Abstract
The minichromosome maintenance (MCM) proteins are essential for DNA replication in eukaryotes. Thus far, all eukaryotes have been shown to contain six highly related MCMs that apparently function together in DNA replication. Sequencing of the entire genome of the thermophilic archaeon Methanobacterium thermoautotrophicum has allowed us to identify only a single MCM-like gene (ORF Mt1770). This gene is most similar to MCM4 in eukaryotic cells. Here we have expressed and purified the M. thermoautotrophicum MCM protein. The purified protein forms a complex that has a molecular mass of ≈850 kDa, consistent with formation of a double hexamer. The protein has an ATP-independent DNA-binding activity, a DNA-stimulated ATPase activity that discriminates between single- and double-stranded DNA, and a strand-displacement (helicase) activity that can unwind up to 500 base pairs. The 3′ to 5′ helicase activity requires both ATP hydrolysis and a functional nucleotide-binding site. Moreover, the double hexamer form is the active helicase. It is therefore likely that an MCM complex acts as the replicative DNA helicase in eukaryotes and archaea. The simplified replication machinery in archaea may provide a simplified model for assembly of the machinery required for initiation of eukaryotic DNA replication.
The minichromosome maintenance proteins (MCMs) are a class of protein that have thus far been specific to eukaryotes (1). Originally identified in yeast as being essential for DNA replication (2), they have since been shown to be required for the initiation of DNA replication in Xenopus (3–5). The binding of the MCMs to chromosomes is a prerequisite for DNA replication initiation and occurs after the origin recognition complex (ORC) and Cdc6 proteins have bound (6–11). Phylogenetic analysis has suggested that all eukaryotes contain six MCM genes (12, 13). Although there are a number of reports on the purification of MCM complexes (4, 14, 15) and subcomplexes containing fewer than six different MCMs (16–19), biochemical analysis has been limited. Sequence comparisons and analysis have suggested that the MCMs may possess a DNA helicase activity (20). Consistent with this, a purified complex containing three of the six human MCM proteins displayed ATPase activity and limited DNA helicase activity (21). More recently, intrinsic DNA helicase and ATPase activity have been shown for a complex containing recombinant mouse MCMs 4, 6, and 7 (22).
The archaea are a group of organisms that comprise a third “domain” of life and contain genetic characteristics of both bacteria and eukaryotes (23). The sequencing of a number of complete archaeal genomes (24–27) has enabled a number of analyses to determine the relatedness of archaeal proteins to those in eukaryotes and bacteria. Interestingly, although more archaeal protein sequences are related to sequences found in bacteria, the majority of proteins required for the processes of DNA replication, transcription, and translation are significantly more related to those found in eukaryotes (28, 29). In the case of the MCMs, we have been able to identify only a single MCM-like sequence in the majority of complete archaeal genomes (24–29). The reduction in the number of MCM genes [as well as a reduction in the number of subunits in other essential replication proteins (30)] suggests that the archaea may provide a simplified model for the mechanism of assembly of the eukaryotic prereplication complex.
Materials and Methods
Expression and Purification of M. thermoautotrophicum MCM (MtMCM).
M. thermoautotrophicum genomic DNA was a kind gift from S. P. Jackson (Wellcome/CRC Institute, Cambridge, U.K.) and T. Darcy and J. Reeve (Ohio State University, Columbus, OH). ORF 1770 was amplified by touchdown PCR into pQE30 (Qiagen, Chatsworth, CA) to yield a construct encoding a fusion protein tagged at the N terminus with RGSHHHHHH (predicted mass, 75.5 kDa, designated WT, plasmid pJC025). This construct was subjected to site-directed mutagenesis to produce an inactivating point mutation in the putative Walker A motif required for ATP hydrolysis (K341E, plasmid pJC055). An N-terminal deletion of 111 aa was cloned into pET-28a (Novagen) to yield a fusion protein with predicted mass of 62.5 kDa, (ΔN, plasmid pJC056). All three proteins were expressed in Escherichia coli. Bacterial lysates were prepared by sonication in buffer T (20 mM Tris, pH 8.0/100 mM NaCl/5% glycerol/0.1% 2-mercaptoethanol). After clarification by centrifugation (15,000 rpm, 20 min) in an SS-34 rotor (Sorvall), the lysate from 2 liters of culture was passed over a 20-ml Talon metal affinity resin column (CLONTECH). The column was washed with buffer T containing 300 mM NaCl/10 mM imidazole and then 500 mM NaCl/40 mM imidazole. The protein was eluted in buffer T containing 100 mM NaCl/300 mM imidazole. The eluate was passed over a 10-ml hydroxyapatite column (Bio-Rad), washed with 20 mM KH2PO4, pH 8.5/50 mM NaCl/5% glycerol/0.1% 2-mercaptoethanol, and eluted with a gradient of 20–500 mM KH2PO4, pH 8.5. Fractions were pooled and supplemented with 5 M NaCl to a final concentration of 2 M NaCl and loaded onto a 15-ml phenyl-Sepharose column (Amersham Pharmacia Biotech). The column was washed with 20 mM Tris, pH 8.5/1 mM EDTA/1 mM EGTA/2 M NaCl/5% glycerol/0.1% 2-mercaptoethanol, and protein was eluted over a 2–0 M NaCl gradient. The sample was diluted 1:1 and passed over a 1-ml Mono Q column (Amersham Pharmacia Biotech), washed with 20 mM Tris, pH 7.5/1 mM EDTA/1 mM EGTA/50 mM NaCl/5% glycerol/0.1% 2-mercaptoethanol, and eluted on a 50–500 mM NaCl gradient. The purified protein was concentrated and stored at −70°C. The ΔN protein was purified as above, substituting Phenyl-5PW (TosoHaas, Montgomeryville, PA) for phenyl-Sepharose.
ATPase Assays.
Reaction mixes containing HDB buffer (50 mM Hepes, pH 7.6/1 mM DTT/0.1 mg/ml BSA/7 mM MgCl2), 203 pmol of ATP containing 2.5 μCi (1 Ci = 37 GBq) of [α-32P]ATP (800 Ci/mmol, ICN), and 331 ng of protein were assembled on ice. Tubes containing DNA were assembled on ice in parallel. Both sets of tubes were preincubated for 5 minutes at 60°C before being mixed and incubated for a further 30 min at 60°C. Reactions were stopped by the addition of EDTA to 10 mM. ATP hydrolysis was visualized by using thin-layer chromatography. The plate was developed in 1 M formic acid/0.5 M LiCl, dried, and exposed to film. The result was quantified on a phosphorimager (Fuji) against standard dilutions of [α-32P]ATP.
Band-Shift Assays.
A 30-base oligomer was labeled by using T4 polynucleotide kinase (PNK) and [γ-32P]ATP. The kinase was heat inactivated and free label was removed by using a Microspin G-25 column (Amersham Pharmacia Biotech). Twenty-microliter reaction mixtures containing HDB buffer, the labeled oligomer, MtMCM protein, and 4 mM nucleotide were assembled on ice before being incubated at room temperature for 10 min. The reaction mixtures were supplemented with 4% glycerol and the products were subjected to electrophoresis on a 2% agarose gel. The gel was dried and exposed to film.
Helicase Assays.
Helicase substrate was made by 5′ (T4 PNK; New England Biolabs) or 3′ (Klenow; New England Biolabs) labeling oligomers that were annealed to pUC118 single-stranded DNA (ssDNA) and purified. Linear substrates to measure polarity were made from a 59-base oligomer that was digested using SmaI (New England Biolabs) after labeling (see scheme in Fig. 3B). To assess processivity, long substrates were made by primer extension of the helicase substrate (outlined above) using Sequenase (Amersham Pharmacia Biotech), dNTPs, and ddGTP in the presence of sodium isocitrate (31). Reaction mixtures containing HDB buffer, substrate, 4 mM ATP (or analogues), and protein were assembled on ice and incubated at 37°C for 30 min. Reactions were terminated with 5 μl of Stop (200 mM EDTA/1% SDS/20% glycerol/bromphenol blue), and products were resolved on nondenaturing 15% acrylamide gels. Gels were fixed, dried, and exposed to film. The displaced oligonucleotide was compared with an equal amount of heat-denatured substrate. Displaced oligonucleotides were quantified on a phosphorimager.
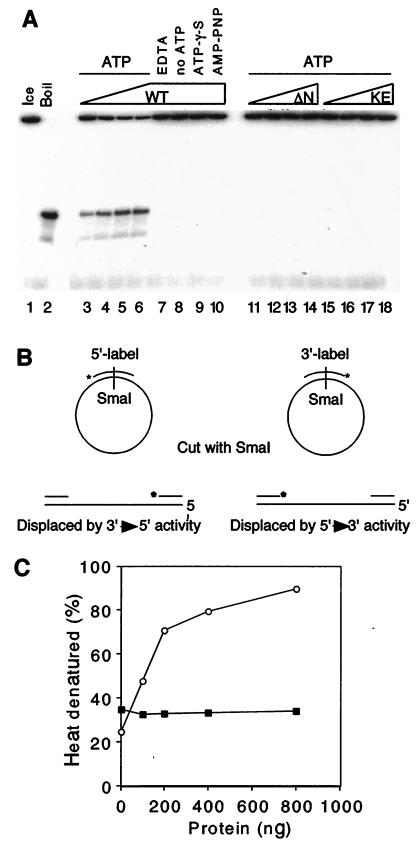
Figure 3.
MtMCM shows an ATP-dependent DNA helicase activity. (A) Reaction mixtures (20 μl) containing 100 ng (lanes 3, 11, 15), 200 ng (lanes 4, 12, 16), 400 ng (lanes 5, 13, 17), or 800 ng (lanes 6–10, 14, 18) of protein were incubated with ≈10 fmol of substrate and 4 mM ATP (lanes 3–7, 11–18), 4 mM ATP[γ-S] (lane 9), 4 mM 5′-[β,γ-imido]triphosphate (AMP-PNP; lane 10), or 20 mM EDTA (lane 7). (B) Oligonucleotides labeled at the 5′ or 3′ ends were annealed to pUC118 ssDNA and digested to produce linear substrates that were used to measure polarity of displacement. (C) MtMCM protein (100, 200, 400, and 800 ng) was incubated with ≈300 fmol of linear substrate in helicase assays. The displaced oligonucleotide was compared with an equal amount of heat-denatured substrate. Displaced 5′-labeled (○) and 3′-labeled (●) oligonucleotides were quantified on a phosphorimager.
Estimation of Native Molecular Mass.
Purified protein (1–1.5 μg) was passed over a 2.4-ml Superose 6 gel-filtration column (Smart PC3.2/30; Amersham Pharmacia Biotech) equilibrated in GF buffer (50 mM Hepes, pH 7.6/150 mM NaCl/1 mM DTT), at 20 μl/min. Fractions (150 μl) were precipitated with deoxycholate/trichloroacetic acid and subjected to SDS/PAGE (stained with Coomassie blue) or assayed for ATPase and helicase activity. Stokes radii were calculated by using Kav (32) from gel-filtration data of samples compared with markers (thyroglobulin, ferritin, aldolase, and bovine serum albumin; 85.0, 61.0, 48.1, and 35.5 Å, respectively). Sucrose gradients (4.8 ml, 20–50%) in GF buffer were spun at 49,000 rpm (no brake, 4°C, 11 hr) in an SW55 rotor (Beckman). Fractions (200 μl) were precipitated, subjected to SDS/PAGE, and compared with markers on parallel gradients. Sedimentation coefficients were calculated from sucrose-gradient sedimentation of samples compared with markers (thyroglobulin, catalase, and aldolase; 19.4, 11.3, and 8.3 × 10−13 sec, respectively). The native molecular masses of MtMCM and its derivatives were calculated as described by Siegel and Monty (33).
Results
wt MtMCM Possesses a DNA-Stimulated ATPase Activity.
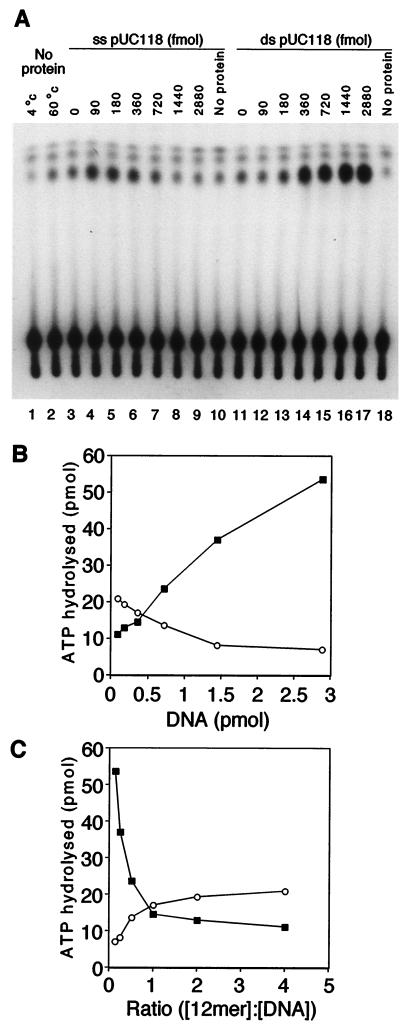
Purified MtMCM (360 fmol of 12-mer) was tested for the ability to hydrolyze ATP in the presence of increasing concentrations of single- or double-stranded DNA (ssDNA and dsDNA, respectively; Fig. 1A). Because this protein was cloned from a thermophilic organism, ATPase assays were performed at the optimal growing temperature of 60°C. Quantification (Fig. 1B) showed that significant ATPase activity was detected only when wild-type protein was incubated in the presence of DNA (Fig. 1A, compare lane 11 to lane 17). ATPase activity was stimulated to a small extent by low concentrations of ssDNA (lanes 4 and 5, 90–180 fmol), but was inhibited as the concentration of ssDNA increased (lanes 6–9, 360-2880 fmol). In contrast, much more ATPase activity was detected in the presence of high dsDNA concentrations (lanes 14–17, 360-2880 fmol). No ATP hydrolysis was detected in reaction mixtures containing the K341E or ΔN mutant proteins (data not shown). Similar results were obtained from performing assays at 37°C, where overall activity was reduced. This suggests, as expected, that the endogenous ATPase activity of MtMCM depends on the Walker A motif. Furthermore, it suggests that the N terminus of the protein is important for ATPase activity, even though this region does not contain the ATPase motif. Fig. 1C presents ATPase activity as a function of the molar ratio of protein to DNA. The protein concentration is calculated assuming that MtMCM forms a double hexamer (see below). The inflection point in the graph suggests that the manner in which ATP is hydrolyzed may change when there is at least one protein complex per DNA molecule. One possible reason for this is that more than one MtMCM complex per DNA molecule prevents movement of the complex and thus blocks ATPase activity.
Figure 1.
MtMCM protein shows DNA-stimulated ATPase activity. (A) ≈360 fmol of 12-mer MtMCM was assayed for ATPase activity in the presence of 90, 180, 360, 720, and 1,440 fmol (lanes 4–8, 12–16) and 2,880 fmol (lanes 9, 10 and 17, 18) of closed circular pUC118 ssDNA or pUC118, respectively. (B) ATPase activity was plotted against the concentration of closed circular single-stranded pUC118 (○) or double-stranded pUC118 (■). (C) The quantified data were replotted as a function of the molar ratio of [protein]/[DNA] (using the same convention as for B). For the purpose of the calculation, MtMCM was assumed to form double hexamers.
DNA-Binding Properties of MtMCM.
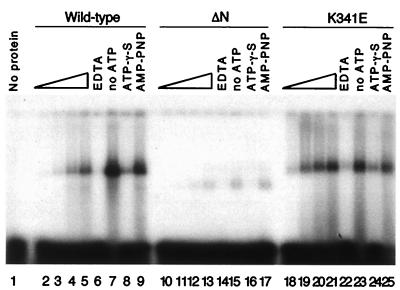
Band-shift experiments performed at 25°C show that wt MtMCM could stably bind to a ssDNA 30-base oligonucleotide (Fig. 2, lanes 2–5, 7–9). Both wt and K341E protein formed complexes with the DNA (lanes 5, 21). The intensity of the shifted band was proportional to the amount of protein added in the presence of hydrolyzable ATP (lanes 2–5 and 18–21). The protein bound most strongly in the presence of Mg2+ alone (lanes 7, 23) or with the nonhydrolyzable analogue adenosine 5′-[β,γ-imido]triphosphate (lanes 9, 25). Interestingly, a second analogue, ATP[γ-S], did not enhance binding of the protein to DNA but showed binding comparable to that seen with hydrolyzable ATP (compare lanes 8, 24 with 5, 21). It is possible that this nucleotide can be hydrolyzed by MtMCM, or produces a steric effect not seen with 5′- [β,γ-imido]triphosphate. Chelation of magnesium ions by EDTA prevented the protein from binding to DNA (lanes 6, 22). The K341E mutant was found to bind to oligonucleotides more efficiently than wt protein (compare lanes 2–5 to 18–21), adding further support to the idea that ATP is dispensable for ssDNA binding and may actually reduce DNA binding. It is interesting to note that, in the absence of ATP, the MtMCM quantitatively shifted all of the ssDNA (lanes 7, 23) whereas, in the presence of ATP, only a small fraction of ssDNA was bound. This might suggest that the ATP binding site and ssDNA interaction site overlap in the MtMCM protein. The ΔN mutant bound DNA to a lesser extent, and resulted in a faster-migrating band, suggesting an incomplete complex (lanes 10–17). Oligonucleotides as small as 12 bases were shifted in the same manner, with no observed sequence dependence. Similar results were seen with dsDNA substrates (data not shown).
Figure 2.
MtMCM requires Mg2+ but not ATP to bind DNA. One hundred nanograms (lanes 2, 10, 18), 200 ng (lanes 3, 11, 19), 400 ng (lanes 4, 12, 20), and 800 ng (lanes 5–9, 13–17, 21–25) of wt and mutant protein were incubated with 20 fmol of labeled oligomer in the presence of 4 mM ATP (lanes 2–6, 10–14, 18–22), 4 mM γ-thio-ATP (ATP[γ-S]; lanes 8, 18, 24), 4 mM 5′-[β,γ-imido]triphosphate (AMP-PNP; lanes 9, 17, 25). Lanes 7, 15, and 23 did not contain ATP, and lanes 6, 14, and 22 were supplemented with an additional 20 mM EDTA to chelate free metal ions.
MtMCM Shows DNA Helicase Activity.
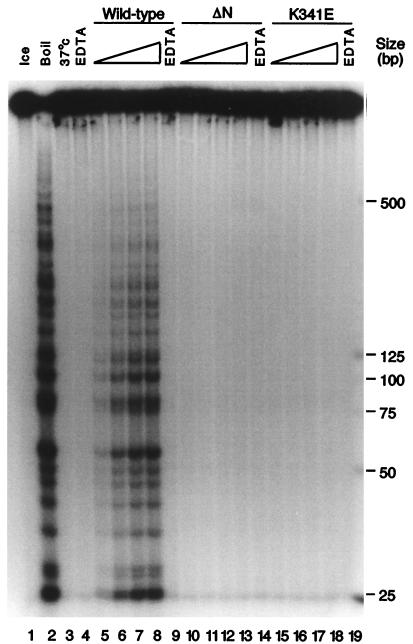
Purified MtMCM was tested for DNA helicase activity by a strand-displacement assay performed at 37°C (34) (Fig. 3A). wt protein was shown to displace a 30-base oligonucleotide annealed to pUC118 ssDNA only under conditions in which ATP hydrolysis could occur (lanes 3–10). Neither of the mutant proteins displayed helicase activity (lanes 11–18). Substrates to determine the polarity of strand displacement were made according to the scheme outlined in Fig. 3B. Strand displacement using the wt protein showed preferential 3′ → 5′ polarity with respect to the potential template strand on the pUC118 ssDNA (Fig. 3C). This is the same preference as reported for simian virus 40 T antigen (35). If an MCM complex really is the replicative helicase, it should be somewhat processive. This has not been the case in previously reported results (21). We attempted to address this point by using a population of substrates possessing displaceable strands of increasing length. We observed displacement of strands up to approximately 500 bases in length on incubation of these substrates with wt MtMCM protein in the absence of additional proteins (Fig. 4, lanes 6–8, 215–870 fmol). MtMCM showed no preference for 3′ or 5′ ssDNA tails as a substrate for helicase activity (data not shown).
Figure 4.
wt MtMCM is capable of displacing up to 500 bp of DNA without additional proteins. ≈10 fmol of long substrate was incubated with 100 ng (lanes 5, 10, 15), 200 ng (lanes 6, 11, 16), 400 ng (lanes 7, 12, 17), or 800 ng (lanes 8, 9, 13, 14, 18, 19) of protein for 30 min at 37°C. Displaced oligonucleotides were separated by PAGE.
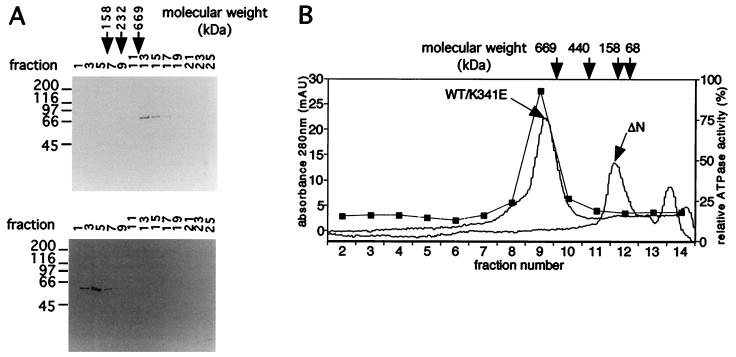
wt MtMCM Forms a Dodecamer.
To determine the size of the active MtMCM complex, purified wt, K341E, and ΔN proteins were subjected to sucrose gradient sedimentation (Fig. 5A) and gel filtration (Fig. 5B). In both cases, wt and K341E protein sedimented and eluted at the same positions (yielding a sedimentation coefficient of 3.0 × 10−13 sec and Stokes radius of 92.1 Å). Native molecular mass calculations using Kav (33) determined that the wt and K341E proteins formed a multimeric complex of some 854 kDa, consistent with the formation of a dodecamer. In contrast, the ΔN mutant protein (yielding a sedimentation coefficient of 22.5 × 10−13 sec and Stokes radius of 47.0 Å) displayed a native molecular mass of some 58 kDa, suggesting a monomeric composition. This suggests that there may be a motif essential for protein multimerization in the N-terminal 111 aa of MtMCM and implies (as the K341E mutant is multimeric) that a functional Walker A motif is not required for multimerization of MtMCM. Both the DNA-stimulated ATPase activity and helicase activity coeluted on gel filtration with the multimeric wt protein (Fig. 5B). These activities, however, were not detected in K341E protein (data not shown). This observation reduces the likelihood that these activities are because of a copurifying contaminant.
Figure 5.
Estimation of the native molecular mass of MtMCM protein. (A) wt and ΔN proteins were sized by sucrose gradient sedimentation, and fractions were subjected to SDS/PAGE and stained with Coomassie blue. (B) Samples were also sized by gel filtration. Gel filtration fractions were assayed for ATPase activity (■). Activity coeluted only with the peak of wt protein.
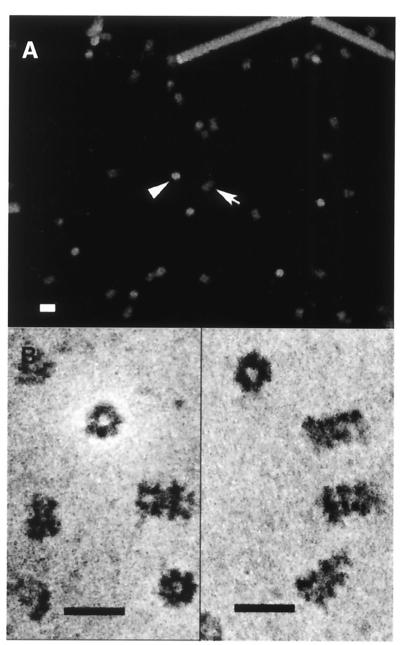
The Dodecamer Form of MtMCM Is a Double Hexamer.
wt MtMCM was examined by scanning transmission electron microscopy (36). The micrographs obtained show two views that combined suggest a cylindrical structure (Fig. 6). Mass measurements of unstained samples (Fig. 6A) yielded estimates of molecular mass of 841.3 ± 45.3 kDa for the end view and 814.2 ± 85.3 kDa for the side view, consistent with our other measurements. This mass is also consistent with the formation of a dodecamer. High-resolution micrographs suggest sixfold symmetry in the protein and hint that the dodecamer is formed from two opposed hexamers (Fig. 6B). A number of other reports on the structure of molecules possessing helicase activity also reveal sixfold symmetry (37–39). In contrast, sizing data for the eukaryotic MCM complexes are more consistent with single hexamers (4, 14, 19, 21). Given our functional data, this finding suggests either that eukaryotic complexes dissociate more readily or that other components are required to yield a functional double-hexamer complex. The RLF-B activity (4, 40) or Cdc45p (9, 11, 41), which are required for assembly of functional MCM complexes onto DNA, may provide such a function.
Figure 6.
wt MtMCM protein forms a double hexamer. (A) Samples of wt protein were subjected to scanning transmission electron microscopy. Separate mass estimates of end (arrowhead) and side views (arrow) were based on unstained sample measurements. (B) Stained samples showed a central pore in the end view and suggested that the complexes consisted of a double hexamer. (Bar = 20 nm.)
Discussion
These data allow us to draw a number of conclusions. wt MtMCM is able to hydrolyze ATP. Stimulation of this activity is greatest in the presence of dsDNA. For stimulation to occur, the protein must be able to form a multimeric complex, which we suggest is a double hexamer. ATP binding or hydrolysis is not required for the binding of MtMCM to ssDNA, suggesting that the ATP hydrolysis is required for a function that occurs after the formation of the dsDNA–MCM complex. One possible requirement for ATP hydrolysis is for the translocation of the protein around the DNA molecule, which may be required for the MCMs to identify an origin marked by ORC. ATP hydrolysis significantly decreases as the number of MCM double hexamers per DNA molecule rises above one, possibly because of the collision of two MCM complexes or topological problems. Other obvious possibilities are that ATP hydrolysis is required for threading of the MCM complex onto the DNA, or for the unwinding of the dsDNA molecule. The lack of helicase activity in the K341E mutant is consistent with ATP hydrolysis being required for helicase activity. Thus, our data extend the initial protein sequence predictions (20) and the helicase data with mammalian MCM proteins (21, 22).
The ΔN mutant data suggest that the first 111 aa of the protein probably contains a region required for multimerization and demonstrates that complex formation is required for helicase activity. Like other helicases (T antigen, E1, DnaB, etc.) (1), the MtMCM seems to function as a hexamer. Interestingly, the 3′ → 5′ polarity of the MtMCM helicase, and the human MCM helicase, on DNA is the same as that of the eukaryotic virus helicases of simian virus 40 (35, 37, 42) and bovine papillomavirus (38, 39). More striking is the observation that MtMCM is a double hexamer in solution, which is analogous to the double hexamer of simian virus 40 T antigen formed at origins and at a replication fork (35, 37, 42, 43). Higher-order complex formation may also be essential for ATP hydrolysis, although both of these activities may be disrupted because of incorrect folding of the protein. The discrepancy in mass measurements between MtMCM complexes and eukaryotic MCM complexes and subcomplexes suggests a double and single hexamer, respectively. The reason for this difference is currently unclear.
It remains to be determined whether MCM proteins promote unwinding of the DNA molecule only at origins of replication or whether the helicase also functions at the DNA replication fork as the principle replicative helicase. It is likely that eukaryotic MCM helicase activity is regulated through the cell cycle. MCM helicase activity may be activated by the presence of proteins such as RP-A and Cdc45p, both of which load onto dsDNA in an MCM-dependent manner (41, 44). A similar activation by RP-A has been observed for simian virus 40 T antigen and bovine papillomavirus E1 (35, 38). Such interactions or activation may be related to the physiological role of MCM helicase activity. For replication fork proteins to be assembled at the origin of replication, unwinding at the origin is probably necessary. This initial melting may require a helicase activity that is distinct from the processive DNA duplex unwinding required during replication fork movement. These helicase activities may be provided by different proteins, and certainly more than one DNA helicase has been identified in eukaryotes that may function during DNA replication (45). At present, it is not clear whether the MCMs provide either one or both of these activities. Many data exist that support the role of MCMs in the initiation of replication (reviewed in ref. 1). Mammalian MCMs seem not to associate with replication forks, and biochemical data for mammalian MCMs show only limited unwinding, consistent with an early melting step (21, 22). Our current data suggest that the MtMCM is capable of more processive unwinding, even without accessory factors. Bell and colleagues (9) report data consistent with yeast MCMs moving with the replication fork, which would support the possibility that the MCMs are the only helicase required for DNA replication.
If the mechanism of archaeal replication faithfully reflects eukaryotic DNA replication, then we would predict that other proteins such as ORC and Cdc6p would be required for origin unwinding to occur. In addition, proteins such as RP-A, RF-C, and PCNA should be required to facilitate processivity. Determining this will allow us to decide whether the archaea can be used as a simplified model for the initiation of eukaryotic replication.
Acknowledgments
Thanks to H. Fearnhead, S. Sepehri, and C. Sanders for helpful discussion and comments on the manuscript, V. Ellison and M. Weinreich for further discussion, and S. P. Jackson, T. Darcy, and J. Reeve for M. thermoautotrophicum genomic DNA. This research was supported by National Institutes of Health Grant GM45436. J.P.J.C. was the recipient of European Molecular Biology Organization and Human Frontier Science Program Postdoctoral Fellowships. The Brookhaven National Laboratory Scanning Transmission Electron Microscope is a National Institutes of Health Supported Resource Center, National Institutes of Health P41-RR01777, with additional support provided by Department of Energy, Office of Biological and Environmental Research.
Abbreviations
- MCM
minichromosome maintenance
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- MtMCM, Methanobacterium thermoautotrophicum MCM
wt, wild type
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.030539597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.030539597
References
- 1.Tye B K. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 2.Maine G T, Sinha P, Tye B K. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 4.Chong J P, Mahbubani H M, Khoo C Y, Blow J J. Nature (London) 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- 5.Madine M A, Khoo C Y, Mills A D, Mushal C, Laskey R A. Curr Biol. 1995;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- 6.Liang C, Weinreich M, Stillman B. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 7.Coleman T R, Carpenter P B, Dunphy W G. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 8.Rowles A, Chong J P, Brown L, Howell M, Evan G I, Blow J J. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 9.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 10.Donovan S, Harwood J, Drury L S, Diffley J F. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 12.Chong J P, Thömmes P, Blow J J. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- 13.Kearsey S E, Maiorano D, Holmes E C, Todorov I T. BioEssays. 1996;18:183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- 14.Adachi Y, Usukura J, Yanagida M. Genes Cells. 1997;2:467–479. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- 15.Richter A, Knippers R. Eur J Biochem. 1997;247:136–141. doi: 10.1111/j.1432-1033.1997.00136.x. [DOI] [PubMed] [Google Scholar]
- 16.Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. J Biol Chem. 1996;271:24115–24122. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- 17.Kimura H, Ohtomo T, Yamaguchi M, Ishii A, Sugimoto K. Genes Cells. 1996;1:977–993. doi: 10.1046/j.1365-2443.1996.840284.x. [DOI] [PubMed] [Google Scholar]
- 18.Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thömmes P, Kubota Y, Takisawa H, Blow J J. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin E V. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishimi Y. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 22.You Z, Komamura Y, Ishimi Y. Mol Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woese C R, Kandler O, Wheelis M L. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 25.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 27.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- 28.Edgell D R, Doolittle W F. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 29.Bernander R. Mol Microbiol. 1998;29:955–961. doi: 10.1046/j.1365-2958.1998.00956.x. [DOI] [PubMed] [Google Scholar]
- 30.Kelly T J, Simancek P, Brush G S. Proc Natl Acad Sci USA. 1998;95:14634–14639. doi: 10.1073/pnas.95.25.14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Desiderio S. In: Transcription Factors: A Practical Approach. Harris E L V, Angal S, editors. Oxford: IRL; 1993. pp. 185–196. [Google Scholar]
- 33.Siegel L M, Monty K J. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 34.Biswas E E, Ewing C M, Biswas S B. Biochemistry. 1993;32:3020–3026. doi: 10.1021/bi00063a013. [DOI] [PubMed] [Google Scholar]
- 35.Goetz G S, Dean F B, Hurwitz J, Matson S W. J Biol Chem. 1988;263:383–392. [PubMed] [Google Scholar]
- 36.Linderoth N A, Simon M N, Russel M. Science. 1997;278:1635–1638. doi: 10.1126/science.278.5343.1635. [DOI] [PubMed] [Google Scholar]
- 37.Mastrangelo I A, Hough P V, Wall J S, Dodson M, Dean F B, Hurwitz J. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 38.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fouts E T, Yu X, Egelman E H, Botchan M R. J Biol Chem. 1999;274:4447–4458. doi: 10.1074/jbc.274.7.4447. [DOI] [PubMed] [Google Scholar]
- 40.Tada S, Chong J P J, Mahbubani H M, Blow J J. Curr Biol. 1999;9:211–214. doi: 10.1016/s0960-9822(99)80092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou L, Mitchell J, Stillman B. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wessel R, Schweizer J, Stahl H. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West S C. Cell. 1996;86:177–180. doi: 10.1016/s0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka T, Nasmyth K. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Budd M E, Choe W C, Campbell J L. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]