Abstract
Pre-mRNA splicing occurs in a large complex spliceosome. The steps of both spliceosome assembly and splicing reaction have been extensively analyzed, and many of the factors involved have been identified. However, the post-splicing intron turnover process, especially in vertebrates, remains to be examined. In this paper, we developed a two-tag affinity purification method for purifying lariat intron RNA–protein complexes obtained from an in vitro splicing reaction. Glycerol gradient sedimentation analyses revealed that there are at least two forms of post-splicing intron complexes, which we named the ‘Intron Large (IL)’ and the ‘Intron Small (IS)’ complexes. The IL complex contains U2, U5 and U6 snRNAs and other protein splicing factors, whereas the IS complex contains no such U snRNAs or proteins. We also showed that TFIP11, a human homolog of yeast Ntr1, is present in the IL complex and the TFIP11 mutant protein, which lacks the interaction domain with hPrp43 protein, caused accumulation of the IL complex and reduction of IS complex formation in vitro. Taken together, our results strongly suggest that TFIP11 in cooperation with hPrp43 mediates the transition from the IL complex to the IS complex, leading to efficient debranching and turnover of excised introns.
INTRODUCTION
Nuclear pre-mRNA splicing takes place in a large RNA–protein complex (RNP) that is termed ‘spliceosome’. Spliceosome is composed of U1, U2, U4/U6 and U5 snRNPs and a large number of non-snRNP proteinaceous splicing factors (1,2). Spliceosome assembly has been extensively analyzed by in vitro splicing systems using extracts from HeLa and Saccharomyces cerevisiae cells (1). In both systems, spliceosome formation proceeds by a sequential assembly of several intermediate complexes. In mammals the intermediate complexes are termed H, E, A, B, B* and C (1).
In the first step, the naked pre-mRNA associates with heterogeneous nuclear ribonucleoproteins (hnRNPs), which are abundant nuclear RNA-binding proteins, to form the H complex. Then, U1 snRNP recognizes and binds to a conserved 5′ splice site in an ATP-independent manner to give the E complex. Subsequently, U2 snRNP stably binds to a branch point sequence in an ATP-dependent manner forming the A complex. Then, preformed tri-snRNP (U4/U6 and U5) joins the A complex giving rise to the B complex. Subsequently, the B complex is activated through RNP rearrangements leading to the displacement or destabilization of U1 and U4 snRNPs from the spliceosome (3,4). The resultant B* spliceosome, which is also called the activated spliceosome, catalyzes the first step reaction, during which the branch-point adenosine attacks the 5′ splice site, generating a cleaved 5′ exon and intron–3′ exon lariat intermediates (3). As a result of the above reaction, the C1 complex is formed and mediates catalysis of the second step of splicing via ATP hydrolysis (1,5). This results in the formation of the C2 complex, which contains ligated exons and excised lariat introns (6). C1 and C2 complexes are often collectively called the ‘C complex’.
In addition to U snRNAs and their specific proteins, a large number of non-snRNP proteins are associated with spliceosome and play essential roles in splicing. In order to learn about the dynamics of the spliceosome's proteome, mass spectrometric (MS) studies of the spliceosomal complexes isolated at defined stages have been performed (1). The complexes analyzed to date include the H, A, BΔU1 (B complex lacking U1 snRNP), B, B* and C1 complexes (3,7–12). The most recent work by Bessonov et al. (10) describes the purification and characterization of fully assembled, active spliceosome C1 complexes that contain the products of the first step of splicing and are capable of carrying out the second-step reaction (exon ligation) without additional protein factors. From these MS analyses, over 100 splicing factors have been identified, and their dynamic association and dissociation during spliceosome formation have been uncovered.
Compared to the extensive analysis of the steps and factors of spliceosome formation, little is known about the post-splicing pathway. Only a few factors involved in the post-splicing process have been identified, almost exclusively by analyses of budding yeast mutants. Upon the completion of the splicing reaction, the spliced RNA (mRNA) is released from the spliceosome, and the remaining post-splicing RNPs containing the lariat intron are disassembled in an active process involving two members of the ATP-dependent DExH box RNA helicase family, Prp22 and Prp43 (13,14). Prp22 is needed for the release of the spliced RNA (14), while Prp43 is necessary for the disassembly of the remaining intron-containing RNP (15,16). Recently it has been shown that two splicing factors, Ntr1 and Ntr2, in the form of a stable dimmer, associate with Prp43. The resultant trimeric complex is termed the ‘NTR complex’ (17). Ntr1 contains a G-patch domain at its N-terminal region, which is responsible for its interaction with Prp43, and this interaction stimulates Prp43′s helicase activity in vitro (17,18). The NTR complex is functional in catalyzing the disassembly of the spliceosome (17). After dissociation of U snRNPs and other splicing factors from the lariat intron, the intron is linearized by a lariat intron debranching enzyme, DBR1, before being degraded (19).
In humans, most of the genes encoded in the nucleus contain multiple introns, which occupy 95% of the protein-coding primary transcripts (20,21). These introns are excised from pre-mRNAs by splicing as a lariat form and get degraded in the nucleus (19). If the excised introns are not processed properly in the nucleus, the splicing factors become trapped on the excised introns and the splicing reaction is negatively affected. The inhibition of intron degradation into mononucleotides might cause a reduction in the amount of free nucleotides, which could result in transcription defects. Moreover, in vertebrates, guide small nucleolar RNAs (snoRNAs), which mediate ribosomal RNAs modifications, are encoded in the introns of some genes. These snoRNAs are known to be processed from the excised introns in coordination with the splicing process (22). The loss of functional snoRNAs leads to production of unmodified rRNAs, which reduces ribosome processivity. Therefore, it is likely that the postsplicing intron turnover process is extremely important in vertebrates.
Although no study has focused on the post-splicing intron turnover process in mammals, the homologs of the factors described above in yeast have been identified in mammals. HRH1 (Human RNA Helicases 1), a human homolog of yeast Prp22 protein, mediates the release of spliced mRNA from the spliceosome (23,24). Homologs of Prp43 have also been identified in mice and humans (25,26). Moreover, human and mouse DBR1 proteins, which show a high sequence homology to the yeast protein, were demonstrated to complement the yeast dbr mutant (27,28). Recently it has been shown that human Ntr1 like protein, which is also called Tuftelin Interacting Protein 11 (TFIP11), interacts with factors involved in DNA double-strand break repair and telomere metabolism (29). However, the function of TFIP11 in splicing has not been investigated. To date, no human homologs of NTR2 have been reported.
In this report, we developed a purification system for excised intron RNPs obtained via in vitro splicing reactions using two affinity tags. Glycerol gradient sedimentation experiments revealed that there are two different forms of post-splicing intron complexes. One is a 40S form with U2, U5 and U6 snRNPs, which was termed the ‘IL (Intron Large) complex’, and the other is a 20S form, which was termed the ‘IS (Intron Small) complex’. We investigated the protein composition of these complexes by MS and western blotting and found stable associations of several human Prp19 complex factors with the IL complex, but none with the IS complex. SF3a/3b proteins, which are 17S U2 snRNP associated factors, did not stably associate with the IL complex although U2 snRNA and its specific protein U2B′′ were present in the complex. The IS complex contained none of the tested U snRNAs and protein factors.
Furthermore, we found that the TFIP11/hNtr1 protein is stably present in the IL complex and that it recruits the hPrp43 protein to the IL complex through interactions mediated by its N-terminal G-patch region. Overexpression of the mutant TFIP11 protein, which lacks the G-patch domain caused the accumulation of IL complex and a reduction in IS complex formation. We also demonstrated that the lariat intron in the IS complex is more susceptible to debranching reaction by hDBR1 than that in the IL complex. Our results strongly suggest that the hPrp43–TFIP11 complex catalyzes the transition of the IL complex to the IS complex leading to efficient debranching and subsequent degradation of the lariat intron during the post-splicing intron turnover process.
MATERIALS AND METHODS
Plasmid construction
For the construction of the pδ-crystallin MS2 plasmid, a DNA fragment containing three copies of the MS2 binding site was generated as described previously (30) although we attached SacI sites at both ends of the fragment. One copy of this fragment was inserted into the SacI site between the 5′ splice site and the branch adenosine of the pSP14-15 plasmid (23). For the construction of the pAd2-MS2 plasmid, three copies of the DNA fragment that contained the SacII site at both ends were inserted into the SacII site of the pAd2 plasmid (31). To prepare plasmids for expressing Flag-tagged proteins in HEK293T cells, full-length TFIP11 and hDBR1 cDNAs were amplified by the PCR reaction from the Marathon ready HeLa cDNA library (Clontech) and were cloned into the EcoRI/XhoI sites and BamHI/XhoI sites of the Flag-pCDNA3 (32), respectively. hDBR1 mutant cDNA (NA) containing a point mutation in its GNHE motif (33) was also prepared by PCR-based mutagenesis using Flag-hDBR1 plasmid as a template. The oligonucleotides used for this mutagenesis were as follows: forward primer: 5′-ATGGGCTCCCCCAATGAAGAGCGT-3′, reverse primer: 5′-GAAGCCTCAAATCATTTGCAAGAG-3′. TFIP11 mutant cDNA (ΔN50) was also prepared by PCR amplification using specific primers (forward primer: 5′-AAAGAATTCCCAAGATGGGCTACGTCCCTGGACGGG-3′ and reverse primer: 5′-AAACTCGAGTCACTTGGCCATGTCGATCAGGCTC-3′) and the Flag-TFIP11 plasmid as a template.
For production of GST-MS2 and GST-hDBR1 protein expression plasmids, PCR-amplified MS2 and hDBR1 cDNAs were inserted between the BamHI and XhoI sites of pGEX6P-1 (GE Healthcare).
For the construction of a His-hPrp43 protein expression plasmid, hPrp43 cDNA, a gift from Dr. Akila Mayeda, was inserted between the NcoI and NotI sites of the pFastBAC HTB plasmid (Invitrogen).
In vitro transcription, splicing and MS2 affinity purification of intron RNP
The in vitro transcription reaction was usually performed at 37°C for 120 min using MEGAscript (Ambion) according to the manufacturer's instructions. The preparation of immobilized pre-mRNA containing a biotin-labeled tag was performed as described previously (34). Briefly, 1 pmol of biotinylated pre-mRNA was incubated with 10 μl of Dynabeads m-280 magnetic streptavidin beads (Invitrogen) in 20 μl of IP1000 buffer [20 mM HEPES-NaOH (pH 7.9), 1000 mM KCl and 0.05% Triton X-100]. Prior to the splicing assay, the immobilized substrate beads were washed twice with 400 μl IP1000 buffer and then with IP150 buffer [20 mM HEPES-NaOH (pH 7.9), 150 mM KCl and 0.05% Triton X-100].
The tethered pre-mRNA was then bound to 1.5 μg of GST-MS2 protein and was incubated with 100 μl of HeLa cell nuclear extracts under splicing conditions (23). The reaction mixture was incubated for 80 min at 30°C while being rocked for 10 s every 10 min using Thermomixer comfort (Eppendorf). After incubation, the resin was precipitated using a magnetic stand to recover the supernatant. Heparin (Sigma) was then added to a final concentration of 0.5 μg/μl, and the resultant mixture was further incubated for 5 min at 30°C. Next, the mixture was incubated with 20 μl of glutathione beads for 1 h at 4°C with rotating, being washed four times with 500 μl IP150 buffer and subsequently eluted with elution buffer (20 mM HEPES-KOH pH7.9, 100 mM KCl, 0.6% sarkosyl, 10% glycerol, 0.1% NP-40, 0.1 m EDTA and 1 mM DTT). For the glycerol sedimentations assays, the supernatant of the biotin-precipitated splicing mixture was subjected to 10–30% glycerol gradient solution (20 mM HEPES-KOH pH7.9, 50 mM KCl, 1.5 mM MgCl2, 1 mM DTT and 10–30% glycerol) and was centrifuged at 40 000 r.p.m. for 3 h with an SW41 rotor (Beckman-Coulter). The fractions were recovered using a Biocomp gradient station (Biocomp).
Antibodies and western blotting
The antibodies used for western blotting are listed below, and they were obtained from the companies shown in parentheses. Anti-Flag M2 antibody (SIGMA); anti- hPrp19, hPrp43 and U5-116k (Bethyl Laboratories, Inc.); anti- Xab2 (GENETYX); anti-U5-220k (Santa Cruz); anti-SF3b130 and SF3a60 (Abcam); anti-U2B′′ (PROGEN); anti-SF3b155 (MBL); and SF3a120 (Synaptic Systems). The antibody against IBP160 was a kind gift from Dr. Tetsuro Hirose. The anti-TFIP11 antibody was kindly provided by Dr. Michael Paine. The anti-hPrp43 antibody was a generous gift from Dr. Reinhard Luhrmann. The anti-hDBR1 antibody was prepared by immunizing two rabbits with GST-tagged hDBR1. Immunization and preparation of antisera were performed by SIGMA.
Western blotting was carried out as described previously (31) except that we used ECL and ECL advance systems (GE Healthcare) for the detection. The signals were obtained and analyzed by a LAS3000mini (Fuji Film, Japan).
Northern blotting analysis
RNA samples were separated by denaturing polyacrylamide gel electrophoresis (PAGE) and were transferred to hybond N+ membranes (GE Healthcare). The oligonucleotides used to prepare probes for U1, U2, U4, U5 and U6 snRNAs are described elsewhere (35). The preparation of [32P]-labeled oligonucleotide probes was carried out with T4 polynucleotide kinase (TOYOBO) as the manufacturer recommends. Hybridization with oligonucleotide DNA probes was performed in PERFECTHYB PLUS (Sigma).
In vitro protein-binding experiments
The TFIP11 and TFIP11ΔN proteins were produced by in vitro transcription–translation using a TNT kit (Promega) in rabbit reticulocyte lysate in the presence of [35S]-methionine (GE Healthcare) according to the manufacturer's instructions. One microgram each of purified His-GST and His-hPrp43 proteins were immobilized on 20 μl Ni–NTA beads (Qiagen) in PBS for 1 h at 4°C. The resin was washed with 500 μl of binding buffer (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 0.1% NP-40 and 20 mM Imidazole). The in vitro translated products were added and incubated with these immobilized proteins at 4°C for 1 hour. After binding, the beads were washed with 500 μl of binding buffer five times, and the bound fraction was analyzed by SDS–PAGE and visualized by fluorography.
Mass-spectrometric analysis
Mass-spectrometric identification of proteins was performed as previously described (36). Briefly, after SDS–PAGE, the proteins were visualized by silver staining and excised separately from gels, followed by in-gel digestion with trypsin (Promega) in a buffer containing 50 mM ammonium bicarbonate (pH 8.0) and 2% acetonitrile overnight at 37°C. Molecular mass analyses of tryptic peptides were performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) using an Ultraflex TOF/TOF (Bruker Daltonics). Proteins were identified by comparing the molecular weights determined by MALDI-TOF/MS and the theoretical peptide masses of proteins registered in the NCBInr.
Immunoprecipitation of RNAs and proteins
For immunoprecipitaion of RNA from the in vitro splicing reaction, 1 μg of anti-hPrp43 antibody (Bethyl), 1 μg of anti-GST antibody (Nakalai Tesque) for mock precipitation or 1 μg of anti-Flag M2 (Sigma) were bound to Protein A-Sepharose beads (GE Healthcare) on a rotating platform for 1 h at 4°C. After the in vitro splicing reaction, the mixture was diluted by a factor of 10 with IP150 buffer, centrifuged at 14 000 r.p.m. for 15 min at 4°C, and the resultant supernatant was used for incubation with the antibody-bound beads for 1 h at 4°C. After the precipitation, the beads were washed five times with 400 μl of IP150 buffer and incubated in 100 μl of HomoMix (50 mM Tris at pH 7.4, 5 mM EDTA, 1.5% SDS, 300 mM NaCl and 1.5 mg/ml Proteinase K) for 30 min at 50°C to elute RNA. Eluted RNAs were extracted with phenol/chloroform and precipitated by ethanol precipitation. The recovered RNAs were analyzed by 6% denaturing PAGE.
For immunoprecipitation of Flag-tagged proteins, nuclear extract from HEK293T cells that were transiently expressing Flag-tagged proteins was prepared (37), and the reaction mixture was incubated with 15 μl of M2 affinity agarose (Sigma). The beads were washed five times with 400 μl of IP150 buffer and eluted with SDS sample buffer. The eluted proteins were analyzed by 12% SDS–PAGE. RNase A (Nakalai Tesque, Japan) was added to a final concentration of 1 μg/ml.
Recombinant protein preparation
GST-MS2 protein was overexpressed in Escherichia coli BL21 (DE3) codonplus cells (Stratagene). Induction was carried out at 20°C overnight. Proteins were purified according to the manufacturer's instructions, and the eluate was then purified with a Mono-Q column (GE Healthcare).
For His-hPrp43 protein, the generation of recombinant bacmids, their transfection into Sf9 cells and purification of his-tagged proteins were performed according to the manufacturer's instructions.
For Flag-tagged proteins, whole-cell lysates from HEK293T cells were prepared from transfected cells as previously described (38). Flag-tagged proteins were purified from whole-cell lysates by using anti Flag-M2 affinity resin (SIGMA) as described previously (39).
RESULTS
The development of an intron RNP purification system from in vitro splicing reaction mixture
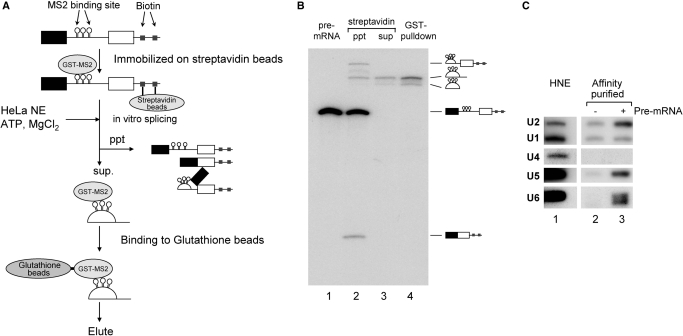
In order to analyze the post-splicing intron turnover pathway, attempts were made to isolate the post-splicing intron complex from the in vitro splicing reaction. We employed a two-affinity tag purification method (Figure 1A). As the first affinity-tag, we used biotin residues. Biotin-labeled oligo DNA was ligated directly onto the 3′ end of a pre-mRNA derived from chicken δ-crystallin gene by the use of T4 DNA ligase as described previously (34). This pre-mRNA also contained three copies of phage MS2 coat protein-binding sites in its intron. The pre-mRNA was tethered to streptavidin magnetic beads and then bound to GST-MS2 protein. The tethered pre-mRNA was incubated with HeLa cell nuclear extracts under splicing conditions, and the beads were precipitated after the incubation. Since pre-mRNA, mRNA and lariat intermediates all contain biotin residues in their 3′ regions, these RNAs were expected to remain bound to the beads. The 5′exon RNA should also be precipitated with the beads through its interaction with the lariat intermediate in the spliceosome (34). Therefore, only the lariat intron that had been released from the spliceosome was anticipated to be in the supernatant (Figure 1A). The intron complex in the supernatant was further purified by a pull-down assay with glutathione beads.
Figure 1.
Purification of the post-splicing intron complex from an in vitro splicing reaction. (A) A schematic representation of the isolation of the intron RNP complex from splicing reaction mixture. Chicken c-crystallin splicing substrate with three MS2 binding loops in its intron (δ-3MS2) was ligated with an oligonucleotide DNA containing biotinylated linker before being immobilized on streptavidin magnetic beads and bound with recombinant GST-MS2 proteins. After immobilization, this pre-mRNA was spliced in HeLa cell nuclear extracts. The reaction was subjected to precipitation using a magnetic stand to remove the pre-mRNA, splicing intermediates and mRNA. The supernatant was subjected to glutathione beads to purify the intron-RNP, which was then eluted with Sarkosyl-containing elution buffer. (B) Analysis of the RNAs recovered from each step of the intron–RNP purification shown in (A). The RNAs were analyzed by 6% denaturing polyacrylamide gel electrophoresis. The structure of each RNA is schematically shown on the right side of the panel. (C) Northern blotting analysis of the spliceosomal U snRNAs in the isolated intron–RNP. HeLa cell nuclear extracts (HNE; lane 1) that contained all five U snRNAs were used as a positive control. The isolated intron–RNPs with (lane 3) or without (lane 2) exogenously added pre-mRNAs were subjected to northern blotting analysis and probed for the five U snRNAs.
The RNA products in each isolation step were analyzed by denaturing PAGE (Figure 1B). The immobilized pre-mRNA was spliced efficiently, and the majority of the splicing products were bound to the beads (Figure 1B, lane 2). In contrast, only lariat intron RNA was detected in the supernatant fraction (lane 3). This lariat RNA was efficiently recovered with glutathione beads (lane 4, about 15% was recovered).
We next analyzed the composition of U snRNAs in this purified fraction. Northern blotting analysis using probes against five spliceosomal U snRNAs revealed that U2, U5 and U6 snRNAs were present in much higher concentrations in the post-splicing intron complex than in the control fraction, which was prepared without the addition of exogenous pre-mRNA (Figure 1C, compare lanes 2 and 3). On the other hand, the levels of U1 and U4 snRNAs in this complex were not higher than those of the control (Figure 1C, lanes 2 and 3). These results are consistent with previous results showing that U1 and U4 snRNPs are not stably associated with late spliceosomes (40). We also employed Adenovirus Major Late (AdML) pre-mRNA (31) in the same system and obtained very similar results (data not shown). Due to these results, we concluded that our method was capable of purifying the post-splicing intron complex that is produced by the in vitro splicing reaction.
Identification of the two forms of lariat–intron RNPs by glycerol density gradient sedimentation analysis
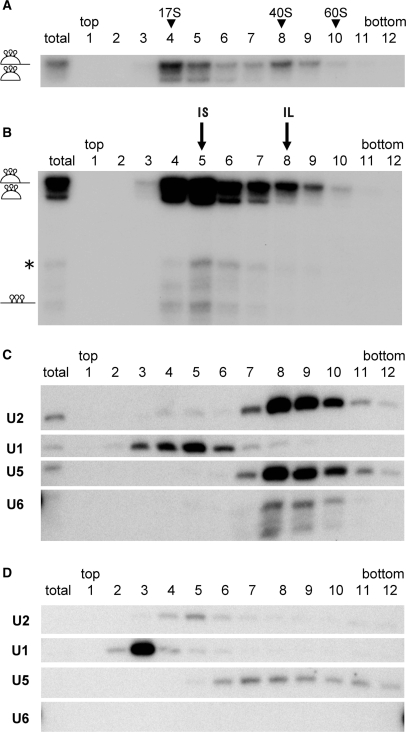
We further analyzed the post-splicing intron complex by glycerol gradient sedimentation. The supernatant of the streptavidin beads precipitation (the same fraction as in Figure 1B, lane 3) was subjected to glycerol gradient centrifugation, and the distribution of the lariat intron across the gradient was analyzed. The results demonstrated that there were at least two peak fractions for the lariat intron (Figure 2A, lanes 4 and 8). The lighter fraction was about 20S, while the heavier fractions was 40S. A lariat intron lacking its 3′ tail was mainly detected in the lighter fraction. These results suggested that two different complexes containing lariat RNA were present in the supernatant of the streptavidin beads precipitation. We designated the two complexes IL and IS complexes.
Figure 2.
Identification of the two major intron-RNPs. (A) Glycerol gradient fractionation (10–30%) of the supernatant of the streptavidin agarose pull-down assay as in Figure 1. The total lane contained 10% of the supernatant before sedimentation. The RNAs from each fraction were recovered and analyzed by 6% denaturing polyacrylamide gel electrophoresis. The fractions that human 17S U2 snRNP, yeast 40S and 60S ribosome migrated to are shown at the top. (B) MS2 affinity purification of intron–RNPs from each fraction. The RNPs were precipitated by glutathione beads through the GST-MS2 binding. The recovered RNAs were analyzed as in (A). The positions of two intron RNA peaks are indicated above as IS and IL. The structure of each RNA is shown on the left of the panel. The lane marked as total contained 10% of the supernatant before sedimentation. (C) Northern blotting analysis of spliceosomal U snRNAs in each glutathione beads purified fraction. The total lane contained a glutathione purified fraction from 10% of the supernatant before sedimentation. (D) Northern blotting analysis of glutathione affinity-purified fractions from a control experiment under the same conditions as (C) except for the absence of pre-mRNA. The exposure time of this membrane to the X-ray film was the same as in (C).
To analyze the composition of the two intron complexes, affinity-selection with glutathione beads was performed with each gradient fraction, and bound intron–RNPs were recovered (Figure 2B). For unknown reasons, the lariat intron RNA was most efficiently recovered from the fifth fraction (Figure 2B, lane 5). The efficiency of GST-MS2 pull down could have been affected by multiple factors. The complex in the fifth fraction might have been more accessible or more stably bound by GST-MS2 protein than that in the fourth fraction. This might mean that two types of complexes exist in fractions 4–6.
Northern blotting analysis revealed that U2, U5 and U6 snRNAs were more concentrated in the IL complex fractions, than in the control fractions (compare Figure 2C with D). On the other hand, the IS complex contained no such U snRNAs. Instead, the IS complex contained some U1 snRNA (Figure 2C, lane 5). However, U1 cannot be a general component of the IS complex since U1 snRNA was not detected in the IS complex when we employed the AdML-MS2 pre-mRNA substrate (Figure S1). Taken together, these results indicate that there were at least two different post-splicing intron complexes in the released supernatant fraction. The heavier IL complex is about 40S and contains U2, U5 and U6 snRNAs, whereas the smaller IS complex is about 20S and is free of spliceosomal U snRNAs.
Identification of protein factors in the lariat–intron RNPs
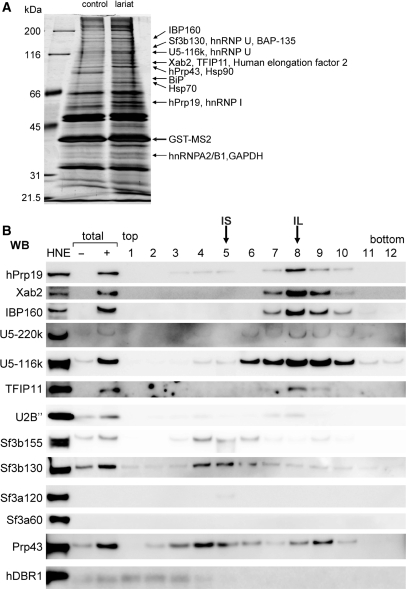
To obtain information regarding the protein composition of the human intron complexes, proteins in the affinity-purified intron complex fraction were separated by SDS–PAGE (Figure 3A, ‘lariat’ lane). Note that the glycerol gradient separation procedure was not employed in this experiment. As a control, gel-purified linear intron RNA was incubated with HeLa nuclear extracts, and the identical affinity selection procedures were performed (Figure 3A, ‘control’ lane). By using this control RNA, we should be able to recognize proteins that bind to the intronic RNA sequence without splicing reaction. Proteins that bind to the lariat–intron through splicing reaction would be anticipated to be enriched in the lariat–intron complex as compared to the linear-intron complex. The protein bands that appeared to be enriched in the ‘lariat’ lane were excised from the gel and analyzed by MS (Figure 3A). Several splicing factors, such as U2 and U5 snRNP specific proteins (SF3b130, U5-116K), hPrp19 complex proteins (IBP160, Xab2, and hPrp19), hPrp43 and some hnRNP proteins (hnRNP A2/B1, I and U) as well as other proteins, were identified (Figure 3A). Among the identified proteins, Tuftelin interacting protein 11 (TFIP11) and hMtr4 (also known as fSAP118) were reported to be present in the spliceosome (1,41), but their roles in splicing have not been clarified. The affinity-purified fraction that was used for the MS analysis should be a mixture of the IL and IS complexes since the glycerol gradient separation procedure was not employed.
Figure 3.
Proteins associated with purified intron-RNP. (A) The protein composition of GST-MS2 affinity-purified intron mRNPs. Proteins from the entire eluate were separated by 10% SDS–PAGE and stained with silver. A subset of the proteins that were concentrated in the lariat intron complex (lane 3) and identified by MS is indicated on the right. Lane 1: molecular weight markers; lane 2: control pull-down experiment performed with an in vitro transcribed linear intron in the presence of GST-MS2. (B) Characterization of purified intron RNPs by western blotting analysis. Intron complexes were separated as in Figure 2, and proteins were subsequently separated by a 5–20% SDS–PAGE gradient, blotted on a PVDF membrane and probed with several antibodies against the proteins shown on the left. The fractions that IS and IL complexes were mainly fractionated into are indicated at the top. HeLa cell nuclear extracts were used as a control (a lane marked HNE). The total lanes contain the supernatant of the MS2 purification step prepared with (+) or without (–) pre-mRNA.
In order to confirm that the identified proteins are indeed associated with post-splicing intron complexes, western blotting analyses were performed using antibodies against some of the identified proteins. The in vitro splicing reaction was carried out with either δ-MS2 (Figure 3B) or AdML-MS2 (Figure S1B) pre-mRNA and the released lariat–intron complexes (supernatant) were separated by glycerol gradient sedimentation. Subsequently, affinity selection by glutathione beads was performed for each glycerol gradient fraction, and the affinity-selected fractions were used for the western blotting analyses. The results indicate that hPrp19 complex proteins such as hPrp19, Xab2 and IBP160 are present in the IL complex. U5 snRNP specific proteins such as U5-220k and U5-116k, and U2 snRNP core protein U2B′′ were also found in the IL complex fraction. This is consistent with the result in Figure 2B, which shows the presence of U2 and U5 snRNAs in the IL complex. On the other hand, no components of the SF3a/3b complex were stably detected in the IL complex (Figure 3B, SF3b155 and 130, SF3a120 and 60), although SF3b130 was identified by MS analysis (Figure 3A). This suggests that the SF3a/3b complex has dissociated prior to IL complex formation (see ‘Discussion’ section). TFIP11 was also detected in the IL complex but not in the IS complex (Figure 3B and Figure S1B). However, the signals of hPrp43 near the IL and IS fractions and of SF3b155 and SF3b130 near the IS fraction were also observed in the control fractions, in which the same procedures were performed in the absence of tagged pre-mRNA (data not shown, see also Figure S1C). Thus, these signals could represent contamination of endogenous complexes unrelated to the IL and IS complexes. It should be pointed out, however, that hPrp43 was detected by the MS analysis (Figure 3A). hPrp43 interacts with TFIP11 and is likely to be targeted transiently at the IL complex as will be shown and discussed later. The hDBR1 protein, which catalyzes the debranching reaction of the lariat intron, was not detected in either complex. These results demonstrate that many of the proteins identified by MS analysis of the lariat–intron complex are associated with the IL complex, whereas the IS complex does not contain any of the spliceosomal U snRNAs or protein factors tested so far.
TFIP11 protein binds specifically to hPrp43 protein and mediates the disassembly of intron RNP in cooperation with hPrp43 protein
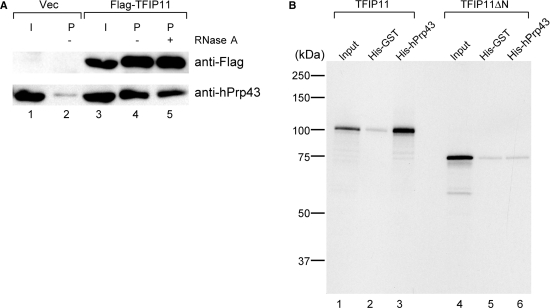
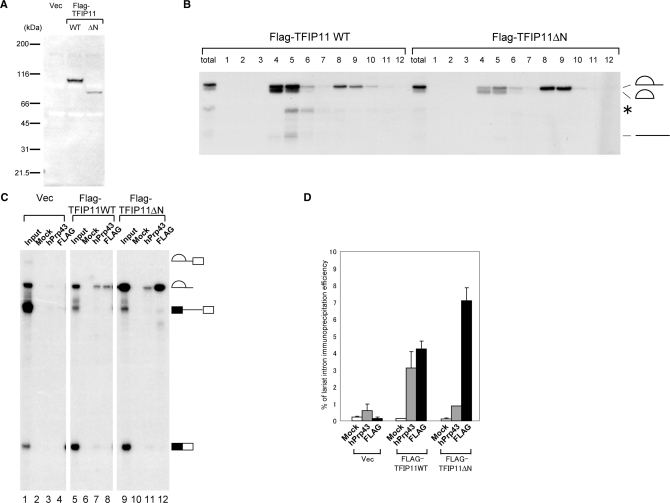
TFIP11, which was identified by MS analysis of the intron complex, was found both in the spliceosome (1,10) and in the IL complex (this study, Figure 3). Since this protein shares relatively high homology (22% identity) with yeast Ntr1 protein and was shown to be able to complement the yeast Δntr1 mutant strain (29), TFIP11 is considered to be a human homolog of yeast Ntr1. The Ntr1 protein associates with Prp43 protein and mediates the disassembly of lariat–intron RNP into U snRNPs and the lariat intron in yeast (17). Therefore using a co-immunoprecipitation experiment, we tested if TFIP11 binds specifically to hPrp43. We transiently expressed Flag-tagged TFIP11 in HEK293T cells and carried out immunoprecipitations with anti-Flag antibody from the whole-cell lysates of the transfected cells. Endogenous hPrp43 protein was efficiently co-precipitated with Flag-TFIP11 (Figure 4A, lane 4). This co-precipitation was not affected by the presence of RNase A (Figure 4A, lane 5), suggesting that TFIP11 and hPrp43 interact with each other through protein–protein interactions. In yeast cells, Ntr1 interacts with Prp43 through one of its domains called a G-patch (17,18). Since TFIP11 also has a G-patch domain at its amino-terminal region, we next sought to determine whether the G-patch domain of TFIP11 is involved in the interaction with hPrp43. We carried out in vitro binding assays with histidine (His)-tagged recombinant hPrp43 and [35S]methionine-labeled, in vitro translated TFIP11 wild-type and mutant proteins (Figure 4B). The TFIP11 wild type bound efficiently to hPrp43 but not to the control protein His-GST (lanes 1–3). When the first 50 amino acids of TFIP11, which correspond to the first half of the G-patch domain, were deleted, the resultant mutant protein, termed TFIP11ΔN, was no longer able to bind to hPrp43 (lanes 4–6). These results strongly suggest that TFIP11 interacts with hPrp43 through the G-patch domain.In analogy with the yeast system, it was plausible to hypothesize that TFIP11 mediates dissociation of U snRNPs and other proteins from the IL complex through its binding to hPrp43, prior to debranching of the lariat intron. To test this hypothesis, we used the TFIP11ΔN protein, which lacks hPrp43-binding domain, since we could expect that it would act as a dominant negative protein. Wild-type or ΔN mutant of TFIP11 was transiently expressed in HEK293T cells by transfection (Figure 5A). Nuclear extracts were prepared from the transfected cells, and in vitro splicing was performed using those extracts with δ-crystallin pre-mRNA as a substrate. When the extract from Flag-TFIP11-expressing HEK293T cells was used, the lariat intron product was most abundantly found in glycerol gradient fractions 4 and 5,which corresponded to the IS complex, but was less present in fractions 8 and 9, which corresponded to the IL complex (Figure 5B, left panel). In contrast, when the nuclear extract from the HEK293T cells that expressed Flag-TFIP11ΔN was used, the lariat intron product in the IS fractions was reduced and that in the IL fractions was increased, suggesting that the IL complex is a precursor of the IS complex and that the transition from IL to IS complexes is slow in this extract (Figure 5B, compare the right and left panels). We also carried out immunoprecipitation assays with anti-Flag tag antibody using the fractionated IL containing complex to examine the association of Flag-tagged TFIP11 proteins with lariat RNA. The results showed that both TFIP11ΔN and the wild-type TFIP11 proteins could co-precipitate with the lariat intron (Figure 5C, lanes 8 and 12, and 5D for quantification). We also used anti-hPrp43 antibody for immunoprecipitation experiments and found that the precipitation efficiency of lariat intron RNA from TFIP11ΔN expressing extract was reproducibly lower than that from the wild-type TFIP11 expressing extract (Figure 5C, lanes 7 and 11, and 5D for quantification), suggesting that hPrp43 is recruited to the intron complex through its interaction with the G-patch domain of TFIP11. These results, taken together, strongly suggest that the TFIP11 protein, in cooperation with hPrp43, mediates the disassembly of the IL complex into IS complex.
Figure 4.
The TFIP11 protein binds to hPrp43 through its amino-terminus. (A) In vivo association of hPrp43 with TFIP11. Immunoprecipitation experiments were performed using anti-Flag M2 agarose from HEK293T whole-cell extracts transfected with either Flag-vector or Flag-TFIP11 and subjected to western blot analyses. The lane marked as ‘I’ contains 10% of the precipitated proteins. (B) The first 50 amino acids of TFIP11 are required for its binding to hPrp43 in vitro. Purified His-GST and His-hPrp43 proteins were immobilized on Ni-NTA beads and incubated with in vitro translated 35S-labeled Flag-TFIP11 wild-type or ΔN mutant protein. Bound proteins were eluted with SDS–containing sample buffer, resolved by 12% SDS–PAGE and detected by autoradiography. An aliquot equivalent to 10% of the Flag-TFIP11 wild-type and ΔN mutant proteins used for binding was run in the lane marked ‘Input’. Molecular mass markers are indicated on the left side of the panel.
Figure 5.
The amino terminus of TFIP11 is required for the transition from the IL complex to the IS complex in vitro. (A) Western blotting of the extracts from the cells transfected with control plasmid (Vec), Flag-TFIP11 wild type (WT), or Flag-TFIP11 ΔN mutant (ΔN). The TFIP11 proteins were detected using a monoclonal antibody against Flag. Molecular mass markers are shown on the left. (B) An in vitro splicing reaction with chicken δ-crystallin pre-mRNA was carried out with nuclear extracts prepared from HEK293T cells expressing either the wild-type or ΔN mutant of Flag-TFIP11 proteins. The supernatant of the streptavidin agarose pull-down assay shown in Figure 1 was separated by 10–30% glycerol gradient. The RNAs in each fraction were recovered and analyzed by 6% denaturing PAGE. The lanes marked as total contain 10% of the reaction mixture used for the sedimentation. The structure of each RNA product is shown schematically on the right. The asterisk indicates the product that is likely to be produced by aberrant splicing. (C) Immunoprecipitation experiments of the RNAs from the IL complex containing fractions. An in vitro splicing reaction was performed and either anti-hPrp43 antibody or anti-Flag antibody bound to protein A-Sepharose was used for immunoprecipitation from the IL complex enriched fraction of either wild-type FLAG-TFIP11 or FLAG-TFIPΔN expressing extracts. Co-precipitated RNAs were analyzed by 6% denaturing PAGE. In the case of mock precipitation (Mock), anti-GST antibody was used. RNA products from 10% of each reaction were present in the lanes marked as ‘Input’. The schematic representation of each RNA is shown on the right of the panel. (D) Quantitation of the lariat intron RNA precipitation results shown in (C). Averages and standard deviations obtained from three independent experiments are shown.
The different sensitivities of lariat intron RNAs in IL and IS complexes to debranching activity
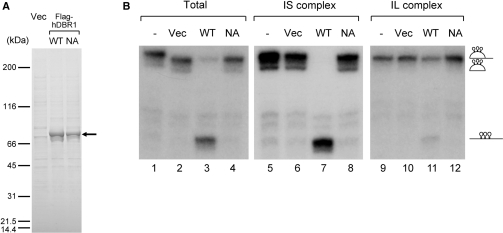
The results so far suggest that the two identified intron complexes are intermediate complexes of the post-splicing intron degradation pathway. It was expected that the lariat intron in the IS complex would be more susceptible to the debranching reaction than that in the IL complex, since U snRNPs and many proteins had already been removed. To test this hypothesis, we carried out an in vitro debranching assay with the two intron complex fractions. For this assay, recombinant Flag-hDBR1 proteins were expressed in HEK293T cells and purified through an anti-Flag M2 antibody column. In addition to the wild-type protein, a mutant hDBR1 protein, which had a substitution at amino-acid residue 84 from asparagine to alanine (N84A), was also prepared (Figure 6A). The equivalent mutation in yeast Dbr1 protein had already been demonstrated to cause the loss of debranching activity (33). When the recombinant wild-type hDBR1 protein was incubated with the intron RNPs immobilized on glutathione beads that were recovered from the supernatant of streptavidin agarose precipitation as in Figure 1B, lane 4, a fraction of the lariat intron was debranched and linearized (Figure 6B, lane 3). On the other hand, neither the mock-purified hDBR1 fraction nor the N84A mutant could cause the debranching of the lariat intron (Figure 6B, lanes 2 and 4). These results indicate that the recombinant Flag-hDBR1 protein has a debranching activity and that the point mutation at N 84 to A caused the loss of this activity. When affinity purified IL and IS complexes on glutathione beads were incubated with the hDBR1 proteins, the lariat intron in the IS complex was efficiently debranched only by the wild-type hDBR1, while the intron in the IL was quite resistant to debranching by the wild-type hDBR1 (Figure 6B, compare lanes 7 and 11). These results demonstrate that the lariat intron in the IS complex, which contains less factors, is more accessible to hDBR1 for its debranching activity. In the IL complex, it is likely that U snRNPs and other proteins identified by MS protect the branch point region of the lariat intron from hDBR1. In summary, our results strongly suggest that the IL and IS complexes are intermediate complexes of post-splicing intron degradation and that the TFIP11–hPrp43 complex mediates the transition of the IL complex to the IS complex before the debranching reaction by hDBR1.
Figure 6.
In vitro debranching assays of the lariat introns from IS and IL complex. (A) The recombinant hDBR1 proteins used for the in vitro debranching assay. Either Flag vector alone, or plasmids encoding the Flag-hDBR1 wild-type or NA mutant were transfected into HEK293T cells, and proteins were purified by anti-Flag M2 resin from whole-cell extracts. Purified proteins were separated by 5–20% gradient SDS–PAGE and detected by Coomassie brilliant blue staining. The position of Flag tagged hDBR1 proteins is indicated by an arrow. (B) In vitro debranching assays of the lariat intron RNAs recovered from IS and IL complexes. In vitro debranching assays with the purified recombinant Flag-hDBR1 proteins shown in (A). Lariat intron RNPs recovered from total supernatant. IS and IL complexes were incubated with Flag-DBR1 proteins. After the reaction, the RNAs were analyzed by 6% denaturing PAGE. The structure of RNAs is demonstrated schematically on the right of the panel.
DISCUSSION
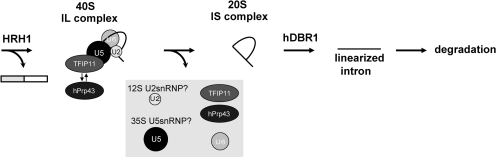
In this study we have established a purification method for the post-splicing lariat intron complex using a two-tag system (Figure 1A). By combining the purification method with glycerol gradient sedimentation analysis, we have found that the excised lariat intron is present in at least two different forms, namely IL (∼40S) and IS (∼20S) complexes (Figure 2). The IL complex contains, besides the excised lariat intron, U2, U5 and U6 snRNPs and many protein factors including hPrp19 complex proteins (hPrp19, Xab2, IBP160) and TFIP11, while the IS complex contains no such U snRNPs or proteins. Our results also suggest that the IL complex is a precursor of the IS complex (Figure 5B). Thus, during the transition from the IL to the IS complexes, all three spliceosomal U snRNAs and many protein factors are dissociated from the IL complex. It is likely from our results that a complex containing TFIP11 and hPrp43 mediates this dissociation. However, how this dissociation process is achieved is not known.
It was reported that U5 snRNP is subjected to drastic remodeling during spliceosome activation and is possibly released after splicing as a form of 35S U5 snRNP containing hPrp19 complex proteins (3). Our IL complex contained both U5 snRNA and hPrp19 complex proteins. Thus, it is plausible to hypothesize that U5 snRNP is released from the IL complex as a form of 35S U5 snRNP (Figure 7). The released 35S U5 snRNP is then converted to 20S U5 snRNP for the next round splicing reactions (3).
Figure 7.
A model for the intron turnover pathway after splicing in the nucleus. Our model for the post-splicing intron turnover pathway. See text for details.
Interestingly, SF3a/3b proteins, which are components of 17S U2 snRNP, were barely detected in the purified IL complex despite the clear presence of U2 snRNA and U2B′′ core protein (Figure 3B and Figure S1). This means that the U2 snRNP in the IL complex is in the 12S form. Recently, it has been demonstrated that the association of SF3a/3b proteins with the spliceosome is destabilized upon B to C transition (10). Our result that SF3a/3b proteins are not stable constituents of the IL complex is consistent with this paper. Taken together, we believe that U2 snRNP is released from the IL complex as a form of 12S U2 snRNP (Figure 7).
While a complex containing TFIP11 and hPrp43 is likely to mediate the IL complex disassembly, our results also suggest that TFIP11 is a stable constituent of the IL complex. However, hPrp43 had a peak in #9 fraction, whereas TFIP11 mainly existed in #8 fraction (Figure 3B). Thus, it is likely that hPrp43 is not a stable constituent of IL complex and targets the IL complex only transiently for the disassembly of the IL complex. This targeting is probably mediated by its interaction with the G-patch domain of TFIP11 (Figures 4 and 5). TFIP11 protein thus acts as a scaffold that recruits hPrp43 protein to the IL complex (Figure 7). However, the hPrp43 protein is reported to be associated with the B spliceosome as a part of 17S U2 snRNP (42). Thus, hPrp43 is thought to be released sometime between the B and IL complexes. When hPrp43 targets the IL complex for disassembly, it has to utilize a different targeting system (TFIP11) since U2 snRNP is no longer in its 17S form in the IL complex. Why such a complicated targeting system is employed is currently unknown. The resultant IS complex serves as an efficient substrate for hDBR1 leading to a complete degradation of the intron (Figure 7).
In comparison to the abundant information about the IL complex in this study, the nature of the IS complex is not well known. In yeast, Ntr1 forms a heterodimer with Ntr2 and functions as a trimer with Prp43 for the disassembly of the post-splicing complex (17,43). The resultant disassembled product is thought to be free lariat intron RNA (17,43). In our case, however, we do not think that the IS complex is devoid of all factors, although it is devoid of all the splicing factors tested in this study. The IS complex migrated as 20S, which is clearly different from the gel-purified free lariat intron (data not shown). It should also be pointed out that no human homolog of Ntr2 has been identified to date. It is still not clear whether humans have a counterpart of yeast Ntr2. A co-immunoprecipitation experiment with TFIP11 may enable us to identify the candidates for human Ntr2.
In this study, we have focused on the post-splicing complexes of general introns. It is of interest to apply our purification strategy to the isolation of post-splicing complexes of particular introns that harbor noncoding RNAs, such as snoRNAs and microRNAs. Of these noncoding RNAs, snoRNAs are known to be produced in coordination with the splicing process (22). Future experiments with these specified pre-mRNAs would help us to gain a comprehensive understanding of post-splicing intron turnover in the nucleus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CREST (to M.O.); JST; grants from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science, and Technology (to R.Y.); and the Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology (SCF) commissioned by the MEXT of Japan (to N.K.). Funding for open access charge: Ministry of Education, Sports, Science and Technology (MEXT) of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Makoto Kitabatake and Ichiro Taniguchi for their suggestions and critical comments about the manuscript. We are grateful to Tetsuro Hirose, Reinhard Luhrmann, and Michael Paine for their generous gifts of antibodies and plasmids.
REFERENCES
- 1.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 2.Will CL, Luhrmann R. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 3.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 4.Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 5.Sawa H, Ohno M, Sakamoto H, Shimura Y. Requirement of ATP in the second step of the pre-mRNA splicing reaction. Nucleic Acids Res. 1988;16:3157–3164. doi: 10.1093/nar/16.8.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirose T, Ideue T, Nagai M, Hagiwara M, Shu MD, Steitz JA. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol. cell. 2006;23:673–684. doi: 10.1016/j.molcel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Luhrmann R. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004;23:2381–2391. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 11.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 14.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 15.Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 16.Arenas JE, Abelson JN. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl Acad. Sci. USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore MJ. Nuclear RNA turnover. Cell. 2002;108:431–434. doi: 10.1016/s0092-8674(02)00645-1. [DOI] [PubMed] [Google Scholar]
- 20.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 21.ConsortiumI.H.GS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 22.Hirose T, Shu MD, Steitz JA. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 23.Ohno M, Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 24.Ono Y, Ohno M, Shimura Y. Identification of a putative RNA helicase (HRH1), a human homolog of yeast Prp22. Mol. Cell Biol. 1994;14:7611–7620. doi: 10.1128/mcb.14.11.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fouraux MA, Kolkman MJ, Van der Heijden A, De Jong AS, Van Venrooij WJ, Pruijn GJ. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA. 2002;8:1428–1443. doi: 10.1017/s1355838202021076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gee S, Krauss SW, Miller E, Aoyagi K, Arenas J, Conboy JG. Cloning of mDEAH9, a putative RNA helicase and mammalian homologue of Saccharomyces cerevisiae splicing factor Prp43. Proc. Natl Acad. Sci. USA. 1997;94:11803–11807. doi: 10.1073/pnas.94.22.11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JW, Kim HC, Kim GM, Yang JM, Boeke JD, Nam K. Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res. 2000;28:3666–3673. doi: 10.1093/nar/28.18.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HC, Kim GM, Yang JM, Ki JW. Cloning, expression, and complementation test of the RNA lariat debranching enzyme cDNA from mouse. Mol. Cells. 2001;11:198–203. [PubMed] [Google Scholar]
- 29.Herrmann G, Kais S, Hoffbauer J, Shah-Hosseini K, Bruggenolte N, Schober H, Fasi M, Schar P. Conserved interactions of the splicing factor Ntr1/Spp382 with proteins involved in DNA double-strand break repair and telomere metabolism. Nucleic Acids Res. 2007;35:2321–2332. doi: 10.1093/nar/gkm127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z, Sim J, Griffith J, Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl Acad. Sci. USA. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 32.Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 33.Salem LA, Boucher CL, Menees TM. Relationship between RNA lariat debranching and Ty1 element retrotransposition. J. Virol. 2003;77:12795–12806. doi: 10.1128/JVI.77.23.12795-12806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trembley JH, Tatsumi S, Sakashita E, Loyer P, Slaughter CA, Suzuki H, Endo H, Kidd VJ, Mayeda A. Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation. Mol. Cell Biol. 2005;25:1446–1457. doi: 10.1128/MCB.25.4.1446-1457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuyama K, Taniguchi I, Okawa K, Ohno M. Factors associated with a purine-rich exonic splicing enhancer sequence in Xenopus oocyte nucleus. Biochem. Biophys. Res. Commun. 2007;359:580–585. doi: 10.1016/j.bbrc.2007.05.144. [DOI] [PubMed] [Google Scholar]
- 37.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kataoka N, Dreyfuss G. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J. Biol. Chem. 2004;279:7009–7013. doi: 10.1074/jbc.M307692200. [DOI] [PubMed] [Google Scholar]
- 39.Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 40.Konarska MM, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 42.Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Luhrmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai RT, Tseng CK, Lee PJ, Chen HC, Fu RH, Chang KJ, Yeh FL, Cheng SC. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell Biol. 2007;27:8027–8037. doi: 10.1128/MCB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.