Abstract
The mechanism by which type-2A topoisomerases transport one DNA duplex through a transient double-strand break produced in another exhibits fascinating traits. One of them is the fine coupling between inter-domainal movements and ATP usage; another is their preference to transport DNA in particular directions. These capabilities have been inferred from in vitro studies but we ignore their significance inside the cell, where DNA configurations markedly differ from those of DNA in free solution. The eukaryotic type-2A enzyme, topoisomerase II, is the second most abundant chromatin protein after histones and its biological roles include the decatenation of newly replicated DNA and the relaxation of polymerase-driven supercoils. Yet, topoisomerase II is also implicated in other cellular processes such as chromatin folding and gene expression, in which the topological transformations catalysed by the enzyme are uncertain. Here, some capabilities of topoisomerase II that might be relevant to infer the enzyme performance in the context of chromatin architecture are discussed. Some aspects addressed are the importance of the DNA rejoining step to ensure genome stability, the regulation of the enzyme activity and of its putative structural role, and the selectively of DNA transport in the chromatin milieu.
INTRODUCTION
Biochemical and structural studies conducted in the past two decades have established the general mechanism by which type-2A topoisomerases transport, in an ATP-dependent manner, one DNA duplex through a transient double-strand break produced in another (1–4). All the enzymes of this class (Bacterial DNA Gyrase, bacterial topoisomerase IV and eukaryotic topoisomerase II) are functional dimers of three structural domains: an ATP-ase or N-terminal domain, and the B′ and A′ domains that constitute the DNA cleavage-rejoining core (3). Before interacting with DNA, type-2A topoisomerases have the shape of an open clamp, in which the N-terminal domains are located at the tips and the A′ domains dimerize at the hinge (5,6). The DNA double helix to be gated (named G-segment) first enters the open clamp and binds to the cleavage-rejoining core (7). Binding of ATP causes the N-terminal domains to interact and the clamp closes. If this closure leads to the capture of a second DNA double helix inside the clamp, a cascade of conformational changes ensues (8). The captured DNA (named T-segment) is transported through a gate transiently opened in the bound G-segment. For this process, the G-segment is cleaved by two trans-estherification reactions that let the 5′ ends of the severed strands covalently linked with a pair of tyrosine residues, one in each half of the topoisomerase (9,10). After crossing the G-segment, the T-segment reaches the cavity delimited by the A′ domains and it is then expelled from the topoisomerase through the side opposite the one it entered, by the A′–A′ dimer interface at the hinge of the closed clamp (8,11). ATP hydrolysis begins during T-segment transport and release of the hydrolytic products allows the N-terminal domains to come apart leaving the enzyme ready for a new reaction cycle.
The DNA transport mechanism of type-2A topoisomerases displays some fascinating traits. One of them is the fine coupling between inter-domainal movements and ATP usage during DNA transport (1,4). The translocation of the T-segment through the complete dimer interface of type-2 topoisomerases demands energetically well-balanced and robust conformational states during the critical DNA cleavage, DNA gating and DNA rejoining steps. Backtracking of the T-segment would cause unnecessary breakages of the G-segment and no net DNA transport. Dissociation of the enzyme halves linked to cleaved DNA would cause chromosomal breaks and rearrangements. The other intriguing aspect of type-2A topoisomerases are the DNA transport preferences that distinguish some enzymes. On the one side, DNA gyrase is specialized to carry out intra-molecular transport. After binding the G-segment, the gyrase–DNA complex enforces the juxtaposition of a contiguous T-segment such that it configures a positive DNA crossing (12,13). As long as this configuration can be settled, DNA gyrase removes positive supercoils and introduce negative ones into circular DNA (14). On the other side, bacterial topoisomerase IV and eukaryotic topoisomerase II can carry out both intra- and inter-molecular DNA transport. Consequently, they can catenate–decatenate, knot–unknot and remove supercoils. These enzymes, however, cannot supercoil DNA alike gyrase. Instead, topoisomerase IV and topoisomerase II reduce the topological complexity of DNA (catenation, knotting and supercoiling) below the levels expected at thermal equilibrium (15). How these enzymes achieve DNA topology simplification is unclear. Several models have been proposed to explain how they assess the global topology of a DNA molecule by their local interactions with DNA segments (16,17).
In parallel to biochemical and structural studies on type-2A topoisomerases, the biological functions of this abundant and essential class of enzymes had also been the focus of attention for long time (18). In eubacteria, DNA gyrase and topoisomerase IV complete different tasks according to their distinct DNA transport preferences. In most eukaryotes, however, a single form of topoisomerase II appears to carry out diverse biological roles (18,19). The major known and essential function of the enzyme is the decatenation of replicated DNA duplexes after the S phase. When two replication forks converge, the last unreplicated segment becomes too short to be unlinked by topoisomerase I or II. The residual intertwines between the parental DNA are then converted to catenane links between the newly replicated daughter duplexes. Removal of such catenanes by topoisomerase II is essential for proper chromosome segregation (recent studies on this topic are refs. 20–22). Another major known cellular activity of topoisomerase II is the relaxation of positive supercoils generated ahead of the replication forks, as well as of oppositely supercoiled domains generated during DNA transcription (23,24). Although eukaryotic topoisomerase I can also fulfil this function, topoisomerase II is likely to be the main relaxase of DNA supercoils inside the cell (25).
In addition to decatenating replicated DNA and relaxing polymerase-driven supercoils, eukaryotic topoisomerase II has been implicated in many other chromosomal processes. A common trait in all these processes is that the topological transformation (if any) catalysed by the enzyme is uncertain. For instance, topoisomerase II is well known to be involved in chromosome condensation, but its precise role in the many steps for assembling morphologically distinct interphase chromosomes and highly condensed metaphase chromosomes remain unclear. At each of these steps, whether a continual DNA transport activity of topoisomerase II is required for chromatin condensation-decondensation or whether the enzyme mainly has a structural role are still controversial questions (26). Other examples of unknown topology conversion are the implications of topoisomerase II in the local regulation of chromatin architecture and in gene expression. Such biological roles became evident after discovering that mammals harbour two topoisomerase II isozymes, IIα and IIβ (27). Both isozymes carry out similar strand-passing activities in vitro (28), though IIα is found to relax positively supercoiled plasmids faster than negatively supercoiled ones (29). Both isozymes are differentially regulated during cell growth and can not compensate for each other in vivo (30–32). On one hand, IIα is only found in proliferating cells, with an expression peak at late S and G2/M phases of the cell cycle (33,34), and it is most likely involved in DNA replication, chromosome condensation–decondensation and sister chromatid segregation (35–37). These functions are quite analogous to those known for the single topoisomerase II form found in budding yeast, fission yeast and drosophila (38–41). On the other hand, IIβ is found in all mammalian cell types but with prominent expression in terminally differentiated cells (42–47). The biological function of topoisomerase IIβ is intriguing. Recent studies uncovered a role in activation–repression of developmentally regulated genes at late stages of differentiation. The enzyme binds to the 5′ region of a number of such IIβ-sensitive genes (48). Topoisomerase IIβ has been also found catalysing a site-specific transient double-stranded DNA break in some gene promoters, which lead to local changes of chromatin architecture and activation of transcription (49). These findings suggest that topoisomerase IIβ, and perhaps the single topoisomerase II form found in yeasts and drosophila, produce critical structural changes in localized chromatin regions to regulate genome transactions.
To find out the range of DNA manipulations that topoisomerase II might catalyse in vivo, it is essential to sum up the capabilities of the enzyme inferred from in vitro studies. Some topoisomerase II activities learned with DNA in free solution that might be relevant for the performance of the enzyme in the context of chromatin architecture and dynamics are surveyed below.
A double-lock rule to open DNA gates and, above all, to close them
Most studies on topoisomerase II mechanism had focussed on the DNA cleavage and gating steps, the DNA transport preferences, and the coupling of inter-domainal movements to ATP usage. However, a less-attended but critical aspect of the enzyme operation that needs to be absolutely effective is the rejoining of the gated DNA. Intracellular DNA is gated by topoisomerase II many million times per cell cycle. Consider, for example, yeast cells deficient in topoisomerase I, in which topoisomerase II alone relaxes the supercoils raised during DNA transcription and reduces to zero the linkage between complementary DNA strands during their replication. Any single DNA gating event not followed by the proper rejoining step will put at risk genome integrity and cell survival. It is reasonable to assume then that most mechanic traits of the enzyme are tuned to ensure rejoining rather than DNA cleavage and DNA transport. Not surprisingly, those compounds affecting the DNA-rejoining step readily produce cell death and have, therefore, prominent pharmacological interest.
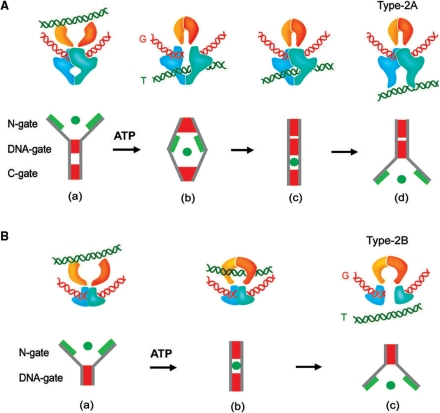
But apart from biochemical targeting, many physiological processes might also threaten the DNA rejoining step. For instance, in contrast to DNA in free solution, intracellular DNA is subjected to considerable twisting, bending and pulling forces generated by DNA tracking ensembles and the conformational dynamics of chromatin fibers. In this environment, holding both severed ends of the gated DNA is essential. Yet, the mechanism of the type-2 topoisomerases implies that none of its dimer interfaces is a permanent interaction. The path of the T-segment involves the sequential crossing of the entrance or N-gate, the DNA gate and the exit or C-gate. Coordination among these three gates is therefore critical to preclude the risk of the two halves of the enzyme coming apart while gating the DNA. In that regard, biochemical studies with yeast topoisomerase II indicate that gate opening and closure follow a double-lock rule: a given gate can open only if the other two are closed (50). Although the N-gate domain is not required for G-segment cleavage, the DNA gate per se is not able to widen unless ATP binds and closes the N-gate (50). Since a captured T-segment cannot be held in the inter-domainal region between the N-gate and the DNA gate, closure of the N-gate and consequent capture of a T-segment is likely to enforce the opening of the DNA gate (50). Once a T-segment has reached the central cavity of the enzyme and before the C-gate opens, the gated G-segment DNA can be rejoined (11,50). Hence, opening of the C-gate is likely to be triggered by the entrapment of the T-segment after being squeezed by the sequential closure of the N-gate and the DNA gate. These inter-domainal couplings maximize the dimer stability, ensure a very transient DNA gating state and enforce unidirectional DNA transport by precluding the backtracking of the T-segment (Figure 1A).
Figure 1.
Mechanical couplings for gate opening and closure in type-2 topoisomerases. (A) In type-2A enzymes (bacterial gyrase, topoisomerase IV, topoisomerase II), the N-gate, DNA gate and C-gate appear to be mechanically coupled with a double-lock rule, such that a given gate can open only if the other two are closed. This coordination minimizes the risk of the two enzyme halves from coming apart while gating the DNA. (a) When the entrance N-gate is open, DNA might be cleaved but the DNA gate is not able to widen. (b) Upon ATP binding and closure of the N-gate, a captured T-segment cannot be held in the inter-domainal region between the N-gate and DNA gate. The T-segment quickly crosses the DNA-gate and reaches the central cavity of the enzyme. (c) The gated DNA is then rejoined. (d) The consequent entrapment of the T-segment enforces its exit by a transient opening of the C-gate. (B) In type-2B enzymes (topoisomerase VI) there is no C-gate. Dimer stability depends on the coordination between the N-gate and DNA gate. (a) When the entrance N-gate is open (no ATP bound) the DNA gate is locked. (b) Upon ATP binding and closure of the N-gate, a captured T-segment is held in the central cavity of the enzyme prior to the aperture of the DNA gate. (c) The captured T-segment crosses then the DNA-gate and exits the complex.
The quaternary couplings during DNA transport in topoisomerase II must be alike in topoisomerase IV and DNA gyrase. However, they might differ from those in the less-extensively studied type-2B topoisomerases (Archea topoisomerase VI and plant topoisomerase VI) (51,52). These enzymes share several mechanistic features with the type-2A topoisomerases, but there are significant structural differences between the two families (53). For T-segment capture, the ATPase domains of type-2B dimerize upon ATP binding alike the N-gate in type-2A enzymes (54). The DNA cleavage-rejoining core of type-2B also contains counterparts of the structural motifs of the DNA-gate of the type-2A enzymes. However, type-2B enzymes do not have an exit- or C-gate (55). The transported duplex is outside the complex right after crossing the DNA gate (Figure 1B). Therefore, dimer stability during the DNA gating step relies mostly in the closure of the N-gate. Type-2A and type-2B topoisomerases should have then distintc couplings between their ATPase and DNA cleavage-rejoining modules. Remarkably, contrary to type-2A topoisomerases (50), the ATPase domains of type-2B enzymes are required for DNA cleavage (56) and the cavity between the two gates of these enzymes can accommodate a captured T-segment before widening the DNA gate (55). The absence of a C-gate in type-2B topoisomerases facilitates perhaps the dissociation of the enzyme halves for some biological tasks. For instance, reduced dimer stability could be operational in Spo11, a type-2B related enzyme that mediates DNA double-strand breaks during recombination (51,57).
Aside of ensuring dimer stability during the DNA cleavage and gating steps, another way to guarantee genome integrity is to not cleave the DNA in the first place. It is conceivable that topoisomerase II discerns whether or not a duplex is suitable for successful rejoining before gating it. For example, if DNA is undergoing extreme tension, its conformation might not be appropriate for catalytic cleavage. In that regard, the recently solved crystal structure of the cleavage-rejoining core of yeast topoisomerase II bound to DNA, reveals a complex producing a 150° DNA bend in the G-segment (58). A sharp bending of the bound G-segment had previously been inferred for bacterial topoisomerase IV and it is postulated to contribute to the capacity of type-2A topoisomerases to simplify DNA equilibrium configurations (59). Bending of the G-segment might be a prerequisite for DNA cleavage, thus protein conformational changes that accompany this deformation create a bipartite catalytic site, in which each DNA strand gets close to a reactive tyrosine and a coordinated magnesium ion (58). Another possible function of such DNA bending is that it would prevent topoisomerase II from gating the duplex in regions undergoing severe stretching forces (Figure 2A).
Figure 2.
Topoisomerase II might discern whether or not a DNA domain is suitable for successful rejoining by bending the G-segment before the gating step. (a) Proper bending of the interacting duplex will be possible as long as the stretching tension along the DNA does not exceed a threshold value (τo). This threshold is determined by the tension (τp) that the enzyme is able to counteract to close the DNA gate. (b) The active deformation of DNA, coupled to ATP binding and hydrolysis, could operate then as a checkpoint to unlock the DNA-gate, as well as a gain of elastic energy to be delivered in the rejoining step.
Recently, oligonucleotides with a pair of fluorophores straddling the DNA cleavage site have been used to observe the dynamics of the DNA gate by FRET (60). The experiment revealed that during steady-state ATP hydrolysis fluorophores repeatedly move over a distance of 20 Å with comparable back and forth rate constants. Whether these displacements reflected opening and closing cycles of the DNA-gate is puzzling in two regards. First, the putative open and closed states are nearly equally probable in a steady state population, whereas all previous biochemical data indicate that in this condition only a small fraction of complexes is in the open DNA-gate state. Second, as mentioned above, the capture of a T-segment is likely required to enforce the opening of the DNA gate. In the FRET experiment, however, no free DNA was available to provide a T-segment. Thus, the observed displacements could reflect mostly bending or twisting distortions of the G-segment coupled to ATP binding and hydrolysis but with no significant widening of the DNA gate. Such active deformation of DNA could operate as a checkpoint to unlock the DNA-gate, as well as a stock of elastic energy to deliver at the rejoining step. By these means, topoisomerase II could discern whether a duplex is suitable for proper rejoining before gating it (Figure 2B).
Gate padlocks to fasten DNA and turn-off topoisomerase activity
Identification of topoisomerase II as one of the main components of the nuclear matrix and metaphase chromosome scaffold (61–63), as well as its preferential interaction to SAR and MAR sequences (64–66), had lead to the proposal of an structural role for the enzyme. The possibility that topoisomerase II could be anchoraging DNA chromosomal loops fit well with the enzyme capabilities to hold DNA through covalent bonds and to interact simultaneously with several DNA segments (67,68). This putative structural role was further supported by the discovery of that, upon binding of a non-hydrolysable ATP analogue, the topoisomerase constituted in vitro a high-salt resistant complex with circular DNA molecules (7,69,70). This observation was essential for unravelling the mechanism of type-2 topoisomerases, reflecting the closure of the N-gate and the subsequent impossibility of the protein ring detaching from a bound G-segment in the absence of a DNA free-end. The topoisomerase–DNA complex was resistant to salt concentrations up to 5 M NaCl or CsCl during several hours (71). Therefore, such a topological interaction was an attractive mechanism to fasten large chromosomal DNA loops, and it seemed more convenient than holding the duplex with covalent links that implied a permanent cleavage of DNA.
In the last decade, however, support for the proposed structural role of topoisomerase II had faded since some observations disputed the requirement of the enzyme to maintain the shape of mitotic chromosomes. Although topoisomerase II is required for mitotic chromosome assembly in vitro, the enzyme could be extracted from in vitro condensed chromosomes with no significant changes in the chromosome conformation (72). Other in vivo studies, which used fluorescently tagged human DNA topoisomerases IIα and IIβ, indicated that neither enzyme is an immobile structural component of the chromosomes (73). Moreover, several studies reported significant mitotic chromosome condensation even after genetic knockdown or knockout of topoisomerase II (30,74–76). Yet, other demonstrations indicated that topoisomerase II remains stably bound in chromosomes assembled in vitro after a round of DNA replication (77). More recent analyses in several species including mammals, chicken, C. elegans and Drosophila, came to agree in that topoisomerase II is not required for initial chromatin compaction and formation of a longitudinal chromosome axis, but that an axial distribution of the enzyme in late prophase to metaphase is real, being dependent on the presence of condensins (76,78,79). On the other side, the existence of a rigid chromosome scaffold, in which DNA loops could be anchored, is also no longer sustained after some morphological and biophysical observations. For instance, depletion of histone H1 results in longer and thinner compacted mitotic chromosomes, and so arguing against a stiff scaffolding structure driving chromosome condensation (80). Also, the elastic response of native metaphase chromosomes and its alteration by endonucleases rule out a contiguous internal non-DNA (e.g. protein) scaffold. The elastic behaviour of mitotic chromosomes suggests instead that they are shaped by isolated DNA-crosslinking elements spaced by few kilo base pairs (81,82).
From the above, the possibility that a highly stable topoisomerase II–DNA complex plays a structural role in chromosome architecture cannot be excluded. The closure of the enzyme around DNA is driven by ATP binding and this conformation could be easily stabilized by some cellular factor. Such factor could interact across the closed N-gate domains, thus behaving like a molecular padlock. Essentially, this is the case with bisdioxopiperazines, such as ICRF-193, that bind to the interface of the dimerized ATP-ase domains of topoisomerase II and preclude N-gate reopening (83,84). The hypothetic existence of gate padlocks could be also considered for the C-gate of topoisomerase II. In this case, stabilisation of the exit gate would be analogous to locking it by means of engineered disulfide bonds (11,50). In such a case, a T-segment would be captured, transported across the gated G-segment and end up entrapped in the central chamber of the topoisomerase between the rejoined DNA-gate and the locked exit-gate. If padlocks close both the C-gate and the N-gate simultaneously, the topoisomerase would become a tight toroidal structure, inside which two DNA segments could be entrapped. This extreme can be observed in vitro by closing the N-gate with a non-hydrolysable ATP analogue in the yeast topoisomerase II mutant having the C-gate locked by disulfide bonds (50). In this condition, the enzyme would firmly hold DNA crossings (Figure 3).
Figure 3.
Gate padlocks can regulate topoisomerase activity and clamp interacting DNA. Analogously to bisdioxopiperazines, such as ICRF-193 that inhibit topoisomerase II activity, closure of the N-gate of topoisomerase II upon ATP binding can be stabilized also by some cellular factor. (A) Such gate padlocks could operate as inhibitors of enzyme binding to chromosomal DNA. (B) If DNA is already bound to the topoisomerase, an N-gate padlock would produce a high salt resistant complex, which might serve to regulate the enzyme activity or operate as a structural element for DNA organization. (C) Padlocks for the C-gate could also exist. If both the N-gate and the C-gate were stabilized in the closed conformation, the topoisomerase could clamp two DNA duplexes, the G-segment and passed T-segment.
In addition to stabilize topoisomerase II–DNA complexes for structural purposes, another plausible function of gate padlocks could be the regulation of the enzyme activity, a subject that remains poorly understood. Topoisomerase II can be phosphorylated in vitro on multiple serine and threonine residues, the majority of which are located in its non-conserved C-terminal region. The plausible effects of phosphorylation in vivo remain controversial, since most studies report that these modifications do not produce significant changes in the DNA transport activity of the enzyme in vitro (85). Topoisomerase II can also undergo poly(ADP-ribosyl)ation in vitro. Whether this modification occurs in vivo and has physiological significance is unclear (86). It is suggested that poly(ADP-ribosyl)ation may play a role in the cellular distribution and degradation the enzyme (87). More recently, topoisomerase II has been shown to undergo SUMO-lation during mitosis (88–90). This modification seems to be a signal for targeting the enzyme to pericentromeric regions of mitotic chromosomes and promotes the decatenation of sister chromatids at anaphase (88–90). The question then is what might prevent intracellular topoisomerase II to hydrolyse ATP, to cleave DNA or to transport DNA when it is not necessary. Gate padlocks could be an effective way to turn-off these enzyme activities. As observed with the topoisomerase inhibitor ICRF-193, locking of the N-gate upon ATP binding can occur in the absence of DNA (83). In this closed conformation, ATP hydrolysis and inter-domainal movements are inhibited (Figure 3). Gate padlocks could therefore be used to transport and stock topoisomerase II inside the cell and let it operate only where needed. So far, no gate padlock has been found in cell extracts or described among the several proteins that might physically interact with topoisomerase II. One reason could be that they do not exist. Another reason, however, is that proper assays to find them have not been conducted.
DNA transport selectivity in the chromatin milieu
Once topoisomerase II is bound to a G-segment, DNA transport depends solely on the probability of finding a potential T-segment properly positioned in the vicinity of the N-gate of the enzyme (91). For naked DNA molecules in free solution, such juxtaposition probability will be determined by the global configuration of the duplex, and also by constrains that the topoisomerase might produce by its local interactions with DNA. This last aspect should account for the enzyme capacity to drive DNA topology below thermal equilibrium configurations and for its chiral preferences (92,93). The question now is whether our current understanding of juxtaposition probability of DNA segments on topoisomerase II, as inferred from in vitro studies, is applicable to intracellular DNA. Although the topological problems inside the cell are equivalent to those portrayed in vitro, the conformational dynamics of DNA is determined by a complex set of variables involving multiple molecular interactions, in addition to the topology of the duplex. For example, although in living yeast cells both DNA topoisomerases I and II are capable to relax plasmid minichromosomes (94), in vitro studies revealed that topoisomerase II relaxes nucleosomal DNA much faster than topoisomerase I (25). Apparently, the DNA cross-inversion mechanism of topoisomerase II is facilitated in nucleosome fibres that might favour the juxtaposition of DNA segments, whereas the DNA strand-rotation mechanism of topoisomerase I might be stalled by twist diffusion barriers imposed by nucleosome organization. How chromatin affects DNA segment juxtaposition and the consequent topoisomerase II activity is complex to envisage. On one hand, juxtaposition can be favoured as DNA is intensely and extensively more folded than in free solution. On the other hand, the number of DNA segments suitable for topoisomerase II manipulation is more reduced since many proteins cover most of the DNA length. To further dissect this problem, three types of DNA juxtaposition should be considered to occur in the intracellular landscape, each one producing a different outcome upon DNA transport by topoisomerase II.
The first type of DNA juxtaposition is that driven by the topology configuration. This is the case of the pre-catenanes and catenane links generated between newly replicated duplexes and the supercoils generated ahead and behind DNA tracking ensembles. These structural complexities are fully removed by topoisomerase II in vivo. One question here is whether such juxtapositions are configured inside the cell in the same way as in naked DNA in vitro, and how such configurations might enhance the DNA transport efficiency of topoisomerase II. For instance, DNA decatenation by yeast topoisomerase II is greatly enhanced by DNA supercoiling in vitro, being catenane links removed much faster than coexisting supercoil crossings (91). Moreover, DNA decatenation is more efficient in negative than in positively supercoiled circles (93). These preferences, however, might not apply to intracellular DNA unless newly replicated duplexes were supercoiled. DNA configurations behind these in vitro preferences are perhaps reproduced in a different way by the chromatin architecture of post-replicative catenanes in vivo. Another dissimilarity between in vitro and in vivo configurations is that intracellular catenane links might produce very tight DNA juxtapositions when chromosome condensation and segregation forces pull apart replicated DNA molecules. In that case, their configuration likely facilitates its removal by topoisomerase II.
In the case of DNA relaxation, yeast topoisomerase II does not have significant preference to invert positive or negative crossings in supercoiled DNA in vitro (91), though this is not the case in relaxed DNA (95). Inside the cell, the enzyme must remove also positive and negative supercoils with comparable efficiency. Otherwise, DNA supercoils of a particular sign would accumulate in yeast mutants deficient in topoisomerase I (96). This situation might be slightly different in mammalian cells, since topoisomerase IIα appears to relax in vitro positive supercoils faster than negative ones, while IIβ displays no such preference (29). To the question of how helical tension configures DNA juxtapositions in vivo, one can assume that DNA is not adopting the same plectoneme folds seen in naked DNA. The nucleosome structure has its intrinsic mode of constraining and adapting to DNA helical stress. Yet, topoisomerase II is found to relax positive helical tension in native nucleosome arrays nearly at the same rate as in naked DNA (25). Without displacement of the histones, DNA juxtaposition appears to be optimized for topoisomerase II activity. In that regard, recent studies indicated that positive helical tension drives a chiral transition of nucleosomes to a meta-stable conformation (reversome) in which H2A–H2B dimers and the H3–H4 tetramer refold to shape a right-handed path for DNA (97). Interestingly, the entry and exit DNA segments of the reversome might configure a positive DNA crossing, which could be inverted by topoisomerase II. Strand passage would readily revert the chiral transition of the nucleosome while relaxing the DNA (Figure 4A).
Figure 4.
Juxtaposition of DNA segments enforced by chromatin architecture. (A) Nucleosomes accommodate to positive helical tension (+) by adopting a meta-stable conformation in which H2A–H2B dimers and the H3–H4 tetramer reorganize and shape a right-handed path for DNA. The entry and exit DNA segments of this nucleosomal conformation might configure an ideal positive DNA crossing to be targeted by topoisomerase II. DNA transport would relax the helical tension and revert the chiral transition of the nucleosome. This scenery might explain why topoisomerase II relaxes helical tension in nucleosome arrays as efficiently as in naked DNA. (B) In absence of DNA helical tension, DNA juxtaposition could be tailored by neighbouring DNA–protein interactions. DNA transport would then result in supercoiling (or knotting). The reaction would be analogous to that of DNA gyrase. The only difference is that protein–DNA interactions enforcing the juxtaposition of a T-segment are established outside rather than inside the topoisomerase–DNA complex.
The second type of DNA juxtaposition is that enforced by the molecular crowding of intracellular DNA. Computer simulations of DNA chains under tough condensation or volume confinement predict that the duplex configuration would equilibrate with many knot and catenane links (98–100). Such equilibrium would be achieved upon random transport of DNA segments, but this extreme is unlikely to occur. First, chromatin architecture might prevent random DNA juxtapositions leading to undesired knots and catenane links. Second, topoisomerase II might keep random knot and catenate links below the equilibrium values, as it does with DNA in free solution. Third, undesired links provoked by molecular crowding will be cleared at the time of chromosome replication and segregation. The fact is, however, than the extent of knots and catenanes driven by molecular crowding of intracellular DNA is unknown. Circular minichromosomes (<10 kb) extracted from yeast cells show negligible catenation and knotting probability (101), but these figures could become significant in larger chromatin domains. In such a case, the question is whether such links might have some biological relevance. For example, knotting of DNA driven by the molecular crowding could contribute to stabilize chromosome condensation.
The third type of DNA juxtaposition is that enforced by the architecture of DNA–protein interactions. Juxtaposition of nearby intra-molecular DNA segments is common in eukaryotic chromatin. Wrapping of DNA around histones brings linker DNA segments in close proximity; and many other DNA-interacting proteins bring together nearby DNA regions by looping or kinking the duplex. The question here is what portion of such juxtapositions not driven by the topology state of DNA can be effective pairs of G- and T-segments. It is conceivable that a juxtaposition is specifically tailored to promote DNA transport by topoisomerase II, so creating a situation analogous to that of DNA gyrase. The only difference would be that protein–DNA interactions enforcing the juxtaposition of a T-segment to configure a positive (or negative) crossing would be established outside rather than inside the topoisomerase-DNA complex (Figure 4B). DNA transport could result then in supercoiling energy that, unless constrained by other interactions, could drive critical structural changes in chromatin micro-domains. Such supercoiling activity could be behind the implication of topoisomerase II in local chromatin condensation-decondensation and gene regulation.
CONCLUSION
In view of our substantial knowledge on topoisomerase II structure and mechanism, as inferred from in vitro studies, many questions need to be now addressed pertaining to how the enzyme performs inside the cell. To the question of how dimer stability is ensured during the DNA gating step, one answer is the double-lock rule that allows any of its three gates to open only if the other two are closed. An additional answer is a checkpoint to unlock the DNA-gate, which might allow topoisomerase II to discern whether a duplex is suitable for rejoining before gating it. To the question of what prevents ATP hydrolysis, DNA cleavage or DNA transport by topoisomerase II when those are not necessary, gate padlocks could be the answer. Gate padlocks might also operate the transition from the catalytic to the putative structural functions of the enzyme. Finally, to the question of what directs DNA transport probability in the chromatin milieu, three kinds of DNA juxtaposition can configure potential pairs of G- and T-segments to be inverted by intracellular topoisomerase II: those enforced by DNA topology configuration, those resulting from molecular crowding, and those customized by chromatin architecture. DNA transport produces a different outcome in each case, so accounting for the multiple biological roles reserved for topoisomerase II. More questions will arise, and the initial answers will be imperfect or quite speculative, as some in this survey. Yet, this exercise is essential to design crucial experiments en route for new discoveries.
FUNDING
Funding for open access charge: Ministerio de Educación y Ciencia de España (Plan Nacional I+D+I) and Generalitat de Catalunya (DURSI).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 2.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 4.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 5.Schultz P, Olland S, Oudet P, Hancock R. Structure and conformational changes of DNA topoisomerase II visualized by electron microscopy. Proc. Natl Acad. Sci. USA. 1996;93:5936–5940. doi: 10.1073/pnas.93.12.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benedetti P, Silvestri A, Fiorani P, Wang JC. Study of yeast DNA topoisomerase II and its truncation derivatives by transmission electron microscopy. J. Biol. Chem. 1997;272:12132–12137. doi: 10.1074/jbc.272.18.12132. [DOI] [PubMed] [Google Scholar]
- 7.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 8.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 9.Lynn R, Giaever G, Swanberg SL, Wang JC. Tandem regions of yeast DNA topoisomerase II share homology with different subunits of bacterial gyrase. Science. 1986;233:647–649. doi: 10.1126/science.3014661. [DOI] [PubMed] [Google Scholar]
- 10.Peng H, Marians KJ. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 11.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc. Natl Acad. Sci. USA. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costenaro L, Grossmann JG, Ebel C, Maxwell A. Small-angle X-ray scattering reveals the solution structure of the full-length DNA gyrase a subunit. Structure. 2005;13:287–296. doi: 10.1016/j.str.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Nöllmann M, Stone MD, Bryant Z, Gore J, Crisona NJ, Hong SC, Mitelheiser S, Maxwell A, Bustamante C, Cozzarelli NR. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat. Struct. Mol. Biol. 2007;14:264–271. doi: 10.1038/nsmb1213. [DOI] [PubMed] [Google Scholar]
- 15.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 16.Maxwell A, Costenaro L, Mitelheiser S, Bates AD. Coupling ATP hydrolysis to DNA strand passage in type IIA DNA topoisomerases. Biochem. Soc. Trans. 2005;33:1460–1464. doi: 10.1042/BST0331460. [DOI] [PubMed] [Google Scholar]
- 17.Bates AD, Maxwell A. Energy coupling in type II topoisomerases: why do they hydrolyze ATP? Biochemistry. 2007;46:7929–7941. doi: 10.1021/bi700789g. [DOI] [PubMed] [Google Scholar]
- 18.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 19.Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 20.Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Cuvier O, Stanojcic S, Lemaitre JM, Mechali M. A topoisomerase II-dependent mechanism for resetting replicons at the S–M-phase transition. Genes Dev. 2008;22:860–865. doi: 10.1101/gad.445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 25.Salceda S, Fernández X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belmont AS. Mitotic chromosome structure and condensation. Current Opin. Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Drake FH, Zimmerman JP, McCabe FL, Bartus HF, Per SR, Sullivan DM, Ross WE, Mattern MR, Johnson RK, Crooke ST, et al. Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J. Biol. Chem. 1987;262:16739–16747. [PubMed] [Google Scholar]
- 28.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase II beta. Bioessays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 29.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 30.Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol. Biol. Cell. 2004;15:5700–5711. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 32.Akimitsu N, Kamura K, Toné S, Sakaguchi A, Kikuchi A, Hamamoto H, Sekimizu K. Induction of apoptosis by depletion of DNA topoisomerase IIalpha in mammalian cells. Biochem. Biophys. Res. Commun. 2003;307:301–307. doi: 10.1016/s0006-291x(03)01169-0. [DOI] [PubMed] [Google Scholar]
- 33.Heck MM, Hittelman WN, Earnshaw WC. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc. Natl Acad. Sci. USA. 1988;85:1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsiang YH, Wu HY, Liu LF. Proliferation-dependent regulation of DNA topoisomerase II in cultured human cells. Cancer Res. 1988;48:3230–3235. [PubMed] [Google Scholar]
- 35.Wood ER, Earnshaw WC. Mitotic chromatin condensation in vitro using somatic cell extracts and nuclei with variable levels of endogenous topoisomerase II. J. Cell Biol. 1990;111:2839–2850. doi: 10.1083/jcb.111.6.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Downes CS, Clarke DJ, Mullinger AM, Gimenez-Abian JF, Creighton AM, Johnson RT. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 37.Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J. Cell Biol. 2003;160:645–655. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: Topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl Acad. Sci. USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 40.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 41.Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol. Cell Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim. Biophys. Acta. 1992;1132:43–48. doi: 10.1016/0167-4781(92)90050-a. [DOI] [PubMed] [Google Scholar]
- 43.Tsutsui K, Okada S, Watanabe M, Shohmori T, Seki S, Inoue Y. Molecular cloning of partial cDNAs for rat DNA topoisomerase II isoforms and their differential expression in brain development. J. Biol. Chem. 1993;268:19076–19083. [PubMed] [Google Scholar]
- 44.Turley H, Comley M, Houlbrook S, Nozaki N, Kikuchi A, Hickson ID, Gatter K, Harris AL. The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. Br. J. Cancer. 1997;75:1340–1346. doi: 10.1038/bjc.1997.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyu YL, Wang JC. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIβ. Proc. Natl Acad. Sci. USA. 2003;100:7123–7128. doi: 10.1073/pnas.1232376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsutsui K, Hosoya O, Sano K, Tokunaga A. Immunohistochemical analyses of DNA topoisomerase II isoforms in developing rat cerebellum. J. Comp. Neurol. 2001;431:228–239. doi: 10.1002/1096-9861(20010305)431:2<228::aid-cne1067>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe M, Tsutsui K, Inoue Y. Differential expressions of the topoisomerase II alpha and II beta mRNAs in developing rat brain. Neurosci Res. 1994;19:51–57. doi: 10.1016/0168-0102(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 48.Lyu YL, Lin CP, Azarova AM, Cai L, Wang JC, Liu LF. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol. Cell Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 50.Roca J. The path of the DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 2004;279:25783–25788. doi: 10.1074/jbc.M402555200. [DOI] [PubMed] [Google Scholar]
- 51.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 52.Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 54.Corbett KD, Berger JM. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J. 2003;22:151–163. doi: 10.1093/emboj/cdg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat. Struct. Mol. Biol. 2007;14:611–619. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- 56.Buhler C, Gadelle D, Forterre P, Wang JC, Bergerat A. Reconstitution of DNA topoisomerase VI of the thermophilic archaeon Sulfolobus shibatae from subunits separately overexpressed in Escherichia coli. Nucleic Acids Res. 1998;26:5157–5162. doi: 10.1093/nar/26.22.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 58.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 59.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smiley RD, Collins TR, Hammes GG, Hsieh TS. Single-molecule measurements of the opening and closing of the DNA gate by eukaryotic topoisomerase II. Proc. Natl Acad. Sci. USA. 2007;104:4840–4845. doi: 10.1073/pnas.0700342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 62.Warburton PE, Earnshaw WC. Untangling the role of DNA topoisomerase II in mitotic chromosome structure and function. Bioessays. 1997;19:97–99. doi: 10.1002/bies.950190203. [DOI] [PubMed] [Google Scholar]
- 63.Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell. 2003;4:467–480. doi: 10.1016/s1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 64.Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 65.Sperry AO, Blasquez VC, Garrard WT. Dysfunction of chromosomal loop attachment sites: illegitimate recombination linked to matrix association regions and topoisomerase II. Proc. Natl Acad. Sci. USA. 1989;86:5497–5501. doi: 10.1073/pnas.86.14.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vassetzky YS, Razin SV, Georgiev GP. DNA fragments which specifically bind to isolated nuclear matrix in vitro interact with matrix-associated DNA topoisomerase II. Biochem. Biophys. Res. Commun. 1989;159:1263–1268. doi: 10.1016/0006-291x(89)92246-8. [DOI] [PubMed] [Google Scholar]
- 67.Zechiedrich EL, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roca J, Berger JM, Wang JC. On the simultaneous binding of eukaryotic DNA topoisomerase II to a pair of double-stranded DNA helices. J. Biol. Chem. 1993;268:14250–14255. [PubMed] [Google Scholar]
- 69.Osheroff N. Eukaryotic topoisomerase II. Characterization of enzyme turnover. J. Biol. Chem. 1986;261:9944–9950. [PubMed] [Google Scholar]
- 70.Roca J. Filter binding assays for topoisomerase-DNA complexes. Methods Mol. Biol. 2001;95:75–80. doi: 10.1385/1-59259-057-8:75. [DOI] [PubMed] [Google Scholar]
- 71.Lindsley JE, Wang JC. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc. Natl Acad. Sci. USA. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J. Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, Mielke C. Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J. Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang CJ, Goulding S, Earnshaw WC, Carmena M. RNAi análisis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. J. Cell Sci. 2003;116:4715–4726. doi: 10.1242/jcs.00797. [DOI] [PubMed] [Google Scholar]
- 75.Sakaguchi A, Kikuchi A. Functional compatibility between isoform a and b of type II DNA topoisomerase. J. Cell Sci. 2004;117:1047–1054. doi: 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- 76.Savvidou E, Cobbe N, Steffensen S, Cotterill S, Heck MM. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J. Cell Sci. 2005;118:2529–2543. doi: 10.1242/jcs.02392. [DOI] [PubMed] [Google Scholar]
- 77.Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J. Cell Biol. 2003;160:645–655. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watrin E, Legagneux V. Contribution of hCAP-D2, a non-SMC subunit of condensin I, to chromosome and chromosomal protein dynamics during mitosis. Mol. Cell Biol. 2005;25:740–750. doi: 10.1128/MCB.25.2.740-750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 2004;166:775–785. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maresca TJ, Freedman BS, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J. Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Poirier MG, Marko JF. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc. Natl Acad. Sci. USA. 2002;99:15393–15397. doi: 10.1073/pnas.232442599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poirier MG, Marko JF. Micromechanical studies of mitotic chromosomes. Curr. Top. Dev. Biol. 2003;55:75–141. doi: 10.1016/s0070-2153(03)01002-0. [DOI] [PubMed] [Google Scholar]
- 83.Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl Acad. Sci. USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc. Natl Acad. Sci. USA. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bakshi RP, Galande S, Muniyappa K. Functional and regulatory characteristics of eukaryotic type II DNA topoisomerase. Crit. Rev. Biochem. Mol. Biol. 2001;36:1–37. doi: 10.1080/20014091074165. [DOI] [PubMed] [Google Scholar]
- 86.Darby MK, Schmitt B, Jongstra-Bilen J, Vosberg HP. Inhibition of calf thymus type II DNA topoisomerase by poly(ADP-ribosylation) EMBO J. 1985;4:2129–2134. doi: 10.1002/j.1460-2075.1985.tb03903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Canitrot Y, de Murcia G, Salles B. Decreased expression of topoisomerase IIbeta in poly(ADP-ribose) polymerase-deficient cells. Nucleic Acids Res. 1998;26:5134–5138. doi: 10.1093/nar/26.22.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takahashi Y, Yong-Gonzalez V, Kikuchi Y, Strunnikov A. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 2006;172:783–794. doi: 10.1534/genetics.105.047167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takahashi Y, Strunnikov A. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma. 2008;117:189–198. doi: 10.1007/s00412-007-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roca J, Wang JC. The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase II. Genes Cells. 1996;1:17–27. doi: 10.1046/j.1365-2443.1996.01001.x. [DOI] [PubMed] [Google Scholar]
- 92.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 93.Roca J. Varying levels of positive and negative supercoiling differently affect the efficiency with which topoisomerase II catenates and decatenates DNA. J. Mol. Biol. 2001;305:441–450. doi: 10.1006/jmbi.2000.4307. [DOI] [PubMed] [Google Scholar]
- 94.Saavedra RA, Huberman JA. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986;45:65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- 95.Trigueros S, Salceda J, Bermudez I, Fernandez X, Roca J. Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J. Mol. Biol. 2004;335:723–731. doi: 10.1016/j.jmb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 96.Trigueros S, Roca J. Failure to relax negative supercoiling of DNA is a primary cause of mitotic hyper-recombination in topoisomerase-deficient yeast cells. J. Biol. Chem. 2002;277:37207–37211. doi: 10.1074/jbc.M206663200. [DOI] [PubMed] [Google Scholar]
- 97.Bancaud A, Wagner G, Conde E, Silva N, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol. Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 98.Arsuaga J, Vázquez M, Trigueros S, Sumners D, Roca J. Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proc. Natl Acad. Sci. USA. 2002;99:5373–5377. doi: 10.1073/pnas.032095099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arsuaga J, Blackstone T, Diao Y, Karadayi E, Saito Y. Knotting of uniform random polygons in confined spaces. J. Phys. A: Math. Gen. 2007;40:11697–11711. [Google Scholar]
- 100.Arsuaga J, Blackstone T, Diao Y, Karadayi E, Saito Y. Linking of uniform random polygons in confined spaces. J. Phys. A: Math. Gen. 2007;40:1925–1936. [Google Scholar]
- 101.Trigueros S, Roca J. Circular minichromosomes become highly recombinogenic in topoisomerase-deficient yeast cells. J. Biol. Chem. 2001;276:2243–2248. doi: 10.1074/jbc.M008930200. [DOI] [PubMed] [Google Scholar]