Abstract
DNA replication across blocking lesions occurs by translesion DNA synthesis (TLS), involving a multitude of mutagenic DNA polymerases that operate to protect the mammalian genome. Using a quantitative TLS assay, we identified three main classes of TLS in human cells: two rapid and error-free, and the third slow and error-prone. A single gene, REV3L, encoding the catalytic subunit of DNA polymerase ζ (polζ), was found to have a pivotal role in TLS, being involved in TLS across all lesions examined, except for a TT cyclobutane dimer. Genetic epistasis siRNA analysis indicated that discrete two-polymerase combinations with polζ dictate error-prone or error-free TLS across the same lesion. These results highlight the central role of polζ in both error-prone and error-free TLS in mammalian cells, and show that bypass of a single lesion may involve at least three different DNA polymerases, operating in different two-polymerase combinations.
Keywords: carcinogenesis, DNA damage, DNA repair, lesion bypass, mutagenesis
Introduction
Despite the existence of efficient DNA repair mechanisms, DNA replication encounters unrepaired lesions, which may lead to arrested progression of replication forks and the formation of replication gaps (Rupp and Howard-Flanders, 1968; Lehmann, 1972; Meneghini, 1976; Friedberg et al, 2006; Lehmann and Fuchs, 2006; Lopes et al, 2006; Mojas et al, 2007). Translesion DNA synthesis (TLS) is a ubiquitous error-prone response to arrested replication forks and gaps, which is conserved from Escherichia coli to humans. TLS is mediated by specialized DNA polymerases that are characterized by low fidelity and an ability to replicate across certain types of damaged sites in template DNA. Such replication can be accurate with respect to the nucleotide composition of the damaged template, or inaccurate, leading to the generation of mutations (Livneh, 2001; Goodman, 2002; Prakash and Prakash, 2002; Lehmann et al, 2007).
TLS in E. coli involves a single major DNA polymerase, DNA polymerase V, (Reuven et al, 1999; Tang et al, 1999), functioning in error-prone TLS across a diversity of DNA lesions (Reuven et al, 1999; Tang et al, 1999, 2000; Maor-Shoshani et al, 2003). E. coli also contains DNA polymerases II and IV, which bypass lesions under specific situations (Napolitano et al, 2000; Jarosz et al, 2006). In Saccharomyces cerevisiae, two genetically distinct TLS pathways have been defined by the DNA polymerases involved. A polη (RAD30)-dependent pathway performs TLS across a TT CPD (thymine-thymine cyclobutane pyrimidine dimer) with high fidelity (Johnson et al, 1999b), and a polζ (REV3 and REV7)-dependent pathway functions in error-prone TLS across UV-induced lesions (Nelson et al, 1996a, 1996b; Lawrence, 2002; Gibbs et al, 2005), and other damages (Xie et al, 2003; Zhao et al, 2006). A key regulatory element in TLS is PCNA, the monoubiquitination of which signals the switch from high-fidelity replication to TLS (Hoege et al, 2002; Stelter and Ulrich, 2003).
The situation in mammalian cells is more complicated, due to the presence of many more low-fidelity DNA polymerases than in E. coli or S. cerevisiae: four specialized Y-family DNA polymerases (compared with two in E. coli and S. cerevisiae), and six additional low-fidelity polymerases that belong to other DNA polymerase families (Hubscher et al, 2000; Goodman, 2002). As in yeast, the monoubiquitination of PCNA appears to have an important regulatory function (Kannouche et al, 2004; Watanabe et al, 2004; Bienko et al, 2005; Bi et al, 2006). Moreover, both monoubiquitination of PCNA, and the efficiency and accuracy of TLS are regulated by the p53-inducible protein p21 (Avkin et al, 2006; Soria et al, 2006) through its interaction with PCNA (Avkin et al, 2006). However, many mechanistic aspects of TLS are still unknown. In the present study, we used kinetics and genetic analysis of TLS based on gene knockout or knockdown, in conjunction with gapped plasmids carrying single site-specific lesions to decipher some of the operation principles of TLS in mammalian cells. Our results indicate that there exist at least three major pathways of TLS in mammalian cells, and that a single polymerase, DNA polymerase ζ (polζ), has a pivotal and irreplaceable role in TLS across a wide variety of DNA lesions in mammalian cells. It functions in both error-free and error-prone TLS, and cooperates with other DNA polymerases in two-polymerase mechanisms. These results highlight the crucial function polζ has in TLS, and provide the first evidence for a two-polymerase mechanism in living mammalian cells.
Results
TLS exhibits distinct classes of kinetics and accuracy in human cells
In this study, we utilized a quantitative TLS assay system based on gapped plasmids carrying site-specific DNA lesions in the single-stranded region opposite to the gap (gap-lesion plasmid) (Avkin et al, 2002, 2004; Adar and Livneh, 2006) (Supplementary Figure 1s). Briefly, cultured cells were transfected with a plasmid mixture comprising a gap-lesion plasmid with a site-specific lesion opposite a gap and a kanR marker, a control gapped plasmid without a lesion (cmR), and an intact carrier plasmid (ampR). After allowing time for TLS-dependent gap filling, plasmids were extracted from the cells using alkaline conditions, such that only fully repaired plasmids remain covalently closed, and those were used to transform an E. coli recA indicator strain, which is defective in TLS. The bacterial cells were plated in parallel on kan-LB plates to select for repaired gap-lesion plasmids, and on cm-LB plates to select for the control plasmid. The extent of TLS was determined from the ratio of kanR/cmR E. coli colonies. To determine the fidelity of TLS, plasmids were extracted from individual kanR colonies and subjected to DNA sequence analysis across the region corresponding to the gap. We demonstrated earlier that this assay system reflects TLS events occurring in the mammalian cell (Avkin et al, 2002), is responsive to the cellular composition of DNA polymerases in a way similar to chromosomal TLS (Avkin et al, 2004; Hendel et al, 2008), and subject to regulation by p53 and p21 through PCNA ubiquitination (Avkin et al, 2006). The plasmids become chromatinized in the cells (data not shown), and may better reflect TLS events occurring at post-replication gaps, rather than replication forks (e.g. behind the fork; Niimi et al, 2008).
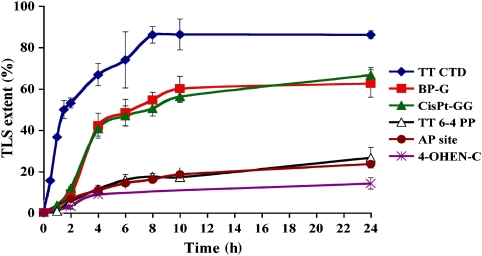
Using the gapped plasmid assay system, we measured the time course of TLS across six diverse, site-specific lesions, to obtain a general view on the kinetics of TLS. The lesions included benzo[a]pyrene-guanine (BP-G; (+)–trans-BPDE-N2-G adduct), a major tobacco smoke-induced DNA lesion; cisplatin-GG (cisPt-GG), an intra-strand adduct formed in DNA by the cancer chemotherapy drug cisplatin; a furanyl AP (apurinic/apyrimidinic (abasic)) site, which is a synthetic analogue of the native AP site generated in DNA by spontaneous hydrolysis; 4-hydroxyequilenin-C (4-OHEN-C), an adduct formed in DNA by metabolites of equilin and equilenin, which are widely used in oestrogen replacement therapy; and TT CPD and thymine-thymine 6-4 photoproduct (TT 6-4 PP), the two main UV-induced DNA lesions. As can be seen in Figure 1 (and Supplementary Tables 1s–6s), the six time course curves fell into three kinetically distinct groups. TLS across the TT CPD was very rapid, and highly efficient, reaching nearly 40% bypass after 1 h, and over 85% after 8 h (Figure 1; Supplementary Table 1s). TLS across the BP-G and cisPt-GG adducts exhibited an initial lag of 1–2 h, after which it proceeded at a fast pace, reaching ∼40% bypass after two additional hours, and ∼60% at 8 h after the lag (Figure 1; Supplementary Tables 2s and 3s). TLS across TT 6-4 PP, the AP site and 4-OHEN-C was slow and relatively inefficient, reaching 14–27% bypass after 24 h (Figure 1; Supplementary Tables 4s–6s). These results are quite remarkable, given that the lesions that share similar kinetics are chemically very different, and have no obvious structural, conformational, or chemical commonalities.
Figure 1.
Kinetics of TLS across six different types of DNA damage in human U2OS cells. Plasmid mixtures containing the indicated gap-lesion plasmid, along with the control and the carrier plasmids, were introduced into human U2OS cells. Following incubation of 0–24 h to allow TLS, the DNA was extracted and used to transform an E. coli indicator strain. The extent of TLS was calculated as described under Materials and methods. Each of the data represents the average of four TLS experiments. The detailed data are presented in Supplementary Tables 1s–6s.
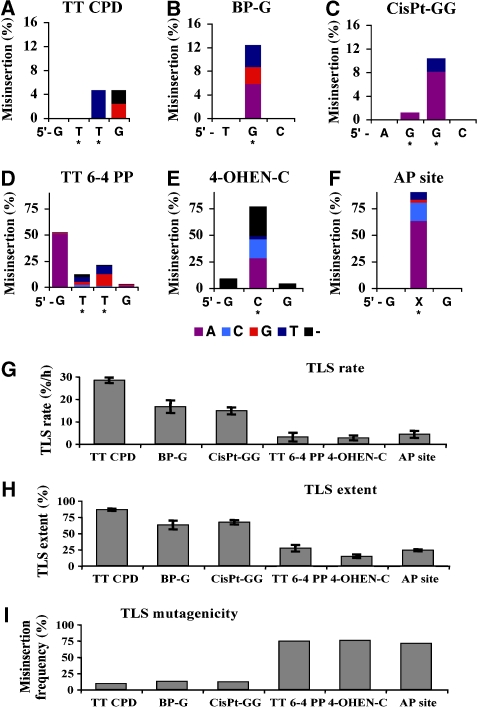
The fidelity of TLS across each lesion was determined using DNA sequence analysis. As can be seen in Figure 2A–F (and Supplementary Tables 7s–12s), each lesion exhibited a unique mutational signature, consistent with previous results (Friedberg et al, 2006), except for 4-OHEN-C, for which no previous mammalian in vivo mutational data existed. TT CPD, BP-G, and cisPt-GG were bypassed accurately in approximately 90% of the TLS events (error frequency of approximately 10%; Figure 2A–C; Supplementary Tables 7s–9s). In contrast, TLS across TT 6-4 PP and 4-OHEN-C was highly mutagenic with error frequencies in the range of 71–75% (Figure 2D and E; Supplementary Tables 10s and 11s). Most of the mutations occurred opposite the damaged template bases; however, for TT CPD, TT 6-4 PP, and 4-OHEN-C, a significant fraction of the mutations were semitargeted to the two nearest bases flanking the lesion (Figure 2A–F; Supplementary Tables 7s–12s). The mutagenicity of TLS across an AP site cannot be defined in the absence of knowledge of the missing base. However, as spontaneous depurination is much faster than depyrimidination, most AP sites generated are at purines. Therefore, any insertion of a purine nucleotide opposite the AP site can be considered functionally mutagenic, providing a minimal estimate of its mutagenicity during TLS. Interestingly, the fidelity of bypass correlated with its speed and efficiency: The rapid and efficient TLS across TT-CPD, BP-G, and cisPt-GG was accurate, whereas the slow and inefficient TLS across TT 6-4 PP, 4-OHEN-C and an AP site was highly mutagenic (Figure 2G–I).
Figure 2.
Mutagenicity of TLS across six different types of DNA damage in human U2OS cells. Individual colonies from the TLS reactions presented in Figure 1 were picked, and their plasmid content was analysed for mutations in the DNA region corresponding to the original site of the lesion. The cumulative height of each column represents the misinsertion frequency opposite the corresponding lesion, whereas the coloured column sections represent specific mutational events, colour coded as shown underneath. The DNA sequence with the damaged bases (marked by stars) is shown in the 5′ → 3′ direction. The detailed DNA sequence data are presented in Supplementary Tables 7s–12s. Results are presented for mutations formed at TT CPD (A), BP-G (B), cisPt-GG (C), TT 6-4 PP (D), 4-OHEN-C (E), and AP site (F). (G) TLS rates, calculated for the linear TLS phase of each lesion from the data presented in Figure 1, namely 0–2 h for TT CPD, TT 6-4 PP, 4-OHEN-C and the AP site, and 2–4 h for BP-G and cisP-GG, are shown. (H) Maximal values of TLS for the six lesions measured at 24 h are shown. The data were taken from Figure 1 and Supplementary Tables 1s–6s. (I) Mutagenic TLS out of total TLS events. For the AP site, insertion of A or G was taken as a mutagenic event. The data were taken from Figures 1 and 2, and Supplementary Tables 1s–12s.
Polζ is involved in both error-free and error-prone TLS
If the kinetically distinct branches of TLS represent distinct pathways, they should exhibit characteristic genetic requirements, particularly of TLS DNA polymerase genes. For the three lesions that were bypassed by rapid and accurate TLS, there is evidence for the involvement of specific DNA polymerases: polη for TLS across TT CPD and cisPt-GG, and polκ for TLS across BP-G. This is based on in vivo studies (Maher et al, 1976; Ogi et al, 2002; Bassett et al, 2004), in vitro results with purified DNA polymerases (Johnson et al, 1999a; Masutani et al, 1999, 2000; Ohashi et al, 2000; Vaisman et al, 2000), as well as studies from our lab using the gap-lesion plasmid assay utilized here (Avkin et al, 2004; Hendel et al, 2008) (and see below). As for the lesions in the slow and mutagenic branch of TLS, based on studies in S. cerevisiae (Haracska et al, 2001; Gibbs et al, 2005) and human cells (Gibbs et al, 1998) we suspected that their bypass involves polζ. We therefore analysed TLS across these three lesions in Rev3L+/+ and Rev3L−/− (the mouse homologue of the yeast REV3 gene, encoding the catalytic subunit of DNA polymerase ζ) mouse embryonic fibroblasts, using the gap-lesion plasmid assay system. As controls, we examined TLS across the three lesions bypassed by the rapid and accurate branches of TLS. In addition, we examined TLS across an artificial lesion in the form of a chain of 12 methylenes inserted into the DNA backbone (M12; Adar and Livneh, 2006).
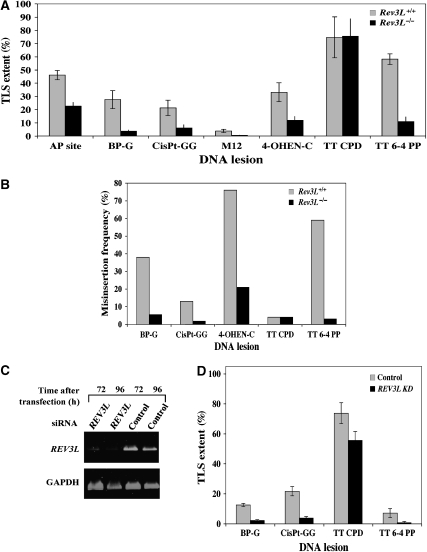
As can be seen in Figure 3A and Supplementary Table 13s, for all lesions except the TT CPD, TLS was significantly reduced in Rev3L−/− cells, compared with Rev3L+/+ cells. This reduction was 7.3-fold for the BP-G adduct, 3.5-fold for the cisPt-GG adduct, 2-fold for the AP site, 2.8-fold for the 4-OHEN-C lesion, 5.3-fold for the TT 6-4 PP, and 7.8-fold for the M12 lesion (Figure 3A; Supplementary Table 13s). Thus, polζ appears to be involved not only in TLS across lesions classified as being bypassed via the slow and mutagenic pathway but also across lesions bypassed by rapid and accurate TLS, except for TT CPD.
Figure 3.
Involvement of Rev3L in TLS in mammalian cells. (A) The TLS assay was performed with Rev3L+/+ or Rev3L−/− MEFs. Cells were assayed for TLS as described under Materials and methods, using the indicated gap-lesion plasmids. Average results of at least four experiments are presented. The detailed data are presented in Supplementary Table 13s. (B) Mutagenicity of the TLS reaction across different lesions in MEF Rev3L+/+ and Rev3L−/− cells. Colonies obtained in the experiments described in (A) were picked, their plasmid contents extracted, and subjected to DNA sequence analysis. The graph shows the percentage of incorrect nucleotides inserted opposite each of the lesions. The statistical significance of the differences in mutagenicity between the two cell types was calculated by the χ2 test, yielding the following P-values: BP-G, 0.0006; cisPt-GG, 0.006; M12, <0.0001; 4-OHEN-C, 0.0002; TT 6-4 PP, <0.0001. The detailed data are presented in Supplementary Tables 14s–18s. (C) RT–PCR of RNA extracted from human U2OS cells transiently transfected with siRNA for REV3L. (D) Extent of TLS in U2OS cells in which REV3L expression was knocked down using specific siRNA. Cells were assayed for TLS as described under Materials and methods, using the indicated gap-lesion plasmids. Average results of at least four experiments are presented. The detailed data are presented in Supplementary Table 21s.
The extent of TLS across several lesions in the mouse cells (Figure 3) was quantitatively different from that obtained in the human cells (Figure 1). This may stem from the difference in organisms and cell types, and the fact that the mice cells were p53 null, a situation under which TLS is deregulated (Avkin et al, 2006). Therefore, to examine whether REV3L is similarly involved in TLS in the human cell line U2OS used in the kinetics analysis (which has a wild-type p53), we analysed TLS across several lesions in U2OS cells in which the expression of REV3L was knocked down using siRNA. As can be seen in Figure 3C, transfection of U2OS cells with REV3L siRNA caused a strong reduction in REV3L mRNA expression, whereas the control siRNA had essentially no effect. We then assayed TLS across four different lesions. As can be seen in Figure 3D and Supplementary Table 21s, when REV3L was knocked down, TLS across TT 6-4 PP, cisPt-GG, and BP-G was decreased compared with the control siRNA. In contrast, TLS across the TT CPD was similar. Thus, similar to the MEF, polζ is involved in TLS in the human cell line U2OS across lesions classified as being bypassed via the slow and mutagenic pathway, as well as the rapid and accurate pathway, with the exception of TT CPD.
Residual TLS in the absence of polζ in murine cells is more accurate
To determine the sequence changes at the lesion caused by the absence of Rev3L, DNA sequence was analysed in descendants of gap-lesion plasmids that were repaired in Rev3L-proficient or Rev3L-deficient cells. As shown in Figure 3B and Supplementary Table 14s, in Rev3L+/+ MEF 62% of TLS events across BP-G were error-free, involving the incorporation of dCMP, consistent with previous results (Avkin et al, 2004). In total, 37% of the TLS events across BP-G were mutagenic, involving insertion of mainly dAMP and dTMP opposite the lesion, although G was also misinserted (Supplementary Table 14s). The major effect in Rev3L−/− MEF was, of course, the strong decrease in TLS, as shown in Figure 3A. DNA sequence analysis of plasmids that were repaired in the Rev3L−/− MEFs showed that a substantial fraction contained large deletions and insertions, indicative of the formation of DSB (27%; Supplementary Table 14s). The absolute frequency of these events was similar in Rev3L−/− and Rev3L+/+ MEFs, indicating that they were unrelated to polζ. Interestingly, residual TLS was more accurate in the absence of Rev3L than in its presence, with only 5.9% mutagenic TLS events. Thus, residual TLS in the absence of Rev3L was very low, but 6.3-fold less mutagenic (P=0.0006; Figure 3B; Supplementary Table 14s).
A similar effect was observed for cisPt-GG adduct (Figure 3B; Supplementary Table 15s): although 13% of the TLS events in Rev3L+/+ cells were error-prone, that is, nucleotides other than CC were inserted opposite cisPt-GG lesions (Supplementary Table 15s), remarkably, all 54 plasmids isolated from Rev3L−/− cells contained the correct sequence (CC) opposite cisPt-GG adducts (Figure 3B; Supplementary Table 15s). This reflects a misinsertion frequency of <1.8% (<1/54 events), indicating that TLS across a cisPt-GG adduct is at least 7.2-fold less mutagenic in the absence of Rev3L (P=0.0062).
Similar phenomena were observed for 4-OHEN-C (Figure 3B; Supplementary Table 16s) where TLS was 3.5-fold less mutagenic in Rev3L−/− cells compared with Rev3L+/+ cells (P=0.0002), and for TT 6-4 PP, where TLS was 20-fold less mutagenic in Rev3L−/− cells compared with Rev3L+/+ cells (P<0.0001) (Figure 3B; Supplementary Table 17s). TLS across TT CPD was accurate in both Rev3L+/+ and Rev3L−/− (Figure 3B; Supplementary Table 18s) similar to the situation in the human cells (Figure 2A; Supplementary Table 7s). We also examined insertion specificity across an AP site and an M12 insert. Regardless of the status of the Rev3L gene most bypass events led to the incorporation of A opposite the AP site, in agreement with previous results (Figure 2; Supplementary Table 19s; Avkin et al, 2002). For the M12 lesion, which represents a severe type of damage, the most pronounced effect was an increase in large deletions of the lesion area in Rev3L−/− cells (89%) compared with Rev3L+/+ cells (38%), indicative of DSB formation followed by NHEJ, possibly due to a difficulty to bypass the lesion (Supplementary Table 20s).
When the mutational spectra were examined in the human cell line U2OS under conditions in which expression of REV3L was knocked down, there was essentially no difference in the accuracy of TLS compared with the control cells (Table I). This may be due to an inherently more accurate TLS in human cells compared with mouse cells, which remained essentially unchanged even when expression of REV3L was knocked down, and TLS inhibited. For example, TLS across BP-G is less accurate in mice cells compared with human cells (Avkin et al, 2004; Table I; Supplementary Table 14s).
Table 1.
DNA sequence analysis of TLS events across BP-G and cisPt-GG in human U2OS cells, in which the expression of specific TLS DNA polymerases was knocked down using siRNA
| siRNA | Control (%) | REV3L (%) | POLK (%) | POLH (%) | POLK +REV3L (%) | POLK+POLH (%) |
|---|---|---|---|---|---|---|
| Nucleotide inserted opposite BP-G | Number of isolates | |||||
| C | 127 (74.3) | 52 (36.1) | 76 (68) | 65(69.9) | 55 (42.3) | 83 (60.1) |
| A | 11 (6.4) | 5 (4.5) | 3 (3.2) | 2 (1.4) | ||
| G | 6 (3.5) | 2 (1.4) | 2 (1.5) | 1 (0.7) | ||
| T | 3 (1.8) | 2 (1.4) | 2 (1.8) | 1 (0.7) | ||
| Deletion/insertion | 24 (14) | 88 (61.1) | 29 (25.9) | 25 (26.9) | 73 (56.2) | 51 (37) |
| Total number | 171 (100) | 144 (100) | 112 (100) | 93 (100) | 130 (100) | 138 (100) |
| TLS clonesa | 147 | 56 | 83 | 68 | 57 | 87 |
| Mutagenic TLS (%)b | 13.6 | 7.1 | 8.4 | 4.4 | 3.5 | 4.6 |
| P=0.2c | P=0.2c | P=0.042c | P=0.037c | P=0.03c | ||
| Nucleotide inserted opposite cisPt-GG | Number of isolates | |||||
| CC | 98 (78.4) | 53 (53) | 60 (87) | 59 (70.2) | 50 (58.1) | 44 (77.2) |
| AC | 15 (12) | 9 (9) | 2 (2.9) | 7 (8.3) | 4 (4.7) | 1 (1.8) |
| TC | 6 (4.8) | 3 (3.6) | ||||
| CT | 1 (0.8) | |||||
| CA | 2 (2.3) | |||||
| Deletion/insertion | 5 (4) | 38 (38) | 7 (10.1) | 15 (17.9) | 30 (34.9) | 12 (21.1) |
| Total isolates | 125 (100) | 100 (100) | 69 (100) | 84 (100) | 86 (100) | 57 (100) |
| TLS clonesa | 120 | 62 | 62 | 69 | 56 | 45 |
| Mutagenic TLS (%)b | 18.3 | 14.5 | 3.2 | 14.5 | 10.7 | 2.2 |
| P=0.006c | P=0.01c | |||||
| aThe number of clones, excluding clones having large deletions or insertions. | ||||||
| bThe percentage of mutagenic TLS is the fraction of events other than insertion of C opposite BP-G, or CC opposite cisPt-GG out of the total number of TLS events. | ||||||
| cP-value is given compared with the results obtained with cells transfected with control siRNA. Analysis was performed using the χ2 test. P-values that reached statistical significance are in bold face type. | ||||||
| U2OS cells were transfected with the indicated siRNAs. After incubation of 72 h, the plasmid mixtures containing the gap-lesion plasmid GP-BP-G1 or GP-cisPt-GG (kanR), along with the control gapped plasmid GP20 (cmR) and the carrier plasmid were introduced into the cells. Following incubation of 8 h to allow TLS, the DNA was extracted and used to transform a recA E. coli indicator strain. GP-BP-G1 or GP-cisPt-GG (kanR) descendents were extracted from kanR colonies and subjected to DNA sequence analysis. Deletions and insertions are taken as non-TLS events. | ||||||
Polζ cooperates with polκ in error-free TLS across BP-G in human cells
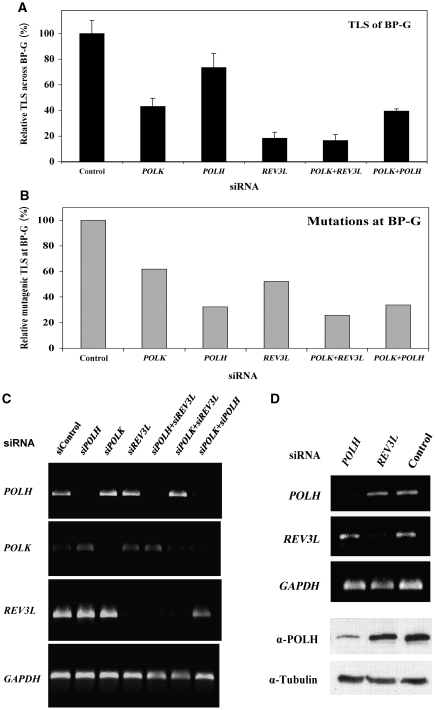
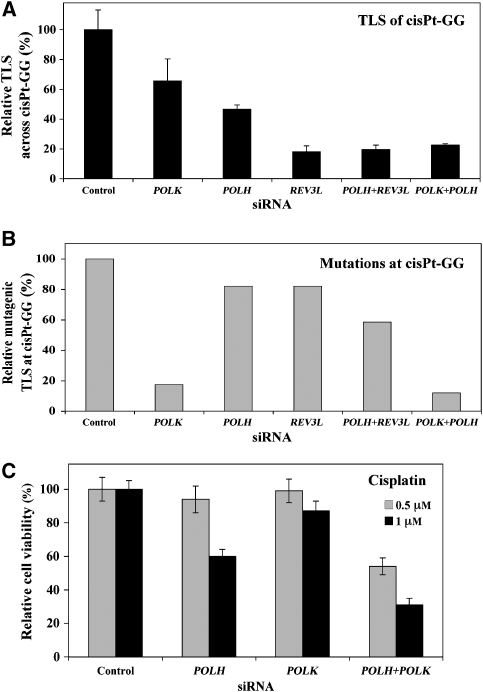
As described above, due to the known role of polκ in TLS across BP-G, the strong dependence on REV3L of TLS across this lesion was unexpected. We therefore decided to further study TLS across this lesion. A possible explanation for these findings was that both polκ and REV3L are involved, perhaps in a two-polymerase pathway (Johnson et al, 2000a). To examine this possibility, we took a gene-knockdown epistasis analysis approach, and assayed TLS in U2OS cells in which the expression of POLK, REV3L, or both was knocked down. As can be seen in Figure 4C, the POLK, POLH, and REV3L siRNAs each specifically knocked down the expression of its target polymerase by approximately 80–90%. Figure 4A and Supplementary Table 22s present the results of TLS across BP-G under these conditions. Knocking down the expression of POLK caused a 2.3-fold decrease in TLS, indicating that at least 57% of the BP-G adducts required POLK for bypass. Knocking down the expression of REV3L caused a stronger 5.5-fold decrease in TLS, indicating that at least 82% of the BP-G adducts required REV3L for bypass. When the expression of both POLK and REV3L was knocked down, TLS was essentially identical to its value in cells where only REV3L was knocked down (Figure 4A; Supplementary Table 22s). To examine whether the combination of any two polymerases would drastically affect TLS across BP-G, we assayed cells in which both POLK and POLH were knocked down. TLS across BP-G in cells in which POLH was knocked down was marginally decreased, whereas when both POLH and POLK were knocked down, TLS was similar to that observed in POLK-knocked-down cells, indicating that polη does not have a major role in the extent of TLS across BP-G (Figure 4A; Supplementary Table 22s). If at least 82% of the BP-G adducts are bypassed by polζ, at most 18% are bypassed by other polymerases, without the involvement of polζ. However, polκ is involved in bypass of 57% of the BP-G adducts. This means that at least 39% of the lesions are bypassed in a TLS pathway that involves the action of both polκ and polζ. TLS became 3.1-fold less mutagenic (P=0.042) when POLH was knocked down (Figure 4B; Table I), whereas when POLK was knocked down, there was a 1.9-fold decrease in mutagenic TLS, but it did not reach statistical significance (P=0.2; Table I). However, the fraction of deletions and insertions increased from 14 to 25.9% (P=0.012; Table I), and a similar increase was observed when POLH was knocked down (Table I). Consistently, when both POLH and POLK were knocked down, the effect was similar to knocking down POLH alone, suggesting that polη is responsible for the majority of error-prone TLS across BP-G (Table I). Knocking down both POLK and REV3L decreased mutagenic TLS across BP-G by 3.9-fold (Figure 4B; Table I; P=0.037).
Figure 4.
Polκ and polζ operate in the same TLS pathway to bypass BP-G in human cells. (A) Relative TLS extent across BP-G in U2OS cells in which the expression of specific TLS DNA polymerases was knocked down. The experiments were performed as described in the legend of Figure 3D, and the detailed data are presented in Supplementary Table 22s. TLS extents were given as percentage relative to TLS assayed with control siRNA, which was 10.9±1.1% (see Supplementary Table 22s). (B) Relative mutagenic TLS across BP-G based on DNA sequence information of plasmids isolated from colonies obtained in the experiments described in (A). The detailed data are presented in Table I. Mutagenic TLS, namely the fraction of nucleotides other than C inserted opposite BP-G, was calculated out of all TLS events, and presented relative to the mutagenic TLS in cells transfected with control siRNA (which was 13.6%; see Table I). (C) RT–PCR of RNA extracted from human U2OS cells transiently transfected with the indicated siRNA. (D) RT–PCR of POLH and REV3L mRNAs (top) and an immunoblot with antibodies against polη (bottom) performed with RNA and protein extracts prepared from U2OS cells transfected with the indicated siRNAs.
Polζ cooperates with polη and polκ in error-free and error-prone TLS, respectively, across cisPt-GG in human cells
The question arises whether lesions additional to BP-G are bypassed by the two-polymerase mechanism. We chose to address this question by analysing TLS across the adduct cisPt-GG using the siRNA-based epistasis approach. This lesion was chosen because there is evidence that polη (Bassett et al, 2004) and REV3L (Wu et al, 2004; Zander and Bemark, 2004) are involved in its bypass; however, as these were experiments performed with different cells and under different conditions it was unknown whether the two operate in the same pathway.
Analysis of TLS across cisPt-GG in cells in which the expression of POLH was knocked down caused a 53% reduction in TLS, whereas knocking down of REV3L caused an 82% decrease (Figure 5A; Supplementary Table 23s). A similar drastic reduction in TLS (80%) was observed when both POLH and REV3L were knocked down. Surprisingly, knocking down the expression of POLK, not known to be involved in TLS across cisPt-GG, caused a 34% decrease, and consistently, knocking down both POLK and POLH caused a 78% reduction in TLS, similar to the effect caused by knocking down REV3L alone (Figure 5A; Supplementary Table 23s). Thus, at least 82% of the cisPt-GG lesions are bypassed by polζ, and a similar fraction is bypassed by polη and polκ combined. Taken together with the other data, this suggests that TLS across cisPt-GG can occur by at least two reactions, each involving two polymerases: one involving polη and polζ, and the other involving polκ and polζ. DNA sequence analysis revealed that the mutagenic TLS decreased by 5.7-fold when POLK was knocked down (Table I). When POLH was knocked down mutagenic TLS remained unchanged but the fraction of deletions/insertions increased 4.5-fold (P<0.001; Table I). Knocking down both POLH and POLK caused both a decrease in mutagenic TLS and an increase in deletions/insertions (Figure 5B; Table I). Taken together, these results suggest that polζ cooperates with polη to carry out error-free TLS, and with polκ to carry out error-prone TLS across cisPt-GG.
Figure 5.
Polζ cooperates with polη and polκ in error-free and error-prone TLS, respectively, across cisPt-GG in human cells. (A) Relative extent of TLS across cisPt-GG in U2OS cells in which the expression of specific TLS DNA polymerases was knocked down. The experiments were performed as described in the legend of Figure 4A, and the detailed data are presented in Supplementary Table 23s. (B) Relative mutagenic TLS across cisPt-GG based on DNA sequence information of plasmids isolated from colonies obtained in the experiments described in (A). The detailed data are presented in Table I. Mutagenic TLS, namely the fraction of nucleotides other than CC inserted opposite cisPt-GG, was calculated out of all TLS events, and presented relative to the mutagenic TLS in cells transfected with control siRNA (which was 18.3%; see Table I). (C) Cisplatin sensitivity of XPA cells in which the expression of POLH and POLK was knocked down. XP12RO cells were transfected with the indicated siRNAs, after which they were treated with 0.5 or 1 μM cisplatin. Results are given relative to the viability of cells transfected with control siRNA (77 and 55% for 0.5 and 1 μM cisplatin, respectively). Cell viability was measured using a luminescence-based assay for measuring ATP.
POLK affects cell viability in response to cisplatin in human XPA cells in which the expression of POLH was knocked down
To examine whether the unexpected involvement of POLK in TLS across the cisPt-GG adduct is relevant to chromosomal TLS, we examined the viability of XPA cells treated with cisplatin. These cells are deficient in nucleotide excision repair, and therefore cell viability in response to cisplatin was expected to be more dependent on DNA damage tolerance mechanisms such as TLS. As can be seen in Figure 5C, cell viability was reduced by 40% in cisplatin-treated (1 μM) cells in which the expression of POLH was knocked down compared with cells transfected with a control siRNA, consistent with the known involvement of POLH in TLS across cisplatin-DNA adducts. Knocking down POLK alone had essentially no effect; however, when both POLH and POLK were knocked down, the viability of cisplatin-treated cells decreased by 70% (3.3-fold decrease; Figure 5C). At a lower cisplatin concentration (0.5 μM) knocking down POLH or POLK alone had no effect on viability, but knocking down both caused a 48% decrease in cell viability (Figure 5C). Although this does not provide direct proof for the involvement of POLK in chromosomal TLS across cisplatin adducts, it is consistent with such a possibility, further strengthening the conclusion from our gap-lesion plasmid assay.
Discussion
A pivotal role for polζ in both error-prone and error-free TLS in mammalian cells
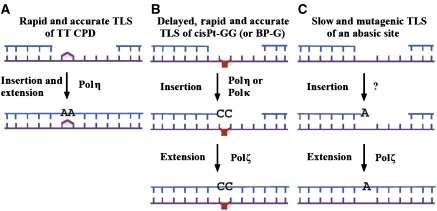
Our results suggest that TLS operates in three distinct pathways: a very fast and accurate pathway that involves polη, a delayed, accurate and fast TLS pathway, which involves two polymerases, including polζ, and a slow and highly mutagenic pathway, which involves polζ (Figure 6). Out of the seven DNA lesions examined, only TT CPD was bypassed in the very fast and accurate TLS pathway (Figure 1). It is well established that polη is involved in TLS across TT CPD (Maher et al, 1976; Johnson et al, 1999a; Masutani et al, 1999), including in our assay (Hendel et al, 2008). The TT CPD was the only lesion analysed, the bypass of which did not require polζ. In fact, it may not need any DNA polymerase except for polη, given its superior catalytic properties and its relatively high fidelity when replicating across a TT CPD (Johnson et al, 2000b; Masutani et al, 2000; McCulloch et al, 2004). These properties are manifested during replication of UV-irradiated cells in culture, as indicated by their ability to divide in the presence of CPD (Spivak and Hanawalt, 1992). Given the evolutionary importance of UV light, the pathway of fast and accurate bypass of TT CPD by polη may represent a special case, which is unique for CPD.
Figure 6.
A model for three pathways of TLS in mammalian cells. TLS across a TT CPD is rapid and accurate, and occurs most likely by a single DNA polymerase, polη (A). TLS across cisPt-GG and BP-G occurs by a two-polymerase TLS pathway, which is rapid and accurate but delayed, and involves a combination of polζ with either polη or polκ (B). TLS across an AP site occurs by a slow and highly mutagenic TLS pathway, which involves polζ, and most likely an additional DNA polymerase (C). See text for details.
Mammalian cells contain no less than 15 DNA polymerases, yet polζ stands out as a key TLS polymerase involved in TLS across all lesions examined, except the TT CPD. Moreover, unlike in the S. cerevisiae paradigm, where polζ appears to be involved only in error-prone TLS, in mammals it is involved in both error-free and error-prone TLS. Although this is somewhat surprising, because the apparent lack of backup to the function of polζ introduces vulnerability into the TLS system, it may explain why polζ is the only TLS polymerase that is essential in mice (Wittschieben et al, 2006; Gan et al, 2008). One possibility for having a common enzyme in several pathways is regulation; however, this possibility remains to be explored.
Discrete two-polymerase combinations define error-free and error-prone TLS across BP-G and cisPt-GG
TLS across two of the lesions studied, BP-G and cisPt-GG involved the action of at least three polymerases: polκ, polη, and polζ. Taking into account the properties of the human polκ and polη, and the putative polζ (based on the S. cerevisiae polζ; Johnson et al, 2000a), the most likely explanation is that these lesions are bypassed in a two-polymerase mechanism: In the first step, polκ or polη inserts a nucleotide opposite BP-G or cisPt-GG, and in the second step, polζ performs the extension past each of the two lesions (Figure 6). Error-prone TLS across BP-G involves primarily polη, as indicated by the decreased mutagenicity of TLS when POLH expression was knocked down (Table I). In the case of cisPt-GG, both polη and polκ are involved in its bypass, and knocking down both causes a reduction of TLS similar to that caused by knocking down polζ. Interestingly, polη and polζ together carry out largely error-free TLS, whereas polκ and polζ lead to a largely error-prone outcome of TLS across cisPt-GG. The involvement of polκ in TLS across cisPt-GG was unexpected, as purified recombinant polκ was reported to be unable to bypass a cisPt-GG lesion (Ohashi et al, 2000; Gerlach et al, 2001). The in vivo activity may be enabled by interaction with auxiliary proteins. Its relevance to chromosomal TLS is indicated by the decreased viability of cisplatin-treated XPA cells in which both POLK and POLH were knocked down, compared with cells in which only POLH was knocked down (Figure 5C). Our results support the two-polymerase model for TLS proposed by L Prakash and S Prakash (Johnson et al, 2000a) based on experiments with purified proteins. To the best of our knowledge, the data presented here provide the first evidence for a two-polymerase TLS mechanism in living mammalian cells.
The fact that BP-G requires two polymerases for bypass in mammalian cells is surprising, because purified polκ completely bypasses BP-G (Zhang et al, 2000; Avkin et al, 2004). Moreover, human purified polκ was shown to be a general extender (Washington et al, 2002), similar to the yeast polζ. Thus, under our conditions, the extender capacity of polκ is not sufficient to catalyse efficient TLS across BP-G, and polζ is needed. The need for polζ in TLS across cisPt-GG is similarly surprising. Human purified polη was shown to bypass cisPt-GG (Vaisman et al, 2000), yet polζ is needed for efficient TLS across it. A possible explanation might be that although polη is efficient and accurate in incorporating a nucleotide opposite the 3′ G of the cisplatin-GG lesion, it is inhibited at the 5′ G (Alt et al, 2007).
Our results are consistent with previous reports demonstrating the involvement of polκ in chromosomal mutation induced by BP in mouse (Ogi et al, 2002; Bi et al, 2005) and of polζ in human cells (Li et al, 2002). Similarly, sensitivity to and mutagenesis by cisplatin in human cells involves polη and polζ (Bassett et al, 2004; Wu et al, 2004). It should be emphasized that the chromosomal studies assayed survival and mutagenesis, not TLS, and therefore accurate TLS, which accounts for most TLS events across BP-G and cisPt-GG, could not be scored. Moreover, any effects observed in assaying chromosomal mutagenesis may have been caused by a combination of effects on excision repair, recombination repair, and checkpoint activation.
Slow and mutagenic polζ-dependent TLS across a wide variety of DNA lesions
Three lesions, TT 6-4PP, 4-OHEN-C, and AP site were bypassed in a slow and highly mutagenic fashion (up to 80% errors). The inferior TLS across these lesions compared with TLS across TT CPD, BP-G, and cisPt-GG may be due to a combination of the severity of the former, namely the large deviation from the normal structure that they force on the DNA, and the lack of an appropriate specialized DNA polymerase. After all, the repertoire of the DNA lesions far exceeds the number of TLS DNA polymerases. At this point, we do not know yet whether any other polymerase is required in this pathway in addition to polζ. TLS across the AP site does not require polη (Avkin et al, 2002) or polκ (unpublished data), but is aphidicolin-sensitive, consistent with the involvement of an additional polymerase, perhaps polδ (Avkin et al, 2002). The big diversity of lesions bypassed by this pathway, including the M12 insert, which represents an artificial and severe type of DNA damage, suggests that this TLS pathway functions as a general-specificity TLS pathway, in charge of dealing with a variety of DNA lesions, including novel ones. Its mutagenic nature indicates that lesions are bypassed at the cost of increased mutation, a price that is apparently not too high given the alternative of a stuck replication fork or persistent gap, which might be broken leading to DSB. The slow rate of this pathway may reflect not only the difficulty of the TLS machinery to deal with these types of lesions but also a negative regulation to enable an alternative mechanism to act. Such a mechanism may involve the Rad5-dependent damage avoidance or homologous recombination repair, which cannot be detected in our assay.
Conclusions
The results presented here suggest that TLS operates in mammalian cells in at least three distinct pathways, of which two are fast and accurate, and one slow and mutagenic. Surprisingly, polζ is involved in both error-free and error-prone TLS, making it a key and bottleneck component of the TLS system in mammalian cells. Moreover, evidence is presented that discrete two-polymerase combinations with polζ dictate error-prone or error-free TLS across the same lesion: polκ and polζ carry out error-free TLS of BP-G, but error-prone TLS of cisPt-GG, whereas polη and polζ carry out error-free TLS of cisPt-GG, but error-prone TLS of BP-G. To the best of our knowledge, this is the first report on the involvement of mammalian polζ in error-free TLS, and the first evidence for a two-polymerase mechanism in mammalian cells.
Materials and methods
Construction of gapped DNA substrates
The construction of gap-lesion plasmids was described earlier (Reuven et al, 1999; Avkin et al, 2002, 2006; Maor-Shoshani et al, 2003; Adar and Livneh, 2006; Hendel et al, 2008), and is further detailed in the Supplementary data, along with the DNA sequences in the vicinity of the lesions.
In vivo TLS assay
The TLS assay was described earlier (Avkin et al, 2004, 2006). Briefly, cells were co-transfected with a mixture containing 50 ng of a gapped-lesion plasmid (kanR), 50 ng of a gapped plasmid without lesion (GP20-cm, cmR), and 10 μg of the carrier plasmid pUC18, using jetPEI/DNA complexes (Polyplus-transfection, Illkirch, France). For kinetics experiments, cells were transfected by electroporation with the Nucleofector™ system (Amaxa GmbH, Köln, Germany), which enables fast delivery. A similar DNA mixture was used, except that it contained 4.9 μg of the carrier plasmid pUC18. The gap-filling kinetics of the normalizing plasmid itself (GP20) was examined by transfecting U2OS cells with a mixture of GP20 (kanR) and a normalizing covalently closed plasmid pSA26 (cmR) (Avkin et al, 2002), and found to be completely filled in less than 30 min (data not shown). The efficiency of gap repair was calculated by dividing the number of transformants obtained from the gap-lesion plasmid (number of colonies on LB-kan plates) by the number of corresponding transformants obtained with the control gapped plasmid GP20-cm (number of colonies on LB-cm plates). Plasmids were extracted from kanR colonies, and the sequence opposite the lesion was determined by automated DNA sequencing analysis in the Biological Services Department at the Weizmann Institute. To obtain values of TLS from values of gap repair, the latter were multiplied by the percentage of TLS events out of the total events, as determined by the DNA sequence analysis.
Knocking down of DNA polymerase genes
The expression of specific DNA polymerase genes was knocked down in U2OS cells by transfection with specific siRNA pools (50 nM) in 6-cm plates. All siRNAs were from Dharmacon: POLK SMARTpool M-021038, REV3L SMARTpool M-006302, POLH SMARTpool M-006454, and siControl non-targeting pool D-001206-14. Transfection was carried out using 12 μl/plate HiPerFect (Qiagen). After 48 h in culture with siRNA, cells were split 1:3, and after additional 24 h the TLS assay was conducted as described above.
Measurement of viability of cisplatin-treated human cells
Human XP12RO (SV40-tranformed XPA) cells were reverse-transfected with on-target plus siRNA (Dharmacon) in a 96-well plate format, and after 24 h the medium was replaced with a fresh medium containing 0.5 or 1 μM cisplatin. Viability was measured 72 h later, using CellTiter-Glo luminescent cell viability assay, which measures ATP levels (Promega).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Richard D Wood (University of Pittsburgh Medical School, Pittsburgh, PA) and Tamar Paz-Elizur (ZL laboratory, Weizmann Institute of Science) for stimulating discussions. ZL is the incumbent of the Maxwell Ellis Professorial Chair in Biomedical Research. This study was supported by grants from the Flight Attendant Medical Research Institute, Florida, USA, the Israel Science Foundation (no. 564/04), and the MD Moross Institute for Cancer Research at the Weizmann Institute of Science to ZL, and by grants from the NIH, USA to NG (CA 099194 and CA112412), ZW (CA92528), ECF (ES11354), and Richard D Wood, University of Pittsburgh (CA098675).
References
- Adar S, Livneh Z (2006) Translesion DNA synthesis across non-DNA segments in cultured human cells. DNA Repair (Amst) 5: 479–490 [DOI] [PubMed] [Google Scholar]
- Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T (2007) Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science 318: 967–970 [DOI] [PubMed] [Google Scholar]
- Avkin S, Adar S, Blander G, Livneh Z (2002) Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci USA 99: 3764–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z (2004) Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells. The role of DNA polymerase κ. J Biol Chem 279: 53298–53305 [DOI] [PubMed] [Google Scholar]
- Avkin S, Sevilya Z, Toube L, Geacintov NE, Chaney SG, Oren M, Livneh Z (2006) p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol Cell 22: 407–413 [DOI] [PubMed] [Google Scholar]
- Bassett E, King NM, Bryant MF, Hector S, Pendyala L, Chaney SG, Cordeiro-Stone M (2004) The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res 64: 6469–6475 [DOI] [PubMed] [Google Scholar]
- Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C (2006) Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol 26: 3527–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Slater DM, Ohmori H, Vaziri C (2005) DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem 280: 22343–22355 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wilder G, Peter M, Lehmann AR, Hofmann K, Dikic I (2005) Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) DNA Repair and Mutagenesis. Washington, DC: ASM Press [Google Scholar]
- Gan GN, Wittschieben JP, Wittschieben BO, Wood RD (2008) DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res 18: 174–183 [DOI] [PubMed] [Google Scholar]
- Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC (2001) Purification and characterization of pol kappa, a DNA polymerase encoded by the human DINB1 gene. J Biol Chem 276: 92–98 [DOI] [PubMed] [Google Scholar]
- Gibbs PE, McDonald J, Woodgate R, Lawrence CW (2005) The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutyl dimer. Genetics 169: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW (1998) A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc Natl Acad Sci USA 95: 6876–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF (2002) Error-prone DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem 71: 17–50 [DOI] [PubMed] [Google Scholar]
- Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L (2001) Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev 15: 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z (2008) Reduced fidelity and increased efficiency of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproducts, in human cells lacking DNA polymerase eta. DNA Repair (Amst) 7: 1636–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hubscher U, Nasheuer HP, Syvaoja JE (2000) Eukaryotic DNA polymerases, a growing family. Trends Biochem Sci 25: 143–147 [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC (2006) A single amino acid governs enhanced activity of DinB DNA polymerases on damaged template. Nature 439: 225–228 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L (1999a) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285: 263–265 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L (1999b) Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science 283: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L (2000a) Eukaryotic polymerases i and z act sequentially to bypass DNA lesions. Nature 406: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L (2000b) Fidelity of human DNA polymerase eta. J Biol Chem 275: 7447–7450 [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR (2004) Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Lawrence CW (2002) Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 1: 425–435 [DOI] [PubMed] [Google Scholar]
- Lehmann AR (1972) Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol 66: 319–337 [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Fuchs RP (2006) Gaps and forks in DNA replication: rediscovering old models. DNA Repair (Amst) 5: 1495–1498 [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM (2007) Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 6: 891–899 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang H, McManus TP, McCormick JJ, Lawrence CW, Maher VM (2002) hREV3 is essential for eror-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat Res 510: 71–80 [DOI] [PubMed] [Google Scholar]
- Livneh Z (2001) DNA damage control by novel DNA polymerases: translesion replication and mutagenesis. J Biol Chem 276: 25639–25642 [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Maher VM, Ouellette LM, Curren RD, McCormick JJ (1976) Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature 261: 593–595 [DOI] [PubMed] [Google Scholar]
- Maor-Shoshani A, Ben-Ari V, Livneh Z (2003) Lesion bypass DNA polymerases replicate across non-DNA segments. Proc Natl Acad Sci USA 100: 14760–14765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Iwai S, Hanaoka F (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase h. EMBO J 19: 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA (2004) Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature 428: 97–100 [DOI] [PubMed] [Google Scholar]
- Meneghini R (1976) Gaps in DNA synthesized by ultraviolet light-irradiated WI38 cells. Biochim Biophys Acta 425: 419–427 [DOI] [PubMed] [Google Scholar]
- Mojas N, Lopes M, Jiricny J (2007) Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev 21: 3342–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J, Fuchs RPP (2000) All three SOS-inducible DNA polymerases (Pol II, Pol IV, and Pol V) are involved in induced mutagenesis. EMBO J 19: 6259–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC (1996a) Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC (1996b) Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science 272: 1646–1649 [DOI] [PubMed] [Google Scholar]
- Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, Green CM, Lehmann AR (2008) Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA 105: 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Shinkai Y, Tanaka K, Ohmori H (2002) Pol kappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA 99: 15548–15553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H (2000) Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev 14: 1589–1594 [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Prakash L (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev 16: 1872–1883 [DOI] [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z (1999) The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA and SSB, and specialized for translesion replication. J Biol Chem 274: 31763–31766 [DOI] [PubMed] [Google Scholar]
- Rupp WD, Howard-Flanders P (1968) Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31: 291–304 [DOI] [PubMed] [Google Scholar]
- Soria G, Podhajcer O, Gottifredi V (2006) P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene 25: 2829–2838 [DOI] [PubMed] [Google Scholar]
- Spivak G, Hanawalt PC (1992) Translesion DNA syhthesis in the dihydrofolate reductase domain of UV-irradiated CHO cells. Biochemistry 31: 6794–6800 [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF (2000) Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, Goodman MF (1999) UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA 96: 8919–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Masutani C, Hanaoka F, Chaney SG (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry 39: 4575–4580 [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash L, Prakash S (2002) Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci USA 99: 1910–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M (2004) Rad18 guides pol eta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 23: 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben JP, Reshmi SC, Gollin SM, Wood RD (2006) Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer Res 66: 134–142 [DOI] [PubMed] [Google Scholar]
- Wu F, Lin X, Okuda T, Howell SB (2004) DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res 64: 8029–8035 [DOI] [PubMed] [Google Scholar]
- Xie Z, Braithwaite E, Guo D, Zhao B, Geacintov NE, Wang Z (2003) Mutagenesis of benzo[a]pyrene diol epoxide in yeast: requirement for DNA polymerase zeta and involvement of DNA polymerase eta. Biochemistry 42: 11253–11262 [DOI] [PubMed] [Google Scholar]
- Zander L, Bemark M (2004) Immortalized mouse cell lines that lack a functional Rev3 gene are hypersensitive to UV irradiation and cisplatin treatment. DNA Repair (Amst) 3: 743–752 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z (2000) Error-free and error-prone lesion bypass by human DNA polymerase κ in vitro. Nucleic Acids Res 28: 4138–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wang J, Geacintov NE, Wang Z (2006) Polh, polz and Rev1 together are required for G to T transversion mutations induced by the (+)- and (−)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells. Nucleic Acids Res 34: 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information