them is supported by experimental evidence. The models are mutually inconsistent in many ways and the topic remains highly controversial [Almawi and Melemedjian, 2002; McKay and Cidlowski, 1999]. The study by Gu et al. [Gu et al., 2006], which demonstrated direct interaction of NF-κB with RXR, provided a potential molecular mechanism for the crosstalk between NF-κB and other nuclear receptors, since RXR is a common partner for many nuclear receptors including CAR, VDR, RAR, TR, LXR and PPARs [Xie and Tian, 2006]. It was also found recently that transrepression of NF-κB target genes by another nuclear receptor, PPARγ, is mediated by SUMOylation of PPARγ [Pascual et al., 2005]. Interestingly, the SXR ligand-binding domain also contains a consensus SUMOylation site, but it is currently unknown whether SUMOylation of SXR is involved in transrepression of NF-κB signaling.
SXR and inflammatory bowel disease (IBD)
As a key regulator of inflammation, activated NF-κB is frequently detected in various inflammatory diseases and tumors [Greten et al., 2004; Karin and Greten, 2005; Pikarsky et al., 2004]. Zhou et al. observed increased pro-inflammatory gene expression in SXR KO mice, which is likely due to the loss of repression of NF-κB by SXR in vivo [Zhou et al., 2006b]. They also found that the small bowel of SXR knockout mice exhibits a prominent, increased chronic inflammatory infiltrate. This histological pattern of a mucosal mononuclear inflammatory infiltrate is reminiscent of that seen in humans with inflammatory bowel diseases such as Celiac disease [Kagnoff, 2005; Wahab et al., 2002].
An independent study conducted by Gonzales et al. also demonstrated that activation of SXR ameliorates experimentally-induced inflammatory bowel disease via inhibition of NF-κB [Shah et al., 2007]. They treated wild-type and SXR knockout mice with the SXR agonist pregnenolone-16α-carbonitrile (PCN) or vehicle while administering 2.5% dextran sulfate sodium (DSS) in drinking water to induce IBD. PCN-treated mice were protected from DSS-induced colitis compared with vehicle-treated mice in wild-type mice, but not in SXR KO mice. PCN treatment did not increase epithelial barrier function; however, it did decrease mRNA levels of several NF-κB target genes in a SXR-dependent manner. Their study clearly demonstrates a protective role for SXR agonists in DSS-induced IBD and suggests that SXR-mediated repression of NF-κB target genes in the colon is a critical mechanism for decreasing the susceptibility of mice to DSS-induced IBD [Shah et al., 2007].
Another study showed that dysregulation of the SXR gene may critically influence intestinal barrier defense and susceptibility to IBD [Langmann et al., 2004]. They further showed, using DNA microarray analyses of nonaffected tissue from IBD patients, that expression of SXR and a known target gene, MDR1, were downregulated in the colon of ulcerative colitis patients. Although the downregulation of MDR1 in IBD patients may be independent of the decreased expression of SXR, these data are consistent with the possibility that dysregulation of SXR in the gut is likely to contribute to the pathophysiology of ulcerative colitis.
In accord with these findings, several clinical studies have found that the SXR gene is associated with susceptibility to inflammatory bowel disease [Dring et al., 2006; Martinez et al., 2007]. Dring et al. showed significant associations of two SXR SNPs (-23585 and -24381) with IBD, Crohn's disease (CD), and ulcerative colitis (UC) in a study of 422 patients with IBD and 350 ethnically-matched controls [Dring et al., 2006]. Two other SNPs (7635 and 8055) were associated with IBD and CD, but not UC. However, a subsequent case-control study failed to replicate the original association in a different population [Ho et al., 2006]. Ho et al. performed a genome-wide association study in total of 387 UC and 328 CD patients, together with 338 healthy controls and did not find any association between SXR and IBD.
Most recently, Martínez et al. analyzed three SXR polymorphisms, including the one most strongly correlated with IBD risk in their initial study (-25385 and the 6 haplotypes), in 365 UC and 331 CD patients compared with 550 ethnically-matched controls [Martinez et al., 2007]. They found that the overall haplotypic distribution showed a significant difference between UC and CD patients. Patients with extensive UC carrying the -25385T allele showed increased susceptibility to IBD compared with healthy subjects. Their data also support the association of the SXR locus with extensive UC and the interaction between SXR and MDR1 genes. Larger studies in other populations may help to clarify the association between SXR and susceptibility to IBD. Further characterization of the effects of SNPs on SXR function in vitro and in vivo will be necessary for a full understanding of what role SXR plays in IBDs.
SXR and cancer
So far, it has become clear that SXR regulates induction of many drug-metabolizing enzymes and drug-transporters. These metabolic enzymes and transporters are involved in biotransformation and clearance of more than 60% of non-prescription and prescription drugs, including widely used anti-cancer agents. Activation of SXR has been associated with clinically important drug-drug interactions. These interactions are even more important in oncology, as cancer patients are typically treated with combinations of anti-cancer agents. It is also very common to give such patients many prophylactic and palliative treatments including anti-emetics, analgesics and glucocorticoids at the same time to relieve symptoms and decrease hypersensitivity.
Many of these drugs, including chemotherapeutic agents themselves, activate SXR. For example, the anti-estrogen, tamoxifen, and its metabolite 4-hydroxy tamoxifen, widely used to treat estrogen-positive breast tumors, are potent activators of SXR [Nagaoka et al., 2006]. Similarly, many taxane and non-taxane microtubule-stabilizing drugs such as taxol, epothilone B and BMS-247550 activate SXR significantly [Mani et al., 2005]. The glucocorticoid dexamethasone, the anti-convulsant phenytoin and the antibiotic rifampicin also activate SXR potently [Luo et al., 2002]. As many of the above-mentioned drugs are given in combination with each other, or other antineoplastic agents, it is clinically relevant that the SXR activators can change the bio-availability and pharmacokinetics of these drugs by inducing their own metabolism and excretion, as well as that of other concomitantly administered drugs.
The induction of metabolism and excretion by anti-cancer agents through SXR has also been associated with drug-resistance or lower efficacy of these agents in cancer. For example, the work done by Baker and coworkers shows that activation of SXR by anti-neoplastic agents in osteosarcoma cell lines induces activity of CYP3A4 and MDR1, which provides a possible mechanism for drug resistance in these cells [Mensah-Osman et al., 2007]. Similarly, activation of SXR in endometrial cancer cell lines by many endocrine-disrupting chemicals has been shown to induce CYP3A4/CYP3A7 expression [Masuyama et al., 2003]. Further studies by this group showed that when SXR expression is downregulated in endometrial cancer cells, the cell growth inhibitory and apoptotic activities of anticancer agents that activate SXR, such as paclitaxel and cisplatin, are significantly enhanced. In contrast, SXR overexpression causes significant decrease in cell growth inhibition and apoptosis mediated by these agents [Masuyama et al., 2007]. Paclitaxel has been identified as a substrate of CYP3A4 and MDR1 [Sparreboom et al., 1997] and the role of MDR1 in mediating paclitaxel resistance in tumors has also been shown [Penson et al., 2004]. Therefore, it was suggested that SXR downregulation might be a way to sensitize these cells to paclitaxel and overcome their resistance to it through the inhibition of drug metabolism and/or transport/efflux of this drug [Masuyama et al., 2007]. Interestingly, these authors also found that there is a significant inverse correlation between SXR expression and estrogen receptor (ER) expression in endometrial tissues. Moreover, they found that downregulation of SXR significantly enhanced the proliferation of endometrial cancer cells in the presence of estradiol. Because estradiol is a substrate for CYP3A4 and an SXR activator, it is plausible that local estrogen levels in endometrial tissue may be controlled by SXR. Thus, SXR downregulation would reduce expression of CYP3A4, thereby increasing local estrogen levels and/or increasing sensitivity of estrogen receptor to estradiol [Cheng et al., 2001].
A similar inverse relationship between SXR and ER expression has also been shown in breast cancer cell lines [Dotzlaw et al., 1999]. Expression of SXR has recently also been found in breast cancer tissues, but not in normal surrounding tissues of the patients [Miki et al., 2006], while other investigators find expression of SXR in both normal and cancerous breast tissue [Dotzlaw et al., 1999]. The link between SXR and ER warrants further evaluation to determine whether SXR modulates estrogen levels only by affecting the metabolism and transport of estradiol, by directly modulating expression of the estrogen receptor itself or through some other mechanism. Similarly, it will be important to determine the relationship between SXR expression and breast cancer and to understand whether activating SXR promotes, or inhibits the growth of breast cancer cells.
Recent studies have shown that expression of the constitutively active form of SXR (VP-SXR) sensitizes transgenic mice and colon cancer cells to the oxidative toxicant paraquat [Gong et al., 2006]. This finding was paradoxical because mice expressing activated SXR have increased GST activity, and GSTs play an important role in detoxification of products from oxidative stress. Thus, a protective effect, rather than apoptosis was expected in mice expressing high levels of GST. Furthermore, another study showed that the survival rate of mice receiving paraquat significantly rose with pretreatment with phenytoin, phenobarbital, or rifampicin, which induced activity in CYP3A, CYP2B, or CYP2C [Shimada et al., 2002]. This study suggested that postmitochondrial fractions play an important role in paraquat detoxication metabolism, and that the combination of CYP induction is highly useful for the survival of paraquat-exposed mice. Interestingly, Gong et al. found that the hepatic activities of two antioxidative enzymes, superoxide dismutase (SOD) and catalase (CAT), are downregulated in VP-SXR mice and in WT mice treated with a SXR agonist, which can potentially explain the paraquat sensitivity [Gong et al., 2006]. SOD and CAT may not be direct transcriptional targets of SXR and the mechanism of SXR-mediated downregulation of SOD and CAT activities remains to be determined. Therefore, the cytotoxic effects of SXR in combination with oxidative stressors such as paraquat needs to be further investigated.
As noted above, the study by Mani and coworkers shows that many microtubule-stabilizing agents activate SXR [Mani et al., 2005]. Interestingly, the compounds which did not cause microtubule stabilization and cytotoxicity also did not activate SXR. Although there is no obvious causal link known between microtubule stabilization and SXR activation, the association suggests that SXR itself may be linked with the cytotoxic effects of these drugs [Mani et al., 2005]. We have found that activation of SXR decreases the growth rate and induces apoptosis in many breast cancer cell lines [Verma et al.]. As SXR is a major regulator of steroid, endobiotic and xenobiotic levels in the body, it will be very important to understand its precise function in cancer tissues, particularly in hormone responsive tumors such as breast, ovary, uterus and prostate.
SXR and endocrine-disrupting chemicals (EDCs)
Considering the possible roles of SXR activation in cancer, and the numerous xenobiotic compounds that are SXR activators, the relationship between SXR and endocrine-disrupting chemicals (EDCs) is very interesting. The term EDCs was coined at a meeting in 1991, chaired by Theo Colborn, in which she first introduced the concept that chemical exposure might be altering reproductive function and fertility in wildlife and humans. This concept was further emphasized in “Our Stolen Future”, which summarized the scientific literature on the potential effects of exposure to persistent chemicals on humans and wildlife [Colborn et al., 1996]. While the doses of individual chemicals required to elicit such effects remain controversial, it is beyond dispute that chemical exposure during sensitive windows of development can cause wide-ranging disturbances in development and physiology. EDCs have been defined as exogenous agents that interfere with the synthesis, secretion, transport, binding, action, or elimination of natural hormones in the body, which are responsible for the maintenance of homeostasis, reproduction, development, and/or behavior [Kavlock et al., 1996].
Some of the most well known examples of EDCs are 17-α ethinylestradiol (the contraceptive pill), dioxins, polychlorinated biphenyls (PCBs), polycyclic aromatic hydrocarbons (PAHs), furans, phenols, organotins (e.g., tributyltin chloride) and various organochlorine pesticides (e.g., DDT and its derivatives, endosulfan, and dieldrin). EDCs can disrupt endocrine function directly by activating or antagonizing the steroid hormone receptors or indirectly by modulating the function of other nuclear receptors involved in metabolism, transport, or elimination of steroid hormones.
The most well-studied effects of EDCs on the endocrine system are those that result from their activity on estrogen or androgen receptors [Kelce et al., 1994; Kelce et al., 1995; Klotz et al., 1996; Sohoni and Sumpter, 1998; Soto et al., 1994; White et al., 1994]. However, there are a variety of other mechanisms by which EDCs can disrupt endocrine function. These include interfering with thyroid and retinoid receptor function, by interfering with activities of other nuclear hormone receptors such as SXR, CAR and PPARγ, and by altering cofactor recruitment (reviewed in [Grun and Blumberg, 2006; Grun and Blumberg, 2007; Tabb and Blumberg, 2006]).
Many EDCs including bisphenol-A, organochlorine and organophosphate pesticides, alkylphenols, phthalates and PCBs have been shown to upregulate CYP3A expression and activate SXR [Coumoul et al., 2002; Hurst and Waxman, 2004; Jacobs et al., 2005; Lemaire et al., 2004; Masuyama et al., 2000; Tabb et al., 2004; Takeshita et al., 2001]. For example, phthalic acid and nonylphenol, the industrial chemicals used in preparation of detergents and perfumes act as xenoestrogens, and also activate SXR-mediated transcription and induce CYP3A1 levels in rat liver [Masuyama et al., 2000].
Similarly, organochlorine pesticides such as endosulfan, chlordane, and dieldrin activate human SXR and induce CYP3A4 transcription [Coumoul et al., 2002]. Since many EDCs can activate SXR and thereby influence their own metabolism as well as the metabolism of other xenobiotic and endobiotic compounds, it will be important to investigate the effects of systemic exposures to EDCs on SXR activity and characterize the in vivo pharmacokinetic effects on other compounds.
Similar to other SXR ligands, some EDCs also have species-specific effects on SXR. For example, bisphenol-A, the monomer for polycarbonate plastics and epoxy resins is a human selective SXR activator [Takeshita et al., 2001]. Highly chlorinated PCBs such as PCB184 and PCB197 are rodent-selective activators, but human-selective SXR antagonists [Tabb et al., 2004]. To our knowledge, PCBs are the first example of ligands acting as agonist on a particular receptor in one species and antagonist in its orthologous receptor in other species.
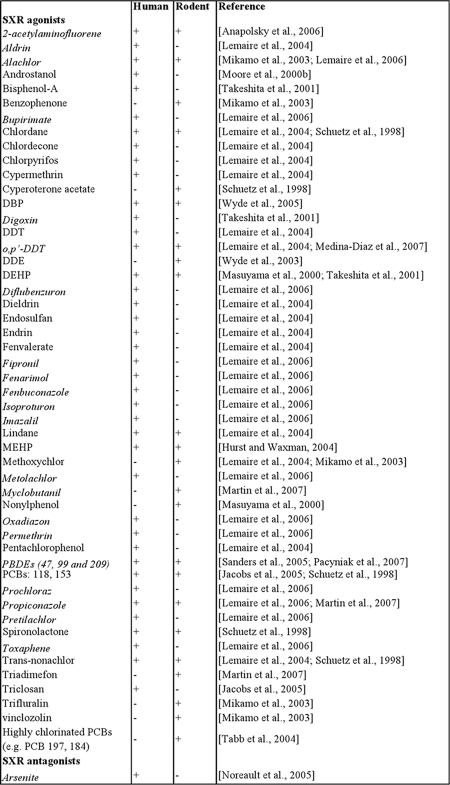
Numerous compounds, many of which are EDCs, can affect the activity of SXR. A current list of other known EDCs reported to alter rodent or human SXR activity is summarized in Table 1. This is an expected response if that SXR functions as a sensor that induces the expression of xenobiotic metabolizing enzymes and transporters, which mediate the body’s response to harmful dietary and environmental compounds. Most of these EDCs are also substrate for CYP action. One implication of this is that activation of SXR and induction of the xenobiotic response will induce metabolism of these chemicals. This may reduce their availability and activity in the body. However, compounds that induce CYPs in human hepatocytes may also lead to pro-carcinogen activation and disrupt normal endocrine function. For example, methoxychlor, an organochlorine pesticide, is not estrogenic in itself, but its metabolism by CYPs converts it to a metabolite that is estrogenically active [Mikamo et al., 2003]. Similarly, some of the hydroxyl metabolites of PCBs are significantly more powerful biologically than their parent compound [Kester et al., 2000; Sandau et al., 2000].
Table 1. EDCs that modulate the activity of human and/or rodent SXR.
The table shows an updated list of EDCs that either activate or inhibit human and/or rodent SXR. EDCs that can modulate SXR activity are indicated by “+” and EDCs that do not modulate SXR activity are indicated by “-”. EDCs that are newly-added in the list after the review done by Kretschmer et al. in 2005 (Kretschmer and Baldwin, 2005) are written in italics.
Inhibition of SXR activity by chemicals such as highly chlorinated PCBs will reduce metabolism of endo- and xenobiotic compounds, as well as the metabolism of the antagonists themselves. Antagonism of SXR activity by EDC could have a high biological impact by decreasing metabolism and elimination (and increasing the blood levels) of endobiotic and xenobiotic compounds in the body. Increased levels of these compounds in the body will increase their potential to affect other hormone receptors (e.g., the estrogen and androgen receptors). Therefore, even low-level exposure to SXR antagonists could lead to a potentially toxic buildup of xenobiotic chemicals in the body. Jacob et al. have suggested an in silico model for testing the ability of various compounds to potentially alter the activity of several nuclear hormone receptors [Jacobs, 2004]. It is more likely that combining in silico approaches with cell-based and in vivo testing of compounds in humanized rodent models will give the most significant and relevant information to predict the effects of environmental compounds on humans. Given that EDCs and other xenobiotic compounds can both activate and antagonize SXR, it will be increasingly important to understand the effects of mixtures in order to predict the effects of real-world chemical exposure.
SXR, FoxO1, FoxA2, and energy homeostasis
FoxO1 and FoxA2 are members of the “forkhead” family of transcription factors that play critical roles in lipid metabolism and gluconeogenesis in the liver [Montminy and Koo, 2004]. FoxO1 promotes gluconeogenesis in liver in the fasted state by activating gluconeogenic genes, such as phosphoenolpyruvate carboxykinase 1 (PEPCK1), glucose-6-phosphatase (G6P) and insulin-like growth factor-binding protein 1. Foxa2 is a key switch that regulates fatty-acid breakdown in the liver during fasting. Long term phenobarbital treatment is known to reduce plasma glucose levels and to improve insulin sensitivity in diabetic patients [Lahtela et al., 1985]. By performing mammalian cell-based two-hybrid screening, Kodama et al. identified FoxO1 as a coactivator to CAR- and SXR-mediated transcription [Kodama et al., 2004]. FoxO1 can directly bind to CAR and SXR in a ligand-dependent manner and facilitate their transcriptional activity. Interestingly, CAR and SXR act as corepressors of FoxO1 and downregulate FoxO1-mediated transcription by preventing its binding to its response elements in target genes. In addition to inhibiting FoxO1 activity, drug-activated SXR and CAR were also shown to inhibit HNF4α transcriptional activity via squelching of PGC1α, thereby repressing transcription of PEPCK1 and G6P [Miao et al., 2006]. These data show that drug and glucose metabolisms, two major liver functions that can be regulated independently, are reciprocally coregulated by crosstalk between xenobiotic sensors and hepatic transcription factors.
When blood glucose levels are low after fasting or prolonged exercise, the liver provides energy to extra-hepatic tissues and organs via β-oxidation and ketogenesis [Eaton et al., 1996]. FoxA2 promotes ketogenesis and β-oxidation by upregulating transcription of genes including mitochondrial 3-hydroxy-3-methylglutartate-CoA synthase 2 (HMGCS2) and carnitine palmitoyltransferase 1A (CPT1A) during fasting or after prolonged exercise [Wolfrum et al., 2004]. FoxA2 is phosphorylated and inactivated by the Akt pathway and decreases lipid metabolism in response to insulin. Treatment with drugs such as phenobarbital can also repress lipid metabolism in an insulin-independent manner [Kiyosawa et al., 2004]. Nakamura et al. reported that SXR can crosstalk with FoxA2 to mediate drug-induced repression of lipid metabolism in fasting mouse livers [Kiyosawa et al., 2004]. Using wild-type and SXR-/- mice, they showed that treatment with PCN reduced steady-state mRNA levels of HMGCS2 and CPT1A in wild-type, but not in SXR-deficient mice. A series of biochemical and cell-based assays demonstrated that SXR directly represses the ability of FoxA2 to induce expression of HMGCS2 and CPT1A. The SXR ligand binding domain can directly interact with DNA-binding domain of FoxA2 and this interaction prevents the binding of FoxA2 to its response elements in target genes. The crosstalk between SXR and FoxO1 and FoxA2 indicates that SXR not only regulates hepatic drug metabolism, but also plays important roles in glucose and energy homeostasis. These results have implications for the design of new drugs to treat insulin resistance and diabetes.
Conclusions
SXR was originally described in 1998 as a xenobiotic sensor that plays important roles in regulating the metabolism and excretion of a large variety of endobiotic, dietary, and xenobiotic chemicals. Its function in drug and xenobiotic metabolism has been extensively studied by many laboratories and much is known about SXR target genes and its interaction with other nuclear receptors. Several new avenues of research have been opened in recent years that have revealed new and mostly unsuspected roles for SXR in inflammation, bone homeostasis, vitamin D metabolism, energy homeostasis and cancer. These results suggest that SXR has a number of important functions in the body that remain to be fully explored. Considering that SXR is activated by such a wide variety of dietary and xenobiotic chemicals, they also suggest new mechanisms through which diet, chemical exposure, and environment ultimately impact heath and disease.
Acknowledgments
We thank N. King for critically reading the manuscript. This work was supported by grants from the US-EPA, DOD and NIH to B.B. S.V. was the recipient of a predoctoral fellowship from the DOD.
Abbreviations
- 1,25(OH)2D3
1,25-dihydroxyvitamin D3
- CAR
constitutive androstane receptor
- CYP
cytochrome P450
- DBD
DNA-binding domain
- EDC
endocrine-disrupting chemicals
- ER
estrogen receptor
- FXR
farnesoid X receptor
- HNF4α
hepatocyte nuclear factor 4α
- LBD
ligand-binding domain
- LXR
liver X receptor
- MDR1
multidrug resistance protein-1
- NR
nuclear receptor
- PAR
pregnane activated receptor
- PCN
pregnenolone 16α-carbonitrile
- PXR
pregnane X receptor
- RIF
rifampicin
- RXR
retinoid X receptor
- SFN
sulforaphane
- SHP
small heterodimer partner
- SNP
single nucleotide polymorphism
- SRC-1
steroid receptor coactivator-1
- SULT
sulfotransferase
- SXR
steroid and xenobiotic receptor
- UGT
UDP-glucuronosyltransferase
- VDR
vitamin D receptor
References
- Aitken A. E., Richardson T. A., Morgan E. T. Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol. 2006;46:123–49. doi: 10.1146/annurev.pharmtox.46.120604.141059. [DOI] [PubMed] [Google Scholar]
- Albermann N., Schmitz-Winnenthal F. H., Z'Graggen K., Volk C., Hoffmann M. M., Haefeli W. E., Weiss J. Expression of the drug transporters MDR1/ABCB1, MRP1/ABCC1, MRP2/ABCC2, BCRP/ABCG2, and PXR in peripheral blood mononuclear cells and their relationship with the expression in intestine and liver. Biochem Pharmacol. 2005;70:949–58. doi: 10.1016/j.bcp.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Almawi W. Y., Melemedjian O. K. Negative regulation of nuclear factor-kappaB activation and function by glucocorticoids. J Mol Endocrinol. 2002;28:69–78. doi: 10.1677/jme.0.0280069. [DOI] [PubMed] [Google Scholar]
- Andress D. L., Ozuna J., Tirschwell D., Grande L., Johnson M., Jacobson A. F., Spain W. Antiepileptic drug-induced bone loss in young male patients who have seizures. Arch Neurol. 2002;59:781–6. doi: 10.1001/archneur.59.5.781. [DOI] [PubMed] [Google Scholar]
- Bachmann K., Patel H., Batayneh Z., Slama J., White D., Posey J., Ekins S., Gold D., Sambucetti L. PXR and the regulation of apoA1 and HDL-cholesterol in rodents. Pharmacol Res. 2004;50:237–46. doi: 10.1016/j.phrs.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Badawy A. H., Shalaby S. A., Abdel Aal S. F. Hydantoin immunosuppression clinical study. J Egypt Soc Parasitol. 1991;21:257–62. [PubMed] [Google Scholar]
- Baes M., Gulick T., Choi H. S., Martinoli M. G., Simha D., Moore D. D. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–52. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaro G. Metabolic and cardiovascular complications of highly active antiretroviral therapy for HIV infection. Curr HIV Res. 2006;4:79–85. doi: 10.2174/157016206775197664. [DOI] [PubMed] [Google Scholar]
- Bertilsson G., Heidrich J., Svensson K., Asman M., Jendeberg L., Sydow-Backman M., Ohlsson R., Postlind H., Blomquist P., Berkenstam A. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci U S A. 1998;95:12208–13. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla S., Ozalp C., Fang S., Xiang L., Kemper J. K. Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem. 2004;279:45139–47. doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. K., Chakrabarti A., Chatterjee S. S. Activity profiles of two hyperforin-containing hypericum extracts in behavioral models. Pharmacopsychiatry. 1998;31 Suppl 1:22–9. doi: 10.1055/s-2007-979342. [DOI] [PubMed] [Google Scholar]
- Blumberg B., Evans R. M. Orphan nuclear receptors--new ligands and new possibilities. Genes Dev. 1998a;12:3149–55. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- Blumberg B., Sabbagh W., Jr., Juguilon H., Bolado J., Jr., van Meter C. M., Ong E. S., Evans R. M. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998b;12:3195–205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch T. M., Deenen M., Pruntel R., Smits P. H., Schellens J. H., Beijnen J. H., Meijerman I. Screening for polymorphisms in the PXR gene in a Dutch population. Eur J Clin Pharmacol. 2006;62:395–9. doi: 10.1007/s00228-006-0108-0. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R., Traber M. G. Vitamin E: function and metabolism. Faseb J. 1999;13:1145–55. [PubMed] [Google Scholar]
- Burt R., Freston J. W., Tolman K. G. The influence of phenobarbital on biotransformation of 25-hydroxycholecalciferol. J Clin Pharmacol. 1976;16:393–8. doi: 10.1002/j.1552-4604.1976.tb02413.x. [DOI] [PubMed] [Google Scholar]
- Carr A., Samaras K., Burton S., Law M., Freund J., Chisholm D. J., Cooper D. A. A syndrome of peripheral lipodystrophy, hyperlipidaemia and insulin resistance in patients receiving HIV protease inhibitors. Aids. 1998a;12:F51–8. doi: 10.1097/00002030-199807000-00003. [DOI] [PubMed] [Google Scholar]
- Carr A., Samaras K., Chisholm D. J., Cooper D. A. Pathogenesis of HIV-1-protease inhibitor-associated peripheral lipodystrophy, hyperlipidaemia, and insulin resistance. Lancet. 1998b;351:1881–3. doi: 10.1016/S0140-6736(98)03391-1. [DOI] [PubMed] [Google Scholar]
- Cheng Z. N., Shu Y., Liu Z. Q., Wang L. S., Ou-Yang D. S., Zhou H. H. Role of cytochrome P450 in estradiol metabolism in vitro. Acta Pharmacol Sin. 2001;22:148–54. [PubMed] [Google Scholar]
- Chiang J. Y. Regulation of bile acid synthesis. Front Biosci. 1998;3:d176–93. doi: 10.2741/a273. [DOI] [PubMed] [Google Scholar]
- Chrencik J. E., Orans J., Moore L. B., Xue Y., Peng L., Collins J. L., Wisely G. B., Lambert M. H., Kliewer S. A., Redinbo M. R. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol. 2005;19:1125–34. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- Chung F. L., Conaway C. C., Rao C. V., Reddy B. S. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis. 2000;21:2287–91. doi: 10.1093/carcin/21.12.2287. [DOI] [PubMed] [Google Scholar]
- Colborn T., Dumanoski D., Meyers J.P. Our Stolen Future. New York: Dutton; 1996. [Google Scholar]
- Conaway C. C., Yang Y. M., Chung F. L. Isothiocyanates as cancer chemopreventive agents: their biological activities and metabolism in rodents and humans. Curr Drug Metab. 2002;3:233–55. doi: 10.2174/1389200023337496. [DOI] [PubMed] [Google Scholar]
- Coumoul X., Diry M., Barouki R. PXR-dependent induction of human CYP3A4 gene expression by organochlorine pesticides. Biochem Pharmacol. 2002;64:1513–9. doi: 10.1016/s0006-2952(02)01298-4. [DOI] [PubMed] [Google Scholar]
- De Bosscher K., Vanden Berghe W., Haegeman G. The interplay between the glucocorticoid receptor and nuclear factor-kappaB or activator protein-1: molecular mechanisms for gene repression. Endocr Rev. 2003;24:488–522. doi: 10.1210/er.2002-0006. [DOI] [PubMed] [Google Scholar]
- Denison M. S., Whitlock J. P., Jr. Xenobiotic-inducible transcription of cytochrome P450 genes. J Biol Chem. 1995;270:18175–8. doi: 10.1074/jbc.270.31.18175. [DOI] [PubMed] [Google Scholar]
- Dotzlaw H., Leygue E., Watson P., Murphy L. C. The human orphan receptor PXR messenger RNA is expressed in both normal and neoplastic breast tissue. Clin Cancer Res. 1999;5:2103–7. [PubMed] [Google Scholar]
- Dring M. M., Goulding C. A., Trimble V. I., Keegan D., Ryan A. W., Brophy K. M., Smyth C. M., Keeling P. W., O'Donoghue D., O'Sullivan M., O'Morain C., Mahmud N., Wikstrom A. C., Kelleher D., McManus R. The pregnane X receptor locus is associated with susceptibility to inflammatory bowel disease. Gastroenterology. 2006;130:341–8; quiz 592. doi: 10.1053/j.gastro.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Dussault I., Yoo H. D., Lin M., Wang E., Fan M., Batta A. K., Salen G., Erickson S. K., Forman B. M. Identification of an endogenous ligand that activates pregnane X receptor-mediated sterol clearance. Proc Natl Acad Sci U S A. 2003;100:833–8. doi: 10.1073/pnas.0336235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I., Lin M., Hollister K., Wang E. H., Synold T. W., Forman B. M. Peptide mimetic HIV protease inhibitors are ligands for the orphan receptor SXR. J Biol Chem. 2001;276:33309–12. doi: 10.1074/jbc.C100375200. [DOI] [PubMed] [Google Scholar]
- Dussault I., Forman B. M. The nuclear receptor PXR: a master regulator of "homeland" defense. Crit Rev Eukaryot Gene Expr. 2002;12:53–64. doi: 10.1615/critreveukaryotgeneexpr.v12.i1.30. [DOI] [PubMed] [Google Scholar]
- Eaton S., Bartlett K., Pourfarzam M. Mammalian mitochondrial β-oxidation. Biochem J. 1996;320 ( Pt 2):345–57. doi: 10.1042/bj3200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echchgadda I., Song C. S., Roy A. K., Chatterjee B. Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin D receptor. Mol Pharmacol. 2004;65:720–9. doi: 10.1124/mol.65.3.720. [DOI] [PubMed] [Google Scholar]
- Eichelbaum M., Burk O. CYP3A genetics in drug metabolism. Nat Med. 2001;7:285–7. doi: 10.1038/85417. [DOI] [PubMed] [Google Scholar]
- Eiselt R., Domanski T. L., Zibat A., Mueller R., Presecan-Siedel E., Hustert E., Zanger U. M., Brockmoller J., Klenk H. P., Meyer U. A., Khan K. K., He Y. A., Halpert J. R., Wojnowski L. Identification and functional characterization of eight CYP3A4 protein variants. Pharmacogenetics. 2001;11:447–58. doi: 10.1097/00008571-200107000-00008. [DOI] [PubMed] [Google Scholar]
- Fahey J. W., Talalay P. Antioxidant functions of sulforaphane: a potent inducer of Phase II detoxication enzymes. Food Chem Toxicol. 1999;37:973–9. doi: 10.1016/s0278-6915(99)00082-4. [DOI] [PubMed] [Google Scholar]
- Faucette S. R., Sueyoshi T., Smith C. M., Negishi M., Lecluyse E. L., Wang H. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther. 2006;317:1200–9. doi: 10.1124/jpet.105.098160. [DOI] [PubMed] [Google Scholar]
- Forman B. M., Tzameli I., Choi H. S., Chen J., Simha D., Seol W., Evans R. M., Moore D. D. Androstane metabolites bind to and deactivate the nuclear receptor CAR-β. Nature. 1998;395:612–5. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Forman B. M., Goode E., Chen J., Oro A. E., Bradley D. J., Perlmann T., Noonan D. J., Burka L. T., McMorris T., Lamph W. W., Evans R. M., Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–93. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- Forrester L. M., Neal G. E., Judah D. J., Glancey M. J., Wolf C. R. Evidence for involvement of multiple forms of cytochrome P-450 in aflatoxin B1 metabolism in human liver. Proc Natl Acad Sci U S A. 1990;87:8306–10. doi: 10.1073/pnas.87.21.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C., Makkonen H., Dunlop T. W., Matilainen M., Vaisanen S., Carlberg C. Identification of pregnane X receptor binding sites in the regulatory regions of genes involved in bile acid homeostasis. J Mol Biol. 2005;346:505–19. doi: 10.1016/j.jmb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Fukuen S., Fukuda T., Matsuda H., Sumida A., Yamamoto I., Inaba T., Azuma J. Identification of the novel splicing variants for the hPXR in human livers. Biochem Biophys Res Commun. 2002;298:433–8. doi: 10.1016/s0006-291x(02)02469-5. [DOI] [PubMed] [Google Scholar]
- Gamet-Payrastre L., Li P., Lumeau S., Cassar G., Dupont M. A., Chevolleau S., Gasc N., Tulliez J., Terce F. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res. 2000;60:1426–33. [PubMed] [Google Scholar]
- Gao X., Talalay P. Induction of phase 2 genes by sulforaphane protects retinal pigment epithelial cells against photooxidative damage. Proc Natl Acad Sci U S A. 2004;101:10446–51. doi: 10.1073/pnas.0403886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner-Stephen D., Heydel J. M., Goyal A., Lu Y., Xie W., Lindblom T., Mackenzie P., Radominska-Pandya A. Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab Dispos. 2004;32:340–7. doi: 10.1124/dmd.32.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaster B., Holroyd J. St John's wort for depression: a systematic review. Arch Intern Med. 2000;160:152–6. doi: 10.1001/archinte.160.2.152. [DOI] [PubMed] [Google Scholar]
- Gong H., Singh S. V., Singh S. P., μ Y., Lee J. H., Saini S. P., Toma D., Ren S., Kagan V. E., Day B. W., Zimniak P., Xie W. Orphan nuclear receptor pregnane X receptor sensitizes oxidative stress responses in transgenic mice and cancerous cells. Mol Endocrinol. 2006;20:279–90. doi: 10.1210/me.2005-0205. [DOI] [PubMed] [Google Scholar]
- Gonzalez F. J. Human cytochromes P450: problems and prospects. Trends Pharmacol Sci. 1992;13:346–52. doi: 10.1016/0165-6147(92)90107-h. [DOI] [PubMed] [Google Scholar]
- Goodwin B., Gauthier K. C., Umetani M., Watson M. A., Lochansky M. I., Collins J. L., Leitersdorf E., Mangelsdorf D. J., Kliewer S. A., Repa J. J. Identification of bile acid precursors as endogenous ligands for the nuclear xenobiotic pregnane X receptor. Proc Natl Acad Sci U S A. 2003;100:223–8. doi: 10.1073/pnas.0237082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B., Hodgson E., Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol. 1999;56:1329–39. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- Greten F. R., Eckmann L., Greten T. F., Park J. M., Li Z. W., Egan L. J., Kagnoff M. F., Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Grun F., Blumberg B. Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology. 2006;147:S50–5. doi: 10.1210/en.2005-1129. [DOI] [PubMed] [Google Scholar]
- Grun F., Blumberg B. Perturbed nuclear receptor signaling by environmental obesogens as emerging factors in the obesity crisis. Rev Endocr Metab Disord. 2007;8:161–71. doi: 10.1007/s11154-007-9049-x. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- Guo G. L., Lambert G., Negishi M., Ward J. M., Brewer H. B., Jr., Kliewer S. A., Gonzalez F. J., Sinal C. J. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem. 2003;278:45062–71. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- Gu X., Ke S., Liu D., Sheng T., Thomas P. E., Rabson A. B., Gallo M. A., Xie W., Tian Y. Role of NF-kappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J Biol Chem. 2006;281:17882–9. doi: 10.1074/jbc.M601302200. [DOI] [PubMed] [Google Scholar]
- Ho G. T., Soranzo N., Tate S. K., Drummond H., Nimmo E. R., Tenesa A., Arnott I. D., Satsangi J. Lack of association of the pregnane X receptor (PXR/NR1I2) gene with inflammatory bowel disease: parallel allelic association study and gene wide haplotype analysis. Gut. 2006;55:1676–7. doi: 10.1136/gut.2006.105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P., Zelko I., Sueyoshi T., Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998;18:5652–8. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinpour F., Ellfolk M., Norlin M., Wikvall K. Phenobarbital suppresses vitamin D3 25-hydroxylase expression: a potential new mechanism for drug-induced osteomalacia. Biochem Biophys Res Commun. 2007;357:603–7. doi: 10.1016/j.bbrc.2007.03.177. [DOI] [PubMed] [Google Scholar]
- Hurst C. H., Waxman D. J. Environmental phthalate monoesters activate pregnane X receptor-mediated transcription. Toxicol Appl Pharmacol. 2004;199:266–74. doi: 10.1016/j.taap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Hustert E., Zibat A., Presecan-Siedel E., Eiselt R., Mueller R., Fuss C., Brehm I., Brinkmann U., Eichelbaum M., Wojnowski L., Burk O. Natural protein variants of pregnane X receptor with altered transactivation activity toward CYP3A4. Drug Metab Dispos. 2001;29:1454–9. [PubMed] [Google Scholar]
- Ichikawa T., Horie-Inoue K., Ikeda K., Blumberg B., Inoue S. Steroid and xenobiotic receptor SXR mediates vitamin K2-activated transcription of extracellular matrix-related genes and collagen accumulation in osteoblastic cells. J Biol Chem. 2006;281:16927–34. doi: 10.1074/jbc.M600896200. [DOI] [PubMed] [Google Scholar]
- Igarashi M., Yogiashi Y., Mihara M., Takada I., Kitagawa H., Kato S. Vitamin K induces osteoblast differentiation through pregnane X receptor-mediated transcriptional control of the Msx2 gene. Mol Cell Biol. 2007;27:7947–54. doi: 10.1128/MCB.00813-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jacobs M. N. In silico tools to aid risk assessment of endocrine disrupting chemicals. Toxicology. 2004;205:43–53. doi: 10.1016/j.tox.2004.06.036. [DOI] [PubMed] [Google Scholar]
- Jacobs M. N., Nolan G. T., Hood S. R. Lignans, bacteriocides and organochlorine compounds activate the human pregnane X receptor (PXR) Toxicol Appl Pharmacol. 2005;209:123–33. doi: 10.1016/j.taap.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Jones G., Strugnell S. A., DeLuca H. F. Current understanding of the molecular actions of vitamin D. Physiol Rev. 1998;78:1193–231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- Jones S. A., Moore L. B., Shenk J. L., Wisely G. B., Hamilton G. A., McKee D. D., Tomkinson N. C., LeCluyse E. L., Lambert M. H., Willson T. M., Kliewer S. A., Moore J. T. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- Jung D., Mangelsdorf D. J., Meyer U. A. Pregnane X receptor is a target of farnesoid X receptor. J Biol Chem. 2006;281:19081–91. doi: 10.1074/jbc.M600116200. [DOI] [PubMed] [Google Scholar]
- Kagnoff M. F. Overview and pathogenesis of celiac disease. Gastroenterology. 2005;128:S10–8. doi: 10.1053/j.gastro.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Kamat J. P., Sarma H. D., Devasagayam T. P., Nesaretnam K., Basiron Y. Tocotrienols from palm oil as effective inhibitors of protein oxidation and lipid peroxidation in rat liver microsomes. Mol Cell Biochem. 1997;170:131–7. doi: 10.1023/a:1006853419214. [DOI] [PubMed] [Google Scholar]
- Karaaslan Y., Haznedaroglu S., Ozturk M. Osteomalacia associated with carbamazepine/valproate. Ann Pharmacother. 2000;34:264–5. doi: 10.1345/aph.19099. [DOI] [PubMed] [Google Scholar]
- Karin M., Greten F. R. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- Kast H. R., Goodwin B., Tarr P. T., Jones S. A., Anisfeld A. M., Stoltz C. M., Tontonoz P., Kliewer S., Willson T. M., Edwards P. A. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem. 2002;277:2908–15. doi: 10.1074/jbc.M109326200. [DOI] [PubMed] [Google Scholar]
- Kavlock R. J., Daston G. P., DeRosa C., Fenner-Crisp P., Gray L. E., Kaattari S., Lucier G., Luster M., Mac M. J., Maczka C., Miller R., Moore J., Rolland R., Scott G., Sheehan D. M., Sinks T., Tilson H. A. Research needs for the risk assessment of health and environmental effects of endocrine disruptors: a report of the U.S. EPA-sponsored workshop. Environ Health Perspect. 1996;104 Suppl 4:715–40. doi: 10.1289/ehp.96104s4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawana K., Ikuta T., Kobayashi Y., Gotoh O., Takeda K., Kawajiri K. Molecular mechanism of nuclear translocation of an orphan nuclear receptor, SXR. Mol Pharmacol. 2003;63:524–31. doi: 10.1124/mol.63.3.524. [DOI] [PubMed] [Google Scholar]
- Kelce W. R., Monosson E., Gamcsik M. P., Laws S. C., Gray L. E., Jr. Environmental hormone disruptors: evidence that vinclozolin developmental toxicity is mediated by antiandrogenic metabolites. Toxicol Appl Pharmacol. 1994;126:276–85. doi: 10.1006/taap.1994.1117. [DOI] [PubMed] [Google Scholar]
- Kelce W. R., Stone C. R., Laws S. C., Gray L. E., Kemppainen J. A., Wilson E. M. Persistent DDT metabolite p,p'-DDE is a potent androgen receptor antagonist. Nature. 1995;375:581–5. doi: 10.1038/375581a0. [DOI] [PubMed] [Google Scholar]
- Kester M. H., Bulduk S., Tibboel D., Meinl W., Glatt H., Falany C. N., Coughtrie M. W., Bergman A., Safe S. H., Kuiper G. G., Schuur A. G., Brouwer A., Visser T. J. Potent inhibition of estrogen sulfotransferase by hydroxylated PCB metabolites: a novel pathway explaining the estrogenic activity of PCBs. Endocrinology. 2000;141:1897–900. doi: 10.1210/endo.141.5.7530. [DOI] [PubMed] [Google Scholar]
- Khogali A. M., Chazan B. I., Metcalf V. J., Ramsay J. H. Hyperlipidaemia as a complication of rifampicin treatment. Tubercle. 1974;55:231–3. doi: 10.1016/0041-3879(74)90050-6. [DOI] [PubMed] [Google Scholar]
- King C. R., Xiao M., Yu J., Minton M. R., Addleman N. J., Van Booven D. J., Kwok P. Y., McLeod H. L., Marsh S. Identification of NR1I2 genetic variation using resequencing. Eur J Clin Pharmacol. 2007;63:547–54. doi: 10.1007/s00228-007-0295-3. [DOI] [PubMed] [Google Scholar]
- Kiyosawa N., Tanaka K., Hirao J., Ito K., Niino N., Sakuma K., Kanbori M., Yamoto T., Manabe S., Matsunuma N. Molecular mechanism investigation of phenobarbital-induced serum cholesterol elevation in rat livers by microarray analysis. Arch Toxicol. 2004;78:435–42. doi: 10.1007/s00204-004-0565-0. [DOI] [PubMed] [Google Scholar]
- Kliewer S. A., Moore J. T., Wade L., Staudinger J. L., Watson M. A., Jones S. A., McKee D. D., Oliver B. B., Willson T. M., Zetterstrom R. H., Perlmann T., Lehmann J. M. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kliewer S. A., Goodwin B., Willson T. M. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- Klotz D. M., Beckman B. S., Hill S. M., McLachlan J. A., Walters M. R., Arnold S. F. Identification of environmental chemicals with estrogenic activity using a combination of in vitro assays. Environ Health Perspect. 1996;104:1084–9. doi: 10.1289/ehp.961041084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama S., Koike C., Negishi M., Yamamoto Y. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol. 2004;24:7931–40. doi: 10.1128/MCB.24.18.7931-7940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano S., Kurose K., Ozawa S., Saeki M., Nakajima Y., Hasegawa R., Komamura K., Ueno K., Kamakura S., Nakajima T., Saito H., Kimura H., Goto Y., Saitoh O., Katoh M., Ohnuma T., Kawai M., Sugai K., Ohtsuki T., Suzuki C., Minami N., Saito Y., Sawada J. Eleven novel single nucleotide polymorphisms in the NR1I2 (PXR) gene, four of which induce non-synonymous amino acid alterations. Drug Metab Pharmacokinet. 2002;17:561–5. doi: 10.2133/dmpk.17.561. [DOI] [PubMed] [Google Scholar]
- Koyano S., Kurose K., Saito Y., Ozawa S., Hasegawa R., Komamura K., Ueno K., Kamakura S., Kitakaze M., Nakajima T., Matsumoto K., Akasawa A., Saito H., Sawada J. Functional characterization of four naturally occurring variants of human pregnane X receptor (PXR): one variant causes dramatic loss of both DNA binding activity and the transactivation of the CYP3A4 promoter/enhancer region. Drug Metab Dispos. 2004;32:149–54. doi: 10.1124/dmd.32.1.149. [DOI] [PubMed] [Google Scholar]
- Kuehl P., Zhang J., Lin Y., Lamba J., Assem M., Schuetz J., Watkins P. B., Daly A., Wrighton S. A., Hall S. D., Maurel P., Relling M., Brimer C., Yasuda K., Venkataramanan R., Strom S., Thummel K., Boguski M. S., Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- Kullak-Ublick G. A. ABC transporter regulation by bile acids: where PXR meets FXR. J Hepatol. 2003;39:628–30. doi: 10.1016/s0168-8278(03)00397-0. [DOI] [PubMed] [Google Scholar]
- Kumar V., Singh P. N., Muruganandam A. V., Bhattacharya S. K. Hypericum perforatum: nature's mood stabilizer. Indian J Exp Biol. 2000;38:1077–85. [PubMed] [Google Scholar]
- Kushad M. M., Brown A. F., Kurilich A. C., Juvik J. A., Klein B. P., Wallig M. A., Jeffery E. H. Variation of glucosinolates in vegetable crops of Brassica oleracea. J Agric Food Chem. 1999;47:1541–8. doi: 10.1021/jf980985s. [DOI] [PubMed] [Google Scholar]
- Laakmann G., Schule C., Baghai T., Kieser M. St. John's wort in mild to moderate depression: the relevance of hyperforin for the clinical efficacy. Pharmacopsychiatry. 1998;31 Suppl 1:54–9. doi: 10.1055/s-2007-979346. [DOI] [PubMed] [Google Scholar]
- Lahtela J. T., Arranto A. J., Sotaniemi E. A. Enzyme inducers improve insulin sensitivity in non-insulin-dependent diabetic subjects. Diabetes. 1985;34:911–6. doi: 10.2337/diab.34.9.911. [DOI] [PubMed] [Google Scholar]
- Lamba J., Lamba V., Strom S., Venkataramanan R., Schuetz E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab Dispos. 2008;36:169–81. doi: 10.1124/dmd.107.016600. [DOI] [PubMed] [Google Scholar]
- Lamba V., Yasuda K., Lamba J. K., Assem M., Davila J., Strom S., Schuetz E. G. PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol. 2004;199:251–65. doi: 10.1016/j.taap.2003.12.027. [DOI] [PubMed] [Google Scholar]
- Landes N., Pfluger P., Kluth D., Birringer M., Ruhl R., Bol G. F., Glatt H., Brigelius-Flohe R. Vitamin E activates gene expression via the pregnane X receptor. Biochem Pharmacol. 2003;65:269–73. doi: 10.1016/s0006-2952(02)01520-4. [DOI] [PubMed] [Google Scholar]
- Langmann T., Moehle C., Mauerer R., Scharl M., Liebisch G., Zahn A., Stremmel W., Schmitz G. Loss of detoxification in inflammatory bowel disease: dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127:26–40. doi: 10.1053/j.gastro.2004.04.019. [DOI] [PubMed] [Google Scholar]
- LeCluyse E. L. Pregnane X receptor: molecular basis for species differences in CYP3A induction by xenobiotics. Chem Biol Interact. 2001;134:283–9. doi: 10.1016/s0009-2797(01)00163-6. [DOI] [PubMed] [Google Scholar]
- Lehmann J. M., McKee D. D., Watson M. A., Willson T. M., Moore J. T., Kliewer S. A. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire G., de Sousa G., Rahmani R. A PXR reporter gene assay in a stable cell culture system: CYP3A4 and CYP2B6 induction by pesticides. Biochem Pharmacol. 2004;68:2347–58. doi: 10.1016/j.bcp.2004.07.041. [DOI] [PubMed] [Google Scholar]
- Li T., Chiang J. Y. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 α-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol. 2005;288:G74–84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- Lim Y. P., Liu C. H., Shyu L. J., Huang J. D. Functional characterization of a novel polymorphism of pregnane X receptor, Q158K, in Chinese subjects. Pharmacogenet Genomics. 2005;15:337–41. doi: 10.1097/01213011-200505000-00009. [DOI] [PubMed] [Google Scholar]
- Linde K., Ramirez G., Mulrow C. D., Pauls A., Weidenhammer W., Melchart D. St John's wort for depression--an overview and meta-analysis of randomised clinical trials. Bmj. 1996;313:253–8. doi: 10.1136/bmj.313.7052.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chen W., Chiang J. Y. PXR induces CYP27A1 and regulates cholesterol metabolism in the intestine. J Lipid Res. 2007;48:373–84. doi: 10.1194/jlr.M600282-JLR200. [DOI] [PubMed] [Google Scholar]
- Luo G., Cunningham M., Kim S., Burn T., Lin J., Sinz M., Hamilton G., Rizzo C., Jolley S., Gilbert D., Downey A., Mudra D., Graham R., Carroll K., Xie J., Madan A., Parkinson A., Christ D., Selling B., LeCluyse E., Gan L. S. CYP3A4 induction by drugs: correlation between a pregnane X receptor reporter gene assay and CYP3A4 expression in human hepatocytes. Drug Metab Dispos. 2002;30:795–804. doi: 10.1124/dmd.30.7.795. [DOI] [PubMed] [Google Scholar]
- Lutjohann D., Hahn C., Prange W., Sudhop T., Axelson M., Sauerbruch T., von Bergmann K., Reichel C. Influence of rifampin on serum markers of cholesterol and bile acid synthesis in men. Int J Clin Pharmacol Ther. 2004;42:307–13. doi: 10.5414/cpp42307. [DOI] [PubMed] [Google Scholar]
- Maheo K., Morel F., Langouet S., Kramer H., Le Ferrec E., Ketterer B., Guillouzo A. Inhibition of cytochromes P-450 and induction of glutathione S-transferases by sulforaphane in primary human and rat hepatocytes. Cancer Res. 1997;57:3649–52. [PubMed] [Google Scholar]
- Mani S., Huang H., Sundarababu S., Liu W., Kalpana G., Smith A. B., Horwitz S. B. Activation of the steroid and xenobiotic receptor (human pregnane X receptor) by nontaxane microtubule-stabilizing agents. Clin Cancer Res. 2005;11:6359–69. doi: 10.1158/1078-0432.CCR-05-0252. [DOI] [PubMed] [Google Scholar]
- Martinez A., Marquez A., Mendoza J., Taxonera C., Fernandez-Arquero M., Diaz-Rubio M., de la Concha E. G., Urcelay E. Role of the PXR gene locus in inflammatory bowel diseases. Inflamm Bowel Dis. 2007;13:1484–7. doi: 10.1002/ibd.20252. [DOI] [PubMed] [Google Scholar]
- Masson D., Lagrost L., Athias A., Gambert P., Brimer-Cline C., Lan L., Schuetz J. D., Schuetz E. G., Assem M. Expression of the pregnane X receptor in mice antagonizes the cholic acid-mediated changes in plasma lipoprotein profile. Arterioscler Thromb Vasc Biol. 2005;25:2164–9. doi: 10.1161/01.ATV.0000183674.88817.fb. [DOI] [PubMed] [Google Scholar]
- Masuyama H., Nakatsukasa H., Takamoto N., Hiramatsu Y. Down-regulation of pregnane X receptor contributes to cell growth inhibition and apoptosis by anticancer agents in endometrial cancer cells. Mol Pharmacol. 2007;72:1045–53. doi: 10.1124/mol.107.037937. [DOI] [PubMed] [Google Scholar]
- Masuyama H., Hiramatsu Y., Kunitomi M., Kudo T., MacDonald P. N. Endocrine disrupting chemicals, phthalic acid and nonylphenol, activate Pregnane X receptor-mediated transcription. Mol Endocrinol. 2000;14:421–8. doi: 10.1210/mend.14.3.0424. [DOI] [PubMed] [Google Scholar]
- Masuyama H., Hiramatsu Y., Kodama J., Kudo T. Expression and potential roles of pregnane X receptor in endometrial cancer. J Clin Endocrinol Metab. 2003;88:4446–54. doi: 10.1210/jc.2003-030203. [DOI] [PubMed] [Google Scholar]
- Ma X., Shah Y., Cheung C., Guo G. L., Feigenbaum L., Krausz K. W., Idle J. R., Gonzalez F. J. The PREgnane X receptor gene-humanized mouse: a model for investigating drug-drug interactions mediated by cytochromes P450 3A. Drug Metab Dispos. 2007;35:194–200. doi: 10.1124/dmd.106.012831. [DOI] [PubMed] [Google Scholar]
- McKay L. I., Cidlowski J. A. Molecular control of immune/inflammatory responses: interactions between nuclear factor-κ B and steroid receptor-signaling pathways. Endocr Rev. 1999;20:435–59. doi: 10.1210/edrv.20.4.0375. [DOI] [PubMed] [Google Scholar]
- Mensah-Osman E. J., Thomas D. G., Tabb M. M., Larios J. M., Hughes D. P., Giordano T. J., Lizyness M. L., Rae J. M., Blumberg B., Hollenberg P. F., Baker L. H. Expression levels and activation of a PXR variant are directly related to drug resistance in osteosarcoma cell lines. Cancer. 2007;109:957–65. doi: 10.1002/cncr.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Fang S., Bae Y., Kemper J. K. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J Biol Chem. 2006;281:14537–46. doi: 10.1074/jbc.M510713200. [DOI] [PubMed] [Google Scholar]
- Mikamo E., Harada S., Nishikawa J., Nishihara T. Endocrine disruptors induce cytochrome P450 by affecting transcriptional regulation via pregnane X receptor. Toxicol Appl Pharmacol. 2003;193:66–72. doi: 10.1016/j.taap.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Miki Y., Suzuki T., Kitada K., Yabuki N., Shibuya R., Moriya T., Ishida T., Ohuchi N., Blumberg B., Sasano H. Expression of the steroid and xenobiotic receptor and its possible target gene, organic anion transporting polypeptide-A, in human breast carcinoma. Cancer Res. 2006;66:535–42. doi: 10.1158/0008-5472.CAN-05-1070. [DOI] [PubMed] [Google Scholar]
- Miki Y., Suzuki T., Tazawa C., Blumberg B., Sasano H. Steroid and xenobiotic receptor (SXR), cytochrome P450 3A4 and multidrug resistance gene 1 in human adult and fetal tissues. Mol Cell Endocrinol. 2005;231:75–85. doi: 10.1016/j.mce.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Montminy M., Koo S. H. Diabetes: outfoxing insulin resistance? Nature. 2004;432:958–9. doi: 10.1038/432958a. [DOI] [PubMed] [Google Scholar]
- Moore L. B., Goodwin B., Jones S. A., Wisely G. B., Serabjit-Singh C. J., Willson T. M., Collins J. L., Kliewer S. A. St. John's wort induces hepatic drug metabolism through activation of the pregnane X receptor. Proc Natl Acad Sci U S A. 2000;97:7500–2. doi: 10.1073/pnas.130155097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. T. Regulation of cytochromes P450 during inflammation and infection. Drug Metab Rev. 1997;29:1129–88. doi: 10.3109/03602539709002246. [DOI] [PubMed] [Google Scholar]
- Murashima M., Watanabe S., Zhuo X. G., Uehara M., Kurashige A. Phase 1 study of multiple biomarkers for metabolism and oxidative stress after one-week intake of broccoli sprouts. Biofactors. 2004;22:271–5. doi: 10.1002/biof.5520220154. [DOI] [PubMed] [Google Scholar]
- Murillo G., Mehta R. G. Cruciferous vegetables and cancer prevention. Nutr Cancer. 2001;41:17–28. doi: 10.1080/01635581.2001.9680607. [DOI] [PubMed] [Google Scholar]
- Myzak M. C., Dashwood W. M., Orner G. A., Ho E., Dashwood R. H. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. Faseb J. 2006;20:506–8. doi: 10.1096/fj.05-4785fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka R., Iwasaki T., Rokutanda N., Takeshita A., Koibuchi Y., Horiguchi J., Shimokawa N., Iino Y., Morishita Y., Koibuchi N. Tamoxifen activates CYP3A4 and MDR1 genes through steroid and xenobiotic receptor in breast cancer cells. Endocrine. 2006;30:261–8. doi: 10.1007/s12020-006-0003-6. [DOI] [PubMed] [Google Scholar]
- Nagengast F. M., Grubben M. J., van Munster I. P. Role of bile acids in colorectal carcinogenesis. Eur J Cancer. 1995;31A:1067–70. doi: 10.1016/0959-8049(95)00216-6. [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Russell D. W. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–62. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Koymans L., Kamataki T., Stegeman J. J., Feyereisen R., Waxman D. J., Waterman M. R., Gotoh O., Coon M. J., Estabrook R. W., Gunsalus I. C., Nebert D. W. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee 1999 . A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–3. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Omdahl J, May B. Vitamin D, 2nd Edition. San Diego: Academic Press; 2004. The 25-hydroxyvitamin D 24-hydroxylase; pp. 85–104. [Google Scholar]
- Ourlin J. C., Lasserre F., Pineau T., Fabre J. M., Sa-Cunha A., Maurel P., Vilarem M. J., Pascussi J. M. The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol Endocrinol. 2003;17:1693–703. doi: 10.1210/me.2002-0383. [DOI] [PubMed] [Google Scholar]
- Owen A., Chandler B., Back D. J., Khoo S. H. Expression of pregnane-X-receptor transcript in peripheral blood mononuclear cells and correlation with MDR1 mRNA. Antivir Ther. 2004;9:819–21. [PubMed] [Google Scholar]
- Ozdemir V., Kalow W., Tang B. K., Paterson A. D., Walker S. E., Endrenyi L., Kashuba A. D. Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics. 2000;10:373–88. doi: 10.1097/00008571-200007000-00001. [DOI] [PubMed] [Google Scholar]
- Pack A. M., Morrell M. J. Epilepsy and bone health in adults. Epilepsy Behav. 2004;5 Suppl 2:S24–9. doi: 10.1016/j.yebeh.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Paolini M., Cantelli-Forti G., Perocco P., Pedulli G. F., Abdel-Rahman S. Z., Legator M. S. Co-carcinogenic effect of β-carotene. Nature. 1999;398:760–1. doi: 10.1038/19655. [DOI] [PubMed] [Google Scholar]
- Parker R. A., Pearce B. C., Clark R. W., Gordon D. A., Wright J. J. Tocotrienols regulate cholesterol production in mammalian cells by post-transcriptional suppression of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 1993;268:11230–8. [PubMed] [Google Scholar]
- Pascual G., Fong A. L., Ogawa S., Gamliel A., Li A. C., Perissi V., Rose D. W., Willson T. M., Rosenfeld M. G., Glass C. K. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature. 2005;437:759–63. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi J. M., Robert A., Nguyen M., Walrant-Debray O., Garabedian M., Martin P., Pineau T., Saric J., Navarro F., Maurel P., Vilarem M. J. Possible involvement of pregnane X receptor-enhanced CYP24 expression in drug-induced osteomalacia. J Clin Invest. 2005;115:177–86. doi: 10.1172/JCI21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunescu E. In vivo and in vitro suppression of humoral and cellular immunological response by rifampicin. Nature. 1970;228:1188–90. doi: 10.1038/2281188a0. [DOI] [PubMed] [Google Scholar]
- Penson R. T., Oliva E., Skates S. J., Glyptis T., Fuller A. F., Jr., Goodman A., Seiden M. V. Expression of multidrug resistance-1 protein inversely correlates with paclitaxel response and survival in ovarian cancer patients: a study in serial samples. Gynecol Oncol. 2004;93:98–106. doi: 10.1016/j.ygyno.2003.11.053. [DOI] [PubMed] [Google Scholar]
- Pikarsky E., Porat R. M., Stein I., Abramovitch R., Amit S., Kasem S., Gutkovich-Pyest E., Urieli-Shoval S., Galun E., Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Ricketts M. L., Boekschoten M. V., Kreeft A. J., Hooiveld G. J., Moen C. J., Muller M., Frants R. R., Kasanmoentalib S., Post S. M., Princen H. M., Porter J. G., Katan M. B., Hofker M. H., Moore D. D. The cholesterol-raising factor from coffee beans, cafestol, as an agonist ligand for the farnesoid and pregnane X receptors. Mol Endocrinol. 2007;21:1603–16. doi: 10.1210/me.2007-0133. [DOI] [PubMed] [Google Scholar]
- Roth A., Looser R., Kaufmann M., Blattler S. M., Rencurel F., Huang W., Moore D. D., Meyer U. A. Regulatory cross-talk between drug metabolism and lipid homeostasis: constitutive androstane receptor and pregnane X receptor increase Insig-1 expression. Mol Pharmacol. 2008;73:1282–9. doi: 10.1124/mol.107.041012. [DOI] [PubMed] [Google Scholar]
- Russell D. W. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–74. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Saini S. P., Sonoda J., Xu L., Toma D., Uppal H., μ Y., Ren S., Moore D. D., Evans R. M., Xie W. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol. 2004;65:292–300. doi: 10.1124/mol.65.2.292. [DOI] [PubMed] [Google Scholar]
- Saito H., Kiyose C., Yoshimura H., Ueda T., Kondo K., Igarashi O. γ-tocotrienol, a vitamin E homolog, is a natriuretic hormone precursor. J Lipid Res. 2003;44:1530–5. doi: 10.1194/jlr.M300061-JLR200. [DOI] [PubMed] [Google Scholar]
- Sandau C. D., Ayotte P., Dewailly E., Duffe J., Norstrom R. J. Analysis of hydroxylated metabolites of PCBs (OH-PCBs) and other chlorinated phenolic compounds in whole blood from Canadian inuit. Environ Health Perspect. 2000;108:611–6. doi: 10.1289/ehp.00108611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saradhi M., Sengupta A., Mukhopadhyay G., Tyagi R. K. Pregnane and Xenobiotic Receptor (PXR/SXR) resides predominantly in the nuclear compartment of the interphase cell and associates with the condensed chromosomes during mitosis. Biochim Biophys Acta. 2005;1746:85–94. doi: 10.1016/j.bbamcr.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Scheinfeld N. Phenytoin in cutaneous medicine: its uses, mechanisms and side effects. Dermatol Online J. 2003;9:6. [PubMed] [Google Scholar]
- Sen C. K., Khanna S., Roy S., Packer L. Molecular basis of vitamin E action. Tocotrienol potently inhibits glutamate-induced pp60(c-Src) kinase activation and death of HT4 neuronal cells. J Biol Chem. 2000;275:13049–55. doi: 10.1074/jbc.275.17.13049. [DOI] [PubMed] [Google Scholar]
- Shafran S. D., Mashinter L. D., Roberts S. E. The effect of low-dose ritonavir monotherapy on fasting serum lipid concentrations. HIV Med. 2005;6:421–5. doi: 10.1111/j.1468-1293.2005.00328.x. [DOI] [PubMed] [Google Scholar]
- Shah Y. M., Ma X., Morimura K., Kim I., Gonzalez F. J. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1114–22. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- Shah S. C., Sharma R. K., Chitle A. R. Rifampicin induced osteomalacia. Tubercle. 1981;62:207–9. doi: 10.1016/0041-3879(81)90008-8. [DOI] [PubMed] [Google Scholar]
- Shimada H., Furuno H., Hirai K., Koyama J., Ariyama J., Simamura E. Paraquat detoxicative system in the mouse liver postmitochondrial fraction. Arch Biochem Biophys. 2002;402:149–57. doi: 10.1016/S0003-9861(02)00059-0. [DOI] [PubMed] [Google Scholar]
- Siest G., Jeannesson E., Marteau J. B., Samara A., Marie B., Pfister M., Visvikis-Siest S. Transcription factor and drug-metabolizing enzyme gene expression in lymphocytes from healthy human subjects. Drug Metab Dispos. 2008;36:182–9. doi: 10.1124/dmd.107.017228. [DOI] [PubMed] [Google Scholar]
- Sohoni P., Sumpter J. P. Several environmental oestrogens are also anti-androgens. J Endocrinol. 1998;158:327–39. doi: 10.1677/joe.0.1580327. [DOI] [PubMed] [Google Scholar]
- Sonoda J., Chong L. W., Downes M., Barish G. D., Coulter S., Liddle C., Lee C. H., Evans R. M. Pregnane X receptor prevents hepatorenal toxicity from cholesterol metabolites. Proc Natl Acad Sci U S A. 2005;102:2198–203. doi: 10.1073/pnas.0409481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell T. C., Forbes I. J. Depression of immune competence by phenytoin and carbamazepine. Studies in vivo and in vitro. Clin Exp Immunol. 1975;20:273–85. [PMC free article] [PubMed] [Google Scholar]
- Sorrell T. C., Forbes I. J., Burness F. R., Rischbieth R. H. Depression of immunological function in patients treated with phenytoin sodium (sodium diphenylhydantoin) Lancet. 1971;2:1233–5. doi: 10.1016/s0140-6736(71)90547-2. [DOI] [PubMed] [Google Scholar]
- Soto A. M., Chung K. L., Sonnenschein C. The pesticides endosulfan, toxaphene, and dieldrin have estrogenic effects on human estrogen-sensitive cells. Environ Health Perspect. 1994;102:380–3. doi: 10.1289/ehp.94102380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparreboom A., van Asperen J., Mayer U., Schinkel A. H., Smit J. W., Meijer D. K., Borst P., Nooijen W. J., Beijnen J. H., van Tellingen O. Limited oral bioavailability and active epithelial excretion of paclitaxel (Taxol) caused by P-glycoprotein in the intestine. Proc Natl Acad Sci U S A. 1997;94:2031–5. doi: 10.1073/pnas.94.5.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearow J. L., Doemeny P., Sera R., Leffler R., Barkley M. Genetic variation in susceptibility to endocrine disruption by estrogen in mice. Science. 1999;285:1259–61. doi: 10.1126/science.285.5431.1259. [DOI] [PubMed] [Google Scholar]
- Squires E. J., Sueyoshi T., Negishi M. Cytoplasmic localization of pregnane X receptor and ligand-dependent nuclear translocation in mouse liver. J Biol Chem. 2004;279:49307–14. doi: 10.1074/jbc.M407281200. [DOI] [PubMed] [Google Scholar]
- Staudinger J. L., Ding X., Lichti K. Pregnane X receptor and natural products: beyond drug-drug interactions. Expert Opin Drug Metab Toxicol. 2006;2:847–57. doi: 10.1517/17425255.2.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., MacKenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., Willson T. M., Koller B. H., Kliewer S. A. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–74. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman C., Robertson G., Coulter S., Liddle C. Feed-forward regulation of bile acid detoxification by CYP3A4: studies in humanized transgenic mice. J Biol Chem. 2004;279:11336–43. doi: 10.1074/jbc.M310258200. [DOI] [PubMed] [Google Scholar]
- Suido H., Tanaka T., Tabei T., Takeuchi A., Okita M., Kishimoto T., Kasayama S., Higashino K. A mixed green vegetable and fruit beverage decreased the serum level of low-density lipoprotein cholesterol in hypercholesterolemic patients. J Agric Food Chem. 2002;50:3346–50. doi: 10.1021/jf0116698. [DOI] [PubMed] [Google Scholar]
- Synold T. W., Dussault I., Forman B. M. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7:584–90. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- Tabb M. M., Kholodovych V., Grun F., Zhou C., Welsh W. J., Blumberg B. Highly chlorinated PCBs inhibit the human xenobiotic response mediated by the steroid and xenobiotic receptor (SXR) Environ Health Perspect. 2004;112:163–9. doi: 10.1289/ehp.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb M. M., Blumberg B. New modes of action for endocrine-disrupting chemicals. Mol Endocrinol. 2006;20:475–82. doi: 10.1210/me.2004-0513. [DOI] [PubMed] [Google Scholar]
- Tabb M. M., Sun A., Zhou C., Grun F., Errandi J., Romero K., Pham H., Inoue S., Mallick S., Lin M., Forman B. M., Blumberg B. Vitamin K2 regulation of bone homeostasis is mediated by the steroid and xenobiotic receptor SXR. J Biol Chem. 2003;278:43919–27. doi: 10.1074/jbc.M303136200. [DOI] [PubMed] [Google Scholar]
- Takai M., Suido H., Tanaka T., Kotani M., Fujita A., Takeuchi A., Makino T., Sumikawa K., Origasa H., Tsuji K., Nakashima M. [LDL-cholesterol-lowering effect of a mixed green vegetable and fruit beverage containing broccoli and cabbage in hypercholesterolemic subjects] Rinsho Byori. 2003;51:1073–83. [PubMed] [Google Scholar]
- Takeshita A., Koibuchi N., Oka J., Taguchi M., Shishiba Y., Ozawa Y. Bisphenol-A, an environmental estrogen, activates the human orphan nuclear receptor, steroid and xenobiotic receptor-mediated transcription. Eur J Endocrinol. 2001;145:513–7. doi: 10.1530/eje.0.1450513. [DOI] [PubMed] [Google Scholar]
- Talalay P., Fahey J. W., Holtzclaw W. D., Prestera T., Zhang Y. Chemoprotection against cancer by phase 2 enzyme induction. Toxicol Lett. 1995;82-83:173–9. doi: 10.1016/0378-4274(95)03553-2. [DOI] [PubMed] [Google Scholar]
- Uno Y., Sakamoto Y., Yoshida K., Hasegawa T., Hasegawa Y., Koshino T., Inoue I. Characterization of six base pair deletion in the putative HNF1-binding site of human PXR promoter. J Hum Genet. 2003;48:594–7. doi: 10.1007/s10038-003-0076-5. [DOI] [PubMed] [Google Scholar]
- Verma S., Tabb M. M., Blumberg B. Activation of the steroid and xenobiotic receptor, SXR, induces apoptosis in breast cancer cells. BMC Cancer. 2009;9:in press. doi: 10.1186/1471-2407-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahab P. J., Meijer J. W., Goerres M. S., Mulder C. J. Coeliac disease: changing views on gluten-sensitive enteropathy. Scand J Gastroenterol Suppl. 2002:60–5. doi: 10.1080/003655202320621472. [DOI] [PubMed] [Google Scholar]
- Wandel C., Witte J. S., Hall J. M., Stein C. M., Wood A. J., Wilkinson G. R. CYP3A activity in African American and European American men: population differences and functional effect of the CYP3A4*1B5'-promoter region polymorphism. Clin Pharmacol Ther. 2000;68:82–91. doi: 10.1067/mcp.2000.108506. [DOI] [PubMed] [Google Scholar]
- Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–53. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- Watkins R. E., Maglich J. M., Moore L. B., Wisely G. B., Noble S. M., Davis-Searles P. R., Lambert M. H., Kliewer S. A., Redinbo M. R. 2.1 A crystal structure of human PXR in complex with the St. John's wort compound hyperforin. Biochemistry. 2003b;42:1430–8. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- Watkins R. E., Davis-Searles P. R., Lambert M. H., Redinbo M. R. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J Mol Biol. 2003a;331:815–28. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- Watkins R. E., Wisely G. B., Moore L. B., Collins J. L., Lambert M. H., Williams S. P., Willson T. M., Kliewer S. A., Redinbo M. R. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–33. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- Waxman D. J. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
- Wei P., Zhang J., Egan-Hafley M., Liang S., Moore D. D. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–3. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Wentworth J. M., Agostini M., Love J., Schwabe J. W., Chatterjee V. K. St John's wort, a herbal antidepressant, activates the steroid X receptor. J Endocrinol. 2000;166:R11–6. doi: 10.1677/joe.0.166r011. [DOI] [PubMed] [Google Scholar]
- White R., Jobling S., Hoare S. A., Sumpter J. P., Parker M. G. Environmentally persistent alkylphenolic compounds are estrogenic. Endocrinology. 1994;135:175–82. doi: 10.1210/endo.135.1.8013351. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Cosme J., Vinkovic D. M., Ward A., Angove H. C., Day P. J., Vonrhein C., Tickle I. J., Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–6. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- Wolfrum C., Asilmaz E., Luca E., Friedman J. M., Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027–32. doi: 10.1038/nature03047. [DOI] [PubMed] [Google Scholar]
- Wrighton S. A., Schuetz E. G., Thummel K. E., Shen D. D., Korzekwa K. R., Watkins P. B. The human CYP3A subfamily: practical considerations. Drug Metab Rev. 2000;32:339–61. doi: 10.1081/dmr-100102338. [DOI] [PubMed] [Google Scholar]
- Xie W., Radominska-Pandya A., Shi Y., Simon C. M., Nelson M. C., Ong E. S., Waxman D. J., Evans R. M. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A. 2001b;98:3375–80. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Barwick J. L., Downes M., Blumberg B., Simon C. M., Nelson M. C., Neuschwander-Tetri B. A., Brunt E. M., Guzelian P. S., Evans R. M. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000a;406:435–9. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- Xie W., Evans R. M. Orphan nuclear receptors: the exotics of xenobiotics. J Biol Chem. 2001a;276:37739–42. doi: 10.1074/jbc.R100033200. [DOI] [PubMed] [Google Scholar]
- Xie W., Barwick J. L., Simon C. M., Pierce A. M., Safe S., Blumberg B., Guzelian P. S., Evans R. M. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 2000b;14:3014–23. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Tian Y. Xenobiotic receptor meets NF-kappaB, a collision in the small bowel. Cell Metab. 2006;4:177–8. doi: 10.1016/j.cmet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Xue Y., Moore L. B., Orans J., Peng L., Bencharit S., Kliewer S. A., Redinbo M. R. Crystal structure of the pregnane X receptor-estradiol complex provides insights into endobiotic recognition. Mol Endocrinol. 2007b;21:1028–38. doi: 10.1210/me.2006-0323. [DOI] [PubMed] [Google Scholar]
- Xue Y., Chao E., Zuercher W. J., Willson T. M., Collins J. L., Redinbo M. R. Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg Med Chem. 2007a;15:2156–66. doi: 10.1016/j.bmc.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Hashizume T., Shuhart M. C., Davis C. L., Nelson W. L., Sakaki T., Kalhorn T. F., Watkins P. B., Schuetz E. G., Thummel K. E. Intestinal and hepatic CYP3A4 catalyze hydroxylation of 1alpha,25-dihydroxyvitamin D(3): implications for drug-induced osteomalacia. Mol Pharmacol. 2006;69:56–65. doi: 10.1124/mol.105.017392. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Talalay P., Cho C. G., Posner G. H. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci U S A. 1992;89:2399–403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kensler T. W., Cho C. G., Posner G. H., Talalay P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc Natl Acad Sci U S A. 1994;91:3147–50. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kuehl P., Green E. D., Touchman J. W., Watkins P. B., Daly A., Hall S. D., Maurel P., Relling M., Brimer C., Yasuda K., Wrighton S. A., Hancock M., Kim R. B., Strom S., Thummel K., Russell C. G., Hudson J. R., Jr., Schuetz E. G., Boguski M. S. The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics. 2001;11:555–72. doi: 10.1097/00008571-200110000-00003. [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhai Y., μ Y., Gong H., Uppal H., Toma D., Ren S., Evans R. M., Xie W. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem. 2006a;281:15013–20. doi: 10.1074/jbc.M511116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Tabb M. M., Nelson E. L., Grun F., Verma S., Sadatrafiei A., Lin M., Mallick S., Forman B. M., Thummel K. E., Blumberg B. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest. 2006c;116:2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Assem M., Tay J. C., Watkins P. B., Blumberg B., Schuetz E. G., Thummel K. E. Steroid and xenobiotic receptor and vitamin D receptor crosstalk mediates CYP24 expression and drug-induced osteomalacia. J Clin Invest. 2006b;116:1703–12. doi: 10.1172/JCI27793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Poulton E. J., Grun F., Bammler T. K., Blumberg B., Thummel K. E., Eaton D. L. The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol Pharmacol. 2007;71:220–9. doi: 10.1124/mol.106.029264. [DOI] [PubMed] [Google Scholar]
- Zhou C., Tabb M. M., Sadatrafiei A., Grun F., Blumberg B. Tocotrienols activate the steroid and xenobiotic receptor, SXR, and selectively regulate expression of its target genes. Drug Metab Dispos. 2004;32:1075–82. doi: 10.1124/dmd.104.000299. [DOI] [PubMed] [Google Scholar]