Abstract
Raf kinase inhibitory protein (RKIP) negatively regulates the MAP kinase (MAPK), G protein-coupled receptor kinase-2, and NF-κB signalling cascades. RKIP has been implicated as a metastasis suppressor for prostate cancer, but the mechanism is not known. Here, we show that RKIP inhibits invasion by metastatic breast cancer cells and represses breast tumour cell intravasation and bone metastasis in an orthotopic murine model. The mechanism involves inhibition of MAPK, leading to decreased transcription of LIN28 by Myc. Suppression of LIN28 enables enhanced let-7 processing in breast cancer cells. Elevated let-7 expression inhibits HMGA2, a chromatin remodelling protein that activates pro-invasive and pro-metastatic genes, including Snail. LIN28 depletion and let-7 expression suppress bone metastasis, and LIN28 restores bone metastasis in mice bearing RKIP-expressing breast tumour cells. These results indicate that RKIP suppresses invasion and metastasis in part through a signalling cascade involving MAPK, Myc, LIN28, let-7, and downstream let-7 targets. RKIP regulation of two pluripotent stem cell genes, Myc and LIN28, highlights the importance of RKIP as a key metastasis suppressor and potential therapeutic agent.
Keywords: let-7, LIN28, metastasis, MAP kinase, raf kinase inhibitory protein
Introduction
Tumour metastasis suppressors are natural regulators of the metastasic process (Massague, 2007). Some of these suppressors prevent progression of tumour cells to metastasis; other suppressors not only block progression, but are also able to reverse the metastatic phenotype. One powerful strategy for both prevention and treatment of aggressive tumours is to identify tumour metastasis suppressors that inhibit the virulent invasive and colonizing properties of metastatic cells and to mimic their mechanism of action.
Raf kinase inhibitory protein (RKIP; also PEBP1), a member of the evolutionarily conserved phosphatidylethanolamine-binding protein family, has been implicated as a suppressor of metastatic progression in an orthotopic murine model using androgen-independent prostate tumour cells (Fu et al, 2003). Although primary prostate tumour growth was unaffected, RKIP inhibited both vascular invasion and lung metastases. RKIP is depleted or deficient in a number of tumours, including prostate, breast, melanoma, hepatocellular carcinoma, and colorectal (Fu et al, 2003, 2006; Schuierer et al, 2004; Hagan et al, 2005; Akaishi et al, 2006; Al-Mulla et al, 2006). Taken together, these results suggest that RKIP may function as a general metastasis suppressor.
Raf kinase inhibitory protein modulates at least three key regulatory pathways in mammalian cells. RKIP inhibits MAP kinase (MAPK) signalling in part by binding to Raf-1, preventing Raf-1 phosphorylation at activating sites (Yeung et al, 1999; Trakul et al, 2005). RKIP phosphorylation at S153 by protein kinase C dissociates RKIP from Raf, enabling MEK and MAPK activation. Phosphorylated RKIP inhibits G protein-coupled receptor kinase-2 (GRK-2)-mediated downregulation of G protein-coupled receptors (GPCRs), thereby mediating cross talk between MAPK and GPCR signalling pathways (Lorenz et al, 2003). RKIP also suppresses NF-κB activation (Yeung et al, 2001), potentiating the efficacy of chemotherapeutic agents (Chatterjee et al, 2004). Finally, RKIP ensures chromosomal integrity by preventing MAPK inhibition of Aurora B kinase and the spindle checkpoint (Eves et al, 2006). Although genomic instability resulting from RKIP loss could contribute to metastatic progression, this mechanism is unlikely to give rise to a common phenotype in multiple tumour types.
Metastasis is a complex process involving a number of steps, including cellular epithelial–mesenchymal transition (EMT), invasion, intravasation into blood or lymph vessels, extravasation from vessels, and metastatic colonization, proliferation, and survival (Massague, 2007). EMT is regulated by the transcription factors Snail, Twist, and Slug, which are themselves regulated by the high mobility group A (HMGA1 and 2) family of non-histone chromatin remodelling proteins (Thuault et al, 2006). Recently, HMGA2 has been shown to be inhibited by the microRNA let-7 (Lee and Dutta, 2007; Mayr et al, 2007).
MicroRNAs are non-coding RNAs of ∼22 nucleotides that regulate key processes in growth and development and have been implicated as tumour oncogenes or suppressors in cancer (Wu et al, 2007). Let-7/miR-98 is an evolutionarily conserved microRNA family that has been implicated as a tumour suppressor of colon and lung cancer, and let-7 loss is associated with breast tumours as well as other less differentiated human cancer cells (Shell et al, 2007; Zhang et al, 2007). However, the signalling cascades that regulate let-7 expression in mammalian cells have not been elucidated.
To examine the mechanism by which RKIP suppresses metastasis, we focused on early and late events in bone metastasis using a breast tumour model. Here, we show that RKIP represses invasion, intravasation and bone metastasis of breast tumour cells in part through a signalling cascade involving inhibition of MAPK, Myc, and LIN28, leading to induction of the microRNA let-7 and downregulation of its targets.
Results
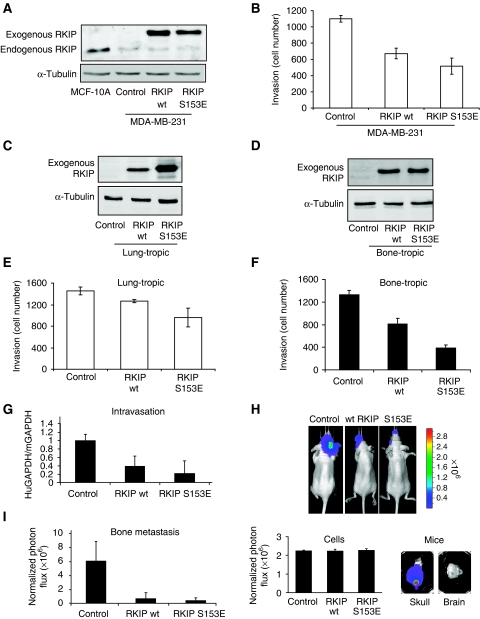
Raf kinase inhibitory protein is expressed in MCF10A mammary gland and MCF-7 cells but is barely detectable in the highly invasive MDA-MB-231 adenocarcinoma cells (Figure 1A, Supplementary Figure 1a and d). To determine the effect of RKIP on invasion, we transduced MDA-MB-231 cells with lentivirus-expressing wild-type (wt) RKIP or an S153E mutant. Phosphorylation at S153 causes RKIP dissociation from Raf-1, and mutating this site results in a more potent MAPK inhibitor (Corbit et al, 2003). The S153E mutant does not function as a phosphomimetic but instead promotes selective inhibition of Raf by preventing phosphorylation and GRK-2 inhibition. As observed previously for prostate cells (Fu et al, 2003), RKIP did not affect breast tumour cell growth in culture or in mice (Supplementary Figure 2a and b). However, stable expression of wt RKIP in MDA-MB-231 cells inhibited invasion without changing E-cadherin or vimentin levels (Figure 1B; Supplementary Figure 1b and c). The S153E mutant induces similar effects at lower expression levels in MDA-MB-231 cells, consistent with Raf-1 as an RKIP target. These results show that RKIP suppresses invasion, an early step in metastasis.
Figure 1.
RKIP regulates breast cancer invasion and metastasis. (A) RKIP is expressed in MCF10A mammary gland and depleted in metastatic MDA-MB-231 cells. MDA-MB-231 cells were stably transduced with wt or S153E RKIP and the lysates immunoblotted with anti-RKIP or anti-tubulin antibody. (B) Wt and S153E RKIP inhibit invasion of MDA-MB-231 cells. Cells were assayed for invasion as described in Materials and methods. Results represent the mean±s.e. for four independent samples (P<0.001 for wt and P=0.002 for S153E RKIP relative to Control). (C, D) Expression of wt and S153E RKIP in lung-tropic (4175) or bone-tropic (1833) breast cancer cells. Cells were stably transduced with wt or S153E RKIP and the lysates were immunoblotted with anti-RKIP or anti-tubulin antibody. All lanes in (C) come from the same gel, but one lane after sample 2 was omitted leading to a composite figure. (E, F) Wt and S153E RKIP inhibit invasion of lung-tropic (4175) or bone-tropic (1833) cells. The 1833 cells stably expressing control vector, wt RKIP, or S153E RKIP were assayed for invasion as described in Materials and methods. Results represent the mean±s.e. for four independent samples (4175) or mean±s.d. for three samples (1833) (P<0.05 for wt RKIP and P<0.05 for S153E RKIP relative to control 4175 cells; P=0.02 for wt RKIP and P<0.001 for S153E RKIP relative to control 1833 cells). (G) Wt and S153E RKIP inhibit intravasation of bone-tropic tumour cells (1833). The 1833 cells stably expressing control vector (six mice), wt RKIP (five mice), or S153E RKIP (five mice) were injected into the mammary fat pad of mice. After 3 weeks, cells isolated from the blood were analysed for GAPDH transcripts derived from human (tumour) or mouse (control). Results represent the mean±s.d. for the animals (P<0.002 for wt RKIP and P<0.001 for S153E RKIP relative to control). (H, I) Wt and S153E RKIP inhibit bone metastases. The 1833 cells expressing luciferase and control vector (six mice), wt RKIP (seven mice), or S153E RKIP (seven mice) were injected into the left ventricle of the mice, and the mice were imaged for luciferase activity after 3 weeks. Representative images show that RKIP wt and S153E greatly reduced bone metastases in skull (H, upper right panel; lower right panel). The 1833 cells stably expressing luciferase have an identical luciferase reporter activity before injection into mice (H, lower left panel). Comparable regions of the mouse skulls were optically imaged and quantified. Results (I) represent the mean±s.d. for the animals (P<0.01 for wt RKIP and P<0.01 for S153E RKIP relative to Control).
MDA-MB-231 cells are a heterogeneous population showing clonal variability in metastatic behaviour (Minn et al, 2005b). To avoid potential confounding effects in studying a mixed population, we introduced wt and S153E RKIP into highly metastatic lung-tropic (4175) and bone-tropic (1833) breast tumour cells derived from MDA-MB-231 by in vivo selection (Kang et al, 2003; Minn et al, 2005a) (Figure 1C and D). As in the parental population, RKIP inhibited in vitro invasion of both 4175 and 1833 cells, and S153E more potently inhibited invasion compared with wt RKIP (Figure 1E and F). As RKIP inhibited invasion more robustly in bone- than in lung-tropic cells by in vitro assays, 1833 cells were used for in vivo experiments and to delineate the mechanism of RKIP inhibition. To test RKIP regulation of invasion in vivo, we determined its effect on tumour cell intravasation from a primary site in a murine orthotopic model. The 1833 cells expressing control vector, wt RKIP, or S153E RKIP were injected into the mammary fat pad. At 3 weeks, cells isolated from the blood were lysed and analysed for human (tumour) and mouse (control) GAPDH transcripts. qRT–PCR quantitation showed that both wt RKIP and S153E RKIP inhibited tumour cell intravasation (Figure 1G).
Invasion and intravasation are necessary early events in the metastatic cascade. To determine if RKIP can also suppress late metastatic events, such as colonization and/or growth at a distant site, we injected luciferase-labelled 1833 cells co-expressing a control vector, wt RKIP, or S153E RKIP directly into the left cardiac ventricle of mice to bypass the intravasation step. Bioluminescence imaging approximately 3 weeks post-injection showed colonization and growth of bone metastatic cells in the skull (a primary osseous site in adult mice) for the control cohort. Analysis of the dissected brain showed no tumour cells, illustrating selective metastasis to the skull. In contrast, RKIP-expressing cells showed a marked decrease in bone metastasis (Figure 1H and I), confirming that RKIP is a suppressor of breast cancer metastasis.
Epithelial–mesenchymal transition is thought to be an important process in tumour invasion and intravasation. HMGA2 is a chromatin remodelling factor that induces transcription factors implicated in EMT and invasion such as Snail (Thuault et al, 2006). Recently, the let-7/miR-98 family of microRNAs was shown to negatively regulate HMGA2 (Lee and Dutta, 2007; Mayr et al, 2007). As let-7 also inhibits MAPK signalling through suppression of Ras (Johnson et al, 2005), the actions of RKIP and let-7 are similar, and we reasoned that they may suppress invasion through common signalling pathways.
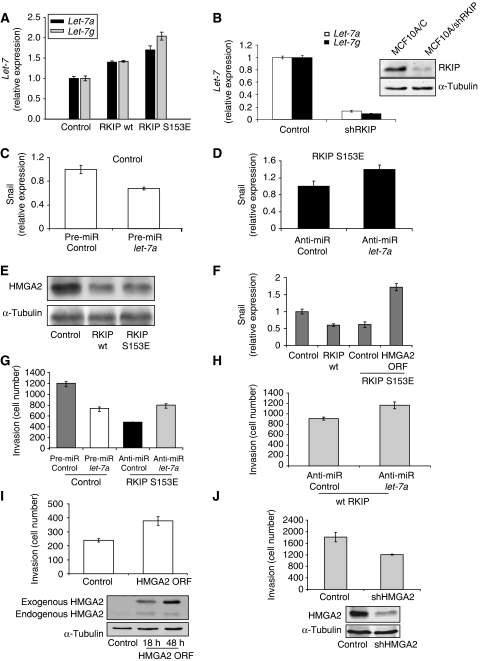
To investigate this possibility, we determined whether let-7 and its targets are influenced by RKIP and mediate RKIP inhibition of invasion. MCF10A cells express higher levels of let-7a than MDA-MB-231 cells, and shRNA depletion of RKIP in MCF10A cells almost completely suppresses let-7a and let-7g expression (Figure 2B, Supplementary Figure 3a). Conversely, 1833 cells transfected with wt RKIP or S153E RKIP exhibit an increase in let-7a and let-7g expression (Figure 2A). As we did not assay the other human let-7 family members with similar binding sites, this induction is probably an underestimate of total let-7 expression. To directly examine the consequences of altering let-7 expression, we transfected 1833 cells with precursor (pre-miR) let-7a. Pre-miR let-7a increased the level of Dicer-processed mature let-7a approximately three-fold, decreased HMGA2 and Snail (Figure 2C, Supplementary Figure 3c and d), and decreased invasion, but proliferation was unaffected (Figure 2G; Supplementary Figure 2c). By contrast, electroporating S153E RKIP or wt RKIP-expressing cells with let-7a anti-miR, an inhibitor of let-7, decreased the level of let-7a expression and increased HMGA2 and Snail (Figure 2D, Supplementary Figure 3e–h). Anti-miR let-7a also promoted invasion (Figure 2G and H) under conditions where proliferation of RKIP-expressing cells was unchanged (Supplementary Figure 2d). Similar to let-7, wt and S153E RKIP inhibited HMGA2 protein and Snail mRNA expression in both MDA-MB-231 and 1833 cells (Figure 2E and F, Supplementary Figure 3b). Furthermore, an HMGA2 cDNA lacking let-7 interaction sites (Lee and Dutta, 2007) restored both Snail expression and invasion in S153E RKIP-expressing 1833 cells (Figure 2F and I), whereas an shRNA for HMGA2 decreased invasion in 1833 cells (Figure 2J). No changes in proliferation rate were observed under the same conditions as the invasion assays (Supplementary Figure 2e and f). In total, these results argue that RKIP inhibits invasion in part through induction of let-7 and inhibition of HMGA2 and Snail.
Figure 2.
RKIP regulates let-7, HMGA2, and Snail. (A) Wt and S153E RKIP increase let-7a and let-7g expression. The 1833 cells expressing control, wt, or S153E were assayed for let-7a or let-7g by qRT–PCR. Results represent the mean±s.d. for three samples (P=0.03 for wt RKIP and P<0.01 for S153E RKIP relative to control). (B) Depletion of RKIP in MCF10A cells suppresses let-7a and let-7g expression. MCF10A cells expressing control vector or shRNA for human RKIP were assayed for let-7a or let-7g by qRT–PCR. Results represent the mean±s.d. for three samples (P<0.0001 for MCF10A/shRKIP relative to MCF10A cells). Cell lysates were immunoblotted with anti-RKIP or anti-tubulin antibodies. (C) Pre-miR let-7a decreases Snail mRNA levels. Snail mRNA isolated from 1833 cells transfected with pre-miR let-7a was quantitated by qRT–PCR. Results represent the mean±s.d. for three samples (P<0.02 for let-7a relative to Control). (D) Anti-miR let-7a increases Snail mRNA levels. Snail mRNA isolated from 1833 cells stably expressing S153E RKIP and transfected with anti-miR let-7a was quantitated by qRT–PCR. Results represent the mean±s.d. for three samples (P=0.03 for anti-miR let-7a relative to Control). (E) RKIP inhibits HMGA2 expression. The 1833 cells expressing vector, wt, or S153E RKIP were lysed and immunoblotted with anti-HMGA2 or anti-tubulin antibody. (F) Snail expression inhibited by RKIP and rescued by let-7-insensitive HMGA2. Snail mRNA was isolated from 1833 cells expressing control vector, wt RKIP, S153E RKIP, or S153E RKIP and HMGA2 lacking the 3′-untranslated region that binds let-7 (ORF). Snail mRNA was quantitated by qRT–PCR. Results represent the mean±s.e. for three independent samples (P<0.001 for wt RKIP and for S153E RKIP relative to control). (G, H) Let-7 regulates invasion. The 1833 cells were transfected with control or pre-miR let-7a, and 1833 cells expressing S153E or wt RKIP were transfected with control or anti-miR let-7a. Cells were assayed for invasion as described in Materials and methods. Results represent the mean±s.e. for four independent samples. (P=0.004 for pre-miR let-7 relative to Control, P=0.008 for anti-miR let-7 relative to Control in S153E RKIP cells, and P=0.025 for anti-miR let-7 relative to Control in wt RKIP cells). (I, J) HMGA2 regulates invasion. The 1833 cells were transfected with either HMGA2 ORF or shRNA for HMGA2 (shHMGA2). Cells were lysed at 18 and 48 h after transfection of HMGA2 ORF or 48 h after transfection of shHMGA2 and immunoblotted with anti-HMGA2 or anti-tubulin antibody. Cells were assayed for invasion as described in Materials and methods. Results represent the mean±s.e. for three independent samples (I: P=0.02 for HMGA2 relative to Control; J: P<0.04 for shHMGA2 relative to Control). Results are representative of at least three independent experiments.
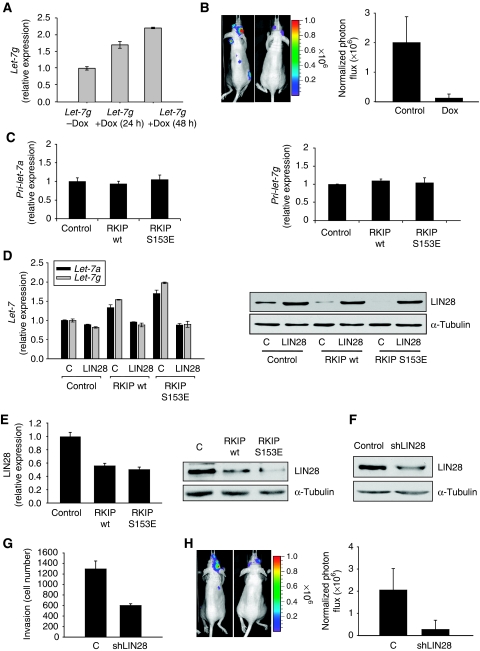
As RKIP is also a metastasis suppressor, we determined whether let-7 similarly represses bone metastasis. To address this question, 1833 cells were transfected with tetracycline-inducible expression vectors for let-7g as described previously (Kumar et al, 2008). Doxycycline did not affect cell growth or invasion in control 1833 cells (data not shown). By contrast, as observed with pre-miR let-7, doxycycline-induced let-7g inhibited cell invasion (Supplementary Figure 4a). Under these conditions (up to 48 h of let-7 induction), no change in cell proliferation was observed (Supplementary Figure 4b), although longer incubation showed some inhibition of cell proliferation. When luciferase-labelled 1833 cells expressing inducible let-7g were injected into the left cardiac ventricle of mice that were subsequently treated with doxycycline, let-7 expression caused a dramatic decrease in bone metastasis (Figure 3A and B). As the in vivo studies take at least 3 weeks, it is possible that the let-7-mediated inhibition is a secondary effect of decreased cell proliferation or increased cell death. In either case, the net effect of let-7 expression, similar to RKIP, is to suppress breast cancer metastasis.
Figure 3.
RKIP induces let-7 through the inhibition of LIN28. (A) Let-7g is induced by doxycycline. The 1833 cells expressing inducible let-7g were treated with 2 μg/ml doxycycline for the indicated times. Results represent the mean±s.d. for three samples (P=0.006 for 24 h and P=0.004 for 48 h treatment relative to Control). (B) Let-7g inhibits bone metastases. The 1833 cells expressing luciferase and either control vector (7 mice) or tet-inducible let-7g (7 mice) were grown in the presence of 2 μg/ml doxycycline for 24 h. Cells were injected into the left ventricle of mice, and 2 days later, mice were administered with drinking water containing 4% sucrose only or 2 mg/ml doxycycyline and 4% sucrose. Mice were imaged for luciferase activity after 3 weeks. Representative images show let-7g greatly reduced bone metastases in skull. Results represent the mean±s.d. for the animals (P=0.001 for let-7g relative to Control). (C) RKIP does not alter primary let-7 expression. The 1833 cells expressing control vector, wt RKIP, or S153E RKIP were lysed. Primary let-7a and let-7g transcripts were analysed by qRT–PCR. Results represent the mean±s.d. for three samples. (D) LIN28 inhibits let-7 expression. The 1833 cells expressing control, wt RKIP, or S153E RKIP were stably transfected with LIN28 expression vector. Let-7a and g transcripts were analysed by qRT–PCR. Left: results represent the mean±s.d. for three samples (P=0.01 and P<0.001 for LIN28 in wt RKIP cells and S153E RKIP cells, respectively, relative to Control); Right: 1833 cells expressing control vector, wt RKIP, or S153E RKIP were lysed and immunoblotted with anti-LIN28 or anti-tubulin antibodies. Results are representative of at least three independent experiments. (E) RKIP decreased LIN28 mRNA in 1833 cells. The 1833 cells expressing control vector, wt RKIP, or S153E RKIP were lysed. LIN28 transcripts were analysed by qRT–PCR. Left: results represent the mean±s.d. for three samples (P=0.003 for wt RKIP and P=0.002 for S153E RKIP relative to Control); right: 1833 cells expressing control vector, wt RKIP, or S153E RKIP were lysed and immunoblotted with anti-LIN28 or anti-tubulin antibodies. Results are representative of at least three independent experiments. (F) shLIN28 downregulates LIN28 expression. The 1833 cells expressing control vector or shLIN28 were lysed and immunoblotted with anti-LIN28 or anti-tubulin antibodies. Results are representative of at least three independent experiments. (G) LIN28 depletion inhibits invasion of 1833 cells. The 1833 cells expressing control vector or shLIN28 were assayed for invasion as described in Materials and methods. Results represent the mean±s.d. for three independent samples (P=0.003 for shLIN28 relative to Control). (H) ShLIN28 inhibits bone metastasis. The 1833 cells expressing luciferase and vector control (seven mice) or shRNA for LIN28 (eight mice) were injected into the left ventricle of mice. Mice were imaged for luciferase activity after 3 weeks. Results represent the mean±s.d. for the animals (P=0.002 for shLIN28 relative to Control).
How does RKIP regulate let-7? Let-7 expression can be controlled at multiple levels, including synthesis of the primary transcript, Drosha processing to the precursor, and Dicer processing to the mature form (Wu et al, 2007). Analysis of one of the three primary let-7a transcripts and the let-7g primary transcript by qRT–PCR showed no increase in response to wt RKIP or S153E RKIP (Figure 3C), indicating that regulation occurs subsequent to primary transcription. Although it is possible that let-7 transcribed from other loci might be subject to transcriptional regulation by RKIP, these data indicate that regulation can occur at the level of Drosha processing to the precursor or Dicer processing to mature let-7.
Recently, LIN28, a regulator of developmental timing in Caenorhabditis elegans that is downregulated by let-7 (Moss et al, 1997; Morita and Han, 2006), was identified as an inhibitor of let-7 primary transcript (pri-miRNA) processing in vitro and in mammalian cells (Newman et al, 2008; Viswanathan et al, 2008). To determine whether LIN28 can regulate let-7 in breast cancer cells, we overexpressed LIN28 in 1833 cells transfected with vector, wt RKIP, or S153E RKIP. LIN28 had little effect in the parental cells, possibly because the LIN28 level in these cells is sufficiently high such that maximal suppression of let-7 to a basal level has already occurred (Figure 3D). By contrast, following induction of let-7a and let-7g expression by RKIP, LIN28 suppressed let-7 back to the basal level (Figure 3D). These data suggest that another regulatory mechanism is responsible for the basal or background let-7 expression levels. Depletion of LIN28 was shown to enhance let-7 expression (Newman et al, 2008; Viswanathan et al, 2008), and LIN28 loss also upregulates let-7 in 1833 cells (Supplementary Figure 4c). To assess whether LIN28 exerts an effect downstream of RKIP to regulate induction of let-7 expression, LIN28 transcripts were analysed by qRT–PCR and immunoblotting. Both wt and S153E RKIP decreased LIN28 mRNA and protein relative to control levels in 1833 cells (Figure 3E). Taken together, these results indicate that RKIP upregulates Drosha or Dicer processing of let-7 via inhibition of LIN28.
If RKIP suppresses invasion and metastasis through a cascade involving inhibition of LIN28, then depletion of LIN28 should mimic RKIP action. To test this hypothesis, we transduced lentivirus expressing shRNA for LIN28 into luciferase-labelled 1833 cells. Loss of LIN28 protein was confirmed by immunoblotting (Figure 3F). Depletion of LIN28 caused a decrease in cell invasion (Figure 3G) but had no effect on cell proliferation (Supplementary Figure 4d). By contrast, transfection of LIN28 into wt RKIP-expressing cells reversed the RKIP-mediated repression and rescued invasion without altering cell growth (Supplementary Figure 4f and g). Finally, injection of the LIN28-depleted 1833 cells into the cardiac ventricle of mice almost completely suppressed bone metastasis (Figure 3H). These data show that LIN28 is required for breast cancer metastasis.
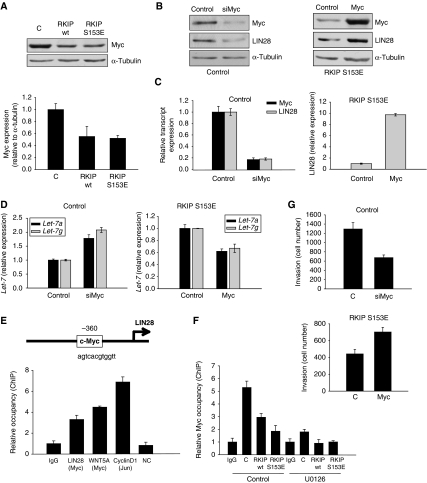
Myc represses the let-7 promoter directly in some cells (Chang et al, 2008), and immunoblotting shows a decrease in Myc protein in RKIP-expressing 1833 cells (Figure 4A). To determine whether Myc can regulate let-7 expression by an alternative mechanism involving LIN28, cells were assayed by immunoblotting and qRT–PCR. Myc depletion by siRNA in 1833 cells decreases LIN28 protein and transcript levels, and overexpression of Myc in 1833 S153E RKIP cells rescues the loss of LIN28 protein and transcripts (Figure 4B and C). In addition, Myc depletion enhances mature let-7 expression in 1833 cells, and Myc overexpression decreases let-7 transcripts in 1833 S153E RKIP cells (Figure 4D). Thus, our data suggest that Myc suppresses let-7 processing in 1833 cells by increasing LIN28 transcripts.
Figure 4.
Myc regulates LIN28 transcription. (A) RKIP downregulates Myc expression. Blot: 1833 cells expressing control vector, wt RKIP, or S153E RKIP were lysed and immunoblotted with anti-Myc or anti-tubulin antibodies. Results are representative of at least three independent experiments. Graph: results represent the mean±s.d. for three independent samples (P=0.03 for wt RKIP and P=0.003 for S153E RKIP relative to Control). (B) Myc regulates LIN28 expression. The 1833 cells transfected with scrambled control or siRNA for Myc (left) or 1833 S153E RKIP cells transfected with a control or a Myc expression vector (right) were lysed and immunoblotted with anti-Myc, anti-LIN28, or anti-tubulin antibodies. Results are representative of at least three independent experiments. (C) Left: Myc regulates LIN28 transcript levels. The 1833 cells were transfected with a control or siRNA for Myc. LIN28 and Myc transcripts were analysed by qRT–PCR 48 h after transfection. Right: 1833 cells expressing S153E RKIP were transfected with a control or Myc expression vector. LIN28 transcripts were analysed by qRT–PCR 48 h after transfection. Results represent the mean±s.d. for three samples (P<0.0001 for siMyc and P<0.0001 for Myc relative to Control). (D) Myc regulates let-7 expression. The 1833 cells were transfected with a control or siRNA for Myc. Let-7a and g transcripts were analysed by qRT–PCR 48 h after transfection. The 1833 cells expressing S153E RKIP were transfected with a control or Myc expression vector. Let-7a and g transcripts were analysed by qRT–PCR 48 h after transfection. Results represent the mean±s.d. for three samples (P<0.01 for siMyc and P<0.01 for Myc relative to Control). (E) Myc regulates LIN28 transcription by binding to its promoter. Schematic representation of LIN28 promoter with the putative Myc-binding site. Chromatin immunoprecipitations (ChIPs) were carried out with anti-Myc antibody and anti-Jun antibody (a positive control for ChIP assay). ChIP was analysed by qRT–PCR, with primers in the LIN28, WNT5A (a positive control), CyclinD1 (a positive control for ChIP assay), and β-globin (a negative control; NC) promoters. Results represent the mean±s.d. for three samples (P<0.004 for LIN28 relative to IgG). (F) RKIP regulates Myc binding to the LIN28 promoter. Chromatin immunoprecipitation (ChIP) were carried out with anti-Myc antibody on the LIN28 promoter. The 1833 cells expressing control vector (C), wt RKIP, or S153E RKIP were treated with 2% serum or 2% serum with U0126 (10 μM) for 2 h after 48 h serum starvation. Results represent the mean±s.d. for three samples (P=0.01 and 0.001 for wt RKIP S153E RKIP, respectively, relative to untreated Control). (G) Myc expression regulates invasion of 1833 cells. Top: 1833 cells transfected with scrambled control or siRNA for Myc were assayed for invasion as described in Materials and methods. Results represent the mean±s.d. for three independent samples (P=0.004 for siMyc relative to Control). Bottom: 1833 S153E RKIP cells transfected with vector control or an expression vector for Myc were assayed for invasion as described in Materials and methods. Results represent the mean±s.d. for three independent samples (P=0.01 for Myc relative to Control).
Examination of the LIN28 promoter shows at least one potential Myc-binding site. To determine whether Myc directly regulates LIN28 transcription by binding to its promoter, we performed quantitative chromatin immunoprecipitation (ChIP) assays. Lysates from 1833 cells were immunoprecipitated with anti-Myc antibody, and the relative association of Myc with the following gene promoters was assessed: LIN28, Wnt5A (a positive control), and β-globin (a negative control) (Figure 4E). As a positive control for the assay, antibody to Jun was used to immunoprecipitate the cyclin D1 promoter. ChIP analysis of cells expressing wt RKIP or S153E RKIP showed that RKIP decreased Myc occupancy of the LIN28 promoter relative to control cells, consistent with the decreased Myc expression (Figure 4F).
If Myc induces LIN28, then Myc depletion should mimic LIN28 depletion by suppressing invasion. As predicted, 1833 cells transfected with siRNA for Myc exhibited decreased invasion relative to control cells (Figure 4G). Conversely, Myc overexpression promoted invasion (Figure 4G). Under the same conditions as the invasion assay, neither Myc depletion nor Myc overexpression affected cell proliferation (Supplementary Figure 5a). However, as Myc is required for cell growth at longer incubation times, it was not possible to assess the function of Myc in regulating metastasis in the absence of major proliferative effects. These results show that Myc binds to the LIN28 promoter to induce its transcription, and RKIP reduces LIN28 expression through suppression of Myc (see scheme in Figure 5).
Figure 5.
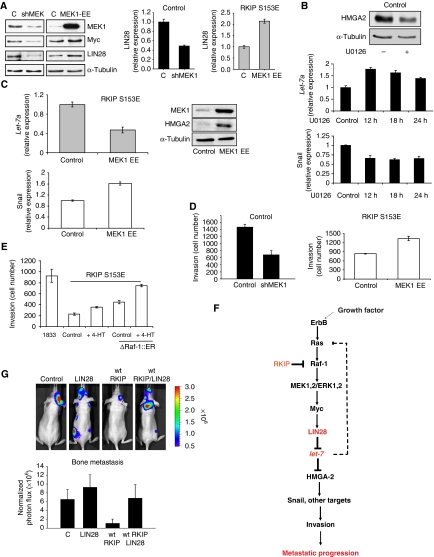
RKIP regulates let-7 in part through the MAPK pathway, and LIN28 reverses the inhibition of bone metastasis by RKIP. (A) MEK1 regulates LIN28 and Myc in 1833 cells. Constitutively, active MEK induces LIN28 and Myc. The 1833 cells expressing S153E RKIP were transfected with control vector (C) or MEK1-EE for 48 h. LIN28 was also assayed by qRT–PCR. MEK1, Myc, lIN28, and α-tubulin expression were measured by western blot. Conversely, stable depletion of MEK by lentiviral shRNA downregulates LIN28 and Myc in 1833 cells. Results represent the mean±s.d. for three samples (P=0.005 for shMEK and P=0.001 for MEK1-EE relative to Control). Immunoblot results are representative of at least three independent experiments. (B) Inhibition of MEK by U0126 induces let-7a and inhibits HMGA2 and Snail. Blot: 1833 cells treated with or without the MEK inhibitor U0126 (10 μM, 12 h) were lysed and immuoblotted with antibodies to HMGA2 or tubulin. Graphs: 1833 cells were treated with 10 μM U0126 for the indicated times, and let-7a was assayed by qRT–PCR. Snail expression was quantitated by qRT–PCR. Results represent the mean±s.d. for three samples (Let-7: P=0.001, 0.01, and 0.03 for 12, 18, and 24 h, respectively, relative to Control; Snail: P=0.01, 0.004, and 0.01 for 12, 18, and 24 h, respectively, relative to Control). Results are representative of at least three independent experiments. (C) Constitutively active MEK inhibits let-7a expression and induces HMGA2 and Snail. The 1833 cells expressing S153E RKIP were transfected with control vector or MEK1-EE for 48 h, and let-7a and Snail were assayed by qRT–PCR. Results represent the mean±s.d. for three independent samples (Let-7a: P<0.02 for MEK1-EE relative to Control; Snail: P=0.01 for MEK1-EE relative to Control). MEK1 and HMGA2 expression were measured by western blot. Results are representative of at least three independent experiments. (D) MEK regulates invasion. Left: 1833 cells expressing either vector control or shRNA for MEK1 were assayed for invasion as described in Materials and methods. Results represent the mean±s.d. for three independent samples (P=0.004 for shMEK1 relative to Control). Right: 1833 cells expressing either vector control or an expression vector for MEK1-EE were assayed for invasion as described in Materials and methods. Results represent the mean±s.d. for three independent samples (P<0.005 for MEK1-EE relative to Control). (E) Inducible Raf kinase rescues the inhibition of invasion caused by S153E RKIP. The 1833 cells expressing S153E RKIP and transfected with control vector or ΔRaf-1:ER were untreated or treated with tamoxifen (4-HT). Results represent the mean±s.e. for three independent samples (P<0.001 for S153E RKIP+ΔRaf-1:ER+HT relative to S153E RKIP Control; P<0.001 for S153E RKIP+ΔRaf-1:ER+HT relative S153E RKIP+ΔRaf-1:ER). (F) Scheme showing the mechanism for RKIP regulation of invasion and metastasis through the MAPK/LIN28/let-7 pathway. (G) LIN28 overcomes the inhibitory effect of wt RKIP on bone metastasis. The 1833 cells expressing luciferase and either control vector or wt RKIP were stably transfected with control vector or LIN28 and injected into the left ventricle of the mice (Control, six mice; LIN28, 5 mice; wt RKIP, six mice; wt RKIP+LIN28, five mice), and mice were imaged for luciferase activity after 3 weeks. Results represent the mean±s.d. for the animals (P=0.001 for wt RKIP relative to Control, and P=0.01 for wt RKIP+LIN28 relative to wt RKIP).
As RKIP is an inhibitor of the Raf/MEK/MAPK signalling cascade, we determined whether MAPK regulates Myc, LIN28, and let-7 in breast epithelial cells. As observed previously for other cell types (Yeung et al, 1999; Trakul et al, 2005), RKIP depletion from MCF10A cells by shRNA upregulates EGF-induced ERK activation, and RKIP expression in 1833 cells downregulates ERK activation (Supplementary Figure 5b and c). Expression of constitutively active MEK (MEK1-EE) enhances Myc and LIN28 expression; conversely, stable depletion of MEK by shRNA or MEK inhibition by the inhibitor U0126 decreases Myc and LIN28 expression (Figure 5A; Supplementary Figure 4e). Consistent with these results, U0126 treatment of control 1833 or RKIP-expressing cells suppresses Myc binding to the LIN28 promoter (Figure 4F). RKIP potentiates U0126-induced repression of Myc binding to the LIN28 promoter, suggesting that RKIP also regulates an MAPK-independent signalling pathway. Finally, suppression of MEK/ERK signalling by U0126 treatment of 1833 cells induces let-7a and inhibits HMGA2 and Snail after 12 h of treatment (Figure 5B). Conversely, constitutively active MEK1-EE expression suppresses let-7a and induces HMGA2 and Snail in 1833 S153E RKIP-expressing cells (Figure 5C). Taken together, these results implicate MEK and ERK1,2 as upstream regulators of Myc, LIN 28, and let-7.
If RKIP downregulates invasion by ERK inhibition, then ERK activation should promote invasion and rescue the RKIP inhibitory phenotype. Consistent with this prediction, MEK1 depletion by shRNA in 1833 cells decreased invasion but had no effect on proliferation (Figure 5D, Supplementary Figure 6a). By contrast, constitutively active MEK1-EE enhanced invasion without altering cell proliferation (Figure 5D; Supplementary Figure 6b). Finally, tamoxifen activation of a stably expressed oestrogen receptor/Raf kinase fusion protein (ΔRaf-1:ER) enhances invasion in 1833 S153E RKIP-expressing cells (Figure 5E). As Raf could theoretically activate non-MEK targets, we confirmed the key function of MAPK in this effect by pretreating the ΔRaf-1:ER cells with the MEK inhibitor U0126 under conditions that do not alter cell proliferation (Supplementary Figure 6c). MEK inhibition blocks the Raf-mediated increase and potentiates RKIP suppression of invasion, consistent with the lack of complete MAPK inhibition by RKIP (Supplementary Figure 6d).
These results support a model whereby RKIP negatively modulates Raf-1/MEK/ERK1,2 activity, leading to the inhibition of Myc and LIN28 and the induction of let-7. Let-7 inhibits HMGA2, and suppression of HMGA2 blocks the induction of Snail transcription and other genes involved in tumour cell invasion and metastatic colonization (Figure 5F). This scheme highlights the function of LIN28 as a key target of RKIP suppression. If RKIP regulates let-7 by inhibiting LIN28 expression in breast tumour cells, then co-transfection of LIN28 should rescue the metastatic phenotype. To test this prediction, we injected luciferase-labelled 1833 cells co-expressing either a control vector or wt RKIP in the presence or absence of stably expressed LIN28 directly into the left cardiac ventricle of mice. Bioluminescence imaging analysis of 1833 cells expressing wt RKIP and LIN28 showed comparable colonization and growth of bone metastasis with that observed for control 1833 cells (Figure 5G), confirming that RKIP suppresses and LIN28 promotes breast cancer metastasis.
Discussion
The results presented herein move from RKIP repression of metastasis and invasion to successive functions for HMGA2 and Snail, let-7, LIN28, Myc, and finally the Raf/MEK/ERK cascade in the RKIP regulatory mechanism (see Figure 5F). These data represent the first direct evidence that let-7 regulates not just tumour growth but also key steps in metastasis. We show that processing of at least some let-7 isoforms is dependent upon LIN28 expression, and that LIN28 is required for bone metastasis by a highly aggressive basal-subtype breast cancer. Finally, we show that MAPK (ERK1,2) negatively regulates let-7 by inducing LIN28 expression through Myc transcription. RKIP regulates this cascade through partial inhibition of both the amplitude and kinetics (data not shown) of ERK activation. As let-7 inhibits Ras (Johnson et al, 2005), an upstream activator of Raf-1, these data implicate let-7 in a positive feedback loop. A study showing negative regulation of RKIP transcription by Snail highlights another potential feedback loop (Beach et al, 2007). RKIP also regulates Myc by an MEK/ERK-independent pathway as illustrated in the ChIP assay, showing Myc regulation of LIN28 transcription. Thus, although this mechanism is depicted as a linear pathway, it is part of a larger network involving feedback regulatory pathways, cross talk, and other RKIP targets.
The MDA-MB-231 metastatic cell line used in this study is a basal-like subtype of breast carcinoma that represents a highly aggressive but small fraction of breast tumours. We chose to focus on an enriched metastatic population derived from this line because we were investigating whether RKIP expression is sufficient to suppress metastatic cells as opposed to preventing progression to a metastatic phenotype. Thus we have not addressed the question in this study of the function of RKIP in tumour progression, particularly with regard to other subtypes, such as invasive luminal breast cancer. It is interesting to note that, in contrast to some other breast cancer subtypes, the MDA-MB-231 cells readily exhibit changes related to EMT (Waerner et al, 2006; Sarrio et al, 2008). Thus, it is most likely that the signalling cascades in these different breast tumour populations and the function of RKIP in their regulation are context specific.
Epithelial–mesenchymal transition is a complex process involving loss of epithelial genes and acquisition of mesenchymal genes. We do not have any evidence that RKIP induces expression of epithelial genes or is sufficient to reverse EMT. For example, unlike miR-200 (Burk et al, 2008; Gregory et al, 2008; Korpal et al, 2008; Park et al, 2008), RKIP does not regulate E-cadherin expression. However, we have observed some inhibition of vimentin expression by the RKIP mutant construct, and we consistently see a decrease in Snail expression, a factor that can contribute to the mesenchymal transition among other functions. Nonetheless, the relatively small change in Snail expression suggests that Snail is not the major mediator of HMGA2 action. We also see consistent reduction in HMGA2, a gene that we have shown to enhance invasion. HMAG2 regulates a number of target genes that contribute to invasion and metastasis independent of Snail (Yun et al., manuscript in preparation). Finally, we do not see a strong morphological change in the cells indicative of transition back to the epithelial phenotype. Therefore, although RKIP may partially counteract EMT by inhibiting some genes required for the mesenchymal phenotype, the evidence is strongest for a function in suppressing invasion and metastasis.
Our results add let-7 to the list of microRNAs that have recently been implicated in breast tumour cell metastasis (Ma et al, 2007; Tavazoie et al, 2008) and represent the first signalling cascade mediated by a tumour metastasis suppressor protein that regulates let-7. The data presented here are supported by a recent report that let-7 inhibits tumorigenesis in a breast cancer stem cell line (Yu et al, 2007a). As either Myc (along with OCT3/4, SOX2, and KLF4) or LIN28 (along with OCT4, SOX2, and NANOG) is sufficient to transform human fibroblasts into pluripotent stem cells (Takahashi et al, 2007; Yu et al, 2007b), our results raise the possibility that RKIP negatively regulates tumour-initiating stem cells. Finally, as a potent inducer of let-7, RKIP represents an important therapeutic target and diagnostic marker.
Materials and methods
Cell culture and reagents
MDA-MB-231 cells were grown in a complete medium consisting of RPMI-1640 supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin and 50 μg/ml streptomycin (Invitrogen Corp., Carlsbad, CA). The bone (1833) and lung (4175) metastatic breast cancer cells were grown in a complete medium consisting of DMEM supplemented with 10% FBS, 50 U/ml penicillin, and 50 μg/ml streptomycin. Cells were treated with EGF (Biomedical Technologies Inc., Stoughton, MA) for 5 min at 50 ng/ml or with U0126 (Fisher, Pittsburgh, PA), used at 10 μM, for 24 h, or as labelled for time-course experiments.
Antibodies specific for vimentin (sc-32322), E-cadherin (sc-21791), α-catenin (sc-7894), N-cadherin (sc-7939), MEK1 (sc-219), and α-tubulin (IgG2a,sc-5286 and IgM, sc-8035) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies for ppERK1/2 (9101) and pMKK1/2 (9121) were purchased from Cell Signaling Technology (Danvers, MA), the antibody for c-Myc (M5546) was purchased from Sigma-Aldrich (St Louis, MO), and the antibody for HMGA2 (59170AP) was purchased from BioCheck Inc. (Foster City, CA). Anti-RKIP antisera used was generated by immunizing rabbits with purified GST-RKIP (α-RKIP). The ΔRaf-1:ER expression vector was generously provided by M McMahon (Samuels et al, 1993). siRNA for Myc (M00328204) and control (D001206405) were purchased from Dharmacon (Lafayette, CO). Pre-miR control (AM17111), pre-miR let-7a (AM17100), anti-miR control (AM17010), and anti-miR let-7a (AM17000) were purchased from Ambion (Austin, TX). LIN28 antibodies were purchased from R&D Systems (Minneapolis, MN).
Generation of stable cell lines
Raf kinase inhibitory protein rescue cell lines were generated by transducing target cells with HA–RKIP wt or HA–RKIP S153E sub-cloned into a pCDH1-CMV-MCS1-EF1-copGFP lentiviral vector (System Biosciences, Mountain view, CA). Stable RKIP depletion in MCF10A cells was achieved using shRNA retroviral vectors as described earlier (Trakul et al, 2005). Stable bone metastatic cells (1833) expressing LIN28 were generated by transducing cells with a pMSCV-puro-IRES-GFP retroviral vector. Cells were transfected using LT-1 transfection reagent (Invitrogen Corp.), using the manufacturer's protocol; 24 h after transfection, cells were selected and maintained in 0.5 μg/ml puromycin. MEK1 knockdown (shMEK1), LIN28 knockdown (shLIN28), and HMGA2 knockdown (shHMGA2) were achieved by transducing the bone metastatic cells (1833) with the respective shRNAs in a pLKO.1 lentiviral vector (Open Biosystems, Huntsville, AL). Cell lines inducible for let-7g expression were constructed by the sequential transductions of lentivirus containing let-7g under the control of a Tet-response element (TRE), and a retroviral vector (pREVTet-On, Clontech) encoding rtTA which in the presence of doxycycline (Dox) binds to TRE to activate transcription (Kumar et al, 2008). In addition, the lentiviral vector encoded puromycin resistance and the retroviral vector encoded neomycin resistance allowing for the selection of stable cell lines at each step.
Immunoblotting
Cells were washed twice with cold phosphate-buffered saline (PBS) on ice and then lysed in cold 1 × SDS-sample buffer. The lysates were immediately boiled at 100°C and then centrifuged at 14 000 g for 10 min. Proteins were resolved on an SDS–PAGE gel before immunoblotting and then detected using either enhanced chemoluminescence western reagents (GE healthcare, Piscataway, NJ) or the membranes were scanned using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). For analysis using the Odyssey System, membranes were probed with an IR dye-tagged secondary antibody (LI-COR Biosciences) rather than an HRP-tagged secondary antibody.
Transient transfection
Cells were transiently transfected using LT-1 transfection reagent (Invitrogen) using the manufacturers' protocol or using the Nucleofector Kit V (Amaxa Biosystems, Gaithersburg, MD) and program X-013 for electroporation.
In vitro cell invasion assay
The invasive capability of the different breast cancer cell lines was evaluated in a 24-well plate using 8-μm-pore-size polycarbonate inserts (BD Biosciences, Bedford, MA). Briefly, the inserts were coated with 50 μg Matrigel basement membrane matrix (BD Biosciences), which was reconstituted in serum-free medium. To assess the ability of cells to invade, 2 × 104 breast cancer cells were seeded on top of the polymerized matrigel in serum-free medium, whereas complete medium (10% FBS) was placed in the lower compartment. The plates were then incubated for 24 h at 37°C, at the end of which the cells were washed with PBS and then the cells on the lower part of the insert were fixed with 4% paraformaldehyde for 10 min. The cells were then stained with Giemsa (1:5) for 15 min, the excess stain was rinsed off with PBS, and cells on top of the insert that had not invaded were scraped off. Invasion was quantified by counting the number of cells in 5 fields at × 100 magnification per filter.
Quantitative ChIP analysis
Chromatin immunoprecipitation was carried out as described previously (Wu et al, 2005). The chromatin fragments were immunoprecipitated with 2 μg of antibodies against either Myc (N-262, Santa Cruz Biotechnology) or Jun (Santa Cruz Biotechnology). Preimmune serum was used as negative controls. The precipitated chromatin was purified by phenol–chloroform extraction and ethanol precipitation. DNA aliquots (2 μl) were analysed by quantitative real-time PCR with the indicated primer pairs. The amounts of products were determined relative to a standard curve generated from a titration of input chromatin. Forward and reverse primers for real-time PCR (5′–3′) ChIP analysis were as follows: LIN28(Myc), GGG AGG GCC CAT TCA TTT C and GGG TCC CCA AAG CAG ATA CA; WNT5A(Myc), GTC GGG AAG TGG TCA AGG TT and AAG TGC CAG AGA CAG ATG CT; CyclinD1(Jun), GTC CCA GGC AGA GGG GAC and CGG CAA TTT AAC CGG GAG A; β-globin promoter as a negative control, AGT GCC AGA AGA GCC AAG GA and CAG GGT GAG GTC TAA GTG ATG ACA; and rRNA as an internal control, ATT AGT CAG CGG AGG AAA AGA AAC and TCG CCG TTA CTG AGG GAA TC.
Animal studies
All animal work was done in accordance with a protocol approved by the Institutional Animal Care and Use Committee. BALB/c female nude mice (Charles River) 6–7 weeks old were used for animal studies. For intravasation assays, cells were orthotopically injected (2 × 106 cells/0.1 ml) into the second mammary fat pad of anaesthetized mice (100 mg ketamine, per kg 10 mg xylazine per kg). The mouse blood was taken from heart and lysed by the addition of RBC lysis buffer (0.155 M NH4Cl, 0.01 M KHCO3, and 0.1 mM EDTA, pH 7.4) at a volume ratio of 3:1 lysis buffer:blood. The samples were incubated for 10 min at 4°C. Cells were collected by centrifugation and resuspended in 1 ml of TRIzol reagent (Invitrogen Corp.). The isolated RNA was analysed for human and mouse GAPDH by qRT–PCR.
For bone metastasis assays, 105 cells in PBS (0.1 ml) were injected into the left ventricle of mice. Mice were imaged for luciferase activity using an IVIS200 Imaging System (Xenogen). Ten minutes before in vivo imaging, mice were anaesthetized with 2% isoflurane and injected intraperitoneally with D-luciferin (100 mg/kg in PBS).
RNA isolation and reverse transcription
mRNA was isolated using TRIzol reagent (Invitrogen Corp.), using the manufacturer's protocol. Briefly, cells were lyzed in TRIzol, followed by a phenol/chloroform (Invitrogen Corp.) extraction to remove impurities. RNA in the aqueous phase was then precipitated using isopropanol, washed with ethanol, and dissolved in DNase–RNase-free water (Invitrogen Corp.). The isolated RNA was then treated with DNase (Turbo DNA-free, Applied Biosystems, Foster City, CA), following the manufacturer's protocol, before being purified using the RNeasy Mini Kit (Qiagen, Valencia, CA). A quantity of 2 μg RNA was then reverse-transcribed using the high-capacity cDNA Reverse Transcription Kit from Applied Biosystems.
For microRNA isolation, we used the mirVana microRNA isolation kit (Applied Biosystems) and followed the manufacturer's protocol. Reverse transcription of 10 ng of isolated microRNA was carried out using the high-capacity cDNA reverse transcription kit (Applied Biosystems), and a specific microRNA primer provided with the TaqMan MicroRNA Assay (Applied Biosystems) was used.
RT–PCR
Quantification of RNA and microRNA was performed by real-time RT–PCR using the Applied Biosystems 7500 Fast Real-Time PCR System. For mRNA, a TaqMan Gene Expression Assay (Applied Biosystems) for the gene of interest was used in conjunction with ABsolute QPCR Low ROX Mix (Thermo Fisher Scientific, Pittsburgh, PA). For microRNA, a TaqMan MicroRNA Assay (Applied Biosystems) was used along with TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). Relative quantification was through the 2−ΔΔCt method. For let-7 analysis, the Primary microRNA primer sequences were as follows (Jiang et al, 2005):
Let-7a-1 forward: CCT GGA TGT TCT CTT CAC TG
Let-7a-1 reverse: GCC TGG ATG CAG ACT TTT CT
Let-7g forward: AGC GCT CCG TTT CCT TTT
Let-7g reverse: CCC CAC TTG GCA GCT G
Cell proliferation assays
Cell proliferation assays were performed using CellTiter-Blue® assay (Promega, Madion, WI) as described by the manufacturer. Briefly, 2000 cells were seeded in a 96 well plate. After 24 h, 20 μl of CellTiter-Blue reagent was added to each well and incubated for 2 h. Fluorescence measurements were used to record data with 560 nm for excitation and 590 nm for fluorescence emission.
Tetracycline induction of let-7g
Cells were treated with 2 μg/ml doxycycline for in vitro studies. For animal studies, 1833 let-7g tet-inducible cells were plated in the presence of 2 μg/ml doxycycline. Twenty-four hours later, cells (105) were injected into the left ventricle of mice. Two days later, mice were administered drinking water containing 4% sucrose only or 2 mg/ml doxycycline and 4% sucrose. Mice were imaged for luciferase activity after 3 weeks.
Statistical analysis
Samples were analysed by the two-sample Student's t test assuming unequal variances (two-tailed). Excel software was used for statistical analysis. P-values were calculated for samples from three independent experiments unless otherwise indicated.
Supplementary Material
Supplementary Figures
Supplementary Figure Legend
Acknowledgments
We thank M Peter for helpful advice and generously sharing his reagents, A Dutta for HMGA2 cDNA lacking let-7 binding sites, and S Gomes for excellent technical assistance. This work was supported by NIH grants NS33858 and CA112310 to MRR, and a gift from the Cornelius Crane Trust for Eczema Research to MRR.
References
- Akaishi J, Onda M, Asaka S, Okamoto J, Miyamoto S, Nagahama M, Ito K, Kawanami O, Shimizu K (2006) Growth-suppressive function of phosphatidylethanolamine-binding protein in anaplastic thyroid cancer. Anticancer Res 26: 4437–4442 [PubMed] [Google Scholar]
- Al-Mulla F, Hagan S, Behbehani AI, Bitar MS, George SS, Going JJ, Garcia JJ, Scott L, Fyfe N, Murray GI, Kolch W (2006) Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J Clin Oncol 24: 5672–5679 [DOI] [PubMed] [Google Scholar]
- Beach S, Tang H, Park S, Dhillon AS, Keller ET, Kolch W, Yeung KC (2007) Snail is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T (2008) A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 9: 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT (2008) Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D, Bai Y, Wang Z, Beach S, Mott S, Roy R, Braastad C, Sun Y, Mukhopadhyay A, Aggarwal BB, Darnowski J, Pantazis P, Wyche J, Fu Z, Kitagwa Y, Keller ET, Sedivy JM, Yeung KC (2004) RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J Biol Chem 279: 17515–17523 [DOI] [PubMed] [Google Scholar]
- Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR (2003) Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem 278: 13061–13068 [DOI] [PubMed] [Google Scholar]
- Eves EM, Shapiro P, Naik K, Klein UR, Trakul N, Rosner MR (2006) Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint. Mol Cell 23: 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Kitagawa Y, Shen R, Shah R, Mehra R, Rhodes D, Keller PJ, Mizokami A, Dunn R, Chinnaiyan AM, Yao Z, Keller ET (2006) Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate 66: 248–256 [DOI] [PubMed] [Google Scholar]
- Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, Keller ET (2003) Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst 95: 878–889 [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601 [DOI] [PubMed] [Google Scholar]
- Hagan S, Al-Mulla F, Mallon E, Oien K, Ferrier R, Gusterson B, Garcia JJ, Kolch W (2005) Reduction of Raf-1 kinase inhibitor protein expression correlates with breast cancer metastasis. Clin Cancer Res 11: 7392–7397 [DOI] [PubMed] [Google Scholar]
- Jiang J, Lee EJ, Gusev Y, Schmittgen TD (2005) Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res 33: 5394–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3: 537–549 [DOI] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283: 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 105: 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A (2007) The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21: 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz K, Lohse MJ, Quitterer U (2003) Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 426: 574–579 [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688 [DOI] [PubMed] [Google Scholar]
- Massague J (2007) Sorting out breast-cancer gene signatures. N Engl J Med 356: 294–297 [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J (2005a) Genes that mediate breast cancer metastasis to lung. Nature 436: 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, Ponomarev V, Gerald WL, Blasberg R, Massague J (2005b) Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest 115: 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Han M (2006) Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J 25: 5794–5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646 [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ML, Weber MJ, Bishop JM, McMahon M (1993) Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human Raf-1 protein kinase. Mol Cell Biol 13: 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J (2008) Epithelial–mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 68: 989–997 [DOI] [PubMed] [Google Scholar]
- Schuierer MM, Bataille F, Hagan S, Kolch W, Bosserhoff AK (2004) Reduction in Raf kinase inhibitor protein expression is associated with increased Ras-extracellular signal-regulated kinase signaling in melanoma cell lines. Cancer Res 64: 5186–5192 [DOI] [PubMed] [Google Scholar]
- Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME (2007) Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA 104: 11400–11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A (2006) Transforming growth factor-beta employs HMGA2 to elicit epithelial–mesenchymal transition. J Cell Biol 174: 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakul N, Menard RE, Schade GR, Qian Z, Rosner MR (2005) Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J Biol Chem 280: 24931–24940 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H (2006) ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10: 227–239 [DOI] [PubMed] [Google Scholar]
- Wu J, Iwata F, Grass JA, Osborne CS, Elnitski L, Fraser P, Ohneda O, Yamamoto M, Bresnick EH (2005) Molecular determinants of NOTCH4 transcription in vascular endothelium. Mol Cell Biol 25: 1458–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Sun M, Zou GM, Chen J (2007) MicroRNA and cancer: current status and prospective. Int J Cancer 120: 953–960 [DOI] [PubMed] [Google Scholar]
- Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W (1999) Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401: 173–177 [DOI] [PubMed] [Google Scholar]
- Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, McFerran B, Wyche J, Kolch W, Sedivy JM (2001) Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation. Mol Cell Biol 21: 7207–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E (2007a) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131: 1109–1123 [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA (2007b) Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA (2007) microRNAs as oncogenes and tumor suppressors. Dev Biol 302: 1–12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Figure Legend