Abstract
The nucleotide sequence of DNA is the repository of hereditary information. Yet, it is now clear that the DNA itself plays an active role in regulating the ability of the cell to extract its information. Basic biological processes, including control of gene transcription, faithful DNA replication and segregation, maintenance of the genome and cellular differentiation are subject to the conformational and topological properties of DNA in addition to the regulation imparted by the sequence itself. How do these DNA features manifest such striking effects and how does the cell regulate them? In this review, we describe how misregulation of DNA topology can lead to cellular dysfunction. We then address how cells prevent these topological problems. We close with a discussion on recent theoretical advances indicating that the topological problems, themselves, can provide the cues necessary for their resolution by type-2 topoisomerases.
INTRODUCTION
DNA has evolved into a stable vehicle for transmitting genes from one generation to the next, providing a remarkably reliable set of instructions for building a cell. The functional elegance of DNA is reflected in the beauty of its double helical structure. Watson and Crick wrote, “It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material (1)”. The concept of one strand of DNA serving as a template explains the inheritance of the genome and explicitly describes a faithful mechanism for DNA replication and repair of a damaged strand. As such, the information encoded by DNA is often considered solely as a linear sequence of nucleotides to be read by cellular proteins. However, considering only the sequence of DNA neglects the unique set of challenges imposed by the mechanical, structural and topological features of double helical DNA, especially considering that it is confined to the cramped cellular space. Incorporating the feedback that the DNA double helix exerts on the proteins that must read the nucleotide sequence is critical for a better understanding of DNA metabolism. In this review, we explore why DNA topology must be maintained, corrected and altered in cells and outline existing models for how topoisomerases may control DNA topology.
CELLULAR CONSEQUENCES OF DNA TOPOLOGY
The topological and conformational consequences of the DNA double helix create an uphill struggle that Sisyphus could appreciate: to be active, DNA must be maintained in a higher energy conformation than relaxed B-form. In this case, topoisomerase acts as Sisyphus to maintain DNA in an underwound, untangled state (2,3; Figure 1). If DNA topology is not maintained in such a state, disaster can result, as recent data demonstrate.
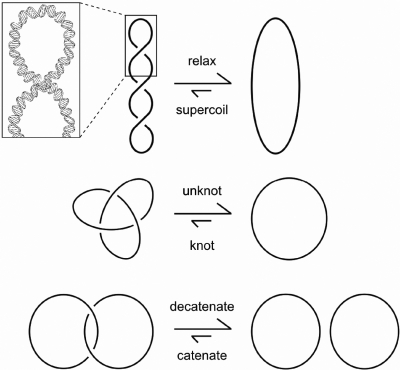
Figure 1.
Biologically relevant topological structures of DNA. Depicted are schematics of the three topological forms of DNA that topoisomerases maintain and modulate. For simplicity, each line represents a double-stranded DNA helix, as shown by the upper left inset. As indicated by the arrow sizes, most type-2 topoisomerases shift the DNA topology equilibrium toward relaxing, unknotting and decatenating. Bacterial DNA gyrase and archaeal reverse gyrase are unique enzymes that introduce supercoils into DNA. This figure is reproduced from (3).
Effects of DNA supercoiling
The canonical image of double helical DNA has the two backbone strands intertwined approximately every 10.5 bp. The processes that read the genetic code, such as semiconservative DNA replication, gene expression and homologous recombination, require access to the internal nucleotide bases. Cells allow access to the code by maintaining the DNA in a homeostatically underwound state (4–6). This helix underwinding means that the linking number (Lk; the number of times the Watson strand wraps around the Crick strand in plane projection) of DNA is less than in the lowest free-energy state. The underwound state can be manifest in two geometric forms: twist and writhe, described by the well-known equation ΔLk = ΔTw + ΔWr (7). Unconstrained, the underwinding causes DNA to buckle around itself, and this has been referred to as negative supercoiling (7).
Negative supercoiling provides the energy for localized, controlled melting of the DNA duplex to allow access of DNA polymerases, RNA polymerases, repair factors and recombinases to the internal nucleotide sequence. In addition to accessing these sequences, many DNA metabolic processes have additional specific DNA conformational requirements. For example, transcriptional regulation, through enhancers, and synapsis during site-specific recombination both require that distant DNA sites come in close physical proximity. Monte Carlo simulations (8) and experimental work (9,10,11) have demonstrated that supercoiling facilitates the synapsis of distant sites by two orders of magnitude. Site-specific recombination further requires that site juxtaposition occurs with a defined spatial arrangement and orientation, which negative supercoiling will also promote (12).
The dependence on negative supercoiling for chromosomal metabolism affords the cell a precise means of regulating DNA metabolism. Similar to signal transduction, the eukaryotic cell cycle and cell fate specification, much of DNA metabolism requires that a cell be fully in one state or another without an intermediate. The state in which a biological process can be on or off, ‘switch-like’, has been of much current interest (13,14). DNA supercoiling appears to be a switch. Site-specific recombination catalyzed by the λ Int protein changes from a low, background level activity with a nearly relaxed plasmid substrate to full activity over a very narrow (<2-fold) increase in negative supercoiling (15). In addition, transcription of a supercoiling-dependent leu500 promoter (4) and other bacterial processes (5) all switch on at the same supercoiling value. This switch-like dependence of supercoiling appears to be true for eukaryotic DNA function as well (6). Accordingly, the control of DNA supercoiling can have important consequences in terms of evolutionary fitness. Escherichia coli grown for 20 000 generations in a glucose-limited medium were found to contain mutations in topA, which encodes for topoisomerase I, or fis, which encodes for a general DNA-binding protein. Either of these mutations cause increased negative supercoiling, which can indicate that evolution favors increased negative supercoiling (16). However, excessive negative supercoiling also can lead to an inhibition of bacterial growth and RNA degradation (17). Evidently, it is very important for a cell to control DNA supercoiling and this feat is accomplished by the combined activities of the topoisomerases.
Effects of DNA catenation
Although the double helix suggested a mechanism for the faithful copying of nucleotide sequence, it also revealed a problem that Max Delbrück recognized over 50 years ago: every link between the parental Watson and Crick strands would be preserved in the daughter molecules following semiconservative DNA replication (18). He suggested an alternate model of discontinuous DNA replication to avoid this linking problem. In the midst of proposing this incorrect model, however, he did propose another mechanism (one he considered less elegant) for solving the linkage problem: an enzyme to break and reseal the DNA strands. The discovery of topoisomerases in the 1970s (19,20) and later experiments on the topology of replication products have confirmed Delbrück's insights and the mechanism is extraordinarily elegant.
Newly replicated DNA is intertwined (21–24). The intertwined DNAs, known as catenanes, or links, have a right-handed (+), parallel structure and are formed by DNA synthesis in vitro and in vivo. It is the job of type-2 topoisomerases, which change Lk by cleaving both DNA strands, passing another duplex through the transient gate, and resealing the gate, to unlink catenated intermediates of DNA replication; type-1 topoisomerases, which cleave only one strand, are not able to unlink catenanes unless the DNA contains nicks or gaps. For E. coli, the decatenating type-2 enzyme is topoisomerase IV (24,25); for metazoans, it is topoisomerase II (21,26,27). The inactivation of topoisomerase IV in E. coli results in highly catenated plasmid reporter molecules, as observed by electron microscopy and high-resolution gel electrophoresis, and nucleoids that failed to segregate (24,25), but does not affect the rate of DNA replication for at least half a dozen doubling times. This result indicates that topoisomerase IV is not needed for replication initiation or elongation, but only for decatenation.
Recent work using fluorescence microcopy has revealed that the separation of replicated genetic loci is impaired in E. coli in the absence of topoisomerase IV (28), which suggests a relationship between DNA catenation and chromosomal cohesion following DNA replication (28,29). Similar trends are apparent in eukaryotic cells. In yeast, the absence of topoisomerase II leads to impaired chromosome segregation and cell death at cytokinesis (30). When a hypomorphic mutant of topoisomerase II replaces the wild-type version, the yeast cell arrests its cell cycle before chromosome segregation at anaphase (31). These findings indicate that the cell is sensitive to catenated chromosomes and that their detection can activate a cell cycle checkpoint to prevent errors in genome transmission. It has been suggested that mammalian cells, with their larger genomes, are exquisitely sensitive to the presence of replication catenanes and contain a catenane checkpoint (32–34). In the presence of a catalytic inhibitor of topoisomerase II, ICRF-193, that does not produce a detectable DNA damage response, mammalian cells arrest prior to anaphase (33). ICRF-193 causes topoisomerase II to clamp stably at a catenane node (35). Thus, the stable binding of type-2 topoisomerase to catenane nodes could initiate a checkpoint response in mammalian cells in the same manner as yeast with a defective topoisomerase II, although it remains a formal possibility that ICRF-193 can induce undetectable levels of DNA damage. The sensitivity of different human cancer cell lines to ICRF-193 is highly variable; lung and bladder cancer cells are much less sensitive than others (34,36). It is possible that a defect in the ability of the cell to detect and resolve chromosomal linkages could lead to aneuploidy, a common occurrence in these tumors, and promote genomic changes associated with cancer development. The story is not so clear, however, as healthy mouse embryonic and human hematopoietic stem cells also appear to have a weakened DNA catenation checkpoint (37). What is clear is that the interplay of DNA linkage and the enzymes that unlink DNA are important for genome stability, cancer and development.
Effects of DNA knotting and additional topological problems originally associated with DNA segregation that may be associated with knots
When two daughter chromosomes are topologically linked, segregation of the genetic material cannot occur normally. However, a DNA molecule tangled in itself, a DNA knot, is also problematic for cells, perhaps even more problematic than catenanes. The biophysical properties of the DNA polymer give it a propensity to become knotted (38–42). This idea makes intuitive sense: the same drive that causes headphone wires and computer cables to become self-entangled (and prompted the wireless forms of these devices) applies also to long, flexible DNA in the cellular space crunch. In spite of a drive to entanglement, cellular DNA rarely is found knotted under physiologically normal conditions. At least three factors keep the DNA unknotted in a cell: (i) type-2 topoisomerases, which remove knots as they form; (ii) the organization of DNA into nucleosomes, which serves not only to compact the bulk of the DNA, but also holds it in an unknotted state. Nucleosomes, then, effectively reduce the length of DNA that must be surveyed by type-2 topoisomerases for knots; and (iii) DNA supercoiling, which suppresses formation of knots, at least for protein-free DNA (43). Early results from simulations indicated that supercoiling promoted DNA knotting and that knots represented a lower free-energy state than DNA supercoils (44). However, in those simulations, as knots formed, supercoils were concomitantly removed. In a cell, as discussed above, DNA negative supercoiling is tightly maintained homeostatically. When such homeostasis is maintained in the simulations, in fact DNA supercoiling is a lower energy conformation than DNA knotting (43).
Why do cells keep their genomes unknotted and what would happen if they did not? Experimentally, this question can be addressed by increasing the activity of processes known to increase DNA knotting or by inhibiting the activity of the type-2 topoisomerase needed to untie the knot. Overexpressing the Hin site-specific recombinase in E. coli, which leads to increased knotting in a plasmid containing its recombination sites, blocked DNA replication and transcription, increased mutation, and led to loss of the replicon (45). The inhibition of topoisomerase IV exacerbated these effects (45). Thus, DNA knotting interferes with genetic metabolism.
Recent experiments have suggested a link between the topological state of the DNA and cellular differentiation in eukaryotes. Differentiated cells, such as Xenopus erythrocytes, have completed and exited the cell cycle. When nuclei from these differentiated cells are added to interphase Xenopus egg extracts, a system that recapitulates a multitude of biological processes, they exhibit only poor DNA replication. If these chromosomes are “reprogrammed” in mitotic extract first, they will undergo very robust DNA replication (46). This reprogramming, known as “replicon resetting”, is topoisomerase II-dependent and accompanies a visible shortening of chromosomal loops (46). It has been suggested that to make way for DNA replication, topoisomerase II must remove chromosomal roadblocks that have appeared in the differentiated cells (46,47). Considering that the DNA from these cells is not yet replicated, it seems unlikely that catenation of linked daughter chromosomes are what causes the roadblock. Similarly, because the block is not removed by the endogenous topoisomerase I, the barrier probably is not caused by an altered DNA supercoiling state, such as an area of localized overwound DNA. We propose that instead of catenanes, knots prevent the facile return of differentiated nuclei to the cell cycle. Although elucidated in metazoans, this phenomenon of a topological resetting of chromosomes to allow complete and efficient DNA replication appears widespread. Yeast cells lacking topoisomerase II protein cannot resolve DNA entanglements and undergo pervasive DNA damage and chromosome missegregation during cytokinesis (48). The expression of a catalytically inactive topoisomerase II, in which the active site tyrosine is replaced with phenylalanine, in this null topoisomerase II background, cannot prevent DNA entanglements from arising. However, the mutant topoisomerase II prevents DNA damage and missegregation by restoring a cell cycle arrest. The resulting DNA entanglements prevent the completion of DNA replication and block entry into mitosis (48). Although the formation of precatenanes during DNA synthesis could be the impediment, these are likely to form behind replication forks. As for the experiments discussed above, one alternative explanation of this finding is that overexpression of catalytically inactive topoisomerase II renders yeast cells unable to unknot DNA, and it is the knots that block progressing DNA replication forks. In support of this alternative interpretation of the data, the electrophoretic migration of reporter plasmids (48) is what would be expected for knotted plasmids.
The consequences of DNA knotting can reach beyond DNA replication. Proper chromatin assembly profoundly influences genetic activity. Using the Xenopus oocyte extract system, researchers found that knotted DNA is an unsuitable substrate for chromatin assembly (49). In a purified transcription system (50) and likely in E. coli (45), RNA polymerases have difficulty transcribing a knotted DNA molecule. DNA knotting might also destabilize the eukaryotic genome. Finally, knotted polymers have a reduced tensile strength compared to unknotted counterparts, and knotted DNA could be more likely to break (45,51).
The brief review above indicates that DNA supercoiling, catenating and knotting, which are the natural topological consequences of storing, replicating, recombining, transcribing and likely also repairing the DNA double helix, can have negative effects on cells. This fact illustrates the cellular need for topoisomerases, particularly for type-2 topoisomerases. How the type-2 topoisomerases carry out their essential roles has been studied extensively since their discovery (19,20). Much about these remarkable enzymes, however, remains a mystery.
MODELS FOR HOW TYPE-2 TOPOISOMERASES UNTANGLE DNA
Whereas the topological state of DNA is a global property, the data discussed above demonstrate that DNA topology affects processes that are occurring on the local level such as replication, transcription and recombination. How does the global topology affect local interactions and, conversely, how do locally acting type-2 topoisomerases guard against detrimental global entanglements of a DNA that is orders of magnitude larger than the enzyme? Type-2 topoisomerases, including prokaryotic DNA gyrase, which introduces negative supercoils into DNA, bind two DNAs (52–56). Experiments with Drosophila topoisomerase II revealed that the enzyme cleaves DNA only after both DNA helices are bound (55). Based upon these findings, a statistical, teleological argument can be made: type-2 topoisomerases discern DNA topology by recognizing helix–helix juxtapositions that exist more frequently in their substrates, overwound (positively supercoiled) and underwound (negatively supercoiled) DNA, as well as in catenanes and knots, than in their products, relaxed and untangled DNA (52,57). Can such a statistical argument explain type-2 topoisomerase function? Not entirely.
Rybenkov et al. found that type-2 topoisomerases, but not type-1 topoisomerases, added to a DNA mixture at equilibrium that consists of a very small fraction of knotted and linked DNAs will change the equilibrium to a state with even less entangled DNA (58). Assuming that the helix–helix juxtapositions were all identical in the equilibrium DNA mix (including relaxed DNA plasmid and sticky-ended linear DNA, which can, with a certain frequency, anneal to form circles and rarely also forms knots when the annealing entraps an entanglement or catenanes when it entraps a DNA plasmid) in the experiment, then it might be expected that the type-2 enzymes should not have altered the equilibrium. Indeed, if DNA juxtapositions are treated as “phantom chains” in which one chain can freely pass through itself or other chains, an idealized equilibrium distribution of knots and catenanes results (59). Because the results of Rybenkov et al. showed that type-2 topoisomerases do not turn DNA into phantom chains, then the straightforward statistical helix–helix juxtaposition model cannot explain the results. Therefore, the researchers suggested that type-2 topoisomerases actively slide along the DNA to trap the catenane or knot nodes and reduce the effective size of the DNA (58). This model, however, was abandoned by the authors for a lack of experimental evidence and in favor of the “active bending model” discussed below. In addition, recent experimental evidence argue against a model that invokes type-2 topoisomerase sliding, as roadblocks had no affect on the ability of a type-2 enzyme to shrink the topoisomer distribution (60). Another three DNA strand model was proposed by Trigueros et al. (61). Their model envisioned a bound, stationary (not sliding) topoisomerase interacting with three DNA segments. The model was proposed to explain the experimental observations that type-2 topoisomerases narrow the distribution of DNA supercoiled topoisomers in an asymmetric way. How this model can address the reduction in knots and catenanes to below equilibrium levels has not been addressed and is hard to envision.
A kinetic proofreading model (62,63) was put forth, in which two sequential type-2 topoisomerase–DNA collisions occurring within a short time interval were required to bring about a segment passage (64). This intriguing model provided rationalization for some of the unknotting and unlinking data, but the model predicted a constant supercoil suppression factor of 2.0 (65), which is inconsistent with experiments showing supercoil suppression factors ranging from approximately 1.4 to 1.8 for several type-2 topoisomerases from different organisms (58,60).
As mentioned above, an active bending model has been put forth that type-2 topoisomerases can actively untangle DNA by bending the DNA gate segment, which, would increase the probability of capturing a second ‘transfer’ segment in knotted or linked molecules relative to the probability in unknotted or unlinked DNA. DNA strand passage is allowed to proceed only along the direction of entry into the hairpin and not in the reverse direction (66,67). In support of this active bending model, Monte Carlo simulations showed that untangling could be achieved if a hairpin is introduced as a preformed conformational kink in the computation (66,67). How to translate the pre-existing kink used in the simulations to a kink actively introduced by the topoisomerases acting on DNA is not obvious. In addition, the experimental evidence for active bending is complicated by the assays used. Interpreting the results from experiments that showed that the addition of type-2 topoisomerase increases ligase-mediated cyclization of short linear DNA is problematic because of the potential that type-2 topoisomerases interact with DNA ends (61,67). Transmission electron microscopy (TEM), which was used to assess bending by type-2 topoisomerases (67), involves flattening and drying a three-dimensional object in buffer into two dimensions and is also difficult to interpret. Therefore, the active bending model remains to be clarified.
Although each of the above models can explain certain aspects of type-2 topoisomerase actions, none of them can account for all of the existing experimental data. For example, how would any of them predict simultaneous supercoiling-independent unknotting and supercoiling-dependent decatenating of plasmids of a few thousand base pairs (15,25,68,69)? Also, all of these models are fairly ‘protein-centric’ in that they seem to ascribe the ability to assess DNA topology solely to the type-2 topoisomerase, which behaves like Maxwell's smart demon (70), cruising the system to gather information, count DNA binding events, and exert its influence onto a passive DNA.
An implicit assumption in the interpretation of the data from Rybenkov et al. was that all of the different helix–helix juxtapositions in the experiment were alike; that there was no difference between the helix–helix juxtaposition formed by two circles near each other to be linked by type-2 topoisomerase and the helix–helix juxtaposition in a knot or a catenane to be unlinked. Is this a valid assumption? Is there nothing at the local level implicit in the DNA–DNA juxtapositions that a type-2 topoisomerase might use to discriminate its substrate from its product? Buck and Zechiedrich stipulated that the local geometric properties of two DNA helices juxtaposing are not the same when the DNA is supercoiled, linked or knotted, compared to when it is linear, relaxed and unknotted (71). Thus, in the experiments of Rybenkov et al., the catenane resulting from the sticky ends of the DNA annealing to encircle another relaxed DNA circle contains a DNA–DNA juxtaposition that is fundamentally distinct from the juxtaposition of two circles that are not linked. Likewise, the juxtaposition at a knot node is distinct from a juxtaposition formed when two segments of one DNA collide. The local geometry of DNA juxtapositions provides significant information about global topology (71) and this may instruct type-2 topoisomerases where to act. Specifically, DNA segments that are linked tend to curve toward each other when they juxtapose, creating hooked juxtapositions whereas segments that are not linked tend to curve away from each other, creating free juxtapositions. Another important distinction between juxtapositions in tangled or untangled DNA is how long they may persist. These observations suggested a simple model for type-2 topoisomerase action; the enzymes will act on hooked, but not free juxtapositions (71, Figure 2).
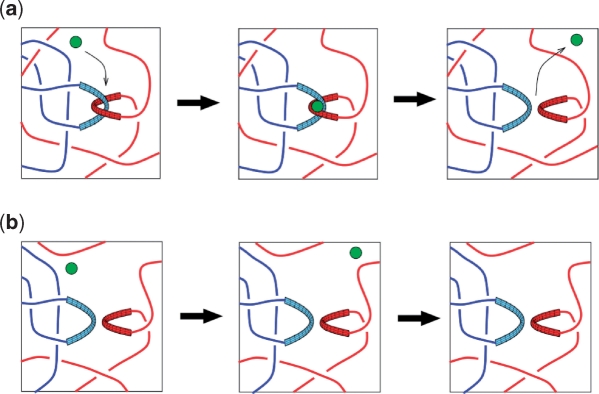
Figure 2.
The hooked juxtaposition hypothesis. The hypothesis put forth by Buck and Zechiedrich stipulates that type-2 topoisomerases unknot and decatenate by selective segment passages at hooked juxtapositions but not at free juxtapositions. Schematized here, as an example, is the hypothesized decatenating mechanism by type-2 topoisomerases of two daughter chromosomes, one red and one blue. Shown is the perspective of the small topoisomerase and the global linkage is hard to ascertain globally. Type-2 topoisomerase (schematically represented by a green circle) catalyzes segment passage specifically at a hooked juxtaposition (top row), which is more likely to occur when the two chromosomes are linked globally. The type-2 enzyme will not act at a free juxtaposition (bottom row), which is more likely to occur when two chromosomes are not linked globally (71). Although depicted here for decatenation, this model is the same for knot-generated juxtapositions as well.
The recent determination of the crystal structure of the DNA binding and cleavage core of yeast topoisomerase II in a complex with a putative gate DNA segment (72), where the DNA segment was strongly bent to 150° lends support to the two models that invoke DNA bending (67,71). Use of the crystal structure to distinguish models, however, must be done with caution because the DNA used to achieve the crystal had unique sequence and structural properties (including two nicks). It is not known whether the bend would exist in DNA without these specific properties or whether the enzyme can actively bend DNA. Whereas type-2 topoisomerases exhibit strong DNA binding and cleavage site preferences (73,74), in general the enzyme must act relatively indiscriminately.
RELATIONSHIP BETWEEN GLOBAL TOPOLOGY AND LOCAL INTERACTIONS
At the same time that experiments are devised to understand type-2 topoisomerase action, it seems reasonable to also consider the question from another perspective. What local parameters of DNA–DNA juxtapositions reflect whether or not DNA is linked, and whether strand passage at certain juxtapositions is more or less likely to link or unlink DNA? The premise of this juxtaposition-centric approach is that distinct conformations of tangled and untangled DNA molecules tend to cause, in a statistical manner, the sites of DNA juxtaposition to adopt different geometries.
Starting with a specific DNA–DNA juxtaposition, exact enumeration or Monte Carlo sampling determines what new conformations would result from segment passage (Figure 3). This method is computationally more efficient than other sampling methods because it ensures that all enumerated or sampled conformations are consistent with the presence of a specific segment juxtaposition. The juxtaposition-centric approach was motivated by a similar constrained lattice conformational enumeration method that also bore on a relationship between local and global properties (75). That earlier study led to the discovery that local helical and sheet-like motifs similar to protein secondary structures tend to be enhanced by the overall conformational compactness of a polymer (76), a trend that was also observed subsequently in tube theory (77).
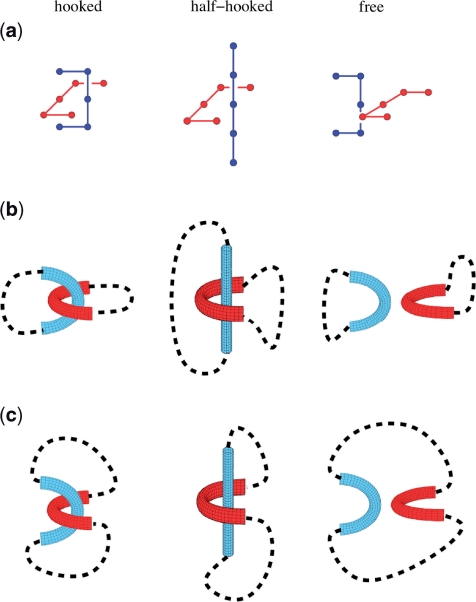
Figure 3.
The juxtaposition-centric computational approach. (a) The hooked, half-hooked and free juxtapositions in the simple cubic lattice (Z3) model. The schematics in (b and c) illustrate how conformational enumeration and sampling are conducted in the juxtaposition-centric approach. The geometry of a preformed juxtaposition (tube-like drawings) remains unchanged during a simulation, while the conformational possibilities of the rest of the chain(s) (dashed curves) are either enumerated exhaustively for short chains or sampled statistically using Monte Carlo techniques for longer chains. The connectivity of the dashed curves to the preformed juxtapositions in (b) are for the studies looking at two separate chains, which consider the decatenating potentials, whereas those in (c) are for one-chain studies for the corresponding unknotting potentials. In addition to the three juxtapositions shown here, the juxtaposition-centric approach has been applied to several thousand lattice juxtapositions (84,85).
Lattice models, with a long productive history in polymer physics (78,79), knot theory (38,80,81) and protein folding (82,83), have been used to study the global topological information contained in segment juxtapositions. More realistic model chains configured in the continuum can also be used in the juxtaposition-centric approach. In addition to the hooked and free juxtapositions (71,84,85), the half-hooked juxtaposition (85) is included in Figure 3a, because of its relevance to the active bending model (67). Two-chain configurations (Figure 3b) and one-chain conformations (Figure 3c) constructed from preformed juxtapositions can be used to study decatenating and unknotting, respectively, by type-2 topoisomerase-like segment passages.
Once these conformations are established, the topological consequences of type-2 topoisomerase-like segment passages at the hooked, half-hooked and free juxtapositions can be investigated (Figure 4a–c). Based on population distributions, a master equation formulation (67,85) was used to determine the steady-state populations of various topological states. These calculations yielded a link (catenane) reduction factor RL (Figure 4d) and a knot reduction factor RK (Figure 4e). Essentially, RL and RK of a juxtaposition type are, respectively, the catenane and knot population at topological equilibrium divided by the corresponding steady-state population resulting from selective segment passages at the given juxtaposition. A high value for these reduction factors means that the steady-state fraction of catenanes or knots is small relative to that at topological equilibrium, and therefore selective segment passage at the given juxtaposition is effective in decatenating or unknotting. On the other hand, a low value (<1) means that selective segment passage at the given juxtaposition tends to tangle rather than untangle. The chain length dependence of link and knot reduction factors for the three juxtapositions is shown in Figure 4d and 4e. Consistent with the idea that global topology is manifested locally, both RL and RK exhibit dramatic dependence on juxtaposition geometry.
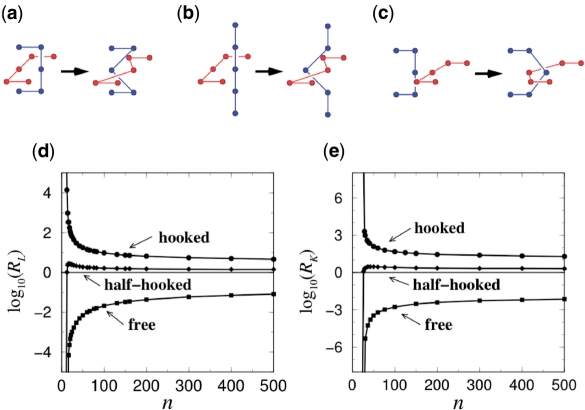
Figure 4.
Effects on catenane and knot populations of segment passage at (a) hooked, (b) half-hooked and (c) free juxtapositions in the simple cubic lattice model. Chain length dependences of (d) link (catenane) reduction factor RL and (e) knot reduction factor RK were computed by determining the link and knot probabilities before and after topoisomerase-like segment passage at the given juxtaposition in configurations of two chains of equal lengths (d) and conformations of a single chain (e). Chain length n is the number of edges in the lattice polygon used to model a ring polymer. Data presented in this figure are identical to that in (84,85; see these references for further computational details).
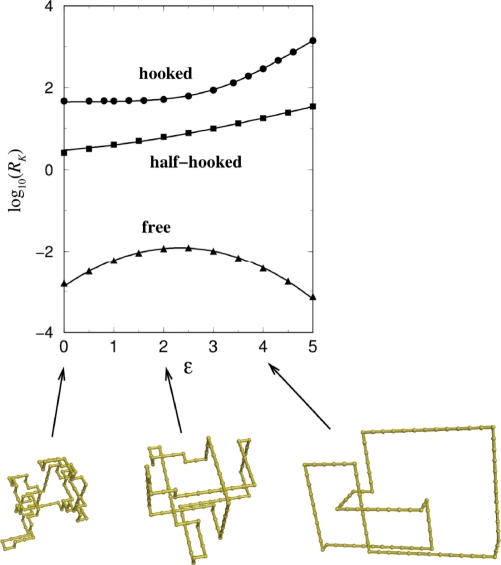
Several features in these plots are noteworthy: (i) segment passage with only the local information of the hooked juxtaposition is effective in decatenating and unknotting and is comparable to the experimental measurements (58). A survey of hundreds of different lattice juxtapositions with a crossing found that the untangling potential correlates with the “hookedness” of the juxtaposition (85). (ii) With RK values of at most ∼3, strand passage at the half-hooked is much less effective than at the hooked juxtaposition. This finding may explain partly why the active bending model (67) predicts much smaller effects than are observed experimentally (58). (iii) Disentangling by selective passage at hooked juxtapositions is even more highly effective for ring polymers of shorter chain length. This trend is consistent with experimental RK values of 90 and 50, respectively, for a 7-kb plasmid, pAB4, and 10-kb bacteriophage, P4 DNA (58). (iv) The juxtaposition-centric computation demonstrates that at any chain length the knot reduction factor (Figure 4e) is higher than the link reduction factor (Figure 4d); but the chain length dependences of the link and knot reduction factors took very similar shapes. This pattern reflects an approximate power-law relationship in the model, RK ≈ (RL)2.0 (85), which mirrors a similar experimental RK ≈ (RL)1.6 scaling for a set of type-2 topoisomerases from different organisms studied by Rybenkov et al. (58). (v) These results with the lattice model are likely general and applicable to DNA juxtapositions. Indeed, a recent study shows that results are the same using the freely jointed chain model (86). The lattice model utilizes a flexible chain to model DNA. However a new calculation made here (Figure 5) indicates that increasing chain stiffness enhances the unknotting potential of the hooked and half-hooked juxtapositions [RK for n = 100 rings increase from ≈50 and ≈2.5 in the original ε = 0 (flexible) model to ≈300 and ≈18 at ε = 4 (stiff), respectively]. (vi) The juxtaposition-centric approach can be extended to treat supercoiling (87) by incorporating a global torsional energy as commonly used in continuum wormlike DNA chain models (88) and considering the writhe of the lattice chains (89) to explain a tightening of steady-state distribution of linking number, i.e. a reduction in <ΔLk2> (Z.L., L.Z. and H.S.C., manuscript in preparation), similar to that observed experimentally for the effect of topoisomerase IV on plasmid DNA (58,60).
Figure 5.
Unknotting effects of strand passage at specific juxtapositions depend on chain stiffness. The plot (top) shows knot reduction factors (RK, in logarithmic scale) resulting from segment passage at the hooked, half-hooked and free juxtapositions (operations are as in Figure 4a–c) as a function of the chain stiffness parameter ε for circular chains with length n = 100. In these model chains, a 180° bond angle is favored by a factor exp(ε) over a 90° bond angle. Thus, chain stiffness increases with ε, with ε = 0 corresponding to the original model from which the results in Figure 4 were obtained. The bottom drawings are representative knotted conformations at ε-values indicated by the arrows.
Considering all of the results obtained from lattice modeling thus far, the hooked juxtaposition hypothesis (Figure 2) and the general juxtaposition-centric approach it inspired afford a coherent rationalization for type-2 topoisomerase action not only in decatenating and unknotting (Figure 4), but in suppressing the equilibrium distribution of supercoils as well. Recently, advances have also been made in extending juxtaposition-centric conformational sampling from lattice to continuum (off-lattice) models. Preliminary studies using the newly developed Monte Carlo sampling techniques (87) showed that selective segment passages at hooked juxtapositions in a more realistic wormlike DNA chain model—like the corresponding operations in lattice models—could indeed achieve significant reduction in knot population comparable to that observed in experiments (Z.L., L.Z. and H.S.C., manuscript in preparation).
PROSPECTS FOR UNDERSTANDING THE IMPACT OF DNA TOPOLOGY ON BIOLOGICAL PROCESSES
As the fundamental principles of local conformational effects of the global topological state are being elucidated experimentally and theoretically, it will be important to clarify how nucleic acid metabolism will alter these properties. Biological processes provide a link between global topology and local DNA structure. The case of DNA knotting is a good example. The shape of a knotted molecule is quite different whether it is loose or tight, and it is likely that the biological consequences of a knot would be influenced by its tightness. Indeed, atomic force microscopy images of knotted plasmids indicate that the knot is normally localized to a small area of the total molecule; thus knots can be tight (90). Theoretical results reveal that knots in DNA tend to localize and remain in that conformation for entropic reasons (91,92). Although knots tighten on their own, there could be an even greater tendency for this to occur in vivo as the DNA is subjected to forces applied by enzymes involved in transcription and replication.
In the cell, a loose knot (Figure 6a) might not be substantially different from an unknotted molecule, whereas a tight knot (Figure 6b) might form a much more formidable impasse to DNA tracking enzymes like RNA and DNA polymerases. This consideration also suggests that even if the molecule is globally knotted, knots can have very transient and localized effects based upon where the knot tightens (Figure 6b). On a chromosome, barriers exist that prevent the diffusion of superhelical tension beyond a ∼50–100 kb loop of DNA (93). It will be interesting to determine whether these blocks to the diffusion of changes in torsional tension are general topological barriers that can prevent the spread of a knot in the chromosome as well. These genomic barriers have been demonstrated to impede large chromosomal movements, as measured by the synapsis of distant chromosomal sites in a site-specific recombination reaction (γδ resolvase) (94), and possibly could also inhibit the movements of DNA that would occur during knot translocation. Although these considerations suggest that knot tightening is detrimental, it is possible that it could be beneficial to the cell. For example, a knot might become localized to an intergenic region of the genome and, thus, not disrupt transcription before a topoisomerase unties it. The processes that affect knot localization should be those that exert force on the DNA, including transcription, replication and segregation. Indeed, both theoretical and experimental evaluations have indicated that sufficient forces exist in vivo to affect the tightness of a knot (95,96).
Figure 6.
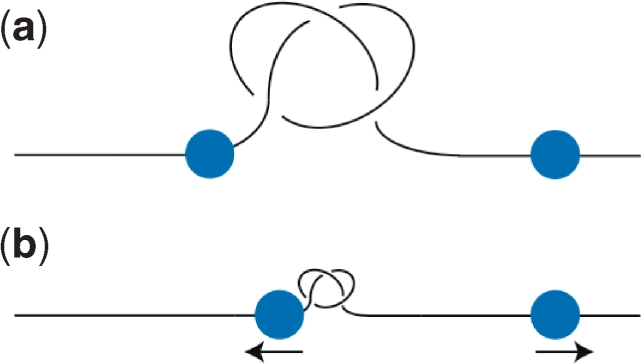
Biological consequences of DNA knotting. Shown is knotted DNA (for simplicity, each line represents a double-stranded DNA helix) and tracking polymerases (ball) with the arrows indicating the direction of force on the DNA. (a) A knot in a loose conformation. (b) A knot pulled in a tighter conformation by polymerases (blue) tracking along the DNA.
Unlike with DNA knots, which have a relatively stable structure and change more in size and location rather than shape, the conformation of DNA supercoils will greatly change as a consequence of nearby biological activities. The folding of chromosomes in cells involves the wrapping of supercoiled DNA around proteins—nucleoid-associated proteins in prokaryotes and the nucleosomal histones in eukaryotes. In this protein-bound state, the supercoiled DNA is constrained and unable to promote DNA activity. In E. coli, ∼40%, and in humans, nearly 100% of bulk chromosomal DNA is constrained in this way (97,98). The transient release of this supercoiling allows high levels of DNA activity, particularly in eukaryotes. Yet, once these unconstrained supercoils are released what form will they take? The highly writhed conformation of plectonemic (interwound) supercoils would promote DNA site juxtaposition, but it is also possible that the DNA inside the cell could be held in an untwisted conformation instead, which would have different physiological properties. What are the effects of negative supercoiling when unwinding of the duplex is maintained, but the juxtaposition of DNA sites is no longer facilitated? How can type-2 topoisomerases regulate topology when the crossover is eliminated? Do they need to? Can the type-1 topoisomerases regulate topology in this case?
Genetic regulation has primarily focused on the role of nucleotide sequence in this process. However, the evolution of DNA as the genetic medium with its inherent set of conformational features has resulted in a feedback of the properties on the regulation of basic chromosomal functions. Furthering our understanding of this feedback will be crucial in completing our understanding of basic genetic mechanisms.
FUNDING
Ministry of Science and Technology of China (2009CB918504 to Z.L.); the Jane Coffin Childs Memorial Fellowship Fund (to R.W.D.); the Natural Sciences and Engineering Research Council of Canada (Discovery Grant 216901 to H.S.C.); H.S.C. also holds a Canada Research Chair in Proteomics, Bioinformatics and Functional Genomics. National Institutes of Health (RO1 AI054830 to L.Z.). Funding for open access charge: Natural Sciences and Engineering Research Council of Canada Discovery Grant 216901.
Conflict of interest statement. None declared.
Acknowledgements
We thank Dr Daniel J. Catanese Jr and Dr Jonathan M. Fogg for providing Figure 1 and Dr Jonathan M. Fogg for critically reading the article.
References
- 1.Watson JD, Crick F.HC. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 3.Fogg JM, Catanese DJ, Randall GL, Swick MC, Zechiedrich L. Differences between positively and negatively supercoiled DNA that topoisomerases may distinguish. Proc. Institute Math. Appl. 2008 (in press) [Google Scholar]
- 4.Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley D.MJ, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 5.Travers A, Muskhelishvili G. DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 6.Travers A, Muskhelishvili G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007;8:147–151. doi: 10.1038/sj.embor.7400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boles TC, White JH, Cozzarelli NR. Structure of plectonemically supercoiled DNA. J. Mol. Biol. 1990;213:931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- 8.Vologodskii A, Cozzarelli NR. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys. J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polikanov YS, Bondarenko VA, Tchernaenko V, Jiang YI, Lutter LC, Vologodskii A, Studitsky VM. Probability of the site juxtaposition determines the rate of protein-mediated DNA looping. Biophys. J. 2007;93:2726–2731. doi: 10.1529/biophysj.107.111245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Embleton ML, Vologodskii AV, Halford SE. Dynamics of DNA loop capture by the Sfil restriction endonuclease on supercoiled and relaxed DNA. J. Mol. Biol. 2004;339:53–66. doi: 10.1016/j.jmb.2004.03.046. [DOI] [PubMed] [Google Scholar]
- 11.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamin KR, Abola AP, Kanaar R, Cozzarelli NR. Contributions of supercoiling to Tn3 resolvase and phage Mu Gin site-specific recombination. J. Mol. Biol. 1996;256:50–65. doi: 10.1006/jmbi.1996.0067. [DOI] [PubMed] [Google Scholar]
- 13.Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc. Natl. Acad. Sci. USA. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crozat E, Philippe N, Lenski RE, Geiselmann J, Schneider D. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics. 2005;169:523–532. doi: 10.1534/genetics.104.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baaklini I, Usongo V, Nolent F, Sanscartier P, Hraiky C, Drlica K, Drolet M. Hypernegative supercoiling inhibits growth by causing RNA degradation. J. Bacteriol. 2008;190:7346–7356. doi: 10.1128/JB.00680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delbrück M. On the replication of deoxyribonucleic acid (DNA) Proc. Natl. Acad. Sci. USA. 1954;40:783–788. doi: 10.1073/pnas.40.9.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase – enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR. Mechanism of action of nalidixic-acid – purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundin O, Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980;21:103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- 22.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers – dissection of the final stages of SV40 DNA-replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 23.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams DE, Shekhtman EM, Zechiedrich EL, Schmid MB, Cozzarelli NR. The role of topoisomerase-IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA-replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 25.Zechiedrich EL, Cozzarelli NR. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;15:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 26.Akimitsu N, Adachi N, Hirai H, Hossain MS, Hamamoto H, Kobayashi M, Aratani Y, Koyama H, Sekimizu K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase II alpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerases II beta and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 28.Wang XD, Reyes-Lamothe R, Sherratt DJ. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 2008;22:2462–2433. doi: 10.1101/gad.487508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bates D. The bacterial replisome: back on track? Mol. Microbiol. 2008;69:1341–1348. doi: 10.1111/j.1365-2958.2008.06378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 31.Andrews CA, Vas AC, Meier B, Gimenez-Abian JF, Diaz-Martinez LA, Green J, Erickson SL, Vanderwaal KE, Hsu WS, Clarke DJ. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes Dev. 2006;20:1162–1174. doi: 10.1101/gad.1367206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Downes CS, Clarke DJ, Mullinger AM, Gimenez-Abian JF, Creighton AM, Johnson RT. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 33.Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol. Cell. 2004;15:977–990. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa T, Hayashita Y, Maeno K, Masuda A, Sugito N, Osada H, Yanagisawa K, Ebi H, Shimokata K, Takahashi T. Identification of decatenation G2 checkpoint impairment independently of DNA damage G2 checkpoint in human lung cancer cell lines. Cancer Res. 2004;64:4826–4832. doi: 10.1158/0008-5472.CAN-04-0871. [DOI] [PubMed] [Google Scholar]
- 35.Germe T, Hyrien O. Topoisomerase II-DNA complexes trapped by ICRF-193 perturb chromatin structure. EMBO Rep. 2005;6:729–735. doi: 10.1038/sj.embor.7400465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doherty SC, McKeown SR, McKelvey-Martin V, Downes CS, Atala A, Yoo JJ, Simpson DA, Kaufmann WK. Cell cycle checkpoint function in bladder cancer. J. Natl.. Cancer Inst. 2003;95:1859–1868. doi: 10.1093/jnci/djg120. [DOI] [PubMed] [Google Scholar]
- 37.Damelin M, Sun YE, Sodja VB, Bestor TH. Decatenation checkpoint deficiency in stem and progenitor cells. Cancer Cell. 2005;8:479–484. doi: 10.1016/j.ccr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Sumners DW, Whittington SG. Knot in self-avoiding walks. J. Phys. A: Math. Gen. 1988;21:1689–1694. [Google Scholar]
- 39.Rybenkov VV, Cozzarelli NR, Vologodskii AV. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl. Acad. Sci. USA. 1993;90:5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arsuaga J, Vazquez M, Trigueros S, Sumners DW, Roca J. Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proc. Natl. Acad. Sci. USA. 2002;99:5373–5377. doi: 10.1073/pnas.032095099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raymer DM, Smith DE. Spontaneous knotting of an agitated string. Proc. Natl. Acad. Sci. USA. 2006;104:16432–16437. doi: 10.1073/pnas.0611320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hickford J, Jones R, du Pont SC, Eggers J. Knotting probability of a shaken ball-chain. Phys. Rev. E. 2006;74:062101. doi: 10.1103/PhysRevE.74.052101. [DOI] [PubMed] [Google Scholar]
- 43.Burnier Y, Dorier J, Stasiak A. DNA supercoiling inhibits DNA knotting. Nucleic Acids Res. 2008;36:4956–4963. doi: 10.1093/nar/gkn467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podtelezhnikov AA, Cozzarelli NR, Vologodskii AV. Equilibrium distributions of topological states in circular DNA: interplay of supercoiling and knotting. Proc. Natl. Acad. Sci. USA. 1999;96:12974–12979. doi: 10.1073/pnas.96.23.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deibler RW, Mann JK, Sumners D.WL, Zechiedrich L. Hin-mediated DNA knotting and recombining promote replicon dysfunction and mutation. BMC Mol. Biol. 2007;8:44. doi: 10.1186/1471-2199-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M. Mitotic remodeling of the replicon and chromosome structure. Cell. 2005;123:787–801. doi: 10.1016/j.cell.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 47.Cuvier O, Stanojcic S, Lemaitre JM, Mechali M. A topoisomerase II-dependent mechanism for resetting replicons at the S-M-phase transition. Genes Dev. 2008;22:860–865. doi: 10.1101/gad.445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Campos A. DNA knotting abolishes in vitro chromatin assembly. J. Biol. Chem. 1996;271:14150–14155. doi: 10.1074/jbc.271.24.14150. [DOI] [PubMed] [Google Scholar]
- 50.Portugal J, Rodriguez-Campos A. T7 RNA polymerase cannot transcribe through a highly knotted DNA template. Nucleic Acids Res. 1996;24:4890–4894. doi: 10.1093/nar/24.24.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arai Y, Yasuda R, Akashi K, Harada Y, Miyata H, Kinosita K, Itoh H. Tying a molecular knot with optical tweezers. Nature. 1999;399:446–448. doi: 10.1038/20894. [DOI] [PubMed] [Google Scholar]
- 52.Zechiedrich EL, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howard MT, Lee MP, Hsieh T.-S, Griffith JD. Drosophila topoisomerase II-DNA interactions are affected by DNA structure. J. Mol. Biol. 1991;217:53–62. doi: 10.1016/0022-2836(91)90610-i. [DOI] [PubMed] [Google Scholar]
- 54.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by topo II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 55.Corbett AH, Zechiedrich EL, Osheroff N. A role for the passage helix in the DNA cleavage reaction of eukaryotic topoisomerase II. A two-site model for enzyme-mediated DNA cleavage. J. Biol. Chem. 1992;267:683–686. [PubMed] [Google Scholar]
- 56.Moore CL, Klevan L, Wang JC, Griffith JD. Gyrase-DNA complexes visualized as looped structures by electron microscopy. J. Biol. Chem. 1983;258:4612–4617. [PubMed] [Google Scholar]
- 57.Osheroff N, Zechiedrich EL, Gale KC. Catalytic function of DNA topoisomerase-II. Bioessays. 1991;13:269–275. doi: 10.1002/bies.950130603. [DOI] [PubMed] [Google Scholar]
- 58.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 59.Sikorav JL, Jannink G. Kinetics of chromosome condensation in the presence of topoisomerases – a phantom chain model. Biophys. J. 1994;66:824–837. doi: 10.1016/s0006-3495(94)80859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stuchinskaya T, Mitchenall LA, Schoeffler AJ, Corbett KD, Berger JM, Bates AD, Maxwell A. J. Mol. Biol. 385:1397–1408. doi: 10.1016/j.jmb.2008.11.056. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trigueros S, Salceda J, Bermudez I, Fernández X, Roca J. Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J. Mol. Biol. 2004;335:723–731. doi: 10.1016/j.jmb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 64.Yan J, Magnasco MO, Marko JF. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- 65.Yan J, Magnasco MO, Marko JF. Kinetic proofreading can explain the suppression of supercoiling of circular DNA molecules by type-II topoisomerases. Phys. Rev. E. 2001;63:031909. doi: 10.1103/PhysRevE.63.031909. [DOI] [PubMed] [Google Scholar]
- 66.Vologodskii AV. RECOMB 98: Proceedings of the Second Annual International Conference on Computational Molecular Biology. New York, USA: Association for Computing Machinery; 1998. Maxwell demon and topology simplification by type II topoisomerases. In; pp. 266–269. [Google Scholar]
- 67.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl. Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 69.Deibler RW, Rahmati S, Zechiedrich EL. Topoisomerase IV alone, unknots DNA in E. coli. Genes Dev. 2001;15:748–761. doi: 10.1101/gad.872301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pulleyblank DE. Of topo and Maxwell's dream. Science. 1997;277:648–649. doi: 10.1126/science.277.5326.648. [DOI] [PubMed] [Google Scholar]
- 71.Buck GR, Zechiedrich EL. DNA disentangling by type-2 topoisomerases. J. Mol. Biol. 2004;340:933–939. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 72.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 73.Masliah G, René B, Zargarian L, Fermandjian S, Mauffret O. Identification of intrinsic dynamics in a DNA sequence preferentially cleaved by topoisomerase II enzyme. J. Mol. Biol. 2008;381:692–706. doi: 10.1016/j.jmb.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 74.Mueller-Planitz F, Herschlag D. DNA topoisomerase II selects DNA cleavage sites based on reactivity rather than binding affinity. Nucleic Acids Res. 2007;35:3764–3773. doi: 10.1093/nar/gkm335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan HS, Dill KA. The effects of internal constraints on the configurations of chain molecules. J. Chem. Phys. 1990;92:3118–3135. [Google Scholar]
- 76.Chan HS, Dill KA. Origins of structure in globular proteins. Proc. Natl. Acad. Sci. USA. 1990;87:6388–6392. doi: 10.1073/pnas.87.16.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maritan A, Micheletti C, Trovato A, Banavar JR. Optimal shapes of compact strings. Nature. 2000;406:287–290. doi: 10.1038/35018538. [DOI] [PubMed] [Google Scholar]
- 78.Orr W.JC. Statistical treatment of polymer solutions at infinite dilution. Trans. Faraday Soc. 1947;43:12–27. [Google Scholar]
- 79.de Gennes P.-G. Scaling Concepts in Polymer Physics. Ithaca, New York: Cornell University Press; 1979. [Google Scholar]
- 80.Soteros CE, Sumners DW, Whittington SG. Entanglement complexity of graphs in Z3. Math. Proc. Cambridge Phil. Soc. 1992;111:75–91. [Google Scholar]
- 81.Matsuda D, Yao A, Tsukahara H, Deguchi T, Furuta K, Inami T. Average size of random polygons with fixed knot topology. Phys. Rev. E. 2003;68:011102. doi: 10.1103/PhysRevE.68.011102. [DOI] [PubMed] [Google Scholar]
- 82.Taketomi H, Ueda Y, Gō N. Studies on protein folding, unfolding and fluctuations by computer simulation. 1. The effect of specific amino acid sequence represented by specific inter-unit interactions. Int. J. Peptide Protein Res. 1975;7:445–459. [PubMed] [Google Scholar]
- 83.Chan HS, Shimizu S, Kaya H. Cooperativity principles in protein folding. Methods Enzymol. 2004;380:350–379. doi: 10.1016/S0076-6879(04)80016-8. [DOI] [PubMed] [Google Scholar]
- 84.Liu ZR, Zechiedrich EL, Chan HS. Inferring global topology from local juxtaposition geometry: interlinking polymer rings and ramifications for topoisomerase action. Biophys. J. 2006;90:2344–2355. doi: 10.1529/biophysj.105.076778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu ZR, Mann JK, Zechiedrich EL, Chan HS. Topological information embodied in local juxtaposition geometry provides a statistical mechanical basis for unknotting by type-2 DNA topoisomerases. J. Mol. Biol. 2006;361:268–285. doi: 10.1016/j.jmb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Burnier Y, Weber C, Flammini A, Stasiak A. Local selection rules that can determine specific pathways of DNA unknotting by type II DNA topoisomerases. Nucleic Acids Res. 2007;15:5223–5231. doi: 10.1093/nar/gkm532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu ZR, Chan HS. Efficient chain moves for Monte Carlo simulations of a wormlike DNA model: Excluded volume, supercoils, site juxtapositions, knots and comparisons with random-Flight and lattice models. J. Chem. Phys. 2008;128:145104. doi: 10.1063/1.2899022. [DOI] [PubMed] [Google Scholar]
- 88.Vologodskii AV, Levene SD, Klenin KV, Frank-Kamenetskii M, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. J. Mol. Biol. 1992;227:1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- 89.Lacher RC, Sumners DW. Data structures and algorithms for the computation of topological invariants of entanglements: link, twist and writhe. In. In: Roe RJ, editor. Computer Simulations of Polymers. New York: Prentice-Hall; 1991. pp. 365–373. [Google Scholar]
- 90.Ercolini E, Valle F, Adamcik J, Witz G, Metzler R, De Los Rios P, Roca J, Dietler G. Fractal dimension and localization of DNA knots. Phys. Rev. Lett. 2007;98:058102. doi: 10.1103/PhysRevLett.98.058102. [DOI] [PubMed] [Google Scholar]
- 91.Katritch V, Olson WK, Vologodskii A, Dubochet J, Stasiak A. Tightness of random knotting. Phys. Rev. E. 2000;61:5545–5549. doi: 10.1103/physreve.61.5545. [DOI] [PubMed] [Google Scholar]
- 92.Grosberg AY, Rabin Y. Metastable tight knots in a wormlike polymer. Phys. Rev. Lett. 2007;99:217801. doi: 10.1103/PhysRevLett.99.217801. [DOI] [PubMed] [Google Scholar]
- 93.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Staczek P, Higgins NP. Gyrase and topo IV modulate chromosome domain size in vivo. Mol. Microbiol. 1998;29:1435–1448. doi: 10.1046/j.1365-2958.1998.01025.x. [DOI] [PubMed] [Google Scholar]
- 95.Bao XR, Lee HJ, Quake SR. Behavior of complex knots in single DNA molecules. Phys. Rev. Lett. 2003;91:265506. doi: 10.1103/PhysRevLett.91.265506. [DOI] [PubMed] [Google Scholar]
- 96.Vologodskii A. Brownian dynamics simulation of knot diffusion along a stretched DNA molecule. Biophys. J. 2006;90:1594–1597. doi: 10.1529/biophysj.105.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J. Mol. Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- 98.Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]