Abstract
Gene expression microarrays can estimate the prevalence of mRNA for thousands of genes in a small sample of cells or tissue. Organ transplant researchers are increasingly using microarrays to identify specific patterns of gene expression that predict and characterize acute and chronic rejection, and to improve our understanding of the mechanisms underlying organ allograft dysfunction. We used microarrays to assess gene expression in bronchoalveolar lavage cell samples from lung transplant recipients with and without acute rejection on simultaneous lung biopsies. These studies showed increased expression during acute rejection of genes involved in inflammation, apoptosis, and T-cell activation and proliferation. We also studied gene expression during the evolution of airway obliteration in a murine heterotopic tracheal transplant model of chronic rejection. These studies demonstrated specific patterns of gene expression at defined time points after transplantation in allografts, whereas gene expression in isografts reverted back to that of native tracheas within 2 wk after transplantation. These studies demonstrate the potential power of microarrays to identify biomarkers of acute and chronic lung rejection. The application of new genetic, genomic, and proteomic technologies is in its infancy, and the microarray-based studies described here are clearly only the beginning of their application to lung transplantation. The massive amount of data generated per tissue or cell sample has spawned an outpouring of invention in the bioinformatics field, which is developing methodologies to turn data into meaningful and reproducible clinical and mechanistic inferences.
Keywords: allograft rejection, lung transplantation, microarray
Lung and heart–lung transplants, introduced into clinical practice in 1981, have now been performed in over 20,000 individuals worldwide for whom effective medical therapy was not available (1). These procedures have been highly beneficial for many recipients, with 2-yr survival rates of approximately 70% and dramatic improvements in quality of life. Despite these remarkable successes, problems remain, and long-term survival rates for lung transplant recipients are considerably lower than those observed in kidney, heart, and liver recipients. This is due, in large part, to the development of chronic allograft rejection despite administration of immunosuppressive medications.
CHRONIC REJECTION AFTER LUNG TRANSPLANTATION
Bronchiolitis obliterans syndrome after lung transplantation is defined as a progressive, irreversible decrease in the FEV1 (2). Bronchiolitis obliterans syndrome is often accompanied by obliterative bronchiolitis, a histologic lesion characterized by inflammation and fibrosis of small airways. Obliterative bronchiolitis is usually considered to be analogous to late graft loss after transplantation of other organs—that is, they represent forms of chronic graft rejection (3). The histologic appearance and bronchoalveolar lavage (BAL) fluid and cell analyses suggest that an inflammatory process is followed by fibroproliferation in the bronchiolar walls and lumens (4).
As traditionally conceptualized, chronic rejection of the lung and other transplanted organs results from two distinct, but linked, processes: an alloimmune response directed against target cells in the organ, followed by a fibroproliferative response that results in irreversible structural changes and impaired function. However, this simple linear model of lung rejection as two discrete entities is almost certainly a vast oversimplification. In reality, chronic rejection is probably better modeled in the same way as other chronic illness states that are dependent on a large number of genetic factors (in this case of both donor and recipient origin), environmental influences, and host–environment interactions. In fact, bronchiolitis obliterans syndrome after lung transplantation has been associated with infections, gastroesophageal reflux disease, and other pathophysiologic processes that are not directly related to the alloimmune response.
The incidence of chronic rejection is highest during the first 2 yr after transplantation, but patients remain at risk indefinitely, and the cumulative risk of bronchiolitis obliterans syndrome may reach 60 to 80% between 5 and 10 yr after transplantation (5, 6). In view of this, it would be quite useful to have a relatively noninvasive, reliable means to identify patients who are at risk for chronic rejection. In addition, no therapies have been identified to date which result in recovery of lung function lost to chronic rejection, and current treatment approaches are aimed at stabilizing lung function. Therefore, identification of molecular markers of early disease, before the development of severe airflow obstruction, is an important research goal. Specifically, identification of patients during the earliest disease stages would allow them to be treated with augmented immune suppression or novel therapies. Conversely, those patients at low risk can be candidates for reduction of immunosuppressive medications.
ACUTE REJECTION IS AN IMPORTANT RISK FACTOR FOR CHRONIC REJECTION
Acute rejection occurs in 35 to 50% of lung transplant recipients, often during the first post-transplant year. Histologically, acute lung rejection is characterized by perivascular and subendothelial mononuclear infiltrates and by lymphocytic bronchitis and bronchiolitis (7). The pathogenesis of acute lung rejection is believed to be similar to acute rejection of other organ allografts: that is, primarily a manifestation of a CD8+ T-cell–mediated cytotoxic reaction initiated by recognition of graft alloantigens by CD4+ and CD8+ T lymphocytes.
Acute rejection, by itself, is usually not life- or graft-threatening, and its abnormal histology can be reversed with treatment. However, it is an important problem because it often is accompanied by irreversible decreases in lung function, and it is an important risk factor for the later development of chronic rejection. In fact, acute lung rejection histology, especially when prolonged, repeated, or severe, has been identified in multiple publications as the strongest known risk factor for chronic rejection (8–10). Recent evidence also indicates that even histologically mild acute rejection is statistically linked to future bronchiolitis obliterans syndrome (11).
THE SEARCH FOR MOLECULAR BIOMARKERS OF ACUTE AND CHRONIC LUNG REJECTION
Biomarkers are cellular, biochemical, molecular (genetic and epigenetic) alterations by which a normal or abnormal process can be recognized or monitored. In this regard, many investigators have attempted to identify diagnostic and predictive biomarkers of acute rejection and chronic rejection. These have included markers in blood, BAL fluid and cells, and exhaled air and breath condensate. “Genomic” approaches have included identification of single nucleotide polymorphisms (SNPs) that are correlated with lung transplant outcomes, and analysis of gene expression products in BAL cells and lung biopsy specimens.
INFLAMMATORY MARKERS IN BAL CELLS DURING ACUTE AND CHRONIC LUNG REJECTION
The lung allograft is ideally suited to study by BAL, which allows repeated sampling of cells and secretions in the bronchiolar microenvironment. Studies of BAL cells have provided insight into the cellular and molecular effectors of local events in the process of acute and chronic lung allograft rejection (12). Several investigators have identified markers of inflammation in BAL fluid and cells during acute rejection, including IFN-γ (13–15), interleukin (IL)-6 (16, 17), IL-4 (13, 18), perforin (19), IL-1, IL-15, and granzyme B (20). Neutrophilia and neutrophil products have also been identified (21).
Markers of fibroproliferation have also been found in BAL cells of lung recipients with bronchiolitis obliterans syndrome. For example, we determined that increased concentrations of platelet-derived growth factor (PDGF) from alveolar macrophages are observed in BAL fluid before irreversible bronchiolar obliteration. Immunochemical and in situ hybridization studies of histologic sections and BAL cells suggested that alveolar macrophages are one cellular source of PDGF (22). BAL cell transforming growth factor (TGF)-β, insulin-like growth factor-1, and IL-12 gene expression are also increased in bronchiolitis obliterans syndrome (17, 23–25). These studies indicated that BAL cells are likely to be an informative source of RNA for use in characterizing acute and chronic lung rejection. However, a lack of sensitivity and specificity has limited the clinical utility of the individual biomarkers that have been identified to date.
MULTIPLE BIOMARKERS CAN IMPROVE DIAGNOSTIC ACCURACY COMPARED WITH SINGLE-GENE ASSAYS
In an attempt to improve the specificity and sensitivity of intragraft gene expression studies for diagnosis of acute rejection, investigators have evaluated the efficacy of using gene expression patterns of several or many genes, rather than of only one gene. This approach was pioneered in renal transplant recipients by Strom and Suthanthiran, who found that the accuracy of the correlation between gene expression and acute rejection histology was enhanced by simultaneous analysis of three CTL markers—Fas ligand, perforin, and granzyme B—in renal biopsy specimens; if any two of these markers were up-regulated, the sensitivity and specificity were 100% for detection of acute rejection histology (26).
USING GENE EXPRESSION MICROARRAYS TO IDENTIFY REJECTION BIOMARKERS
The biology of acute and chronic rejection, almost certainly involves a wide spectrum of molecules (proteins, RNA) that play a role in inflammation, chemoattraction, apoptosis, T-cell activation and proliferation, fibrosis, signal transduction, and effector functions. To better understand the orchestration of these events, an assay that can effectively gauge all of these processes simultaneously would be optimal. Although measurement at the protein level would be most biologically relevant, measurement at the RNA level can be a reasonable surrogate and can be done on a genomic scale. Mapping and sequencing of whole genomes have led to the advent of assays that can directly take advantage of this information to simultaneously measure mRNA levels of tens of thousands of genes in a sample. Two methods of assaying gene expression at a genomic level are cDNA and oligonucleotide microarrays (27, 28). Both of these technologies work by immobilizing nucleotide sequences onto a surface and reading intensities of fluorescent molecules conjugated to mRNA complementary to sequences attached to the array (28, 29).
MICROARRAY GENE EXPRESSION ANALYSIS IN HUMAN TRANSPLANT RECIPIENTS
Microarrays have been used in numerous contexts by the transplant community in recent years. Gene expression signatures have been explored for diagnosis and prediction of rejection events after kidney, liver, and heart transplant. Expression patterns discriminating acute-rejection state after kidney transplant have been found in biopsy samples and peripheral blood (30–32). Expression patterns in biopsy samples suggest that there are subgroups of acute-rejection profiles with differences in immune activation, cellular proliferation, and B-cell infiltrates, and that patients with chronic allograft nephropathy, a renal transplant correlate of obliterative bronchiolitis, have an expression profile that is distinguishable from the normal and acute-rejection profiles (31). Microarray analysis has also been used to establish an expression profile for early prognosis of renal chronic allograft rejection from amplified biopsy RNA samples (33–35). Acute rejection and cytomegalovirus (CMV) infection were analyzed together in liver transplant recipients, and genes associated with T-cell activation, adhesion, and apoptosis were found to be up-regulated in both conditions (36). An association between anemia and acute kidney rejection has been explored by gene expression profiling of peripheral blood, suggesting a collection of hemoglobin synthesis and erythropoiesis genes in common (37). Self-protection in ABO-incompatible kidney transplant recipients was analyzed by microarray comparison of ABO-compatible renal graft biopsies to ABO-incompatible graft biopsies, and a set of 440 probe sets, including SMADs, protein tyrosine kinases, tumor necrosis factor-α, and mucin, was found to be significant (38). Acute rejection has also been monitored by gene expression profiling in liver (39, 40) and heart (41–43) transplant recipients through analysis of biopsies and peripheral blood, respectively.
Our group has investigated gene expression patterns in BAL cells after lung transplantation. We reported that microarray analysis of BAL cells could identify genes and gene expression patterns indicative of acute rejection of the lung allograft (44). In this work, statistical methods were used to identify genes on the microarray whose expression levels were correlated, positively or negatively, with the histopathologic findings on simultaneously obtained transbronchial lung biopsies. Biopsies were graded from 0 to 4 for “A” (perivascular inflammation) and “B” (lymphocytic bronchiolitis) scores, according to standardized nomenclature (7). To simplify the numerous possible combinations of scores, samples were assigned to one of two groups based on the sum of the A and B scoring. The “no rejection” group included samples with a combined sum of A and B scores of 0 or 1 (A0B0, A1B0, and A0B1) and the “acute rejection” samples with a sum of greater than 1 (e.g., A2B1).
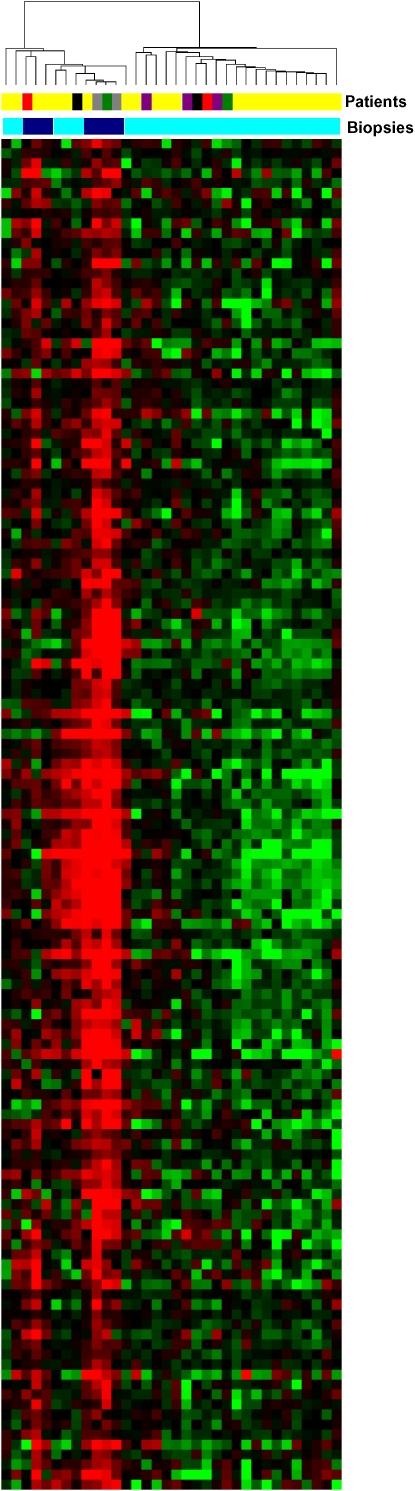
A software package developed at Stanford University, Significance Analysis of Microarrays (SAM), was used to find a set of transcripts that were differentially expressed between subjects in the two groups (45). Because of the large number of simultaneous comparisons, we controlled for false discovery rate (FDR) rather than type I error rate in selecting interesting genes. At an FDR of 0.001, a group of 135 transcripts was identified, and visualized by two-dimensional hierarchical clustering (Figure 1). Based on the clustering algorithm, we separated the samples into two major groups. One group included all seven acute-rejection samples and six no-rejection samples. The second group included the majority (21 of 27) of the no-rejection samples, and no acute-rejection samples. The five subjects represented by more than one BAL sample afforded the opportunity to compare the importance of acute-rejection status in sorting samples obtained from specific individuals. In general, samples sorted on the basis of the corresponding acute-rejection score, and not on the basis of the subject from whom the sample came.
Figure 1.
Cluster analysis of gene expression patterns of bronchoalveolar lavage (BAL) cells from lung transplant recipients with and without acute rejection. Microarray analysis of human BAL cell samples from subjects with no rejection (A+B score 0 or 1) and those with acute rejection (A+B score > 1). RNA from BAL cells was isolated and applied to Affymetrix U133A microarrays (Affymetryx, Inc., Santa Clara, CA) according to a set of standard procedures. Expression levels for 22,283 transcripts, representing 13,267 unique Entrez gene IDs, were estimated using algorithms from Microarray Suite 5.0 and GeneData Expressionist Refiner 4.0 (GeneData, Basel, Switzerland) to preprocess, normalize, and summarize probe level information. SAM (45) clustered the samples into two main groups: one which included all of the acute-rejection samples and the other which included only no-rejection samples. Subjects A and B each contributed one acute-rejection sample and one no-rejection sample; each of these samples clustered with their appropriate groups according to rejection status. Subject C contributed two acute-rejection samples, each of which clustered with other acute-rejection samples. Conversely, subject D had three no-rejection samples that all clustered within the no-rejection group. Subject E contributed two no-rejection samples: one grouped with the majority of no-rejection samples, but one clustered with the acute-rejection samples. Patients: Yellow = individual patients; red = patient A; green = patient B; gray = patient C; purple = patient D; black = patient E. Biopsies: light blue = no rejection; dark blue = acute rejection. Genes: red = up-regulated genes; green = down-regulated genes. Reprinted by permission from Reference 76.
Microarray analysis is particularly well suited for discovering complex changes in gene expression profiles under different conditions. Inherent within these complex changes is that knowledge of the dynamics of a single gene or even a small group of genes may be insufficient to understand the process occurring due to the change in conditions. A greater understanding of the overall process may be facilitated by seeing whether any pathways, networks, or groups of genes with related function are up-regulated or down-regulated as a whole. Organization of genes into pathways, functional groups, or networks is the critical step in such an analysis. Once this organization is established, it is then possible to design a metric to assign significance to a pathway, network, or group.
The Gene Ontology Consortium has provided a dynamic vocabulary of three independent gene ontology (GO) categories—molecular function, biological process, and cellular component—arranged into hierarchically nested nodes (46). Within a node, the hypergeometric distribution can be used to determine if there are more members of that node represented in a set of genes than would be expected by chance. MAPPFinder is a freely available application that uses either GO categories or curated pathways to implement the hypergeometric distribution and find pathway or GO nodes that are overrepresented by a given set of genes (47). Significant GO groups or pathways can then be visualized with the GenMAPP application, which can overlay expression information on top of pathways or GO groups to color code individual genes with statistical significance across experimental conditions.
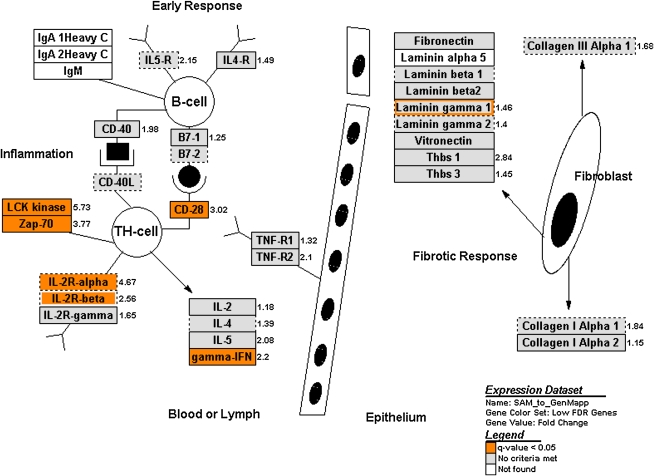
We used GenMAPP to categorize our BAL cell microarray results. A relatively large group of transcripts (FDR < 0.05; n = 885) was used in the GenMAPP analysis to develop a more robust picture of pathway activation. Pathways overrepresented by this set of transcripts included TGF-β signaling, inflammatory response, apoptosis, nucleotide G-protein–coupled receptors, peptide G-protein–coupled receptors, the Wnt family of signaling molecules, and other sets of related cytokines and chemokines. Several of these pathways are expected to be involved in acute rejection biology, whereas others represent novel observations worthy of further investigation (Figure 2).
Figure 2.
Relationship of genes significantly changed in acute rejection to process pathways. The relationship of genes significantly changed in acute rejection (as compared with no rejection) to different biological pathways was investigated using GenMAPP, a computer application designed to visualize gene expression data through preconfigured or custom-developed biological pathways and groupings of genes (47). For this analysis, a delta value of 1.05, which identified 885 genes with a false discovery rate (FDR) of 4.63%, was used to include a larger pool of candidate genes. Six of the 52 preconfigured pathways showed significant changes in expression of some of their component genes. These included pathways for transforming growth factor-β signaling, inflammatory response, apoptosis, nucleotide G-protein–coupled receptors, peptide G-protein–coupled receptors, and the Wnt family of signaling molecules. The inflammatory response pathway is shown here. Genes highlighted in orange are significantly changed; those highlighted in gray are not significantly changed; those in white were not represented on the Affymetrix Human U133A GeneChip. Reprinted by permission from Reference 76.
MICROARRAY GENE EXPRESSION ANALYSIS OF NONHUMAN TRANSPLANT MODELS
Analysis of human samples is limited by heterogeneity among patients and an inability to obtain samples at specific stages in the development of disease. To address these limitations, the effects of various interventions on grafted organs in mouse and rat models have been assessed using expression microarrays. The overall dynamics of gene expression after cardiac transplant in mice (48–51) and rats (52, 53) have been assessed. A collection of cytotoxic T-lymphocyte–associated transcripts leading to the development of tubulitis was found in mouse kidney allografts (54). Tolerated and rejected islet grafts were compared in a diabetic mouse model (55). Xenotransplantation of rat hearts in mouse recipients has been analyzed using a murine microarray (56). Ischemia–reperfusion injury has been explored in rat models of both cardiac (57, 58) and kidney (59) transplant.
Standard immunosuppression of cardiac transplantation was analyzed in a heterotopic rat model (60–62). Analyses of the gene expression profile changes of an experimental immunosuppressant on rat kidney allografts (63), anti-CD80 and anti-CD86 antibodies on mouse cardiac allografts (64), and anti-CD40 and anti-LFA antibodies (65) on mouse tracheal allografts have been tested to explore gene expression changes associated with attenuation of rejection in these models. A group of chemokine genes was found to be up-regulated in IFN-γ–knockout cardiac recipient mice compared with wild-type mice (66), and a set of genes was associated with IFN regulatory factor-1–deficient mouse cardiac recipients (67). Peripheral blood from rat liver allografts with FK506-induced tolerance was compared with peripheral blood of syngeneic liver recipients (68). Genes involved with tolerance regulation in cardiac allografts from allochimeric therapy have been identified as well (69, 70).
Our group has performed microarray analysis of gene expression in a well-characterized murine model of obliterative airway disease that reproduces characteristic features of obliterative bronchiolitis (71). This is an attractive model because the evolution of the process is more rapid and the heterogeneity involved with human samples is avoided. We used this model system to test the hypothesis that there are sequential, stereotypic gene expression patterns that reflect pathophysiologic events in the grafts and in the graft-infiltrating cells. Gene expression dynamics at three distinct time points were studied, which correlated with the previously identified evolution of the lesion: an initial stage of ischemia-induced injury; re-epithelialization; and an innate immune response followed by an adaptive immune response, which includes both cell-mediated and humoral components (72, 73).
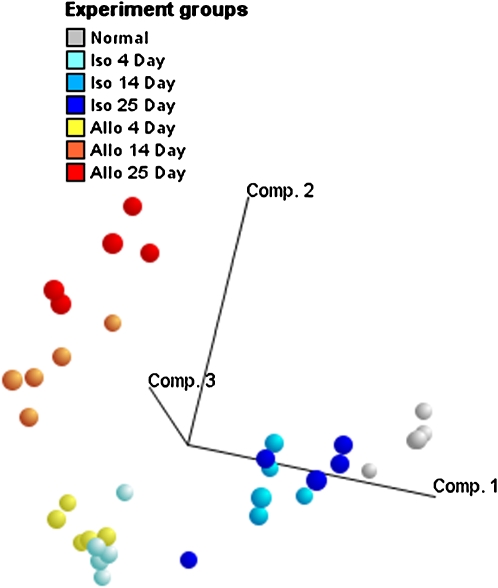
Tracheal grafts from BALB/c or C57B6 mice were transplanted into BALB/c recipients. Isografts from BALB/c donors have identical major histocompatibility (MHC) class I and II phenotypes to the recipients, whereas allografts from C57B6 mice were incompatible for both MHC class I and II antigens. Isografts and allografts were compared with each other at Days 4, 14, and 25, and with untransplanted BALB/c tracheas, which represent baseline gene expression. Comparisons with untransplanted tracheas using both hierarchical clustering and principal components analysis (Figure 3) indicated that, although both gene expression patterns from isografts and allografts change substantially from untransplanted tracheas early after transplantation, isografts revert back to the baseline pattern, whereas allografts continue to differ substantially.
Figure 3.
Comparison of gene expression patterns of isograft and allograft tracheas harvested at 4, 14, and 25 d after heterotopic transplantation. Principal components analysis was performed on gene expression measurements of all allografts, isografts, and untransplanted tracheas for the most variant transcripts across the different samples. Principal components analysis is a data-reduction technique that derives an ordered set of orthogonal components, each of which is a linear combination of the original variables, and explains the most amount of variability that has not been accounted for by prior components. The first three principal components of the most variant genes on the microarray, based on the coefficient of variation (SD/mean; n = 3,554), were plotted in three dimensions to visually assess the degree to which graft day and type explain the largest sources of variation in the data. The first three principal components accounted for 15.3, 10.7, and 6.9% of the variation in the data, respectively. Normal, untransplanted tracheas (gray) were closest to 14- and 25-d isografts when plotted according to the first three principal components, suggesting that isografts revert back to the normal baseline gene expression of the mouse trachea. Comp. = component. Reprinted by permission from Reference 77.
At the earliest time point after transplantation, although differing from the untransplanted tracheas, isografts and allografts had few gene expression differences from each other. Using SAM to compare isografts and allografts pairwise at all three time points, there were only 24 transcripts differentially expressed at Day 4 compared with over 1,000 genes at both Days 14 and 25, at the same level of statistical significance (FDR < 0.05).
Implementing a gene-wise linear modeling approach, a collection of 1,677 transcripts was found to be associated with graft type, or graft type and time interaction. Visualization using two-dimensional hierarchical clustering revealed several clusters of interest. A small number of genes were specifically more highly expressed at the 14-d point. Genes with high Day 14 expression included granzymes C and E, regenerating islet-derived 1, proprotein convertase subtilisin/kexin type 5, indoleamine-pyrrole 2,3 dioxygenase, ubiquitin D and IFN-γ, and an expressed sequence tag. In addition to a set of transcripts with high expression at both Days 14 and 25, including CD3, CD8, and T-cell receptor genes, there also was a set of transcripts that stood out for high levels of expression in 25-d allografts. These transcripts principally comprised genes related to the humoral immune response. A cluster of genes with generally higher allograft expression included many genes that were found differentially expressed at all time points in the SAM analysis. A large cluster of transcripts exhibited lower expression levels at both Days 14 and 25 in the isografts compared with allografts on those days. These clusters included epithelial cell markers and many genes with GO classifications associated with epithelial cell processes and functions. Although the particular genes found in the mouse overlap differed from the gene list found in the human microarray studies, overrepresentation of genes associated with CD8+ T-cell immune function was clearly observed in both studies; these included granzymes, T-cell receptor genes, CD3, and IFN-γ.
LIMITATIONS AND FUTURE DIRECTIONS
The application of new genetic, genomic, and proteomic technologies is in its infancy, and the microarray-based studies described here are clearly only the beginning of their application to human disease, including lung transplantation. The massive amount of data generated per tissue or cell sample has spawned an outpouring of invention in the bioinformatics field, which is still in the process of developing methodologies to turn data into meaningful clinical and mechanistic inferences. In parallel to the evolution of analytic methods, the underlying technology is also in evolution. For example, although the available microarray platforms all may accurately reflect the relative quantities of specific mRNA species in a sample, they are likely to yield different numerical results, thus limiting the ability to compare work done in different laboratories (74). For this and other reasons, microarrays are presently probably best suited as “screening tools,” which can estimate the quantity of individual and groups of gene products. Precise quantitation for diagnostic purposes is more accurately performed using real-time polymerase chain reaction or related technologies.
Important future directions for this research fall into several general areas. First, the number of patients and patient samples in studies needs to be increased greatly to improve statistical validity. This will require the establishment of collaborative study groups that share biological samples from hundreds of subjects (75). These larger studies will likely use high-throughput, low-cost technologies, such as real-time reverse transcriptase–polymerase chain reaction, to study genes that have been identified in microarray-based experiments.
Second, expression microarrays of BAL cells only describe one dimension in the chain of events between donor and recipient genotypes and clinical phenotypes. To optimize our ability to characterize patients in good and poor prognostic groups, information regarding gene expression will need to be integrated with clinical, genetic, genomic, and proteomic information.
Third, our studies to date have focused on correlation of gene expression with the concurrent histologic diagnosis of “acute rejection” versus “no rejection” (i.e., diagnostic biomarkers). Although promising, these studies require additional refinement. For example, we have excluded subjects with bronchopulmonary infections at the time of RNA sampling, which would undoubtedly influence gene expression. To apply these studies to “real world” patients, we will have to understand and correct for the influence of infections and other confounding factors on gene expression.
Finally, although the identification of diagnostic biomarkers of acute rejection is important, an equally important goal is to identify patterns of gene expression that predict poor outcomes, such as bronchiolitis obliterans syndrome and obliterative bronchiolitis (i.e., predictive biomarkers). This will require monitoring the evolution of gene expression patterns in individual subjects, and correlating these patterns with the development of the outcomes of interest. Identifying these predictive biomarkers will help realize the goal of identifying patients who are or are not at risk for poor outcomes, and intervening before their development.
Supported by NIH PPG 5PO1-AI50162-02 (principal investigator: M.I.H.) and NIH Pulmonary Training Grant 5T32HL07741 (principal investigator: David Ingbar).
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Trulock EP, Edwards LB, Taylor DO, Boucek MM, Keck BM, Hertz MI. Registry of the International Society for Heart and Lung Transplantation: twenty-second official adult lung and heart-lung transplant report—2005. J Heart Lung Transplant 2005;24:956–967. [DOI] [PubMed] [Google Scholar]
- 2.Estenne M, Maurer JR, Boehler A, Egan JJ, Frost A, Hertz M, Mallory GB, Snell GI, Yousem S. Bronchiolitis obliterans syndrome 2001: an update of the diagnostic criteria. J Heart Lung Transplant 2002;21:297–310. [DOI] [PubMed] [Google Scholar]
- 3.Estenne M, Hertz MI. Bronchiolitis obliterans after human lung transplantation. Am J Respir Crit Care Med 2002;166:440–444. [DOI] [PubMed] [Google Scholar]
- 4.Neuringer IP, Chalermskulrat W, Aris R. Obliterative bronchiolitis or chronic lung allograft rejection: a basic science review. J Heart Lung Transplant 2005;24:3–19. [DOI] [PubMed] [Google Scholar]
- 5.Cooper JD, Patterson GA, Trulock EP. Results of single and bilateral lung transplantation in 131 consecutive recipients. Washington University Lung Transplant Group. J Thorac Cardiovasc Surg 1994;107:460–470. [PubMed] [Google Scholar]
- 6.Valentine VG, Robbins RC, Berry GJ, Patel HR, Reichenspurner H, Reitz BA, Theodore J. Actuarial survival of heart-lung and bilateral sequential lung transplant recipients with obliterative bronchiolitis. J Heart Lung Transplant 1996;15:371–383. [PubMed] [Google Scholar]
- 7.Yousem S, Berry G, Cagle P, Chamberlain D, Husain A, Hruban R, Marchevsky A, Ohori N, Ritter J, Stewart S, et al. Revision of the 1990 working formulation for the classification of pulmonary allograft rejection: Lung Rejection Study Group. J Heart Lung Transplant 1996;15:1–15. [PubMed] [Google Scholar]
- 8.Bando K, Paradis IL, Similo S, Konishi H, Komatsu K, Zullo TG, Yousem SA, Close JM, Zeevi A, Duquesnoy RJ. Obliterative bronchiolitis after lung and heart-lung transplantation: an analysis of risk factors and management. J Thorac Cardiovasc Surg 1995;110:4–13. [DOI] [PubMed] [Google Scholar]
- 9.Husain AN, Siddiqui MT, Holmes EW, Chandrasekhar AJ, McCabe M, Radvany R, Garrity ER. Analysis of risk factors for the development of bronchiolitis obliterans syndrome. Am J Respir Crit Care Med 1999;159:829–833. [DOI] [PubMed] [Google Scholar]
- 10.Kroshus TJ, Kshettry VR, Savik K, John R, Hertz MI, Bolman RM III. Risk factors for the development of bronchiolitis obliterans syndrome after lung transplantation. J Thorac Cardiovasc Surg 1997;114:195–202. [DOI] [PubMed] [Google Scholar]
- 11.Khalifah AP, Hachem RR, Chakinala MM, Yusen RD, Aloush A, Patterson GA, Mohanakumar T, Trulock EP, Walter MJ. Minimal acute rejection after lung transplantation: a risk for bronchiolitis obliterans syndrome. Am J Transplant 2005;5:2022–2030. [DOI] [PubMed] [Google Scholar]
- 12.Slebos DJ, Postma DS, Koeter GH, van der Bij W, Boezen M, Kauffman HF. Bronchoalveolar lavage fluid characteristics in acute and chronic lung transplant rejection. J Heart Lung Transplant 2004;23:532–540. [DOI] [PubMed] [Google Scholar]
- 13.Iacono A, Dauber J, Keenan R, Spichty K, Cai J, Grgurich W, Burckart G, Smaldone G, Pham S, Ohori NP, et al. Interleukin 6 and interferon-gamma gene expression in lung transplant recipients with refractory acute cellular rejection: implications for monitoring and inhibition by treatment with aerosolized cyclosporine. Transplantation 1997;64:263–269. [DOI] [PubMed] [Google Scholar]
- 14.Moudgil A, Bagga A, Toyoda M, Nicolaidou E, Jordan SC, Ross D. Expression of gamma-IFN mRNA in bronchoalveolar lavage fluid correlates with early acute allograft rejection in lung transplant recipients. Clin Transplant 1999;13:201–207. [DOI] [PubMed] [Google Scholar]
- 15.Ross DJ, Moudgil A, Bagga A, Toyoda M, Marchevsky AM, Kass RM, Jordan SC. Lung allograft dysfunction correlates with gamma-interferon gene expression in bronchoalveolar lavage. J Heart Lung Transplant 1999;18:627–636. [DOI] [PubMed] [Google Scholar]
- 16.Magnan A, Mege JL, Reynaud M, Thomas P, Capo C, Garbe L, Meric B, Badier M, Bongrand P, Viard L. Monitoring of alveolar macrophage production of tumor necrosis factor-α and interleukin-6 in lung transplant recipients. Marseille and Montreal Lung Transplantation Group. Am J Respir Crit Care Med 1994;150:684–689. [DOI] [PubMed] [Google Scholar]
- 17.Magnan A, Mege JL, Escallier JC, Brisse J, Capo C, Reynaud M, Thomas P, Meric B, Garbe L, Badier M, et al. Balance between alveolar macrophage IL-6 and TGF-β in lung-transplant recipients. Marseille and Montreal Lung Transplantation Group. Am J Respir Crit Care Med 1996;153:1431–1436. [DOI] [PubMed] [Google Scholar]
- 18.Whitehead BF, Stoehr C, Wu CJ, Patterson G, Burchard EG, Theodore J, Clayberger C, Starnes VA. Cytokine gene expression in human lung transplant recipients. Transplantation 1993;56:956–961. [DOI] [PubMed] [Google Scholar]
- 19.Clement MV, Legros-Maida S, Israel-Biet D, Carnot F, Soulie A, Reynaud P, Guillet J, Gandjbakch I, Sasportes M. Perforin and granzyme B expression is associated with severe acute rejection: evidence for in situ localization in alveolar lymphocytes of lung-transplanted patients. Transplantation 1994;57:322–326. [DOI] [PubMed] [Google Scholar]
- 20.Shi R, Yang J, Jaramillo A, Steward NS, Aloush A, Trulock EP, Alexander PG, Suthanthiran M, Mohanakumar T. Correlation between interleukin-15 and granzyme B expression and acute lung allograft rejection. Transpl Immunol 2004;12:103–108. [DOI] [PubMed] [Google Scholar]
- 21.Tikkanen J, Lemstrom K, Halme M, Pakkala S, Taskinen E, Koskinen P. Detailed analysis of cell profiles in peripheral blood, bronchoalveolar lavage fluid, and transbronchial biopsy specimens during acute rejection and CMV infection in lung and heart-lung allograft recipients. Transplant Proc 1999;31:163–164. [DOI] [PubMed] [Google Scholar]
- 22.Hertz MI, Henke CA, Nakhleh RE, Harmon KR, Marinelli WA, Fox JM, Kubo SH, Shumway SJ, Bolman RM III, Bitterman PB. Obliterative bronchiolitis after lung transplantation: a fibroproliferative disorder associated with platelet-derived growth factor. Proc Natl Acad Sci USA 1992;89:10385–10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charpin JM, Stern M, Grenet D, Israel-Biet D. Insulinlike growth factor-1 in lung transplants with obliterative bronchiolitis. Am J Respir Crit Care Med 2000;161:1991–1998. [DOI] [PubMed] [Google Scholar]
- 24.Charpin JM, Valcke J, Kettaneh L, Epardeau B, Stern M, Israel-Biet D. Peaks of transforming growth factor-beta mRNA in alveolar cells of lung transplant recipients as an early marker of chronic rejection. Transplantation 1998;65:752–755. [DOI] [PubMed] [Google Scholar]
- 25.Meloni F, Vitulo P, Cascina A, Oggionni T, Bulgheroni A, Paschetto E, Klersy C, D'Armini AM, Fietta A, Bianco AM, et al. Bronchoalveolar lavage cytokine profile in a cohort of lung transplant recipients: a predictive role of interleukin-12 with respect to onset of bronchiolitis obliterans syndrome. J Heart Lung Transplant 2004;23:1053–1060. [DOI] [PubMed] [Google Scholar]
- 26.Strom TB, Suthanthiran M. Prospects and applicability of molecular diagnosis of allograft rejection. Semin Nephrol 2000;20:103–107. [PubMed] [Google Scholar]
- 27.Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet 1999;21:10–14. [DOI] [PubMed] [Google Scholar]
- 28.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet 1999;21:20–24. [DOI] [PubMed] [Google Scholar]
- 29.Eberwine J. Amplification of mRNA populations using aRNA generated from immobilized oligo(dT)-T7 primed cDNA. Biotechniques 1996;20:584–591. [DOI] [PubMed] [Google Scholar]
- 30.Akalin E, Hendrix RC, Polavarapu RG, Pearson TC, Neylan JF, Larsen CP, Lakkis FG. Gene expression analysis in human renal allograft biopsy samples using high-density oligoarray technology. Transplantation 2001;72:948–953. [DOI] [PubMed] [Google Scholar]
- 31.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, Salvatierra O. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med 2003;349:125–138. [DOI] [PubMed] [Google Scholar]
- 32.Flechner SM, Kurian SM, Head SR, Sharp SM, Whisenant TC, Zhang J, Chismar JD, Horvath S, Mondala T, Gilmartin T, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant 2004;4:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherer A, Krause A, Walker JR, Sutton SE, Seron D, Raulf F, Cooke MP. Optimized protocol for linear RNA amplification and application to gene expression profiling of human renal biopsies. Biotechniques 2003;34:546–544, 556. [DOI] [PubMed] [Google Scholar]
- 34.Scherer A, Krause A, Walker JR, Korn A, Niese D, Raulf F. Early prognosis of the development of renal chronic allograft rejection by gene expression profiling of human protocol biopsies. Transplantation 2003;75:1323–1330. [DOI] [PubMed] [Google Scholar]
- 35.Donauer J, Rumberger B, Klein M, Faller D, Wilpert J, Sparna T, Schieren G, Rohrbach R, Dern P, Timmer J, et al. Expression profiling on chronically rejected transplant kidneys. Transplantation 2003;76:539–547. [DOI] [PubMed] [Google Scholar]
- 36.Inkinen K, Lahesmaa R, Brandt A, Katajamaa M, Halme L, Hockerstedt K, Lautenschlager I. DNA microarray-based gene expression profiles of cytomegalovirus infection and acute rejection in liver transplants. Transplant Proc 2005;37:1227–1229. [DOI] [PubMed] [Google Scholar]
- 37.Chua MS, Barry C, Chen X, Salvatierra O, Sarwal MM. Molecular profiling of anemia in acute renal allograft rejection using DNA microarrays. Am J Transplant 2003;3:17–22. [DOI] [PubMed] [Google Scholar]
- 38.Park WD, Grande JP, Ninova D, Nath KA, Platt JL, Gloor JM, Stegall MD. Accommodation in ABO-incompatible kidney allografts: a novel mechanism of self-protection against antibody-mediated injury. Am J Transplant 2003;3:952–960. [DOI] [PubMed] [Google Scholar]
- 39.Tannapfel A, Geissler F, Witzigmann H, Hauss J, Wittekind C. Analysis of liver allograft rejection related genes using cDNA-microarrays in liver allograft specimen. Transplant Proc 2001;33:3283–3284. [DOI] [PubMed] [Google Scholar]
- 40.Sreekumar R, Rasmussen DL, Wiesner RH, Charlton MR. Differential allograft gene expression in acute cellular rejection and recurrence of hepatitis C after liver transplantation. Liver Transpl 2002;8:814–821. [DOI] [PubMed] [Google Scholar]
- 41.Horwitz PA, Tsai EJ, Putt ME, Gilmore JM, Lepore JJ, Parmacek MS, Kao AC, Desai SS, Goldberg LR, Brozena SC, et al. Detection of cardiac allograft rejection and response to immunosuppressive therapy with peripheral blood gene expression. Circulation 2004;110:3815–3821. [DOI] [PubMed] [Google Scholar]
- 42.Shulzhenko N, Yambartsev A, Goncalves-Primo A, Gerbase-DeLima M, Morgun A. Selection of control genes for quantitative RT-PCR based on microarray data. Biochem Biophys Res Commun 2005;337: 306–312. [DOI] [PubMed] [Google Scholar]
- 43.Morgun A, Shulzhenko N, Silva IDCG, Rampim GF, Chinellato AP, Borra RC, Gerbase-DeLima M. Differentially expressed genes in cardiac transplant biopsies and in mixed lymphocyte culture. Transplant Proc 2002;34:471–473. [DOI] [PubMed] [Google Scholar]
- 44.Gimino VJ, Lande JD, Berryman TR, King RA, Hertz MI. Gene expression profiling of bronchoalveolar lavage cells in acute lung rejection. Am J Respir Crit Care Med 2003;168:1237–1242. [DOI] [PubMed] [Google Scholar]
- 45.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet 2002;31:19–20. [DOI] [PubMed] [Google Scholar]
- 48.Christopher K, Liang Y, Mueller TF, DeFina R, He H, Haley KJ, Exley MA, Finn PW, Perkins DL. Analysis of the major histocompatibility complex in graft rejection revisited by gene expression profiles. Transplantation 2004;78:788–798. [DOI] [PubMed] [Google Scholar]
- 49.Christopher K, Mueller TF, Liang Y, Finn PW, Perkins DL. Modulation of gene expression by alloimmune networks following murine heart transplantation. Mol Genet Genomics 2004;271:687–696. [DOI] [PubMed] [Google Scholar]
- 50.Christopher K, Mueller TF, Ma C, Liang Y, Perkins DL. Analysis of the innate and adaptive phases of allograft rejection by cluster analysis of transcriptional profiles. J Immunol 2002;169:522–530. [DOI] [PubMed] [Google Scholar]
- 51.Christopher K, Mueller TF, DeFina R, Liang Y, Zhang J, Gentleman R, Perkins DL. The graft response to transplantation: a gene expression profile analysis. Physiol Genomics 2003;15:52–64. [DOI] [PubMed] [Google Scholar]
- 52.Saiura A, Mataki C, Murakami T, Umetani M, Wada Y, Kohro T, Aburatani H, Harihara Y, Hamakubo T, Yamaguchi T, et al. A comparison of gene expression in murine cardiac allografts and isografts by means DNA microarray analysis. Transplantation 2001;72:320–329. [DOI] [PubMed] [Google Scholar]
- 53.Stegall M, Park W, Kim D, Kremers W. Gene expression during acute allograft rejection: novel statistical analysis of microarray data. Am J Transplant 2002;2:913–925. [DOI] [PubMed] [Google Scholar]
- 54.Einecke G, Melk A, Ramassar V, Zhu LF, Bleackley RC, Famulski KS, Halloran PF. Expression of CTL associated transcripts precedes the development of tubulitis in T-cell mediated kidney graft rejection. Am J Transplant 2005;5:1827–1836. [DOI] [PubMed] [Google Scholar]
- 55.Berg T, Wu T, Levay-Young B, Heuss N, Pan Y, Kirchhof N, Sutherland DE, Hering BJ, Guo Z. Comparison of tolerated and rejected islet grafts: a gene expression study. Cell Transplant 2004;13:619–629. [PubMed] [Google Scholar]
- 56.Saiura A, Sugawara Y, Harihara Y, Sata M, Hamakubo T, Kodama T, Makuuchi M. Gene expression profile during acute rejection in rat-to-mouse concordant cardiac xenograft by means of DNA microarray. Transpl Int 2002;15:535–540. [DOI] [PubMed] [Google Scholar]
- 57.Amberger A, Schneeberger S, Hernegger G, Brandacher G, Obrist P, Lackner P, Margreiter R, Mark W. Gene expression profiling of prolonged cold ischemia and reperfusion in murine heart transplants. Transplantation 2002;74:1441–1449. [DOI] [PubMed] [Google Scholar]
- 58.Stegall MD, Park WD, Kim DY, Covarrubias M, Khair A, Kremers WK. Changes in intragraft gene expression secondary to ischemia reperfusion after cardiac transplantation. Transplantation 2002;74: 924–930. [DOI] [PubMed] [Google Scholar]
- 59.Yoshida T, Kurella M, Beato F, Min H, Ingelfinger JR, Stears RL, Swinford RD, Gullans SR, Tang SS. Monitoring changes in gene expression in renal ischemia-reperfusion in the rat. Kidney Int 2002;61:1646–1654. [DOI] [PubMed] [Google Scholar]
- 60.Erickson L, Crews G, Pan F, Kobayashi M, Jiang H. Gene expression profiling of acute rejection and mixed lymphocyte reaction with tacrolimus immunosuppression. Transplant Proc 2002;34:1385–1386. [DOI] [PubMed] [Google Scholar]
- 61.Erickson LM, Pan F, Ebbs A, Kobayashi M, Jiang H. Microarray-based gene expression profiles of allograft rejection and immunosuppression in the rat heart transplantation model. Transplantation 2003;76:582–588. [DOI] [PubMed] [Google Scholar]
- 62.Erickson LM, Yang XF, Pan F, Kobayashi M, Jiang H. Gene expression profiles of rat heart allografts with immunosuppression. Transplant Proc 2001;33:562–566. [DOI] [PubMed] [Google Scholar]
- 63.Fisniku O, Pan F, Wynn C, Erickson LM, Crews G, Jang MS, Sudo Y, Tamura K, Kobayashi M, Benediktsson H, et al. Protective effects of PG490-88 on chronic allograft rejection by changing intragraft gene expression profiles. Transplant Proc 2005;37:1962–1964. [DOI] [PubMed] [Google Scholar]
- 64.Matsui Y, Saiura A, Sugawara Y, Sata M, Naruse K, Yagita H, Kohro T, Mataki C, Izumi A, Yamaguchi T, et al. Identification of gene expression profile in tolerizing murine cardiac allograft by costimulatory blockade. Physiol Genomics 2003;15:199–208. [DOI] [PubMed] [Google Scholar]
- 65.Murakawa T, Kerklo MM, Zamora MR, Wei Y, Gill RG, Henson PM, Grover FL, Nicolls MR. Simultaneous LFA-1 and CD40 ligand antagonism prevents airway remodeling in orthotopic airway transplantation: implications for the role of respiratory epithelium as a modulator of fibrosis. J Immunol 2005;174:3869–3879. [DOI] [PubMed] [Google Scholar]
- 66.Saiura A, Kohro T, Yamamoto T, Izumi A, Wada Y, Aburatani H, Sugawara Y, Hamakubo T, Taniguchi T, Naito M, et al. Detection of an up-regulation of a group of chemokine genes in murine cardiac allograft in the absence of interferon-gamma by means of DNA microarray. Transplantation 2002;73:1480–1486. [DOI] [PubMed] [Google Scholar]
- 67.Erickson L, Crews G, Pan F, Fisniku O, Jang MS, Wynn C, Kobayashi M, Jiang H. Unique gene expression profiles of heart allograft rejection in the interferon regulatory factor-1-deficient mouse. Transpl Immunol 2004;13:169–175. [DOI] [PubMed] [Google Scholar]
- 68.Fujino M, Kitazawa Y, Kawasaki M, Funeshima N, Kimura H, Nakajima T, Saito H, Li XK. Differences in lymphocyte gene expression between tolerant and syngeneic liver grafted rats. Liver Transpl 2004;10:379–391. [DOI] [PubMed] [Google Scholar]
- 69.Liu D, Shen XD, Fang Z, Gao F, Semiletova N, Cao MJ, Busuttil RW, Kupiec-Weglinski JW, Ghobrial RM. Identification of early tolerance regulator genes induced by allochimeric therapy using microarray-based genomewide scan. Transplant Proc 2005;37:1942–1943. [DOI] [PubMed] [Google Scholar]
- 70.Lee BP, Mansfield E, Hsieh SC, Hernandez-Boussard T, Chen W, Thomson CW, Ford MS, Bosinger SE, Der S, Zhang ZX, et al. Expression profiling of murine double-negative regulatory T cells suggest mechanisms for prolonged cardiac allograft survival. J Immunol 2005;174:4535–4544. [DOI] [PubMed] [Google Scholar]
- 71.Hertz MI, Jessurun J, King MB, Savik SK, Murray JJ. Reproduction of the obliterative bronchiolitis lesion after heterotopic transplantation of mouse airways. Am J Pathol 1993;142:1945–1951. [PMC free article] [PubMed] [Google Scholar]
- 72.Kelly KE, Hertz MI, Mueller DL. T-cell and major histocompatibility complex requirements for obliterative airway disease in heterotopically transplanted murine tracheas. Transplantation 1998;66:764–771. [DOI] [PubMed] [Google Scholar]
- 73.Jaramillo A, Fernandez FG, Kuo EY, Trulock EP, Patterson GA, Mohanakumar T. Immune mechanisms in the pathogenesis of bronchiolitis obliterans syndrome after lung transplantation. Pediatr Transplant 2005;9:84–93. [DOI] [PubMed] [Google Scholar]
- 74.Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet 2006;22: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilkes DS, Egan TM, Reynolds HY. Lung transplantation: opportunities for research and clinical advancement. Am J Respir Crit Care Med 2005;172:944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gimino VJ, Lande JD, Berryman TR, King RA, Hertz MI. Gene expression profiling of bronchoalveolar lavage cells in acute lung rejection. Am J Respir Crit Care Med 2003;168:1237–1242. [DOI] [PubMed] [Google Scholar]
- 77.Lande JD, Dalheimer SL, Mueller DL, Hertz MI, King RA. Gene expression profiling in murine obliterative airway disease. Am J Transplant 2005;5:2170–2184. [DOI] [PubMed] [Google Scholar]