Abstract
Chloroplast RNA metabolism is integrated into wider gene regulatory networks. To explore how, we performed a chloroplast genome-wide expression analysis on numerous nuclear Arabidopsis mutants affected in diverse chloroplast functions and wild-type plants subjected to various stresses and conditions. On the basis of clustering analysis, plastid genes could be divided into two oppositely regulated clusters, largely congruent with known targets of nucleus- and plastid-encoded RNA polymerases, respectively. Further eight sub-clusters contained co-transcribed and functionally tightly associated genes. The chloroplast transcriptomes could also be classified into two major groups comprising mutants preferentially affected in general plastid gene expression and other chloroplast functions, respectively. Deviations from characteristic expression profiles of transcriptomes served to identify novel mutants impaired in accumulation and/or processing of specific plastid RNAs. Expression profiles were useful to distinguish albino mutants affected in plastid gene expression from those with defects in other plastid functions. Remarkably, biotic and abiotic stressors did not define transcriptionally determined clusters indicating that post-transcriptional regulation of plastid gene expression becomes more important under changing environmental conditions. Overall, the identification of sets of co-regulated genes provides insights into the integration of plastid gene expression into common pathways that ensures a coordinated response.
Key words: chloroplast transcriptome, Arabidopsis mutants, cluster analysis, expression profiling, macroarray and microarray analysis
1. Introduction
Plastid genes are embedded in regulatory networks that enable adaptive and developmentally flexible chloroplast biogenesis. Coordination of plastid and nuclear gene expression on both transcriptional and post-transcriptional levels is important for chloroplast function.1–4
The activity of the plastid-encoded RNA polymerase (PEP) depends on nucleus-encoded sigma factors, which are mainly involved in global, tissue-specific, as well as environmentally and developmentally dependent regulation of transcription.5–11 Moreover, two newly acquired nucleus-encoded phage-type RNA polymerases (NEP, represented by RpoTp and RpoTmp) are involved in the regulation of plastid transcription, adding a further layer of complexity to chloroplast RNA metabolism.12–17
Another important characteristic of chloroplast gene regulation is the predominance of post-transcriptional control.18–20 Although nuclear mutants in higher plants often show pleiotropic phenotypes, there is increasing evidence that numerous nuclear genes for chloroplast proteins control post-transcriptional processing and stabilization of individual plastid transcripts.1,21–25
DNA array technologies of chloroplast transcriptomes have so far largely been restricted to individual conditions and/or single mutations affecting chloroplast functions.20,26–29 Moreover, in contrast to in-depth studies on nuclear gene expression, relatively little genome-wide information is available regarding the co-regulated expression of groups of plastid genes. Since the expression of nuclear and chloroplast genes must be tightly coupled, comparative array-based analysis of both genomes should provide a framework for the understanding of the integrated gene regulatory network. Affymetrix 22K ATH1 is an Arabidopsis oligonucleotide array containing more than 22 500 probe sets and is currently well used for global evaluation of gene expression from plants grown under various conditions. In order to elucidate the interactive networks and to identify novel Arabidopsis mutants impaired in chloroplast gene expression, we have established macroarrays and complemented our results with data acquired using the Affymetrix 22K ATH1 array for expression profiling.30
Distinct expression profiles highlight clusters of plastid genes that are potential targets for concerted nuclear control.
2. Materials and methods
2.1. Sources of Arabidopsis mutants, phenotypes and plant growth conditions
Mutant plants were characterized on the basis of their color and the maximum quantum yield (Fv/Fm) of photosystem II (Supplementary Table S1).31 Wild-type (WT) and mutant seeds were surface-sterilized before plating on an MS medium supplemented with 15 g sucrose/L. Plates were incubated at 4°C for 2 days in the dark, and then placed in a climate chamber under continuous light (60 µmol photons m−2 s−1) at 22°C.
2.2. RNA isolation and gel blot analysis
Total RNA was extracted from frozen plant tissues using TRIzol® Reagent according to the manufacturer’s (Invitrogen, Karlsruhe, Germany) instructions. Northern analysis was performed using radio-labeled DNA probes as described.24
2.3. Preparation of macroarray filters
Ninety-four probes for genes encoding plastid proteins, tRNAs and rRNAs were amplified from DNA of WT plants (accession Columbia) using gene-specific oligonucleotides (data available upon request). Intron-containing genes were amplified with the Titan One Tube RT–PCR Kit (Roche, Mannheim, Germany). Size and quality of PCR products purified with PCR Purification Kit (Qiagen, Hilden, Germany) were checked by electrophoresis on 1.2% agarose gels, and three different dilutions (30.0, 7.5 and 1.87 ng/µL) were prepared. Probes were spotted onto 11.9 cm x 7.8 cm positively charged nylon membranes (Hybond™-N+Amersham Pharmacia Biotech, Munich, Germany) using a 96-pin tool (0.4 mm pins) with a BioGrid Spotting Device Roboter (BioRobotics, Boston, USA) as described.27,32 Each probe was spotted 20 times in duplicate giving 1.25, 5 or 20 ng per spot (Supplementary Fig. S1A and B). The spotted DNA was denatured (1.5 M NaCl, 0.5 M NaOH), neutralized (0.5 M Tris–HCl pH 7.2, 1 M NaCl) and fixed with UV light (120 mJ, 302 nm, UV Stratalinker 1800, Stratagene, La Jolla, USA).
2.4. Hybridization of labeled cDNAs to macroarray filters
Before hybridization, macroarray filters were incubated at 65°C for 1 h in 10 ml of buffer (0.25 M Na2HPO4 pH 7.2, 7% SDS). [α-32P] dCTP-labeled cDNAs were synthesized at 50°C for 1 h, with hexanucleotide primers (Roche) and 20 µg of total RNA as template, using SuperScript™ III RNase H− Reverse Transcriptase (Invitrogen). After inactivation of the transcriptase at 70°C for 20 min, the labeled cDNAs were incubated at 37°C for 20 min with RNase H (Invitrogen) to remove RNA. cDNAs were purified on MicroSpin™ G-25 columns (Amersham Pharmacia Biotech) and used for hybridization (12 h at 65°C in hybridization buffer). Filters were washed separately at 65°C for 20 min each in three different washing buffers (2× SSC, 0.1% SDS; 1.0× SSC, 0.1% SDS; 0.5× SSC, 0.1% SDS).
2.5. Normalization and statistical analysis
To maximize the precision of array data, probes were spotted in three different concentrations and in duplicate. The radioactive images were scanned with an FLA-3000 phosphoimager (Fuji, Tokyo, Japan), and the AIDA Image Analyzer (3.52) software was used for background correction and normalization of the signals. The mean value of three selected background dots within each sub-grid was used for background subtraction. Background-corrected hybridization signals were normalized using R/MAANOVA version 0.98.8 implemented in the R program (www.r-project.org).33 The robust locally weighted regression (LOWESS) method in R/MAANOVA was applied for normalization.34 After performing standard t-tests, adjusted P-values for each gene were calculated from ratios of six individual spots using a web-based microarray analysis toolbox (http://nbc11.biologie.uni-kl.de).
2.6. Microarray data analysis
ATH1 (22 k) expression data from Arabidopsis thaliana were obtained from Genevestigator, GEO and AtGenExpress databases, using data for mutants and a variety of biological conditions.35,36 For identification of co-expressed genes located within operons, the MAS 5.0 method is recommended.37 Therefore, all microarray data were normalized by the MAS 5.0 method using Simpleaffy implemented in the R program.38 Expression profiles of 79 plastid genes were selected for cluster analysis. Fold changes were first converted to log2 and expressed relative to the mean value for normalization. Several clustering methods (hierarchical, SOM, K-means and Terrain clustering) were implemented using the program Genesis.39–41 The latter Terrain clustering method was recently successfully applied in DNA microarray experiments of Caenorhabditis elegans. Co-regulated genes were grouped together and visualized in a three-dimensional expression map that displays correlations of gene expression profiles as distances in two dimensions and gene density in the third dimension.42
3. Results
3.1. Plant growth and mutant phenotypes
Unless otherwise indicated, leaves from 3-week-old WT and mutant plants grown in a climate chamber (Percival, Iowa, USA) under continuous light (60 µmol photons m−2 s−1) were used for our analyses. Salient information on mutants, their sources and phenotypic characteristics including photosynthetic parameters is given in Supplementary Table S1, together with details of the growth and stress conditions employed. Most of the genes affected in the mutants studied are essential for photoautotrophic growth. Such mutants survived only when grown on medium supplemented with sucrose. The mutants exhibited three major phenotypes: albino mutants arrested at an early stage of chloroplast development, hcf (high chlorophyll fluorescence) mutants with impaired photosynthetic electron transport capacity and yellow to pale-green lines with defects in various, often unknown, chloroplast functions (Supplementary Table S1).43
3.2. Construction of plastid macroarrays
DNA macroarrays bearing probes for genes encoding all plastid proteins, ribosomal RNAs and 11 tRNAs were constructed for comprehensive expression analyses of various Arabidopsis mutants affected in chloroplast development and function, and WT plants grown under various environmental conditions (Supplementary Fig. S1A and B). Mutant names and gene functions are listed in Supplementary Table S2. In order to ensure statistical reliability for the evaluation of expression levels, three different concentrations of each gene probe were spotted in duplicate onto filters, background subtraction for each gene grid was performed, P-values were calculated taking into account all spots and selected experiments were repeated several times (Supplementary Fig. S1). Moreover, scatter-plot analyses were performed routinely (Supplementary Fig. S1C).
3.3. Expression profiling of plastid genes from plants grown under various biological conditions
In addition to non-photosynthetic mutants, different WT tissues (stems, leaves and flowers), and WT plants exposed to various hormones (gibberellin, abscisic acid, auxin and cytokinin), herbicides 3-(3′, 4′-dichlorphenyl)-1,1-dimethylurea (DCMU) and methylviologen (N,N′-dimethyl-4,4′-bipyridinium, MV), biological stresses (heat, light, cold and dark) or treated with sucrose for defined times, were used to study plastid gene expression patterns (Supplementary Tables S1 and S2).
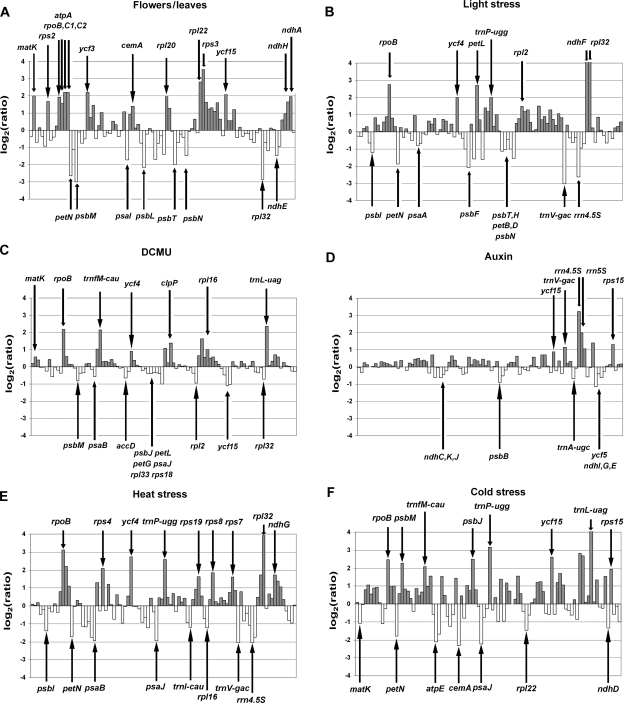
In the flower, some photosynthetic genes tended to be down-regulated and non-photosynthetic genes were preferentially up-regulated when compared with leaves (Fig. 1A). Most of the chosen stress conditions induced significant deviations from the expression pattern observed under standard conditions (Fig. 1B–F). Similarly, excess light, which induces photoinhibition and formation of reactive oxygen species, caused both a reduction in photosynthetic gene expression and an induction of genes required for RNA and protein synthesis in the chloroplast (Fig. 1B). Compared with light stress, DCMU, preventing reduction of plastoquinone, had comparable but milder effects on the expression of plastid genes (Fig. 1C).44 Auxin plays an essential role in the coordination of numerous developmental processes in the plant life cycle.45 Although auxin treatment had a strong bleaching effect, the expression of most plastid genes was not significantly changed indicating that translational and post-translational processes are prevalent to cope with the hormone treatment. However, the 4.5S and 5S ribosomal RNAs were much more abundant following treatment with auxin (Fig. 1D). In contrast, several other stress conditions, including heat (Fig. 1E), cold (Fig. 1F) and sucrose depletion, significantly affected the expression of numerous plastid genes (Supplementary Table S2). Interestingly, differential expression of many plastid genes was more pronounced in WT plants exposed to various physical stress conditions (e.g. light, heat and cold) than in several non-photosynthetic mutants [e.g. hcf145 and cyt160 (cyt b6f mutant)] (Figs. 1 and 2).24
Figure 1.
Changes in plastid transcript levels in tissues and leaves exposed to the indicated stressors. (A–F) Log2-transformed fold changes in plastid RNA levels were determined in flowers when compared with leaves (A), and in leaves subjected to high light stress (B), DCMU treatment (C), auxin treatment (D), heat (E) and cold stress (F) when compared with untreated control leaves. High expression ratios are indicated by arrows. Detailed information on the stress conditions employed is given in Supplementary Table S1. In all six histograms, genes are listed according to their positions on the plastid chromosome.
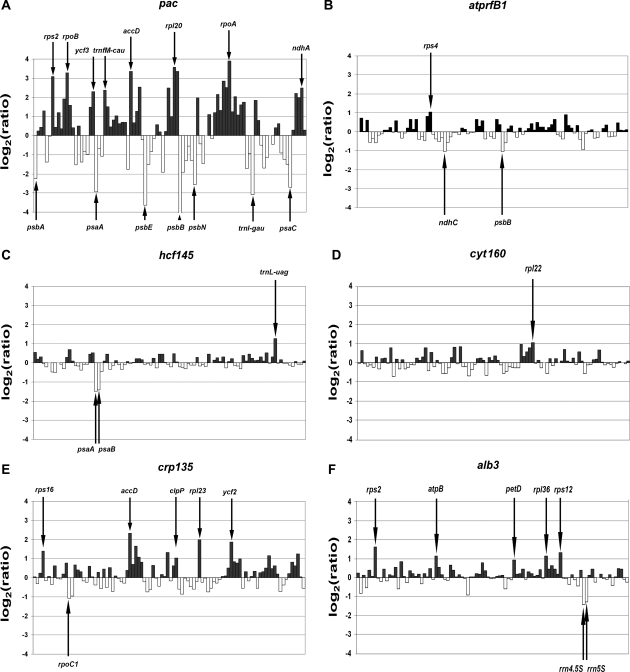
Figure 2.
Plastid transcript levels in six representative nuclear mutants compared with WT. (A–F) Log2-transformed fold changes in RNA levels in each mutant are expressed relative to WT. Up-regulated genes have positive, down-regulated genes negative values. Significantly deviating expression ratios are indicated by arrows. Adjusted P-values for each gene are listed in Supplementary Table S2. (A) pac, (B) atprfB1, (C) hcf145, (D) cyt160, (E) crp135 and (F) alb3.
3.4. Cluster analyses of plastid genes based on the analysis of 89 transcriptomes
For all data sets, we calculated P-values as a measure of the significance of the differential expression of plastid genes in 75 mutants, 3 WT tissues and WT plants placed under 12 different stresses (see previous section) relative to levels in WT plants grown under standard conditions. Filtering of genes with P-values below 0.05 or showing more than 1.5-fold expression changes did not significantly affect clustering of co-expressed plastid genes. Therefore, we included all data sets in our clustering analysis to permit a more comprehensive and subtle expression analysis of the plastid genome.
K-means clustering of all 89 transcriptomes identified two groups, I and II, which displayed essentially opposite expression patterns (Fig. 3A). Most of the mutants in group I (∼70%) exhibited an albino phenotype. The majority of genes up-regulated in group I had non-photosynthetic functions, whereas most down-regulated genes coded for photosynthetic proteins.
Figure 3.
Expression profiles of 94 plastid genes in 89 transcriptomes. (A) Transcript levels in 75 mutants and in WT plants exposed to 14 different biological conditions were determined by macroarray analysis. Fold change values were transformed to log2 and normalized relative to the mean value of genes and experiments. Non-hierarchical K-means clustering (K=2) was performed as described in the ‘Results’. Fold changes close to, higher and lower than the mean values are represented by black, red and green colors, respectively. Co-expressed plastid genes were distributed into two major clusters A and B, which were further divided into eight classes (A–H). Cluster A (green bar) and cluster B (red bar) contain each four classes. Detailed information can be found in Table 1 and Supplementary Table S3. (B) Average expression views of plastid genes in each class show eight distinct expression patterns of plastid genes in 89 transcriptomes. The colors used correspond to the classes in Fig. 3A. The mean expression pattern within each gene class is shown by the black line. The x- and y-axes represent the 89 transcriptomes and log2-transformed fold changes of plastid genes, respectively. The order of the transcriptomes is according to Fig. 3A. (C) Average expression views of plastid genes in clusters A (green) and B (red). The order of the 89 transcriptomes is identical to that shown in panel A. (D) Here, expression profiles were used to cluster the 89 transcriptomes rather than genes using non-hierarchical terrain clustering as described in the ‘Methods’. The terrain map is reminiscent of a model of a complex mountain ridge and illustrates the correlation of the 89 transcriptomes in three dimensions. The appearing clusters reflect individual mountains of specific size and shape depending on the number of and correlation between genes in that cluster, respectively. Peak height corresponds to the density of transcriptomes, denoted by red, yellow and green colors. The white cube on each peak indicates an individual transcriptome or a group of transcriptomes and neighboring peaks have similar expression profiles. The arrows indicate the two distinct transcriptome groups.
Group II comprises transcriptomes of WT plants exposed to a variety of stress conditions and mutants affected in various chloroplast functions. Mutants of group II often showed a pale or hcf phenotype. The behavior of the albino mutants in group II (∼21%), like alb3 and vipp1, deviates from the group I signature typical for most albino mutants.46,47 Unlike those in group I, the albino mutants in group II did not show marked differential expression of plastid genes. Interestingly, early arrest of chloroplast development and the acquisition of an albino phenotype are not necessarily correlated with the expression signature of group I. Most mutants in group II exhibited similar expression patterns, showing less striking expression changes and a smaller number of significantly differentially expressed genes than mutants of group I (Fig. 3A and Supplementary Table S2), which is consistent with primary defects in the gene expression system in mutants of group I. The data also imply that related genetic defects affecting chloroplast functions and resulting in comparable phenotypes exhibit similar expression patterns, like those of albino or hcf mutants, which are characteristic for mutants in group I or II, respectively.
K-means clustering identified two major gene clusters, A and B, of similar sizes that exhibited opposite patterns of expression (Fig. 3A and Table 1). Cluster A contains 30 genes involved in gene expression, 6 ATP synthase and 6 NDH genes. Cluster B comprises genes for 31 components of linear and cyclic electron transport, rbcL and 16 non-photosynthetic genes.
Table 1.
Functional categories and distribution of plastid genes within the two clusters shown in Fig. 3
| Functional categories | Cluster A | Cluster B |
|---|---|---|
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| NADH dehydrogenase | ndhA, ndhB, ndhC, ndhH, ndhK, ndhJ | ndhD, ndhE, ndhF, ndhG, ndhI |
| Cytochrome b6f | petA, petB, petD, petG, petL, petN | |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbTc, psbZ | |
| Ribosomal proteins | rpl14, rpl16, rpl20, rpl36, rps11, rps12, rps15, rps16, rps18, rps2, rps4, rps7, rps8 | rpl2, rpl22, rpl2, rpl32, rpl33, rps14, rps19, rps3 |
| RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| rRNAs | rrn23S, rrn4.5S | rrn16S, rrn5S |
| tRNAs | trnA-ugc, trnF-gaa trnfM-cau, trnI-cau trnK-uuu, trnL-uag trnP-ugg | trnE-uuc, trnI-gau trnR-acg, trnV-gac |
| Unknown | ycf15, ycf2 | ycf1 |
| Others | ycf3, ycf4, ccsA/ycf5, ycf10, accD, clpP | matK, rbcL |
Remarkably, most Arabidopsis genes known to be transcribed by the NEP are present in cluster A, whereas genes transcribed preferentially by the PEP are found in cluster B.17,48,49 With the exception of rrn16S and ycf1, Arabidopsis genes that contain both NEP and PEP promoters are found in cluster A. In accordance with this, the expression pattern of group I mutants is indicative of a loss of the PEP activity since the expression profile is almost identical to mutants affected in the PEP activity.12,27 Therefore, most group I mutants are affected, either primarily or secondarily, in general RNA metabolism.
K-means clustering further sub-classified plastid genes into eight distinct, co-regulated and often functionally associated gene classes; classes A–D are found in cluster A and classes E–H in cluster B (Fig. 3B and Supplementary Table S3). The averaged expression ratios for each class provide a differential expression view (Fig. 3B and Supplementary Table S3). Clustered genes generally derived from the same polycistronic operons (e.g. psbB-, psbD-, atpB-, petG, atpA-, ndhC- and the ribosomal S10 operons) (Supplementary Tables S3 and S4). There are also numerous exceptions, like rbcL/accD, psbE/psbF, psaB/rps14, petA/ycf10 and others. In these cases it is likely that processed monocistronic transcripts of a polycistronic transcription unit undergo different half-lives so that their levels do not correlate with co-transcriptional control. This has also been confirmed genetically since nuclear mutants exist that are specifically affected in the stability of individual RNA segment intervals of polycistronic precursors.1,2,21–24
Co-expression of ndhH and ndhA in cluster A and association of ndhI, ndhG, ndhE and ndhD expressed farther downstream of the DNA strand of the former two in cluster B may designate a hitherto unidentified internal PEP promoter within the ndhH operon (Table 1).
The sizes and membership of clusters that emerge from clustering analysis are known to strongly depend on the program and parameters utilized. We therefore performed clustering analyses with additional methods to confirm the identification of co-regulated genes, using self-organizing map (SOM) and hierarchical clustering methods.50 We confirmed the existence of two distinct oppositely expressed gene clusters (A and B), and both methods assign essentially the same sets of genes to the two clusters (Fig. 3C, Table 1). Each peak in the terrain map represents a transcriptome and peak heights reflect the dynamics of gene expression. The two-dimensional orientations and distances between peaks, which reflect relationships between transcriptomes, indicate that members of group I are quite unrelated to those of group II (Fig. 3D). The biological significance of the terrain cluster analysis is the existence of two distinguishable groups of related transcriptomes and data obtained are consistent with SOM and hierarchical clustering methods (Fig. 3D).
According to the original expression data clustering is largely based on an opposite expression of genes in albino mutants of group I relative to gene members of group II. Notably, clustering of transcriptomes of WT plants adapted to various detrimental conditions failed to assign two main clusters of transcriptomes and genes (data not shown) indicating that post-transcriptional control of plastid gene expression prevails under changing environmental conditions. This finding is consistent with data obtained by using microarry expression profiles (see below).
3.5. Identification of mutants affected in the chloroplast RNA metabolism
Most mutants of group I exhibiting pleiotropic effects are affected in general plastid gene expression and most likely in plastid RNA metabolism. To distinguish mutants primarily affected in plastid RNA metabolism from those perturbed in other plastid functions, we used the mutants pac,51 atprfB122,52 and hcf14524 as controls, which are known to be primarily affected in plastid RNA degradation and processing. The data obtained from the macroarray analysis of these mutants are in good agreement with those obtained previously by northern and quantitative RT–PCR analysis (Fig. 2).22,24,51 For example, the expression of most photosynthetic genes was severely reduced (>2.5-fold) but many genes encoding ribosomal proteins, sub-units of the RNA polymerase, ycf3 and accD were significantly up-regulated (>2.5-fold) in pac (Fig. 2A).51 Severely reduced expression of UGA stop codon-containing psbB and ndhC/K/J transcripts could be confirmed in the atprfB1 mutant defective in the expression of a ribosomal release factor (Fig. 2B).22,52 The nucleus-encoded factor HCF145 is involved in stabilization of the tricistronic psaA–psaB–rps14 transcript in Arabidopsis.24 Accordingly, the histogram for the hcf145 mutant shows that, although the expression of most genes was not severely altered, the abundance of psaA and psaB transcripts was drastically reduced (Fig. 2C), confirming for the first time the high specificity of the effect of this mutation on mRNA metabolism.24 Similarly, the severe defect in accumulation of the cytochrome complex in the mutant cyt160 (data not shown) did not result in the differential expression of most plastid genes (Fig. 2D). In contrast, the newly identified mutant crp135 (chloroplast RNA processing) showed highly elevated levels of some non-photosynthetic genes and down-regulation of 16S and the two autocatalytically cleaved 23S rRNA species a and b, whereas general differential expression of plastid genes was less prominent (Fig. 2E).
Our analyses identified 25 mutant lines in which at least 10 genes showed at least a fourfold change in expression level compared with the WT. In 43 mutants, including the pac control and the T-DNA insertion (ins) mutants ins20, ins24 and ins25, the number of genes showing significant differential expression (>2.5-fold change) was greater than 10. In six mutants, levels of only a few transcripts were significantly altered in either direction, whereas in the control mutant hcf145, only the psaA–psaB–rps14 transcript showed a marked decrease (Supplementary Table S2). In the remaining mutants, such as vipp1 and alb3, the degree of up- and down-regulation was less striking (<2.5-fold) suggesting that the expression profile in these mutants essentially reflects secondary or milder primary effects (Fig. 2F). Indeed, the plastid proteins VIPP1 and ALB3 have been shown to be responsible primarily for the assembly of membranes and thylakoid complexes, respectively.47,48
Since almost all chloroplast transcriptomes responded rather sensitively to mutations and displayed complex expression patterns, the characteristic expression signatures of groups I and II or simple quantification of changes in transcript levels could not be used to identify specific defects in chloroplast RNA metabolism. Instead, we selected mutants in groups I and II that deviated from the general expression signature for their group, e.g. mutants in groups I and II showing down-regulation especially of co-transcribed genes in clusters A and B, respectively, or vice versa. In addition to mutants of group I generally affected in plastid RNA abundance, this approach allowed the identification of 14 specific RNA metabolism mutants: the three control mutants, pac, hcf145 and hcf109, and 11 new mutants, including crp135 (Supplementary Table S5).
Exemplarily, one putative RNA metabolism mutant, crp135 (Fig. 2E), has been selected for preliminary northern analysis to check its deviated expression atypical for group II members. Although the crp135 transcriptome clustered in group II, non-photosynthetic genes, such as clpP and accD, were up-regulated several fold and 16S rRNA was severely down-regulated when compared with the WT (Fig. 4). This is in good agreement with the array data (Fig. 2E). Furthermore, the mutant also showed severe RNA processing defects, indicating a functional link between mRNA processing and abundance. This demonstrates that the array-based approach is well suited to identifying nuclear genes relevant for regulating the general RNA metabolism (group I mutants) and specific plastid RNA transcripts.
Figure 4.
Northern analysis of the plastid genes clpP and accD in WT and crp135. Each lane was loaded with 10 µg of total leaf RNA isolated from 3-week-old mutant and WT seedlings that had been grown on sucrose-supplemented agar medium. Staining shows equal loading of RNAs and reduced levels of plastid rRNAs (23Sa, 23Sb, 16S) when compared with cytoplasmic rRNAs (25S, 18S). The numbers on the left indicate RNA sizes in bases. clpP (caseinolytic protease); accD (carboxyltransferase beta sub-unit of the acetyl-CoA carboxylase).
3.6. Use of Affymetrix microarrays for the evaluation of plastid gene expression in various Arabidopsis mutants
ATH1 microarrays of A. thaliana represent probes of ∼24 000 nuclear and 79 plastid genes.53 As has shown before the hybridization signals of plastid genes were generally several-fold higher than or comparable to that of even highly expressed nuclear genes confirming that the ATH1 microarray data are useful for evaluating plastid gene expression.30
We collected expression data of plastid genes from 136 different transcriptomes associated with mutants that are mostly affected in non-chloroplast functions, such as morphogenesis of plant organs and signaling pathways. Similar to stress conditions, these data did not allow plastid transcriptomes and genes to be classified into two groups and two NEP and PEP determined clusters, respectively (Supplementary Fig. S2 and Table S6). Nevertheless, in accordance with the operon organization of plastid genes, 35 out of all 79 protein-coding genes co-transcribed in 14 polycistronic units were identified as being co-expressed (Supplementary Table S7). Hierarchical clustering identified nine groups of plastid transcriptomes (I–IX) and six co-regulated plastid gene clusters (A–F) (Supplementary Table S8). The largest differences in the average expression views were between clusters A and B (Supplementary Fig. S2B). Taken together, these results demonstrate that genetic defects unrelated to the chloroplast do not impose discernible plastid gene expression profiles or identify new clusters of plastid genes that are under common transcriptional control.
3.7. Use of microarrays for the evaluation of plastid gene expression under various stress conditions
Fold changes in plastid gene expression were calculated from data for plants subjected to 83 stress conditions, including various abiotic, biotic, pathogen, chemical, nutrient, hormone and light stresses.35 It appeared that plastid transcriptomes and genes clustered according to whether or not different stress conditions affected the chloroplast (Fig. 5 and Supplementary Table S9). Again, two groups of generally oppositely regulated transcriptomes, I and II, could be defined. In group I, the stressors included biotic stresses, light (all spectral qualities tested, except UV-B) and various chemicals, whereas group II includes UV-B light, abiotic, nutrient, and hormone stresses. Six clusters (A–F) of co-regulated genes emerged (Supplementary Table S10). Although genes in clusters A–D encode quite heterogeneous functions, clusters E and F contain genes with almost exclusively non-photosynthetic (19 out of 21 genes) and photosynthetic (14 out of 18 genes) functions, respectively (Supplementary Table S10). Genes present in clusters E and F generally show a comparable expression behavior like a down-regulation under biotic stress and an up-regulation after hormone treatment but are oppositely regulated predominantly under different light conditions. This indicates that genes containing NEP and PEP promoters are preferentially light-dependent down- and up-regulated, respectively.
Figure 5.
Expression map of 79 plastid genes under 83 various conditions generated from Genevestigator. (A) Hierarchical clustering identified six co-regulated gene clusters as illustrated by different color bars. Up-regulated, down-regulated and unchanged gene expressions are labeled by red, green and black colors, respectively. (B) The average expression views of plastid genes in each identified cluster are shown. The mean expression pattern within each cluster is shown by black color. The x- and y-axes represent 83 different stress conditions and log2-transformed fold changes of plastid genes, respectively.
Expression of genes in clusters A and B was generally highly induced in group I but reduced in group II (Fig. 5A and B). Transcript levels for genes in clusters C and D were relatively high under various hormone stress conditions, but low under chemical stresses. The average expression views for clusters A–D displayed severe up- and down-regulation of plastid genes, whereas those for clusters E and F did not show significant alterations in plastid gene expression (Fig. 5B). Again, 14 groups of co-expressed and often co-transcribed genes were identified, indicating that co-transcriptional processes contribute to controlling the abundance of transcripts originating from single operons (Supplementary Table S11). However, in accordance with the macroarray data using solely various detrimental conditions, biotic and abiotic effects did not lead to clustering in two major oppositely regulated and transcriptionally determined gene associations, again indicating a prevailing post-transcriptional control under changing environmental conditions rather than a transcriptional control.
4. Discussion
4.1. Hierarchical clustering of plastid mutant transcriptomes identifies two distinguishable expression signatures and novel mutants impaired in RNA metabolism
K-means and hierarchical clustering of transcriptomes of mutants affected in chloroplast functions led to the definition of two groups, I and II. Mutants that fell into group I mostly exhibited an albino phenotype with loss of photosynthetic capability. Group II consists preferentially of expression profiles of hcf and pale green mutants, and patterns characteristic of various tissues or induced by particular environmental conditions. Transcriptomes of group I were generally oppositely regulated relative to those of group II (Fig. 3). The plastid gene expression profile of tobacco PEP mutants revealed that most genes for photosynthesis and ribosomal RNAs were down-regulated and those for ribosomal proteins and RNA polymerase were relatively up-regulated.27 Interestingly, the signature of group I transcriptomes closely resembled not only the expression pattern of the PEP mutants in tobacco, but also those of plastid gene expression mutants in Arabidopsis and WT lines treated with lincomycin which inhibits plastid translation and therefore expression of the PEP.3,12,27,54,55 Several genes in tobacco, which were shown to be transcribed preferentially by the NEP (rpl33, ndhF) or the PEP (ndhA) appeared otherwise to contain most likely PEP and NEP promoters in Arabidopsis, respectively (Table 1), indicating plant-specific strategies for regulation of plastid gene expression consistent with a high diversity of plastid promoters in the two lineages.12,16,27
Genes transcribed by the NEP and the PEP were generally severely up- and down-regulated in group I, respectively. Therefore, group I mutants are expected to be primarily affected in plastid gene expression at various levels, allowing preferential or exclusive transcription of plastid genes by the NEP. The data indicate that the activity of the NEP is prevalent in those mutants and under those conditions where expression of the PEP is decreased or limited. The important roles of NEP and PEP for gene expression in plastids in all tissues during plant development is consistent with the assumption that both polymerases are active in non-photosynthetic tissue and important for the early development of the chloroplast during germination.56 The appearance of several albino mutants found in group II indicates that they are not primarily impaired in plastid gene expression. Examples include alb3 and vipp1, mutants known to have defects at the post-translational level.46,47 This contrasts previous assumptions that albino plastids always exhibit strong, pleiotropic aberrations in plastid gene expression.1
Reduced levels of plastid 16S and/or 5S rRNAs in mutants of group I imply a severe drop in translation rates and consequently a general loss of transcripts synthesized by the PEP. This makes it difficult to define the primary cause of the lesions in gene expression simply by comparing transcript levels. Instead of changes in levels and diversity of gene expression, deviations from the expression profiles that are generally characteristic for members of groups I and II may provide a more secure basis for identifying mutants primarily affected in the accumulation of specific plastid RNAs. Among the 75 mutant lines analyzed, eight and six bona fide specific plastid RNA metabolism mutants have been identified by this means in groups I and II, respectively (Supplementary Table S5), among them the control mutants hcf145, pac and atprfB1. Representative northern analysis of crp135 finally confirmed the specific defect in plastid RNA metabolism (Fig. 2E). Detailed molecular analysis of identified mutants and gene mapping approaches are in progress.
4.2. Analysis of plastid genes based on various transcriptomes of WT and mutants defective in chloroplast functions
Genes previously confirmed to be preferentially transcribed by the NEP and the PEP, respectively, were present in clusters A and B, respectively (Table 1).17,48 It appears that plastid gene expression in mutants impaired in translation and/or chloroplast gene expression is mainly under control of the NEP activity, and, to a lesser degree, under post-transcriptional control. Furthermore, NEP and PEP control different sets of genes in opposite senses. The mutant transcriptomes unequivocally demonstrate the predominance of transcriptional control mechanisms in albino mutants of group I presumably all directly or indirectly affected in the expression of the PEP (Fig. 2). Therefore, clustering analysis identified presumably all plastid promoters, which are preferentially transcribed by the two polymerase types.
In contrast, as revealed by macroarray and microarray data, clustering of plastid genes of WT plants exposed to various stress and exogenic conditions, which also affect the chloroplast, did not identify two major transcriptionally determined gene clusters, which behave oppositely (Fig. 5A and Supplementary Table S9). Only two out of the six clusters (E and F) that emerged preferentially contain genes transcribed by the NEP and the PEP, respectively. Cluster E and F genes are only oppositely expressed under changing light conditions, indicating that changes in light quantity and quality significantly induce the transcriptional regulation of photosynthetic gene expression mediated predominantly by the activity of the PEP. However, post-transcriptional events are shown to be major determinants of plastid transcript abundance under most environmental changes in Arabidopsis, providing sufficient fine-tuning for adaptation to environmental changes. A similar situation has been proposed previously for some plastid genes in other organisms.57–59 In contrast, transcriptional control by the PEP and post-transcriptional regulation collapses in mutants that are generally and severely affected in chloroplast gene expression, as is the case for the numerous albino mutants of group I with prevalent NEP-induced transcription (Fig. 3A). This might also be the reason why the degree and diversity of expression changes were often less pronounced in mutants affected in chloroplast functions than in WT plants exposed to various stresses (Figs 1 and 2).
Post-transcriptional RNA modifications are relevant not only for the control of transcript abundance but also for the generation of spliced, edited, endo- and exonucleolytically cleaved plastid transcripts in order to generate translation-competent mRNAs.21,60,61 Processing of plastid primary transcripts seems to be especially relevant to both transcript abundance and the translatability of individual gene segments of polycistronic mRNAs to fine tune regulation of gene expression independently of transcriptional control. This is supported by our findings that clustering of plastid genes is less prominent determined by transcriptional control under various biological conditions.
4.3. Conclusion
In summary, the present report provides a rich source of information with which to investigate the involvement of the chloroplast and the roles of yet unknown nuclear genes in the management of gene expression in this organelle, especially under conditions of abiotic and biotic stresses. The data allude to defined expression programs regulating plastid functions in response to changing environmental conditions. We show that expression profiles can be used to monitor the functional state of the plant and to identify mutants in plastid RNA metabolism, which deviate from the general expression response. Especially the acquisition of a large number of nuclear genes by de novo synthesis and horizontal gene transfer has significantly increased the complexity of plastid RNA metabolism. The high frequency of plant-specific genes that are important for chloroplast RNA homeostasis demonstrates that transcript regulation has shaped the emergence of plant-specific expression systems for components of the plastid.
Supplementary data
Supplementary data are available online at www.dna.research.oxfordjournals.org.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (SFB TR1 Project B2) to JM.
Supplementary Material
Acknowledgements
We would like to thank Klaus Apel, Ekkehard Neuhaus, Ute Vothknecht, and Peter Westhoff for providing Arabidopsis mutants.
Footnotes
Edited by Katsumi Isono
References
- 1.Barkan A., Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. doi: 10.1016/s0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- 2.Rochaix J. D. Posttranscriptional control of chloroplast gene expression. From RNA to photosynthetic complex. Plant Physiol. 2001;125:142–144. doi: 10.1104/pp.125.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray J. C., Sullivan J. A., Wang J. H., Jerome C. A., MacLean D. Coordination of plastid and nuclear gene expression. Phil. Trans. R. Soc. Lond. B. 2002;358:135–145. doi: 10.1098/rstb.2002.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura T., Schuster G., Sugiura M., Sugita M. Chloroplast RNA-binding and pentatricopeptide repeat proteins. Biochem. Soc. Trans. 2004;32:571–574. doi: 10.1042/BST0320571. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka K., Oikawa K., Ohta N., Kuroiwa H., Kuroiwa T., Takahashi H. Nuclear encoding of a chloroplast RNA polymerase sigma subunit in a red alga. Science. 1996;272:1932–1935. doi: 10.1126/science.272.5270.1932. [DOI] [PubMed] [Google Scholar]
- 6.Allison L. A. The role of sigma factors in plastid transcription. Biochimie. 2000;82:537–548. doi: 10.1016/s0300-9084(00)00611-8. [DOI] [PubMed] [Google Scholar]
- 7.Privat I., Hakimi M. A., Buhot L., Favory J. J., Lerbs-Mache S. Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Mol. Biol. 2003;51:385–399. doi: 10.1023/a:1022095017355. [DOI] [PubMed] [Google Scholar]
- 8.Ichikawa K., Sugita M., Imaizumi T., Wada M., Aoki S. Differential expression on a daily basis of plastid sigma factor genes from the moss Physcomitrella patens. Regulatory interactions among PpSig5, the circadian clock, and blue light signaling mediated by cryptochromes. Plant Physiol. 2004;136:4285–4298. doi: 10.1104/pp.104.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanamaru K., Tanaka K. Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response in higher plants. Biosci. Biotechnol. Biochem. 2004;68:2215–2223. doi: 10.1271/bbb.68.2215. [DOI] [PubMed] [Google Scholar]
- 10.Favory J. J., Kobayshi M., Tanaka K., et al. Specific function of a plastid sigma factor for ndhF gene transcription. Nucleic Acids Res. 2005;33:5989–5999. doi: 10.1093/nar/gki908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zghidi W., Merendino L., Cottet A., Mache R., Lerbs-Mache S. Nucleus-encoded plastid sigma factor SIG3 transcribes specifically the psbN gene in plastids. Nucleic Acids Res. 2007;35:455–464. doi: 10.1093/nar/gkl1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajdukiewicz P. T., Allison L. A., Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedtke B., Börner T., Weihe A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science. 1997;277:809–811. doi: 10.1126/science.277.5327.809. [DOI] [PubMed] [Google Scholar]
- 14.Hedtke B., Legen J., Weihe A., Herrmann R. G., Börner T. Six active phage-type RNA polymerase genes in Nicotiana tabacum. Plant J. 2002;30:625–637. doi: 10.1046/j.1365-313x.2002.01318.x. [DOI] [PubMed] [Google Scholar]
- 15.Emanuel C., Weihe A., Graner A., Hess W. R., Börner T. Chloroplast development affects expression of phage-type RNA polymerase in barley leaves. Plant J. 2004;38:460–472. doi: 10.1111/j.0960-7412.2004.02060.x. [DOI] [PubMed] [Google Scholar]
- 16.Swiatecka-Hagenbruch M., Emanuel C., Hedtke B., Liere K., Börner T. Impaired function of the phage-type RNA polymerase RpoTp in transcription of chloroplast genes is compensated by a second phage-type RNA polymerase. Nucleic Acids Res. 2007;36:758–792. doi: 10.1093/nar/gkm1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liere K., Börner T. Transcription of plastid genes. In: Grasser K., editor. Regulation of Transcription in Plants. 1st Ed. Vol. 291. Oxford: Blackwell Publishing; 2006. pp. 184–223. [Google Scholar]
- 18.Monde R. A., Schuster G., Stern D. B. Processing and degradation of chloroplast mRNA. Biochimie. 2000;82:573–582. doi: 10.1016/s0300-9084(00)00606-4. [DOI] [PubMed] [Google Scholar]
- 19.Bollenbach T. J., Schuster G., Stern D. B. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 2004;75:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T., Furuhashi Y., Hasegawa K., et al. Array-based analysis on tobacco plastid transcripts: preparation of a genomic microarray containing all genes and all intergenic regions. Plant Cell Physiol. 2003;44:861–867. doi: 10.1093/pcp/pcg101. [DOI] [PubMed] [Google Scholar]
- 21.Felder S., Meierhoff K., Sane A. P., et al. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meurer J., Lezhneva L., Amann K., et al. A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell. 2002;14:3255–3269. doi: 10.1105/tpc.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lezhneva L., Meurer J. The nuclear factor HCF145 affects chloroplast psaA–psaB–rps14 transcript abundance in Arabidopsis thaliana. Plant J. 2004;38:740–753. doi: 10.1111/j.1365-313X.2004.02081.x. [DOI] [PubMed] [Google Scholar]
- 25.Schmitz-Linneweber C., Williams-Carrier R. E., Williams-Voelker P. M., Kroeger T. S., Vichas A., Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson B., Stern D. B., Higgs D. C. Microarray analysis confirms the specificity of a Chlamydomonas reinhardtii chloroplast RNA stability mutant. Plant Physiol. 2005;137:534–544. doi: 10.1104/pp.104.053256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legen J., Kemp S., Krause K., Profanter B., Herrmann R. G., Maier R. M. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 2002;31:171–188. doi: 10.1046/j.1365-313x.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 28.Nagashima A., Hanaoka M., Motohashi R., Seki M., Shinozaki K., Kanamaru K., Takahashi H., Tanaka K. DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Biosci. Biotechnol. Biochem. 2004;68:694–704. doi: 10.1271/bbb.68.694. [DOI] [PubMed] [Google Scholar]
- 29.Kahlau S., Bock R. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell. 2008;20:856–874. doi: 10.1105/tpc.107.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhingra A., Bies D. H., Lehner K. R., Folta K. M. Green light adjusts the plastid transcriptome during early photomorphogenic development. Plant Physiol. 2006;142:1256–1266. doi: 10.1104/pp.106.088351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber U. Detection of rapid induction kinetics with a new type of high-frequency modulated chlorophyll fluorimeter. Photosynth. Res. 1986;9:261–272. doi: 10.1007/BF00029749. [DOI] [PubMed] [Google Scholar]
- 32.Mrácek J., Greiner S., Cho W. K., et al. Construction, database integration, and application of an Oenothera EST library. Genomics. 2006;88:372–380. doi: 10.1016/j.ygeno.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 33.Wu H., Kerr M. K., Cui X. Q., Churchill G. A. MAANOVA: a software package for the analysis of spotted cDNA microarray experiments. In: Parmigiani G., Garret E., Irizarry R., Zeger S., editors. The Analysis of Gene Expression Data, 1st Ed., Vol. 14. New York: Springer; 2003. pp. 313–341. [Google Scholar]
- 34.Cleveland W. S., Devlin S. J. Locally weighted regression: an approach to regression analysis by local fitting. JASA. 1988;83:596–610. [Google Scholar]
- 35.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR, Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett T., Edgar R. Mining microarray data at NCBI’s gene expression omnibus (GEO) Methods Mol. Biol. 2006;338:175–190. doi: 10.1385/1-59745-097-9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harr B., Schlötterer C. Comparison of algorithms for the analysis of Affymetrix microarray data as evaluated by co-expression of genes in known operons. Nucleic Acids Res. 2006;34:e8. doi: 10.1093/nar/gnj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson C. L., Miller C. J. Simpleaffy: a BioConductor package for Affymetrix quality control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- 39.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soukas A., Cohen P., Socci N. D., Friedman J. M. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;15:963–980. [PMC free article] [PubMed] [Google Scholar]
- 41.Sturn A., Quackenbush J., Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 42.Kim S. K., Lund J., Kiraly M., Duke K., Jiang M., Stuart J. M., Eizinger A., Wylie B. N., Davidson G. S. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 43.Meurer J., Meierhoff K., Westhoff P. Lolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta. 1996;198:385–396. doi: 10.1007/BF00620055. [DOI] [PubMed] [Google Scholar]
- 44.Yamagishi A., Fork D. C. Photoreduction of QA, QB, and cytochrome b-559 in an oxygen-evolving photosystem II preparation from the thermophilic cyanobacterium Synechococcus sp. Arch. Biochem. Biophys. 1987;15:124–130. doi: 10.1016/0003-9861(87)90477-2. [DOI] [PubMed] [Google Scholar]
- 45.Teale W. D., Paponov I. A., Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 46.Ossenbühl F., Göhre V., Meurer J., Krieger-Liszkay A., Rochaix J. D., Eichacker L. A. Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell. 2004;16:1790–1800. doi: 10.1105/tpc.023226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aseeva E., Ossenbühl F., Sippel C., et al. Vipp1 is essential for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol. Biochem. 2007;45:119–128. doi: 10.1016/j.plaphy.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Shiina T., Tsunoyama Y., Nakahira Y., Khan M. S. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 2005;244:1–68. doi: 10.1016/S0074-7696(05)44001-2. [DOI] [PubMed] [Google Scholar]
- 49.Kuhn K., Bohnem A. V., Liere K., Weihe A., Börner T. Arabidopsis phage-type RNA polymerases: accurate in vitro transcription of organellar genes. Plant Cell. 2007;19:959–971. doi: 10.1105/tpc.106.046839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamayo P., Slonim D., Mesirov J., et al. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA. 1999;96:2907–2892. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meurer J., Grevelding C., Westhoff P., Reiss B. The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 1998;258:342–351. doi: 10.1007/s004380050740. [DOI] [PubMed] [Google Scholar]
- 52.Meurer J., Berger A., Westhoff P. A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell. 1996;8:1193–1207. doi: 10.1105/tpc.8.7.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hennig L., Menges M., Murray J. A., Gruissem W. Arabidopsis transcript profiling on Affymetrix GeneChip arrays. Plant Mol. Biol. 2003;53:457–465. doi: 10.1023/B:PLAN.0000019069.23317.97. [DOI] [PubMed] [Google Scholar]
- 54.Pfalz J., Liere K., Kandlbinder A., Dietz K. J., Oelmüller R. pTAC2,-6 and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koussevitzky S., Nott A., Mockler T. C., et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;4:715–719. [PubMed] [Google Scholar]
- 56.Demarsy E., Courtois F., Azevedo J., Buhot L., Lerbs-Mache S. Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol. 2006;142:993–1003. doi: 10.1104/pp.106.085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng X. W., Tonkyn J. C., Peter G. F., Thornber J. P., Gruissem W. Post-transcriptional control of plastid mRNA accumulation during adaptation of chloroplasts to different light quality environments. Plant Cell. 1989;1:645–654. doi: 10.1105/tpc.1.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tonkyn J. C., Deng X. W., Gruissem W. Regulation of plastid gene expression during photooxidative stress. Plant Physiol. 1992;99:1406–1415. doi: 10.1104/pp.99.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gruissem W., Tonkyn J. Control mechanisms of plastid gene expression. CRC Critical Rev. Plant Biol. 1993;12:19–55. [Google Scholar]
- 60.Barkan A., Walker M., Nolasco M., Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirose T., Sugiura M. Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J. 1997;16:6804–6811. doi: 10.1093/emboj/16.22.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.