Abstract
In the yeast, Saccharomyces cerevisiae, oligosaccharyl transferase (OT), which catalyzes the transfer of dolichol-linked oligosaccharide chains to nascent polypeptides in the endoplasmic reticulum, consists of nine nonidentical membrane protein subunits. Genetic and biochemical evidence indicated these nine proteins exist in three subcomplexes. Three of the OT subunits (Ost4p, Ost3p, and Stt3p) have been proposed to exist in one subcomplex. To investigate the interaction of these three membrane proteins, initially we carried out a mutational analysis of Ost4p, which is an extraordinarily small membrane protein containing only 36 amino acid residues. This analysis indicated that when single amino acid residues in a region close to the luminal face of the putative transmembrane domain of Ost4p were changed into an ionizable amino acid such as Lys or Asp, growth at 37°C and OT activity measured in vitro were impaired. In addition, using immunoprecipitation techniques and Western blot analysis, we found that with these mutations the interaction between Ost4p, Ost3p, and Stt3p was disrupted. Introduction of Lys or Asp residues at other positions in the putative transmembrane domain or at the N or C terminus of Ost4p had no effect on disrupting subunit interactions or impairing the activity of OT. These findings suggest that a localized region of the putative transmembrane domain of Ost4p mediates in stabilization of the interaction with the two other OT subunits (Ost3p and Stt3p) in a subcomplex in the endoplasmic reticulum membrane.
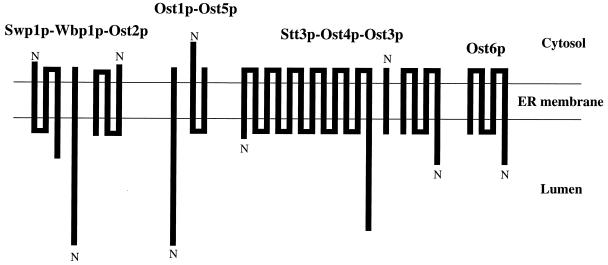
Oligosaccharyl transferase (OT) is a membrane-associated enzyme complex that catalyzes the N-glycosylation of proteins. In the past decade it has become clear that in both higher eukaryotes and yeast this enzyme is a complex consisting of multiple, nonidentical membrane protein subunits residing in the endoplasmic reticulum (ER) (see recent review, ref. 1). An understanding of the mechanism of interaction of these membrane proteins with each other offers a special challenge because of the difficulty of studying their potential interactions in a two-phase system, the hydrophobic environment of the bilayer, and the aqueous environment surrounding the ER membrane. In the case of yeast OT, the situation is especially complex because this enzyme has nine nonidentical membrane proteins with 31 putative transmembrane segments. Some of the OT subunits (Ost1p, Wbp1p, and Swp1p) have very short cytosolic domains, others (Ost5p and Ost2p) have short luminal domains, and one of the subunits that is a mini-membrane protein (Ost4p) has extremely short hydrophilic domains on both sides of the ER membrane. Genetic and biochemical studies (1, 2) have suggested that these nine OT subunits exist in three subcomplexes, as shown in Fig. 1. It is of interest to note that each of these three subcomplexes contains at least one subunit with a short luminal domain and another with a short cytosolic domain, an observation that suggested that the interaction between these OT subunits through their extramembranous domains would be unlikely. In addition, a yeast two-hybrid study using the luminal domains of the OT subunits of Saccharomyces cerevisiae has failed to reveal direct interactions between the luminal domains of these subunits (H.P. and W.J.L., unpublished observations). Based on these observations we hypothesized that the transmembrane domains of these OT subunits would be involved in forming a functional enzyme complex.
Figure 1.
Model for subcomplexes of OT based on the studies of R. Gilmore and M. Aebi and their coworkers (see ref. 1).
One obvious role of transmembrane domains is to anchor proteins in membranes. However, studies on MHC class II molecules, CD8 molecules, the T cell receptor, the human high affinity receptor for Ig E, and glycophorin A have revealed that, in addition to transmembrane domains' structural role in membrane spanning, specific interactions between the transmembrane domains are essential for the assembly and proper sorting of these proteins (3–9). From these studies the idea that transmembrane domains participate in membrane protein oligomerization has emerged. Such studies on membrane protein oligomerization often have been carried out by using truncated chimera transmembrane domains prepared from membrane proteins. For example, truncated constructs of transmembrane domains of glycophorin A have been very useful in studying its mode of dimerization (10). Nevertheless, there is still much to be learned about how membrane proteins oligomerize in vivo in the membrane.
In an earlier study, a novel component of OT, Ost4p, was described (10). Ost4p may be one of the smallest structural membrane proteins that has been discovered in eukaryotic cells. It contains only 36 amino acid residues, of which 21 are predicted to be in the membrane (see topology prediction of membrane proteins at http://www.biokemi.su.se/∼server/toppred2/). OST4 is not an essential gene for cell growth at 25°C, and ost4 mutations exhibit temperature sensitivity for growth. Because of these properties of Ost4p and its small mass, Ost4p is a particularly good model to study transmembrane helix interactions and their possible role in interaction between membrane proteins in vivo.
It has been demonstrated by coimmunoprecipitation (11) and blue native electrophoresis (12) that all nine OT subunits form a complex. However, little is known about the forces involved in the physical interactions between these subunits in the membrane. Karaoglu et al. (11) have reported that when the OT complex containing one OT subunit with a hemagglutinin (HA) epitope fused to its C terminus, namely Ost3HAp, was coimmunoprecipitated by using an anti-HA antibody in the presence of a high concentration of detergent, only Stt3p, Ost4p, and Ost3HAp were found in the immunoprecipitate; all of the other subunits apparently had dissociated from the complex. Based on these observations, it was proposed that these three subunits form a subcomplex. In current studies we have used a mutagenesis approach to ask whether Ost4p is involved in oligomerization with these two proteins, Stt3p and Ost3p, and if so, where the interaction occurs in the primary structure of Ost4p. The results indicate that Ost4p contains a localized region in the transmembrane domain that, when mutagenized to contain single amino acids with ionizable side chains such as Lys or Asp, causes impairment in growth and OT activity measured in vitro. Coimmunoprecipitation revealed that the Ost4p with Lys or Asp residue mutants in this localized region cause the interaction with both Ost3p and Stt3p to be disrupted. Mutations to Lys or Asp elsewhere in the transmembrane domain and at either of the hydrophilic extramembranous domains had no such effect. These findings strongly suggest that the association of Ost3p and Stt3p with Ost4p in the subcomplex of OT is mediated by a very limited region of the transmembrane domain of Ost4p.
Experimental Procedures
Strains and Genetic Methods.
A previously generated ost4Δ strain, JCY11 (MATa ade2 ura3 his3 trp1 leu2 can1 ost4Δ∷URA3), was used in this study (10). To generate the chromosomally integrated OST3 gene with a myc epitope in a background of JCY11, the methods and a plasmid, pFA6a-13MYC-His3Mx6, obtained from Longtine et al. (13) were used. This strain is denoted as LY1 (MATa ade2 ura3 his3 trp1 leu2 can1 ost4Δ∷URA3 with 13 copies of c-myc incorporated into the C terminus of Ost3p). Plasmids encoding wild-type OST4 and various ost4 mutants were transformed into either JCY11 or LY1 by using standard techniques (14).
Plasmid Construction and Mutagenesis.
For expression of Ost4p and various ost4p mutants in the same genetic background, a plasmid containing a triose phosphate isomerase (TPI) promoter, and either wild-type OST4 or ost4 mutants were transformed into JCY11. The TPI promoter region was subcloned into pRS306 by using PstI–SphI. A 1-kb PstI–SphI fragment containing a TPI promoter was subcloned into pRS306 (pRS306TPI). A XhoI–XbaI fragment from pRS306TPI was subcloned into pRS315 (pHP84). Using primers AT1 (5′-GGGGATCCAAGCAAAAGA-3′) and AT2 (5′-CCTCTAGATGTAACCACTTTAG-3′), a BamHI–XbaI OST4 ORF was subcloned by yeast colony PCR and inserted into pRS316 (pHP41). Using AT1 and AT2, a BamHI–XbaI OST4 fragment from pHP41 was subcloned into pHP84 (pHP85). For random mutagenesis, the procedure modified from that of Spee et al. (15) was used. The PCR contained primers AT1 and AT2, 3 mM MgCl2, 0.2 mM MnCl2, 200 μM dITP, 10 μM of one nucleotide, and 200 μM of the each three other nucleotides. pHP85 was used as a template. The PCR-mutated BamHI–XbaI ost4 ORF was inserted into pHP84. A collection of plasmids containing pHP84-randomly mutagenized ost4 was transformed into ost4 null strain (JCY11). Yeast transformants were replica-plated and grown at 25°C and 37°C. A plasmid from the yeast strains that did not grow at 37°C was isolated, sequenced, and transformed back to JCY11. Site-directed mutagenesis was carried out as described (16). To construct pHP84HA, the triple HA sequence was subcloned by using XbaI–NotI. (A XbaI site was generated at the 5′ end, a BamHI site in HA sequence was destroyed, and a NotI site was generated at the 3′ end of the triple HA sequence.) Stop codons of wild-type OST4 or ost4 mutants were destroyed by PCR using primer 5′-CGTCTAGAGGTGTAACCACTTGAG-3′ and, after digestion with BamHI and XbaI, the construct was inserted into pHP84HA. A plasmid carrying wild-type OST4HA or mutant ost4HA was transformed into a strain carrying OST3myc/ost4Δ (LY1), and these transformants were used for coimmunoprecipitation.
Spotting Assay for Growth.
To determine the growth difference between yeast transformants carrying ost4p mutants, the same number of cells (5 × 106 cells) were collected after the strains had been grown to early log phase in −leu media at 25°C. Then 10 μl of serial 1:10 dilutions of the cells were plated on −leu plates and incubated at 37°C for 2 days.
Determination of OT Activity.
The activity of OT in N-glycosylation was carried out by paper chromatography as described (17) except that after addition of the substrate peptide, 3H-Ac-Asn-bpa-Thr-Am, samples were incubated at 37°C for 20 min. The activity of OT was expressed as the amount of labeled glycopeptide formed (in cpm) per unit of protein per unit time.
Protein Expression.
To measure the level of the amount of Ost4p or various ost4p mutants expressed, yeast transformants carrying plasmids with OST4HA or mutant ost4HA were grown to stationary phase in 5 ml of −leu media at 25°C. An equivalent level of cells based on OD600 units was collected by centrifugation, washed with 5 ml of distilled water, and resuspended in 250 μl of SDS/PAGE sample buffer containing protease inhibitors. After addition of 250 μl of acid-washed glass beads, samples were vortexed at high speed four times for 45-sec intervals with 30 sec on ice in between each vortexing. Cell lysates were collected by centrifugation for 5 min and used for analysis by SDS/PAGE with 12.5% gels. The amount of protein expressed was estimated by Western blotting using mouse anti-HA antibody (Babco, Richmond, CA).
Coimmunoprecipitation and Western Blotting.
Coimmunoprecipitation was performed as described (11) except that membrane proteins were solubilized with buffer containing 10 mM Hepes (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, and protease inhibitors. After solubilization of membrane proteins, samples were incubated at 37°C for 20 min. Rabbit anti-myc antibody was purchased from Santa Cruz Biotechnology, and anti-Stt3p antibody was obtained from Satoshi Yoshida (Kirin Brewery Co., Kanagawa, Japan) and used for immunoprecipitation. The immunocomplex of Ost3mycp or Stt3p was washed three times with 1 ml of 50 mM Tris⋅Cl, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, and 0.2% SDS and once with 1 ml of 50 mM Tris⋅Cl, 150 mM NaCl, and 5 mM EDTA, and then analyzed by SDS/PAGE followed by Western blotting with anti-HA antibody.
Results
The disruption of the OST4 gene has been shown to cause underglycosylation of secretory and membrane proteins, and cells lacking this gene were found to be viable at 25°C, but not at 37°C (10). It has been proposed by Karaoglu et al. (11) that Ost4p forms a subcomplex with Stt3p and Ost3p (Fig. 1). In addition, genetic interaction with Ost6p, which is a structural homolog of Ost3p, has been observed (1, 12). However, little is known about the mode of interaction of these proteins or the function of Ost4p. To study how Ost4p interacts with the other protein subunits that exist in its subcomplex with Stt3p and Ost3p, we took a mutational approach using Ost4p. In our initial studies we carried out random mutagenesis of Ost4p and searched for mutant strains that were impaired in growth and OT activity. Surprisingly, considering the small size of the protein (36 aa) we found that many of the mutations in Ost4p had no phenotype. However, several of the mutations that (i) occurred in the hydrophobic domain of Ost4p and (ii) introduced an ionizable amino acid residue (Asp or Lys) resulted in strains that no longer grew at the elevated temperature and showed reduced OT activity assayed in vitro. For example, strains carrying mutations M18K or V23D of ost4p exhibited these characteristics. These results suggested that introduction of ionizable residues might disrupt tight hydrophobic packing of helices in the membrane. This idea was further supported by the finding that mutants with changes in these same positions in the polypeptide chain to nonpolar residues, M18L and V23G, grew as well as wild-type Ost4p.
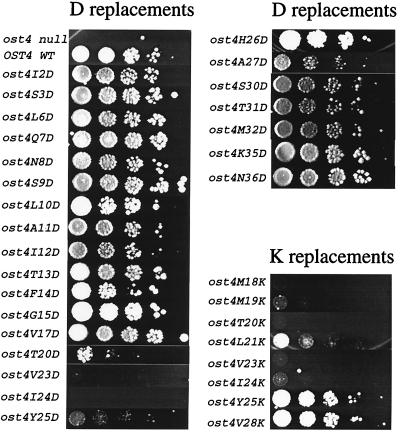
Given these findings, we undertook a more systematic approach and made a series of single amino acid changes to Asp or Lys in residues 2–36 in Ost4p by site-directed mutagenesis. The effect of these mutations on growth, assessed by spotting assays, is shown in Fig. 2. Interestingly, we found that changes to Asp at amino acid positions 16 and 18–24 produced strains that exhibited defects in growth at 37°C. We also examined Lys mutants instead of Asp and found a similar effect on this amino acid. A summary of the growth phenotype of all of the mutant strains that were characterized as discussed above is presented in Fig. 3. Three observations are of particular interest. First, mutations that introduced an ionizable group (Asp, Glu, or Lys) in the hydrophilic N or C terminus of Ost4p had no effect on growth. Second, these residues severely affected growth when introduced into the hydrophobic domain. Third, this negative effect was limited to amino acid residues 18–24. However, as shown, changes to a variety of other amino acids in the 18–24 regions had no significant negative effect on growth.
Figure 2.
Comparison of growth phenotype of strains carrying ost4p mutants with a Asp or Lys replacement at the indicated amino acid positions. Cultures of wild-type (WT), ost4 null, and mutant strains were diluted serially and spotted on plates. After 2 days the growth of colonies at 37°C was compared.
Figure 3.
Summary of growth effect on strains bearing mutations at amino acid residues 2–36. Although the presence of Asp at positions 16, 18, 19, 21, and 22 had a negative effect on growth, these mutant strains were excluded from further study because they behaved like the ost4 null because their level of protein expression was far lower than that of wild-type Ost4p. In all cases Lys and Asp mutants exhibited the same effect except for residue 25. The reason for this difference is not clear.
Based on the fact that ost4 null strain grew poorly and exhibited reduced OT activity, it was important to determine whether the ost4p mutants were expressed at levels comparable to wild-type Ost4p. To do this we used the epitope tagging approach with each of the ost4p mutants. Initially we prepared a construct encoding for a triple HA tag that was linked to the C-terminal end of the wild-type OST4 gene. Then, the plasmid having the OST4HA was transformed into a ost4 null strain. Its growth was found to be the same as strains having the wild-type OST4 sequence. In addition, the OT activity of the transformant with OST4HA was found to be 90–95% of the strain with the wild-type gene of OST4 (data not shown). Therefore, tagging OST4 with HA did not impair the growth or the OT activity. With this result in hand we then tagged all of the ost4p mutants with HA to investigate their level of expression in the cell.
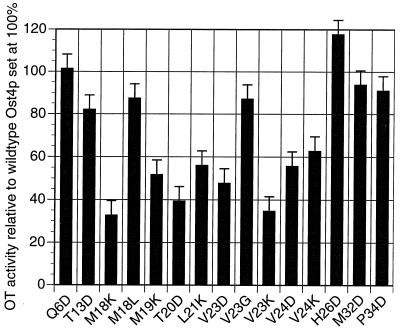
All ost4HA mutants were transformed into the ost4 null strain, cell lysates were prepared from each transformant, and immunoblotting was done by using an anti-HA antibody. It was found that all of the ost4p mutants were expressed, although the level of their expression varied. In the case of some of the Asp mutants, I16D, M18D, M19D, L21D, and I22D, protein expression was very low. Thus, the poor growth (and lowered OT activity) of these mutant strains was similar to that of the ost4 null strain. Consequently these mutants were no longer studied and are not shown in Fig. 2. In all cases when we introduced a Lys instead of an Asp residue in these positions and checked for the growth and OT activity we found that these mutants also exhibited impaired growth and OT activity similar to that observed with the Asp mutants. However, of crucial importance is that we found that the level of expression on the HA-tagged form of these Lys mutant strains was comparable to the expression of wild-type Ost4HAp. Thus, in this case, the phenotypes of all of the Lys mutant strains exhibiting reduced growth at 37°C (Fig. 2) and reduced OT activity measured in vitro (Fig. 4), was caused by the mutation per se, not by a reduction in the synthesis of the mutant protein.
Figure 4.
Measurement of the OT activity of lysates of the indicated mutant strains by using an in vitro assay of the N-glycosylation of a labeled peptide. Values shown are relative to 100% for the strain bearing wild-type Ost4p; shown are the average of three independent experiments and the SD.
Next, all of the strains having ost4 mutant genes were tested for their OT activity. As shown in Fig. 4, single amino acid changes to either Asp or Lys at positions 18–24 of Ost4p showed reduced OT activity measured in vitro. Compared with the wild-type Ost4p, strains containing mutations in this region exhibited a 30–50% decrease in OT activity. In contrast, mutations to nonpolar amino acids in the “sensitive” region of the putative transmembrane domain (M18L and V23G) or mutation to charged residues of any of the other amino acids outside this region of the transmembrane domain (T13D and H26D) had no severe effect on growth. In addition, we found that mutation of single amino acids residue in both the N- and C-terminal extramembranous domains (Q6D, M32D, and P34D) had no effect on OT activity.
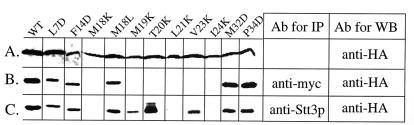
We then asked whether the mutation that impaired growth and OT activity might be a consequence of an alteration in the interaction between Ost4p, Ost3p, and Stt3p. Karaoglu et al. (11) have demonstrated that after detergent solubilization eight of the OT subunits are coimmunoprecipitated by using Ost3HAp, but when the immunocomplex of OT was treated with a higher concentration of detergent, only Ost4p and Stt3p remain associated with Ost3HAp. Based on this observation it was proposed that Ost4p forms a subcomplex with these two other subunits. Therefore, we took a coimmunoprecipitation approach to study the effect of ost4 mutations on complex formation with Ost3p and Stt3p. First, we prepared strains that had Ost3p epitope tagged with c-myc and Ost4p tagged with HA. Cell lysates from a strain carrying Ost3mycp and various ost4HAp mutants were prepared. The expression of ost4HAp mutants was assessed by Western blotting with anti-HA antibody and found to be approximately equal to that of the wild-type strain with a fused HA epitope (Fig. 5A). Next, coimmunoprecipitation using anti-myc antibody to precipitate Ost3mycp was performed and the immunoprecipitated complex was subjected to SDS/PAGE and then analyzed by Western blotting with anti-HA antibody. As shown in Fig. 5B, wild-type Ost4HAp was found to have precipitated with the Ost3mycp. In addition, strains containing ost4p mutants, L7D, F14D, M18L, M32D, and P34D that did not exhibit reduced growth or in vitro OT activity were found to be precipitated with Ost3mycp. However, ost4p mutants that showed severe defects in growth and OT activity (M18K, M19K, T20K, L21K, and I24K) were not immunoprecipitated with Ost3 mycp. These data clearly indicate that mutations that introduce Lys or Asp in residues 18–24 of Ost4p result in impaired interaction of this subunit with Ost3p.
Figure 5.
(A) Western blot analysis (WB) using anti-HA antibody to test the expression of various ost4HAp mutants in cell lysates. The wild-type (WT) strain contains Ost4HAp. (B) Effect of mutations of various amino acid residues of Ost4p on their ability to interact with Ost3p. Strains with Ost3 mycp and the indicated amino acid mutation in Ost4HAp were lysed, treated with detergent as described in Experimental Procedures, and then immunoprecipitated (IP) with anti-myc antibody. Then the immunoprecipitates after SDS/PAGE were assessed for the presence of the mutated ost4HAp by using Western blot with anti-HA antibody. (C) Effect of mutations of ost4p on their ability to interact with Stt3p. In this case, immunoprecipitation was performed by using polyclonal antibody to Stt3p. The immunoprecipitates after SDS/PAGE were assessed for the presence of the various ost4HAp mutants by Western blotting with anti-HA antibody.
To test the effect of these ost4p mutants in a subcomplex formation with Stt3p, the other OT subunit known to be present in this subcomplex (11), the OT complex was immnoprecipitated with anti-Stt3p antibody, and the immunoprecipitate, after SDS/PAGE, was analyzed by Western blotting with anti-HA antibody. Interestingly, whereas all of the HA-tagged ost4p mutants that exhibited reduced growth and OT activity (M18K, M19K, T20K, L21K, V23K, and I24K) caused a disruption of binding to Ost3p, interaction with Stt3p was impaired only when the mutation in Ost4p occurred in position 18, 21, or 24 (Fig. 5C).
Discussion
Now that all nine of the different subunits of yeast OT have been cloned and sequenced, a major challenge is to understand the function of these transmembrane protein subunits and how they interact with each other in the ER. In this context it is of interest to point out that most membrane-anchored glycosyl transferases that have been characterized are far less complex in terms of subunit composition. All glycosyl transferases would be expected to have two recognition domains: one that interacts with the activated sugar donor, and the other that binds the sugar acceptor. In the case of the classical glycosyl transferases the donor is a sugar activated in the form of a sugar nucleotide and the sugar acceptor is either another sugar on the polypeptide or the hydroxyl group of a Ser or Thr residue in a polypeptide. In the case of OT, the donor is an oligosaccharide linked in an activated form via a pyrophosphate bridge to the lipid, dolichol, and the acceptor is the amido N of Asn that is part of the sequence Asn-X-Ser/Thr. Although the recognition requirements for OT activity might be expected to be somewhat higher than that of a “standard” glycosyl transferase, it is completely unclear why the N-glycosylation process catalyzed by OT requires nine different subunits for optimal enzyme activity.
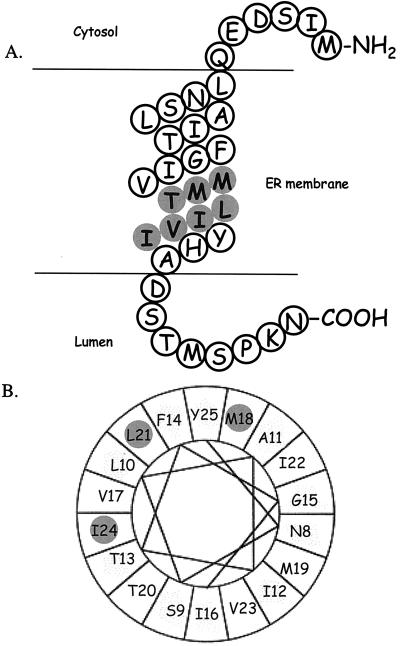
Recently we have shown that one of the subunits, Ost1p, has the property of recognizing a -Asn-X-Ser/Thr- glycosylation site in a peptide (18) and, presumably, in nascent polypeptide chains. In the current study we have undertaken to study another subunit, Ost4p, a mini-membrane protein that is a component of a subcomplex of OT that contains, in addition, Stt3p and Ost3p (11). Because Ost4p is an extraordinarily small subunit of OT it was feasible to consider a comprehensive mutation approach to investigate how it interacts with other components of the OT complex. Initially random mutagenesis led us to focus on mutations with an ionizable residue, either Asp or Lys, in the transmembrane domain of Ost4p. Accordingly we prepared single site mutations to Asp at each of the 36 amino acid residues of Ost4p except residue 1, the initiator methionine. We also prepared a number of Lys mutants. Some of these mutant strains showed low levels of protein expression in the cell. Because the absence of Ost4p, i.e., as the case in the ost4 null strain, results in impaired growth and reduced N-glycosylation assayed in vitro, we excluded all of the ost4p mutants that were underexpressed from further consideration. However, even without these mutants it became clear that there was a positional effect of introducing an ionizable amino acid in Ost4p. As shown in Fig. 6A, assuming that the core of Ost4p is in the form of an α-helix passing through the lipid bilayer, it is obvious that introduction of a ionizable residue only has a negative effect on growth and peptide glycosylation activity when it is located at positions 18–24. Because it is known by epitope tagging and proteolysis experiments that the C terminus of Ost4p faces the lumen of the ER (N. Dean, personal communication; H.K. and W.J.L., unpublished observations) it is clear that the domain of the helix that is sensitive to introduction of an ionizable residue is located near the luminal face of the ER membrane. It is of interest to point out that mutations of Ost4p that caused the disruption of interaction with Stt3p when analyzed by a helical wheel analysis of the sequence, namely residues 18, 21, and 24, all are located in one face of the helical wheel (Fig. 6B). This finding suggests that they may be in direct contact with one of the many transmembrane helices of Stt3p. In contrast, residues 19, 20, and 23, which are located at the opposite side of the helical wheel from residues 18, 21, and 24 may be involved in interaction with Ost3p. One explanation that mutations in residues 18, 21, and 24 of Ost4p also showed reduced interaction with Ost3p is that even though these residues are not in direct contact with Ost3p, the disruption of interaction between Ost4p and Stt3p might cause destabilization of the interaction between Ost4p and Ost3p. Alternatively, there may be more than one copy of Ost4p present in the OT complex, mediating the assembly and stability of Ost3p and Stt3p. Even though it is still premature to describe a detailed model of how Ost4p is organized with other subunits of OT in the ER membrane, our results indicate that residues 18–24 in the transmembrane domain of Ost4p are involved in interaction with two of the other OT subunits, Ost3p and Stt3p. In the future, we hope to obtain a detailed picture of where in these two subunits the interaction with Ost4p occurs.
Figure 6.
(A) Model of Ost4p in a α-helical conformation in the ER membrane. The positions indicated by filled circles are those that exhibited impaired growth at 37°C, reduced OT activity, and disrupted interaction with Ost3 mycp. (B) A helical wheel analysis of the Ost4p transmembrane domain. The filled circles indicate ost4p mutants that exhibited impaired growth at 37°C, reduced OT activity, and disrupted interaction with Stt3p.
Acknowledgments
We thank Dr. S. Yoshida for a generous gift of anti-Stt3p antibody and the Lennarz lab members for their insightful comments on this work. This study was supported by National Institutes of Health Grant GM33185 to W.J.L.
Abbreviations
- OT
oligosaccharyl transferase
- ER
endoplasmic reticulum
- HA
hemagglutinin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040556797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040556797
References
- 1.Knauer R, Lehle L. Biochim Biophys Acta. 1999;1426:259–273. doi: 10.1016/s0304-4165(98)00128-7. [DOI] [PubMed] [Google Scholar]
- 2.Spirig U, Glavas M, Bodmer D, Reiss G, Burda P, Lippuner V, te Heesen S, Aebi M. Mol Gen Genet. 1997;256:628–637. doi: 10.1007/s004380050611. [DOI] [PubMed] [Google Scholar]
- 3.Cosson P, Lankford S P, Bonifacino J S, Klausner R D. Nature (London) 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- 4.Cosson P, Bonifacino J S. Science. 1992;258:659–662. doi: 10.1126/science.1329208. [DOI] [PubMed] [Google Scholar]
- 5.Hennecke S, Cosson P. J Biol Chem. 1993;268:26607–26612. [PubMed] [Google Scholar]
- 6.Letourneur F, Hennecke S, Demolliere C, Cosson P. J Cell Biol. 1995;129:971–978. doi: 10.1083/jcb.129.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manolios N, Bonifacino J S, Klausner R D. Science. 1990;249:274–277. doi: 10.1126/science.2142801. [DOI] [PubMed] [Google Scholar]
- 8.Rutledge T, Cosson P, Manolios N, Bonifacino J S, Klausner R D. EMBO J. 1992;11:3245–3254. doi: 10.1002/j.1460-2075.1992.tb05402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacKenzie K R, Engelman D M. Proc Natl Acad Sci USA. 1998;95:3583–3590. doi: 10.1073/pnas.95.7.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi J H, Roos J, Dean N. J Biol Chem. 1996;271:3132–3140. doi: 10.1074/jbc.271.6.3132. [DOI] [PubMed] [Google Scholar]
- 11.Karaoglu D, Kelleher D J, Gilmore R. J Biol Chem. 1997;272:32513–32520. doi: 10.1074/jbc.272.51.32513. [DOI] [PubMed] [Google Scholar]
- 12.Knauer R, Lehle L. J Biol Chem. 1999;274:17249–17256. doi: 10.1074/jbc.274.24.17249. [DOI] [PubMed] [Google Scholar]
- 13.Longtine M S, McKenzie A, 3rd, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Guthrie C, Fink G R. Methods Enzymol. 1991;194:1–251. [Google Scholar]
- 15.Spee J H, de Vos W M, Kuipers O P. Nucleic Acids Res. 1993;21:777–778. doi: 10.1093/nar/21.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, Lennarz W J. Glycobiology. 2000;10:51–58. doi: 10.1093/glycob/10.1.51. [DOI] [PubMed] [Google Scholar]
- 18.Yan Q, Prestwich G D, Lennarz W J. J Biol Chem. 1999;274:5021–5025. doi: 10.1074/jbc.274.8.5021. [DOI] [PubMed] [Google Scholar]