Abstract
neuroD is a member of the family of proneural genes, which function to regulate the cell cycle, cell fate determination and cellular differentiation. In the retinas of larval and adult teleosts, neuroD is expressed in two populations of post-mitotic cells, a subset of amacrine cells and nascent cone photoreceptors, and proliferating cells in the lineages that give rise exclusively to rod and cone photoreceptors. Based on previous studies of NeuroD function in vitro and the cellular pattern of neuroD expression in the zebrafish retina, we hypothesized that within the mitotic photoreceptor lineages NeuroD selectively regulates aspects of the cell cycle. To test this hypothesis, gain and loss-of-function approaches were employed, relying on the inducible expression of a NeuroDEGFP fusion protein and morpholino oligonucleotides to inhibit protein translation, respectively. Conditional expression of neuroD causes cells to withdraw from the cell cycle, upregulate the expression of the cell cycle inhibitors, p27 and p57, and downregulate the cell cycle progression factors, Cyclin B1, Cyclin D1, and Cyclin E2. In the absence of NeuroD, cells specific for the rod and cone photoreceptor lineage fail to exit the cell cycle, and the number of cells expressing Cyclin D1 is increased. When expression is ectopically induced in multipotent progenitors, neuroD promotes the genesis of rod photoreceptors and inhibits the genesis of Müller glia. These data show that in the teleost retina NeuroD plays a fundamental role in photoreceptor genesis by regulating mechanisms that promote rod and cone progenitors to withdraw from the cell cycle. This is the first in vivo demonstration in the retina of cell cycle regulation by NeuroD.
Keywords: cell cycle, transgenic, neurogenesis, cyclins
Introduction
Cascades of transcriptional regulators determine cell fates and control cellular differentiation (Livesey and Cepko, 2001; Gowan et al., 2001; Vetter and Brown, 2001; Hatakeyama et al., 2001; Akagi et al., 2004; Van Raay and Vetter, 2004; Yan et al., 2005; Wang and Harris, 2005; Hevner et al., 2006; Sugimori et al., 2007). Members of the bHLH class of proneural regulatory proteins act as molecular links connecting withdrawal from the cell cycle, cell fate determination and cellular differentiation (Bertrand et al., 2002; Chae et al., 2004; Yan et al., 2005; Sugimori et al., 2007). The mechanisms by which bHLH proteins link these developmental events are being actively investigated (Logan et al., 2005; Liu et al., 2008). Due to the functional similarity of bHLH proteins, elucidating the role of one member of this family may help identify principal mechanisms that underlie bHLH function. NeuroD is a basic helix-loop-helix transcription factor, which was originally identified as a molecule that functions to regulate the cell cycle, determine neuronal fates and control neuronal differentiation (Lee et al., 1995; Lee, 1997; Farah et al., 2000). Within persistently mitotic cellular lineages, more recent evidence indicates a common role for NeuroD, directly linking cell cycle withdrawal with terminal differentiation. For example, in all persistently mitotic regions in the adult central nervous system, neuroD is expressed in late stage progenitors and appears to be essential for their terminal differentiation (Miyata et al., 1999; Schwab et al., 2000; Pleasure et al., 2000; Lee et al., 2000; Bedard and Parent, 2004; Hevner et al., 2006; see also Naya et al., 1997; Mutoh et al., 1998; Schonhoff et al.,2004).
The retina is an informative model for studying gene function within the central nervous system (Stenkamp, 2007). In the retinas of larval and adult teleosts, neuroD is expressed in two populations of postmitotic cells, amacrine cells and nascent cone photoreceptors, and in proliferating cells in the lineages that give rise exclusively to rod or cone photoreceptors (Hitchcock and Kakuk-Atkins, 2004; Ochocinska and Hitchcock, 2007). Mice share aspects of the teleost pattern of expression. In embryonic mice, neuroD is rarely expressed in retinal progenitors, however it is expressed in nascent cones and in these cells functions to regulate opsin selection (Liu et al., 2008). In contrast to fish and mice, in the avian retina, neuroD is expressed in multipotent progenitors and is determinative for photoreceptor cell fates (Yan and Wang, 1998; Yan and Wang, 2000; Yan and Wang, 2004). In frogs, NeuroD promotes the differentiation of amacrine cells (Kanekar et al., 1997; Moore et al., 2002).
Based on previous studies of NeuroD function in vitro and the cellular pattern of neuroD expression in the zebrafish retina (Ochocinska and Hitchcock, 2007), we hypothesized that within the mitotic photoreceptor lineages NeuroD selectively regulates aspects of the cell cycle. To test this hypothesis, we generated zebrafish transgenic for NeuroDEGFP fusion protein under control of the zebrafish heat shock 70/4 promoter (Halloran et al., 2000; Xiao et al., 2003) for conditional gain-of-function experiments, and we used morpholino oligoncleotides to knock down protein synthesis for loss-of-function experiments. Proliferation and photoreceptor genesis was evaluated using BrdU labeling, proliferation markers and cell type-specific markers. Potential down-stream effectors of the cell cycle were evaluated by in situ hybridization.
The results show that NeuroD functions in rod and cone progenitors to promote these cells' exit from the cell cycle, and suggests that this is accomplished by regulating the expression of cell cycle control genes. Further, when expressed ectopically in multipotent retinal progenitors, NeuroD promotes the genesis of rod photoreceptors and inhibits the genesis of Müller glia.
Materials and Methods
Experimental Animals
Zebrafish were housed at 28.5°C on a 14/10 hr light/dark cycle. Embryos were collected after natural spawns, developed at 28.5°C, and staged by hours post fertilization (hpf) (see Kimmel et al., 1995). Protocols for all procedures using animals were approved by the University Committee for the Use and Care of Animals (UCUCA) at the University of Michigan and conform to NIH guidelines.
Transgene Construct
A pHsp70/4: neuroDegfp construct was made by inserting the PCR-amplified open reading frame of zebrafish neuroD (Korzh et al., 1998) between the SalI and SacII restriction sites in the pHsp70/4:EGFP vector (provided by John Kuwada, University of Michigan [Halloran et al., 2000]). The following PCR primers were used to add the SalI and SacII restriction sites and remove the neuroD stop codon: forward: 5′-GGGGTCCCAAGAAGAAGAAG-3′; reverse: 5′-TAAGGGGTCCGTCAAATGAG-3.′ Following subcloning, the insert was sequenced to rule out potential errors in the PCR.
Transgenic Lines
Plasmid DNA was isolated for injection, linearized at the SacII restriction site, 5′ to the Hsp70/4 promoter, and diluted at 50ng/μl in 1× Danieau buffer (Nasevicius and Ekker, 2000) and 0.25% phenol red. Embryos at the one- to four-cell stage were viewed at 40× magnification, and 2ng of DNA was injected into blastomeres, delivered by glass micropipettes and a Picospritzer (General Valve Corporation, Picospritzer II, Fairfield, NJ). Injected embryos were raised to sexual maturity and pair-wise crosses were made to identify fish showing germline transmission of the transgene. To identify founders (F0), embryos from these crosses were subjected to heat-shock and assayed for EGFP fluorescence, which was then validated by PCR with genomic DNA using primers for the sequence encoding the enhanced green fluorescent protein. Proteins driven by the heatshock promoter are constitutively expressed in the lens (Blechinger et al., 2002), therefore, F1 transgenic embryos were identified at 72hpf for green lenses. F1 transgenic fish were raised to adulthood and mated with wild-type (wt) fish to generate hemizygous progeny. The F2 progeny were raised to sexual maturity and pair-wise crosses of these animals resulted in F3 progeny with a 1:2:1 ratio of wt:hemizygous:homozygous (data not shown).
For the gain-of-function experiments, embryos were harvested from a cross of hemizygous, F2 parents, heat shocked at the indicated ages and, based on fluorescence intensity, sorted into wt, hemizygous and homozygous groups. Hemizygous animals were discarded, and all comparisons were made between wt and homozygous embryos.
Southern Blot analysis
To estimate the copy number of the integrated plasmid in the three lines of Tg(Hsp70/4:nrdegfp) fish, genomic DNA from the F3 generation was digested with SalI, which releases the plasmid from the genomic DNA, and the DNA was subjected to Southern blot analysis. The DNA was separated by agarose gel electrophoresis and transferred to nylon membranes. Blots were hybridized overnight at 65°C with a 200bp fragment from the C-terminal region of NeuroD labeled with [alpha-32P]dCTP and diluted in standard buffer. To estimate copy number, the optical density of bands on the film for endogenous neuroD and the transgene were compared. Band intensity was quantified by computer-assisted image analysis (Adobe Photoshop CS2, Adobe System Incorporation, USA).
Antibodies
Antibodies against zebrafish NeuroD were generated by ZYMED Laboratories (Invitrogen) against the C-terminal, non-conserved region of zebrafish neuroD (amino acids 332-350). Serum containing NeuroD antibodies (zNrd) was affinity purified, and antibody specificity was evaluated by Western analysis (data not shown).
Western blot analysis
Levels of NeuroD protein in experimental and control animals were analyzed by Western blotting. Proteins were extracted from pools of 150-200 embryos by lysing the embryos in buffer with protease inhibitors (Complete Mini, Roche, Mannheim, Germany). Protein concentrations were quantified using a BCA Protein Assay Kit (Pierce, Rockford, IL) and a Perkin-Elmer Lamba Bio 20 spectrophotometer. Proteins were separated in a 10% SDS-PAGE gel and transferred to a nitrocellulosemembrane (Sigma-Aldrich, St. Louis, MO). The membrane was blocked overnight and incubated with the zNrd antibodies diluted at 1:1000 and anti-GFP antibodies (Chemokine) diluted at 1:10,000. Horseradish peroxidase-coupled anti-rabbit IgG secondary antibodies (Sigma-Aldrich, St. Louis, MO) were diluted 1:1000. Chemiluminescence and film (ECL detection system; AmershamBiosciences, Arlington Heights, IL Kodak Chemiluminescence BioMax Film; Rochester, N.Y.) were used to visualize the immunolabeled proteins.
BrdU labeling
Bromodeoxyuridine (BrdU; Sigma, St. Louis, MO) was used to label mitotically active cells. Embryos were systemically labeled with BrdU by soaking them for 20 minutes in 5mM BrdU in embryo rearing solution containing 15% DMSO (Ochocinska and Hitchcock, 2007).
Immunohistochemistry and TUNEL
All immunohistochemistry protocols were performed as described previously (Hitchcock et al., 1996). Omitting primary antibodies served as negative controls. In the absence of primary antibodies, no staining was observed. Ganglion cells were labeled using the monoclonal antibody, zn-12 (The Zebrafish International Resource Center, Eugene, OR; catalog #072103). Amacrine cells were labeled using a monoclonal antibody against rat syntaxin (Monoclonal Anti-Syntaxin Clone HPC-1; Sigma; catalog #S0664). Müller glia were labeled using a monoclonal antibody against glutamine synthetase (GS; Chemicon, Temecula, CA, catalog #MAB305). Red/green cone photoreceptors were labeled using the monoclonal antibody, zpr-1 (formerly Fret43 [Larison and Bremiller, 1990]; The Zebrafish International Resource Center, Eugene, OR; catalog #092502). Rod photoreceptors were labeled using the monoclonal antibody, zpr-3 (formerly Fret11 [Schmitt and Dowling, 1996]; The Zebrafish International Resource Center, Eugene, OR; catalog #011604). Cells in the M-phase of the cell cycle were labeled with using a polyclonal antibody against phosphohistone H3 (Upstate Biotechnology, Lake Placid, NY). Proliferating cells were also labeled using a monoclonal antibody against Proliferating Cell Nuclear Antigen (PCNA; SIGMA, San Louis, MO; product #8825). BrdU was detected using either mouse (Becton Dickinson Immunocytochemistry Systems, San Jose, CA; catalog #347580) or rat (Abcam; Cambridge, MA; catalog #ab6326-250) monoclonal antibodies. All primary antibodies, unless otherwise noted, and secondary antibodies conjugated to fluorescent labels were diluted 1:200. The In Situ Cell Death Detection Kit, TMR red (Roche) was used to label apoptotic cells.
In situ hybridization
In situ hybridization was performed using digoxigenin-labeled riboprobes, synthesized as previously described (Hitchcock et al., 2001). In situ hybridization with whole embryos was performed according to Westerfield (2000). Briefly, embryos were fixed in 4% paraformaldehyde, dehydrated in methanol and stored at −20°C. Embryos were then returned to room temperature, rehydrated, fixed in 4% paraformaldehyde, permeabilized with 0.1 M proteinase K, fixed a second time in 4% paraformaldehyde, treated with acetic anhydride, washed in PBS with 1% Tween and pre-hybridized in hybridization buffer for 1-2 hours. The prehybridization solution was then removed and 200ng of probe in 80μl of hybridization solution was added to the embryos and hybridized overnight at 55°C. The next day, the embryos were washed and the digoxigenin was detected using antibodies conjugated to alkaline-phosphatase (Roche Diagnostics; Indianapolis, IN; catalog #12930020) and a colorimetric reaction with 4-nitrobluetetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP; Roche Molecular Biochemicals, Indianapolis, IN) as the enzymatic substrate. The color reaction was allowed to proceed for approximately 60 minutes and stopped with PBS. Embryos were transferred to single concavity slides (Tri-Ess Sciences, Inc., Burbank, CA) and coverslipped for inspection and photomicroscopy. In situ hybridization with single probes on sections was performed as previously described (Hitchcock et al., 2001). For each probe, two hundred nanograms of probe in 80μl of hybridization solution was placed onto sections, coverslipped, and hybridized overnight at 55°C. The next day, the sections were washed and digoxigenin was immunolabeled using an alkaline phosphatase-conjugated antibody and visualized with NBT/BCIP.
Cell counts
For embryos at 48hpf, labeled cells were counted in one section per animal taken through the optic nerve. Labeled cells were counted in the entire retina and the number of cells was divided by retinal area to normalize for differences in the area of the section. For embryos at 72hpf, labeled cells were counted in 2-3 sections per animal taken through the optic nerve. For each section, the number of labeled cells was divided by retinal area to normalize for differences in the area of the section. The data were then averaged for each animal. The standard deviation was calculated for control and experimental groups, and a two-sample t-test was used to determine significance.
For experiments tracking cell fates following induction of NeuroD among cells in the CMZ, wt and transgenic animals were exposed to BrdU at 48hpf, treated with heat shock and sacrificed at 72hpf. Cells were counted in three sections per animal taken through the optic nerve. For each section, the cohort of BrdU-labeled cells adjacent to the CMZ was identified, and within the BrdU-positive cohort, specific cell types were identified (see Results) and counted. Ratios of the different cell types, e.g. ganglion cells to amacrine cells, were calculated for control and experimental animals, and a test of interactions for proportions was used to identify statistically significant differences (Altman and Bland, 2003) in the proportion of individual cell types.
Morpholino oligonucleotides and microinjections
Morpholino oligonucleotides, complementary to the translation start site, (-13 to +12) of zebrafish neuroD mRNA sequence (GenBank accession number: AF036148) or containing a 5-bp mismatch (control morpholino), were synthesized by Gene Tools, LLC (Cowallis, OR). The sequences of the morpholinos were: neuroD-atg: 5′-TGACTTCGTCATGTCGGAACTCTAG-3′ and neuroD-MM: 5′ TGAgTTgGTCATcTCGcAACTgTAG-3′. Morpholinos were diluted in 1× Danieau buffer (Nasevicius and Ekker, 2000) at 1mg/ml. Embryos were injected with 5ng at the 2- to 8-cell stage.
Photography
All photomicroscopy was performed using a Nikon DMX 1200 digital camera and a Nikon Eclipse E800 microscope. Digital overlays and figures were assembled in Adobe Photoshop CS2.
Results
Transgenic lines
The expression of the pHsp70/4:neuroDEGFP construct and intracellular trafficking of the fusion protein was validated both in cell lines and transient transgenic embryos (supplemental Fig. S1-S3). To generate transgenic lines (see Halloran et al., 2000), embryos were injected, and 150 were raised to adulthood and screened for germline transmission. Based on this screen, three lines were identified and propagated. Southern blotting of digested DNA from each line identified genes encoding both the transgene and endogenous neuroD (supplemental Fig. S4). Southern blots showed that for each line approximately 50 copies of the transgene had inserted per haploid genome. This is within the range previously described for other transgenic lines (Xiao et al., 2003). After being subjected to heat shock, embryos from the first line had robust fluorescence in the retina, embryos from the second line had only a few fluorescent cells in the retina and embryos from the third line had no fluorescent cells in the retina but did have EGFP-positive cells in the olfactory placode. The transgenic line with robust induced fluorescence in the retina was used for the experiments described here. Western analysis using the zNrd antibodies demonstrated that following heat shock the NeuroD fusion protein was stable for at least 24 hours (data not shown).
neuroD fusion protein is functional in vivo
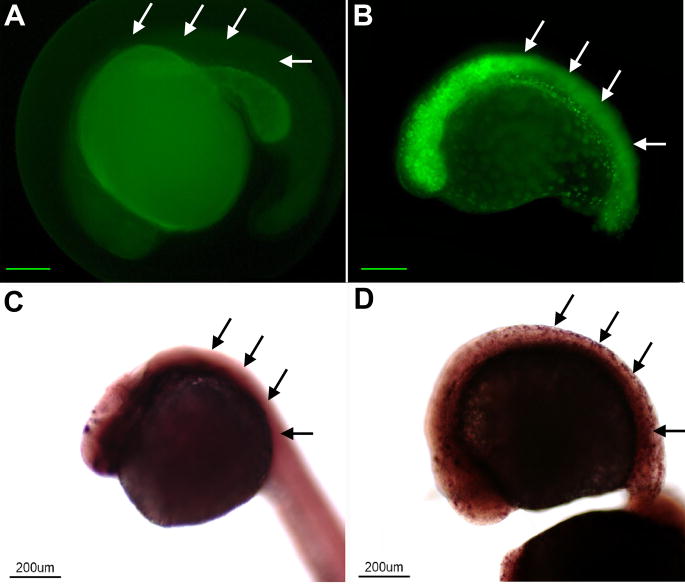
To determine if the NeuroDEGFP fusion protein was functional in vivo, the expression of islet-1, a putative downstream target of NeuroD (see Wang et al., 2000), was examined in wt and transgenic embryos. Embryos received heatshock at 15hpf and were assayed at 25hpf for EGFP fluorescence, neuroD expression and islet-1 expression. Several observations emerged from these early experiments. First, as evidenced by fluorescence microscopy, heat shock induced expression of the fusion protein throughout the embryo (Fig. 1 A, B). Second, prior to 24hpf, induction of the fusion protein arrested subsequent growth. This is likely a consequence of the effects of induced neuroD on the cell cycle (see below). Third, ectopic neuroD mRNA was present throughout the embryo (data not shown). Fourth, following heat shock, islet-1 was ectopically expressed throughout the embryo (Fig.1 C, D). At 25hpf, islet-1 is normally expressed in discrete tissues, including the epiphysis, forebrain, hindbrain, neural tube, pancreatic bud, pharyngeal arch, retina, spinal cord, and trigeminal ganglion (Thisse et al., 2004). In transgenic animals, in addition to the normal pattern of expression, islet-1 was expressed throughout the embryo, including the epidermis. These results demonstrate that the NeuroD fusion protein was functional in vivo.
Figure 1. The NeuroD fusion protein is functional in vivo.
Panels A (wt) and B (transgenic) illustrate embryos from the same clutch treated by heat shock at 15hpf and sacrificed at 25hpf. In the Tg(Hsp70/4:neuroDegfp) EGFP is expressed throughout the body (cf. arrows in A and B, respectively). Panels C (wt) and D (transgenic) illustrate embryos from the same clutch treated by heat shock and processed for in situ hybridization for islet-1. In the transgenic embryo (D) islet-1 gene expression is induced throughout the body. Scale bars equal 200μm in A and B.
Gain of function: neuroD induces mitotic cells to withdraw from the cell cycle
To test the hypothesis that NeuroD regulates mitotic activity among photoreceptor progenitors, embryos were subjected to heat shock at 24hpf, labeled with BrdU at 48hpf and sacrificed immediately afterward. As expected, the retinas of wt fish contained numerous BrdU-positive cells, both in the circumferential marginal zone and scattered throughout the inner and outer nuclear layers (Fig. 2 A). In the retinas of transgenic fish, the vast majority of cells were EGFP-positive (Fig.2 B). However, in contrast to wt fish, the retinas of transgenic fish contained few BrdU+ cells (Fig.2 C). The absence of BrdU+ cells was especially striking for the circumferential marginal zone (CMZ), a proliferative neuroepithelium at the junction of the retina and iris (cf. 2 A and Fig. 2 C; arrows). The few BrdU+ cells present in the transgenic retina lacked EGFP (Fig. 2D) and virtually all the EGFP+ cells lacked BrdU.
Figure 2. Induced neuroD promotes exit from the cell cycle exit in vivo, but does not cause cell death.
Panel A illustrates a retina from a wt embryo treated with heat shock at 24hpf and labeled with BrdU and sacrificed at 48hpf. Note the extensive BrdU labeling throughout. Panel B is a retina from a transgenic embryo treated with heat shock at 24hpf and labeled with BrdU and sacrificed at 48hpf. Panel C is the retina illustrated in B, showing the BrdU labeling. Panel D is the digital overlay of panels B and C. Panel E illustrates histograms comparing the number of phosphohistone H3 immunopositive cells in the retinas of wt and transgenic embryos. Panel F illustrates histograms comparing the number of TUNEL-positive cells in the retinas of wt and transgenic embryos. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; CMZ, circumferential marginal zone. Scale bar equals 50 μM.
The number of proliferating cells was quantified using an antibody against phosphohistone H3. This analysis showed there were significantly fewer pH3-positive cells in transgenic retinas (n=12) following heat shock compared to wt controls (n=12; Fig. 2E). Taken together, data from the BrdU and phosphohistone H3 labeling show that synthesis of NeuroD in retinal progenitors prevents these cells from entering S-phase of the cell cycle and may have exited completely from the cell cycle.
An alternative explanation for the reduction in the number of proliferating cells, however, is that, following heat-shock, dividing cells do not exit the cell cycle, but rather undergo apoptosis. This was tested in the same cohort of embryos using a TUNEL assay for apoptotic cells. Counts of TUNEL-positive cells showed that following heat shock there was no increased cell death in the retinas of transgenic fish (Fig. 2F). In fact, the data reveal there were significantly fewer TUNEL+ cells in the retinas of the transgenic animals. These data show that the decrease in BrdU labeling following the induction of NeuroD fusion protein is not a result of increased cell death.
To investigate the potential mechanism by which NeuroD may promote withdrawal from the cell cycle, in situ hybridization was performed to evaluate the expression of genes encoding the cyclin inhibitors p27 and p57 and genes encoding the cell cycle progression factors, CyclinD1, CyclinB, and CyclinE. Animals were heat-shocked at 24hpf and sacrificed at 48hpf. For each probe, retinas from wt (control) and transgenic (experimental) animals were mounted and processed on the same microscope slide. In wt animals, p27 and p57 are expressed in an irregular distribution within the retina, reflecting the mixed populations of dividing and differentiated cells at this age. Cells within the CMZ, which exclusively contains dividing cells, do not express these cell cycle inhibitors (Fig. 3 A, C). This was most noticeable for p57. In transgenic animals, the expression of both p27 and p57 was significantly increased, as indicated by the intensity of reaction product within each cell, and each probe uniformly labeled cells in the CMZ (Fig. 3 B, D). A converse pattern of expression was observed for the cyclin genes. In control retinas probed for Cyclin D1, B and E, the cellular expression of these genes was largely confined to the dividing cells of the CMZ and adjacent, immature retina (Fig. 3 E, G, I). In contrast, in the retinas of transgenic animals, the expression of the cyclin genes was reduced or absent (Fig. 3 F, H, J). These data show that induced NeuroD protein results in an increase in the expression of the cell cycle inhibitors, p27 and p57, and a concomitant decrease in the expression of the cell cycle promoters, Cyclins D, B and E. These results indicate that the expression of neuroD modulates the expression of these cell cycle-control genes.
Figure 3. NeuroD increases the expression of cyclin inhibitors and inhibits the expression of cyclins.
Panels A, C, E, G and I illustrate retinas from wt embryos heat-shocked at 24hpf, sacrificed at 48hpf and processed for in situ hybridization with probes for p27, p57, Cyclin D1, Cyclin B and Cyclin E, respectively. Panels B, D, F, H and J are transgenic embryos treated similarly. Scale bars equal 50 μM.
Gain of function: NeuroD promotes the genesis of rod photoreceptors and inhibits gliogenesis
bHLH transcription factors, including NeuroD, have a fundamental property in that they function to promote neurogenesis and inhibit gliogenesis (Vetter and Brown, 2001; Van Raay and Vetter, 2004; Yan et al., 2005; Wang and Harris, 2005; Moore et al., 2002; Hevner et al., 2006; Sugimori et al., 2007). Following heatshock with transgenic zebrafish, NeuroD fusion protein was expressed ectopically in multipotent retinal progenitors of the CMZ. This allowed us to test whether or not ectopic NeuroD promotes neurogenesis among multipotent cells. In this experiment, wt and transgenic embryos were subjected to heat-shock at 24hpf or 48hpf and analyzed 24 hours later, at 48hpf or 72hpf, respectively, with a panel of antibody markers. At 48hpf there are few differentiated cells in the retina, and in transgenic animals that received heatshock at 24hpf, there were no additional neurons produced (data not shown). These results show that the induced expression of NeuroD in the early retinal neuroepithelium is not sufficient to generate neurons or photoreceptors.
One explanation for these data is that in the early embryo, regardless of the mitotic state of a cell, the retinal environment is not permissive for cellular differentiation. To circumvent this potential limitation, a similar assay was performed 24hrs later, a time when the retinal environment is permissive for neurogenesis (Raymond et al., 1995). For this experiment, zebrafish were exposed to systemic BrdU at 48hpf then subjected to heat-shock. In this experiment, BrdU was used to mark and follow the cohort of multipotent cells within the CMZ. The fate of these cells was then evaluated at 72hpf. Further, because of the known developmental asymmetry in the retina at 72hpf (Hyatt et al., 1996; Schmitt and Dowling, 1999), cells generated from the dorsal and ventral marginal zones were analyzed separately. The number of cells labeled with each cell type-specific marker was counted. To normalize for the slight differences in retinal growth as a consequence of the induced NeuroD expression, ratios of the various cell types were calculated and compared in a pair-wise manner.
The pair-wise comparisons showed that between control (n=5) and transgenic animals (n=5) there were no statistically-significant differences in the proportion of ganglion, amacrine and cone photoreceptors generated from the CMZ (supplemental Fig. S5). In contrast, in transgenic animals there was a significantly higher proportion of rod photoreceptors in the dorsal retina (Fig. 4A) and a significantly lower proportion of Müller glia in ventral retina (Fig. 4B). This dorso-ventral asymmetry in cell types likely reflects the developmental asymmetries present in the retina at this age. Nonetheless, these data suggest that ectopic expression of neuroD in multipotent retinal progenitors is sufficient to promote the genesis of rod photoreceptors and concomitantly inhibit the genesis of Müller glia.
Figure 4. NeuroD promotes photoreceptor genesis and inhibits gliogenesis.
Panel A illustrates histograms of the ratio of rods to all other cell types generated at the dorsal CMZ. Panel B illustrates histograms of the ratio of Müller glia to all other cell types generated at the ventral CMZ. Asterisks indicate a statistically significant difference as calculated by a pooled two-sample t-test (p<0.05). CMZ – central marginal zone.
Loss of function: effects on development and morpholino specificity
As a second, independent test of our hypothesis, the synthesis of NeuroD was selectively inhibited by injecting antisense morpholino oligonucleotides at the 2-8-cell stage and cell proliferation and differentiation was assayed at 72hpf. Initial comparisons of control and morphant embryos showed there were no apparent differences in gross development and morphology (Fig. 5 A-C; see Corey and Abrams, 2001). The size and shape of the body and size of the eyes was comparable between control and experimental animals. The specificity of the atg-morpholinos was tested by Western blot analysis and in the Tg(pHsp70/4:neuroDEGFP) line. In Western blots, the zNrd antibody recognizes a band at 30kD, and this protein is markedly reduced in morphant embryos (Fig. 5 D). As a second test of morpholino specificity, embryos generated from transgenic animals, were randomly separated into two groups, injected with either mismatch (Fig. 5 E, F) or neuroD-atg morpholinos (Fig. 5 G, H), respectively, subjected to heat shock at 24hpf and photographed at 48hpf. EGFP expression was then qualitatively assayed for each group. A subset of the embryos (presumptive transgenics) injected with the mismatch morpholino showed robust EGFP fluorescence (Fig. 5 E, F). In contrast, embryos injected with the neuroD-atg morpholino uniformly lacked EGFP fluorescence or (more rarely) contained a few EGFP+ cells (Fig. 5 G, H). Together, these data demonstrate that the neuroD-atg morpholinos specifically block translation of NeuroD protein in vivo.
Figure 5. Morpholino oligonucleotides do not alter gross development, and specifically inhibit neuroD synthesis.
Panels A-C illustrate wt, mismatch control and morphant embryos, respectively, at 72hpf. Note the similarity in body shape and eye size. Panel D is a Western blot of proteins from wt and morphant embryos. Panels E and F illustrate Tg(Hsp70/4:neuroDegfp) embryos injected with either mismatch (E, F) or atg (G,H) morpholinos, treated with heat shock and 24hpf and photographed at 48hpf. Note the absence of EGFP fluorescence in the morphant embryos (G, H).
Loss of function: NeuroD is required for photoreceptor progenitors to exit the cell cycle
In contrast to the gain-of-function experiments, where NeuroD was induced in all cells, in the loss-of-function experiments, NeuroD translation is inhibited only in cells that normally synthesize this protein. Since NeuroD is expressed in mitotically active cells of the rod and cone lineages, these studies can directly test the consequence of removing NeuroD in photoreceptor progenitors. Embryos from wild-type crosses were divided into control (uninjected or injected with mismatch morpholinos) and experimental groups (injected with neuroD-atg morpholinos). Following the injections, each group was exposed to a brief systemic pulse of BrdU at 72hpf and sacrificed immediately afterwards. As expected for animals at 72hpf, in both control and morphants, the retinas were laminated, and cell-type specific markers showed that neurons within the inner layers developed normally (supplemental Fig. S6). In control animals (n=12), BrdU+ cells were present in the CMZ and largely absent in the inner and outer nuclear layers (Fig. 6 A, B). However, in striking contrast to controls, in the retinas from morphants (n=12), numerous BrdU-positive cells were present throughout the outer nuclear layer, and individual cells and clusters of cells were present in the inner nuclear layer (Fig. 6 C). The number of mitotically active cells was quantified using antibody labeling for BrdU and phosphohistone H3. These counts showed there are significantly more BrdU and pH3-positive cells in morphant retinas (n=6) compared to wt controls (n=6; Fig. 6 D, E). These data show that NeuroD is required for cells in the photoreceptor lineages to exit the cell cycle, and in the absence of NeuroD, these cells continue to proliferate. Retinas from control and morphant embryos were analyzed at 72hpf with probes for mRNA encoding cell cycle regulatory proteins p27 and CyclinD1. In contrast to the gain-of-function experiments, there was no apparent difference in p27 expression between retinas in control and morphant embryos (Fig. 7 A, B). This likely reflects the inability to identify the small proportion of the total cells in the retinas of morphant embryos that remain in the cell cycle and do not express this gene. In contrast, the number and distribution of cells expressing Cyclin D1 was markedly altered in the morphants. At 72hpf, CyclinD1 is normally expressed only in cells in the CMZ (Fig. 7 C). In retinas from morphant embryos, the expression of CyclinD1 in the CMZ remains unchanged, but CyclinD1 is upregulated in cells in the inner and outer nuclear layers (Fig. 7 D). These data show that the absence of NeuroD results in the upregulation of CyclinD1 expression in retinal layers that contain excess dividing cells.
Figure 6. Cells fail to withdraw from the cell cycle following knock down of neuroD.
Panels A, B and C illustrate retinas form wt, mismatch control and morphant fish, respectively, that were labeled with BrdU and sacrificed at 72hpf. Left-hand panels illustrate nuclear staining with bisbenzimide; middle panels illustrate BrdU labeling; right-hand panels are the respective digital overlays. L – lens. Panel D is histograms comparing the number of BrdU-labeled cells in control and morphant retinas. Panel E is histograms comparing the number of cells immunopositive for phosphohistone H3 in control and morphant retinas. Asterisks, p<0.001.
Figure 7. Cyclin D1 is upregulated in the absence of neuroD.
Panels A and C illustrate wt retinas at 72hpf processed for in situ hybridization for p27 and Cyclin D1, respectively. Panels B and D illustrate morphant retinas processed on the same microscope slides. L – lens.
Photoreceptors differentiation recovers by 7 days post fertilization in NeuroD morphant retinas
As an additional test of NeuroD function, the population of mitotically active cells in the morphant retina was tracked between 72hpf and 7 days post fertilization (dpf), which is 3 or 4 days after the time when morpholino-dependent translation inhibition is lost (Malicki, 2000; Malicki et al., 2002; Pujic et al., 2006). Control and morphant animals were exposed to BrdU at 72hpf and assayed either immediately afterward or at 7dpf for BrdU incorporation and markers of cones (zpr1) or rods (zpr3). Among the cohort of morphants examined at 72hpf, there were no cells that expressed markers of differentiated photoreceptors (Fig. 8 A-B and C-D). In contrast, by 7dpf NeuroD morphants expressed proteins characteristic of mature photoreceptors (Fig. 8 F). Furthermore, these photoreceptors (along with other cells within the inner nuclear layer) were co-labeled with BrdU (Fig. 8F inset). These results show that at 72hpf cells destined to migrate to the outer nuclear layer fail to exit the cell cycle, however they retain their identities as photoreceptor progenitors. These results further demonstrate that neuroD is required for photoreceptor progenitors to exit the cell cycle, and serve, secondarily, as proof for the specificity of the NeuroD morpholinos.
Figure 8. Photoreceptor differentiation is absent in morphant retinas at 72hpf and recovers in morphant retinas by 7 days post fertilization.
Rows A and B illustrate retinas from wt embryos at 72hpf stained for cone and rod photoreceptors, respectively. Rows C and D are retinas from morphants at 72hpf stained for cone and rod photoreceptors, respectively. Row E illustrates the retina from a wt animal labeled with BrdU at 72hpf, sacrificed at 7days post fertilization (dpf) and labeled antibodies against BrdU (green) and the cone specific marker, zpr1 (red). Row F illustrates the retina from a morphant labeled with BrdU at 72hpf, sacrificed at 7dpf and labeled with antibodies against BrdU (green) and the cone-specific marker, zpr1 (red). All left-hand panels illustrate nuclear staining with bisbenzimide. The middle panels are stained with markers for cones (zpr1) or rods (zpr3). The right-hand panels are digital overlays (E3 and F3). The scale bar in panel D1 corresponds to panels A-D and equals 50μm. The scale bar in panel F1 corresponds to panels E-F and equals 50μm. L – lens; wt – wild-type embryos; MO-ATG – embryos injected with translation-blocking morpholinos.
Discussion
In the developing and adult retina of teleosts, neuroD is expressed in mitotic progenitors that give rise exclusively photoreceptors (Hitchcock and Kakuk-Atkins, 2004; Ochocinska and Hitchcock, 2007), and this allowed us to evaluate the function of NeuroD in a mitotic lineage dedicated to a single cell type. The results show that in photoreceptor progenitors NeuroD initiates cell cycle exit. The most parsimonious mechanism is direct, cell autonomous regulation of the expression of cell cycle control genes. However, we cannot exclude the possibility that NeuroD functions indirectly or via non-cell autonomous mechanisms. Nonetheless, these results provide the first in vivo evidence for the role of NeuroD in controlling the cell cycle of neural progenitors in the retina. Our data also suggest that NeuroD does not control cell fates. However, when NeuroD is ectopically expressed in retinal progenitors, a circumstance that normally does not occur in the teleost retina, it promotes the genesis of rod photoreceptors and inhibits the genesis of Müller glia, corroborating the previously demonstrated general role of bHLH proteins in promotoing neurogenesis over gliogenesis.
Progression through all of the phases of the cell cycle is under the control of cyclin–CDK (cyclin-dependent kinase) complexes, and the activity of these cyclin:CDK complexes is regulated by CDK inhibitors, including the CIP/KiP family (Soprano and Giordano, 2003; Dehay and Kennedy, 2007). Whereas cyclin–CDK complexes positively drive progression of the cell cycle, CDK inhibitors negatively regulate progression through the cell cycle by binding to and inactivating cyclin–CDKs (Dyer and Cepko, 2001a,b; Soprano and Giordano, 2003; Dehay and Kennedy, 2007). The cyclin inhibitors are regulated at the transcriptional and post-translational levels, and studies have shown that NeuroD upregulates the expression of CDK inhibitors p21, p27, and p57 (Naya et al., 1997; Mutoh et al., 1998; Farah et al., 2000; Schonhoff et al., 2004; present results). In vitro, forced expression of NeuroD induces dividing cells to withdraw from the cell cycle, which is preceded by elevated expression of the cyclin-dependent kinase inhibitor p27Kip1 (Farah et al., 2000). In enteroendocrine cells, NeuroD induces cell cycle arrest with a concomitant increase in p21 expression, another inhibitor of cyclin-dependent kinases (Naya et al., 1997; Mutoh et al., 1998; Schonhoff et al., 2004). In the retina p27(Kip1) is part of the molecular mechanism that controls the decision of multipotent central nervous system progenitors to withdraw from the cell cycle (Levine et al., 2000; Dyer and Cepko, 2001a, b; Ohnuma et al., 2002; Cunningham et al., 2002). Based on its expression pattern in photoreceptor progenitors (Ochocinska and Hitchcock, 2007) and the data presented here, we suggest that in teleosts NeuroD influences the expression of a network of cell cycle regulatory genes to promote cell cycle exit among photoreceptor progenitors. Specifically, NeuroD may coordinate cell cycle arrest by upregulating the expression of p27 and p57, which, in turn, leads to the downregulation of the expression of cyclins D, B and E.
Previous studies have shown that other transcription factors also regulate cell cycle progression and neurogenesis in the retina. Prox1 is required for horizontal cell genesis and controls progenitor cell proliferation (Dyer, 2003; Dyer et al., 2003), whereas Math5 is required for ganglion cell genesis and controls cell cycle progression (Kanekar et al., 1997; Le et al., 2006). Prox1 expression promotes the upregulation of both p27 and p57 in the retina (Dyer, 2003, Dyer et al., 2003). Math5-/- retinal cells show aberrant p27/Kip1 expression and an inability to become fully postmitotic (Kanekar et al., 1997; Le et al., 2006). These data are consistent with the characteristics of bHLH regulatory proteins acting as a molecular link connecting withdrawal from the cell cycle, cell fate determination, and differentiation (Bertrand et al., 2002; Chae et al., 2004; Yan et al., 2005; Sugimori et al., 2007). A recent study has shown that Meis1, a vertebrate homolog of the TALE-class homeodomain transcription factor Homothorax (Htx), is required for cell cycle progression of mulitpotent progenitors in the retina and functions by regulating the expression of CyclinD1 (Bessa et al., 2008). It is interesting to note that the bHLH transcription factor, MyoD, directly interacts with the Pbx/Meis complex (Tapscott, 2005), suggesting that in the retina similar mechanisms controlling the function of bHLH transcription factors are conserved.
In the embryonic and adult retina of teleosts, NeuroD is transiently and briefly expressed in postmitotic cone photoreceptors. Based on this, we speculated previously that NeuroD might regulate early aspects of cone maturation (Ochocinska and Hitchcock, 2007; see also Liu et al., 2008). Our data are consistent with this speculation. In NeuroD morphants at 72hpf, a distinct outer nuclear layer is present, but cells there do not express markers of differentiated cones. If the outer nuclear layer harbors postmitotic photoreceptors, the absence of cone markers could be interpreted to show that, in the absence of NeuroD, these cells fail to differentiate. This is an attractive interpretation, however, we cannot distinguish this from the possibility that, in the absence of NeuroD, the outer nuclear layer consists only of dividing photoreceptor progenitors. Alternatively, based on the gain-of-function results, we are tempted also to speculate that in newly postmitotic cones NeuroD functions to hold these cells in a post-mitotic state. Cell cycle arrest is an active process (Herrup and Yang, 2007), and the transient expression of NeuroD in nascent cones may block re-entry into the cell cycle as differentiation proceeds.
In the present study, induced expression of NeuroD in the CMZ, which contains retinal progenitors that normally do not express this gene (see Ochocinska and Hitchcock, 2007), promoted rod genesis and inhibited the genesis of Müller glia. This result is consistent with the general function of bHLH transcription factors, which in various neural tissues function to promote neurogenesis and inhibit gliogenesis (Sun et al., 2001; Morrow et al., 1999; Tomita et al., 2000). In contrast, in the loss-of-function experiments, the absence of NeuroD in photoreceptor progenitors did not result in a change in cell fate. Following knock down of NeuroD, photoreceptor progenitors fail to exit the cell cycle, however, these cells migrate normally, form an outer nuclear layer and express markers of differentiated photoreceptors when NeuroD function is allowed to recover. These observations show that in the absence of NeuroD, these cells retain their identity as photoreceptor progenitors and that additional mechanisms, independent of NeuroD, must specify their fates. Together, these data suggest that in the vertebrate retina (and brain), the function of NeuroD is context specific. When expressed in cells that are competent to generate multiple cell types, NeuroD functions as a determination factor, whereas when expressed in cells whose fates are constrained, NeuroD functions more narrowly to regulate exit from the cell cycle or discrete steps in cellular differentiation.
Supplementary Material
Panel A is a cartoon illustrating subcloning of the Hsp70/4:neuroDEGFP construct. The Hsp70/4:neuroDegfp construct was assembled by subcloning the 1kb coding region of the zebrafish neuroD gene into the Hsp70/4:EGFP vector. Panels B1-3 illustrate the localization of EGFP in the cytoplasm of HEK293 cells transfected with the empty Hsp70/4:EGFP construct. Panels C1-3 illustrate localization of EGFP in the nucleus of HEK293 cells transfected with the Hsp70/4:neuroDegfp construct. Left-hand panels illustrate GFP fluorescence; Middle panels illustrate nuclear staining with bisbenzimide; right-hand panels are digital overlays.
Panel A illustrates a transient transgenic embryo treated with heat shock at 24hpf and photographed at 48hpf. Note the mosaic of GFP-labeled cells. Panel B is an uninjected control treated with heat shock. Panel C illustrates a section through the head and retina of a transient transgenic embryo at 48hpf. The enclosed area is illustrated at higher magnification (and rotated slightly) in D. Note the nuclear localization of the GFP. D1 – Nomarski illumination; D2 – GFP fluorescence; D3 – nuclear staining with bisbenzimide; D4 – digital overlay D2 and D3. The scale bar in A represents 200μm for panels A and B and 50μm for panel C.
This panel illustrates in situ hybridization for neuroD in a transient transgenic embryo (see Fig. S2) treated with heatshock at 48hpf and sacrificed at 96hpf. Note the numerous labeled cells throughout the retina and head. Note the ectopic neuroD-expressing cells in the CMZ. CMZ – circumferential germinal zone. L – lens. Scale bar equals 50μm.
Lanes 1-3 correspond to the three Tg(Hsp70/4:neuroDegfp) lines. Lane 3 corresponds to the line selected for this study. Note the bands at 5kb, which represent endogenous neuroD, and the intense bands at 12kb, which represent the transgene. The left-most lane contains molecular weight markers.
Panels G and J are illustrated in Figure 5.
Panels A and B are wt embryos at 72hpf immunostained with zn12 and HPC-1 antibodies, respectively. Panels C and D are neuroD morphants treated similarly. Note that the inner retinal layers of the morphant retinas contain differentiated neurons.
Acknowledgments
The authors thank Laura Kakuk-Atkins for technical assistance, Dr. Deborah Stenkamp for reading an earlier version of this manuscript and members of the Hitchcock lab for useful discussions. This work was supported by NIH grants R01 EY07060 and P30 EY07003 (PFH); T32 EY 13934 and T32 DC00534 (MJO); and a Senior Scientific Investigator Award from Research to Prevent Blindness, Inc. (PFH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman DG, Bland JM. Interaction revisited: the difference between two estimates. BMJ. 2003;326:219. doi: 10.1136/bmj.326.7382.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard A, Parent A. Evidence of newly generated neurons in the human olfactory bulb. Brain Res Dev Brain Res. 2004;151:159–168. doi: 10.1016/j.devbrainres.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–30. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bessa J, Tavares MJ, Santos J, Kikuta H, Laplante M, Becker TS, Gómez-Skarmeta JL, Casares F. Meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development. 2008;135:799–803. doi: 10.1242/dev.011932. [DOI] [PubMed] [Google Scholar]
- Blechinger SR, Evans TG, Tang PT, Kuwada JY, Warren JT, Jr, Krone PH. The heat-inducible zebrafish hsp70 gene is expressed during normal lens development under non-stress conditions. Mech Dev. 2002;112:213–5. doi: 10.1016/s0925-4773(01)00652-9. [DOI] [PubMed] [Google Scholar]
- Chae JH, Stein GH, Lee JE. NeuroD: the predicted and the surprising. Mol Cells. 2004;18:271–88. [PubMed] [Google Scholar]
- Corey DR, Abrams JM. Morpholino antisense oligonucleotides: tools for investigating vertebrate development. Genome Biol. 2001;2:1015. doi: 10.1186/gb-2001-2-5-reviews1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JJ, Levine EM, Zindy F, Goloubeva O, Roussel MF, Smeyne RJ. The cyclin-dependent kinase inhibitors p19(Ink4d) and p27(Kip1) are coexpressed in select retinal cells and act cooperatively to control cell cycle exit. Mol Cell Neurosci. 2002;19:359–374. doi: 10.1006/mcne.2001.1090. [DOI] [PubMed] [Google Scholar]
- Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. p27Kip1 and p57Kip2 regulate proliferation in distinct retinal progenitor cell populations. J Neurosci. 2001a;21:4259–4271. doi: 10.1523/JNEUROSCI.21-12-04259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001b;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- Dyer MA. Regulation of proliferation, cell fate specification and differentiation by the homeodomain proteins Prox1, Six3, and Chx10 in the developing retina. Cell Cycle. 2003;2:350–357. [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucuc HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001;128:1313–1322. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Macdonald RE, VanDeRyt JT, Wilson SW. Antibodies against Pax6 immunostain amacrine and ganglion cells and neuronal progenitors, but not rod precursors, in the normal and regenerating retina of the goldfish. J Neurobiol. 1996;29:399–413. doi: 10.1002/(SICI)1097-4695(199603)29:3<399::AID-NEU10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Otteson DC, Cirenza PF. Expression of the insulin receptor in the retina of the goldfish. Invest Ophthalmol Vis Sci. 2001;42:2125–2129. [PubMed] [Google Scholar]
- Hitchcock P, Kakuk-Atkins L. The basic helix-loop-helix transcription factor neuroD is expressed in the rod lineage of the teleost retina. J Comp Neurol. 2004;477:108–117. doi: 10.1002/cne.20244. [DOI] [PubMed] [Google Scholar]
- Hyatt GA, Schmitt EA, Fadool JM, Dowling JE. Retinoic acid alters photoreceptor development in vivo. Proc Natl Acad Sci USA. 1996;93:13298–13303. doi: 10.1073/pnas.93.23.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekar S, Perron M, Dorsky R, Harris WA, Jan LY, Yan YN, Vetter ML. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron. 1997;19:981–994. doi: 10.1016/s0896-6273(00)80391-8. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Korzh V, Sleptsova I, Liao J, He J, Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev Dyn. 1998;213:92–104. doi: 10.1002/(SICI)1097-0177(199809)213:1<92::AID-AJA9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Larison KD, Bremiller R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development. 1990;109:567–576. doi: 10.1242/dev.109.3.567. [DOI] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006;295:764–778. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lee JE. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- Lee JK, Cho JH, Hwang WS, Lee YD, Reu DS, Suh-Kim H. Expression of neuroD/BETA2 in mitotic and postmitotic neuronal cells during the development of nervous system. Dev Dyn. 2000;217:361–367. doi: 10.1002/(SICI)1097-0177(200004)217:4<361::AID-DVDY3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Levine EM, Close J, Fero M, Ostrovsky A, Reh TA. p27(Kip1) regulates cell cycle withdrawal of late multipotent progenitor cells in the mammalian retina. Dev Biol. 2000;219:299–314. doi: 10.1006/dbio.2000.9622. [DOI] [PubMed] [Google Scholar]
- Logan MA, Steele MR, Van Raay TJ, Vetter ML. Identification of shared transcriptional targets for the proneural bHLH factors Xath5 and XNeuroD. Dev Biol. 2005;285:570–83. doi: 10.1016/j.ydbio.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Liu H, Etter P, Hayes S, Jones I, Nelson B, Hartman B, Forrest D, Reh TA. NeuroD1 regulates expression of thyroid hormone receptor 2 and cone opsins in the developing mouse retina. J Neurosci. 2008;28:749–756. doi: 10.1523/JNEUROSCI.4832-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Malicki J. Harnessing the Power of Forward Genetics: Analysis of Neuronal Diversity and Patterning in the Zebrafish Retina. Trends in Neuroscience. 2000;23:531–541. doi: 10.1016/s0166-2236(00)01655-6. [DOI] [PubMed] [Google Scholar]
- Malicki JJ, Pujic Z, Thisse C, Thisse B, Wei X. Forward and reverse genetic approaches to the analysis of eye development in zebrafish. Vision Res. 2002;42:527–533. doi: 10.1016/s0042-6989(01)00262-0. [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KB, Schneider ML, Vetter ML. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron. 2002;34:183–195. doi: 10.1016/s0896-6273(02)00666-9. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- Mutoh H, Fung BP, Naya FJ, Tsai MJ, Nishitani J, Leiter AB. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA. 1998;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochocinska MJ, Hitchcock PF. Dynamic expression of the basic helixloop-helix transcription factor neuroD in the rod and cone photoreceptor lineages in the retina of the embryonic and larval zebrafish. J Comp Neurol. 2007;501:1–12. doi: 10.1002/cne.21150. [DOI] [PubMed] [Google Scholar]
- Ohnuma S, Hopper S, Wang KC, Philpott A, Harris WA. Co-ordinating retinal histogenesis: early cell cycle exit enhances early cell fate determination in the Xenopus retina. Development. 2002;129:2435–2446. doi: 10.1242/dev.129.10.2435. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Collins AE, Lowenstein DH. Unique expression patterns of cell fate molecules delineate sequential stages of dentate gyrus development. J Neurosci. 2000;20:6095–6105. doi: 10.1523/JNEUROSCI.20-16-06095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujic Z, Omori Y, Tsujikawa M, Thisse B, Thisse C, Malicki J. Reverse genetic analysis of neurogenesis in the zebrafish retina. Dev Biol. 2006;293:330–347. doi: 10.1016/j.ydbio.2005.12.056. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Curran GA. Developmental patterning of rod and cone photoreceptors in embryonic zebrafish. J Comp Neurol. 1995;359:537–550. doi: 10.1002/cne.903590403. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Comparison of topographical patterns of ganglion and photoreceptor cell differentiation in the retina of the zebrafish, Danio rerio. J Comp Neurol. 1996;371:222–234. doi: 10.1002/(SICI)1096-9861(19960722)371:2<222::AID-CNE3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. J Comp Neurol. 1999;404:515–536. [PubMed] [Google Scholar]
- Schonhoff SE, Giel-Moloney M, Leiter AB. Minireview: Development and differentiation of gut endocrine cells. Endocrinology. 2004;145:2639–2644. doi: 10.1210/en.2004-0051. [DOI] [PubMed] [Google Scholar]
- Schwab MH, Bartholomae A, Heimrich B, Feldmeyer D, Druffel-Augustin S, Goebbels S, Naya FJ, Zhao S, Frotscher M, Tsai MJ, Nave KA. Neuronal basic helix-loop-helix proteins (NEX and BETA2/NeuroD) regulate terminal granule cell differentiation in the hippocampus. J Neurosci. 2000;20:3714–3724. doi: 10.1523/JNEUROSCI.20-10-03714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano KJ, Giordano A. Cell Cycle Inhibitors in Cancer Therapy: Current Strategies. New Jersey: Humana Press; 2003. [Google Scholar]
- Stenkamp DL. Neurogenesis in the fish retina. Int Rev Cytol. 2007;259:173–224. doi: 10.1016/S0074-7696(06)59005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M. Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development. 2007;134:1617–1629. doi: 10.1242/dev.001255. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000;19:5460–5472. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raay TJ, Vetter ML. Wnt/frizzled signaling during vertebrate retinal development. Dev Neurosci. 2004;26:352–358. doi: 10.1159/000082277. [DOI] [PubMed] [Google Scholar]
- Vetter ML, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin Cell Dev Biol. 2001;12:491–498. doi: 10.1006/scdb.2001.0273. [DOI] [PubMed] [Google Scholar]
- Wang X, Wan H, Korzh V, Gong Z. Use of an IRES bicistronic construct to trace expression of exogenously introduced mRNA in zebrafish embryos. Biotechniques. 2000;29:814–816. 818, 820. doi: 10.2144/00294st09. [DOI] [PubMed] [Google Scholar]
- Wang JC, Harris WA. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol. 2005;285:101–115. doi: 10.1016/j.ydbio.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish book: A guide for the Laboratory Use of Zebrafish (Brachydanio rerio) Eugene: Institute of Neuroscience, University of Oregon; 2000. [Google Scholar]
- Xiao T, Shoji W, Zhou W, Su F, Kuwada JY. Transmembrane sema4E guides branchiomotor axons to their targets in zebrafish. J Neurosci. 2003;23:4190–4198. doi: 10.1523/JNEUROSCI.23-10-04190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan RT, Wang SZ. NeuroD induces photoreceptor cell overproduction in vivo and de novo generation in vitro. J Neurobiol. 1998;36:485–496. [PMC free article] [PubMed] [Google Scholar]
- Yan RT, Wang SZ. Expression of an array of photoreceptor genes in chick embryonic retinal pigment epithelium cultures under the induction of neuroD. Neuroscience Letters. 2000;280:83–86. doi: 10.1016/s0304-3940(99)01003-4. [DOI] [PubMed] [Google Scholar]
- Yan RT, Wang SZ. Requirement of neuroD for photoreceptor formation in the chick retina. Invest Ophthalmol Vis Sci. 2004;45:48–58. doi: 10.1167/iovs.03-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan RT, Ma W, Liang L, Wang SZ. bHLH genes and retinal cell fate specification. Mol Neurobiol. 2005;32:157–171. doi: 10.1385/MN:32:2:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panel A is a cartoon illustrating subcloning of the Hsp70/4:neuroDEGFP construct. The Hsp70/4:neuroDegfp construct was assembled by subcloning the 1kb coding region of the zebrafish neuroD gene into the Hsp70/4:EGFP vector. Panels B1-3 illustrate the localization of EGFP in the cytoplasm of HEK293 cells transfected with the empty Hsp70/4:EGFP construct. Panels C1-3 illustrate localization of EGFP in the nucleus of HEK293 cells transfected with the Hsp70/4:neuroDegfp construct. Left-hand panels illustrate GFP fluorescence; Middle panels illustrate nuclear staining with bisbenzimide; right-hand panels are digital overlays.
Panel A illustrates a transient transgenic embryo treated with heat shock at 24hpf and photographed at 48hpf. Note the mosaic of GFP-labeled cells. Panel B is an uninjected control treated with heat shock. Panel C illustrates a section through the head and retina of a transient transgenic embryo at 48hpf. The enclosed area is illustrated at higher magnification (and rotated slightly) in D. Note the nuclear localization of the GFP. D1 – Nomarski illumination; D2 – GFP fluorescence; D3 – nuclear staining with bisbenzimide; D4 – digital overlay D2 and D3. The scale bar in A represents 200μm for panels A and B and 50μm for panel C.
This panel illustrates in situ hybridization for neuroD in a transient transgenic embryo (see Fig. S2) treated with heatshock at 48hpf and sacrificed at 96hpf. Note the numerous labeled cells throughout the retina and head. Note the ectopic neuroD-expressing cells in the CMZ. CMZ – circumferential germinal zone. L – lens. Scale bar equals 50μm.
Lanes 1-3 correspond to the three Tg(Hsp70/4:neuroDegfp) lines. Lane 3 corresponds to the line selected for this study. Note the bands at 5kb, which represent endogenous neuroD, and the intense bands at 12kb, which represent the transgene. The left-most lane contains molecular weight markers.
Panels G and J are illustrated in Figure 5.
Panels A and B are wt embryos at 72hpf immunostained with zn12 and HPC-1 antibodies, respectively. Panels C and D are neuroD morphants treated similarly. Note that the inner retinal layers of the morphant retinas contain differentiated neurons.