Abstract
Leukemias with MLL rearrangements are characterized by high expression of the homeobox gene MEIS1. In these studies, we knocked down Meis1 expression by shRNA lentivirus transduction in murine Mll-AF9 leukemia cells. Meis1 knockdown resulted in decreased proliferation and survival of murine Mll-AF9 leukemia cells. We also observed reduced clonogenic capacity and increased monocytic differentiation. The establishment of leukemia in transplantation recipients was significantly delayed by Meis1 knockdown. Gene expression profiling of cells transduced with Meis1 shRNA showed reduced expression of genes associated with cell cycle entry and progression. shRNA-mediated knockdown of MEIS1 in human MLL-fusion gene leukemia cell lines resulted in reduced cell growth. These results show that MEIS1 expression is important for MLL-rearranged leukemias and suggest that MEIS1 promotes cell-cycle entry. Targeting MEIS1 may have therapeutic potential for treating leukemias expressing this transcription factor.
Introduction
Leukemias with MLL gene rearrangements occur most frequently in infants and as secondary malignancies and are associated with poor outcomes. Several studies have demonstrated that both human and murine MLL-rearranged leukemias have high expression of HOXA9 and MEIS1.1–5 Recently, we found that the expression of 5′ Hox-a genes and Meis1 in murine hematopoietic cells is proportional to the level of transformation, suggesting that the MLL-fusion gene induced overexpression of these genes is central to the pathogenesis of leukemia.6 Wong et al have recently shown that retrovirally expressed MLL-fusion genes are incapable of transforming Meis1−/− murine fetal liver cells.7 In the current study, we report on Meis1 inhibition in murine Mll-AF9 knockin leukemia. The knockin model closely mimics the human disease because each cell contains a single copy of the gene, expressed from the endogenous promoter and the mice develop myeloid leukemia.8 A cell line, derived from a leukemic Mll-AF9 knockin mouse with high Meis1 expression, was used in the current study.9 We found that Meis1 is required for the growth and survival of Mll-AF9 leukemia cells in vitro and for growth of leukemia in vivo. Gene profiling data suggested mechanisms by which Meis1 might mediate these growth- and survival-promoting effects in this cell line. Finally, we show that human MLL-fusion gene leukemia cell lines also require MEIS1 for growth.
Methods
Cell culture
The 4166 cell line was established from a leukemic Mll-AF9 knockin mouse. The human cell lines were obtained from ATCC (Manassas, VA).10
Lentivirus shRNAs
Lentivirus short hairpin RNA (shRNA) clones were obtained from OpenBiosystems (Huntsville, AL); details are provided in the “Lentivirus” section in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). For ease of description, these clones were designated M23, M24, M25, M26, and M27. The manually designed construct was labeled M1456.
Cell-cycle and apoptosis analysis by flow cytometry
The 4166 cells were transduced with Meis1 shRNA at multiplicities of infection (MOI) of 10 to 100, and the cells were harvested at days 2 through 5 after transduction. Nuclei were stained with propidium iodide (PI), and analysis of nuclear DNA content was performed using the CellQuest-Pro software (BD Biosciences, San Jose, CA). Apoptosis was detected using the CaspaTag caspase activity kit (Millipore, Billerica, MA) as described previously.11
Western blotting and immunohistochemistry
Western blotting was performed using anti-Meis1 antibody (Upstate Biotechnology, Charlottesville, VA) with antiactin (Sigma-Aldrich, St Louis, MO) used as a loading control. Cell-surface marker expression was determined by immunohistochemistry as previously described.9
Myeloid colony-forming assay
Cells were cultured in methylcellulose medium under myeloid conditions, and colonies were counted and scored at 7 days as previously described.9
Transplantations
For in vivo experiments, 4166 cells were transduced with M26 and control vector at an MOI of 100. Transduced cells were grown in 4166 medium overnight without puromycin selection. Transduced or untreated 4166 cells combined with wild-type bone marrow cells were transplanted via tail vein injection into irradiated (8 Gy) C57BJ/6 mice (105 4166 cells + 5 × 105 wild-type bone marrow cells/mouse; 9 mice in each of the M26, control virus, and untreated groups). All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Minnesota.
Gene expression analysis
RNA extraction and processing and data analysis were performed as in “Supplemental materials” in Document S1. Real-time quantitative reverse-transcribed polymerase chain reaction (RT-PCR) was performed as before.6 The data from this analysis can be found on the Gene Expression Omnibus under accession number GSE14101.12
Results
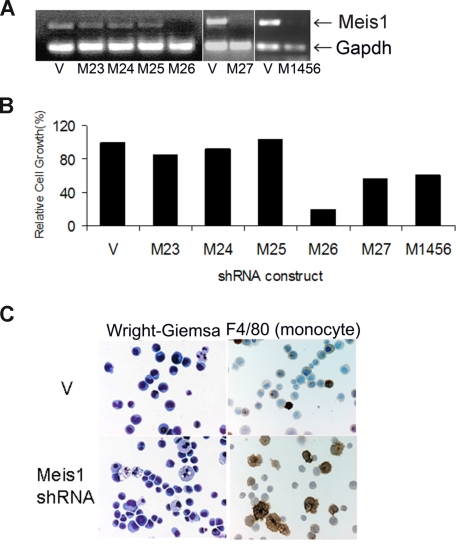
We first studied the effects of Meis1 knockdown in cell line 4166, established from a leukemic Mll-AF9 mouse. Of all the shRNA clones studied, 3 (M26, M27, and M1456) showed consistent Meis1 knockdown associated with significant inhibition of cell growth (Figure 1A,B). M26 was the most efficient and was therefore chosen for further studies. Knockdown of Meis1 expression was observed by real-time quantitative RT-PCR (P < .01, Figures S1A,B) and immunoblotting (Figure S1C). Meis1-negative murine cell lines (C1498 and BaF3) showed no significant change in growth when transduced with M26 or vector control virus (Figure S1D). These results demonstrate that the 4166 cell line is specifically sensitive to Meis1 knockdown by 3 shRNAs.
Figure 1.
shRNA lentivirus-mediated Meis1 knockdown inhibits cell growth and promotes differentiation in murine Mll-AF9 leukemia. The 4166 cells were transduced for 5 days with the various lentivirus constructs and cultured in the presence of puromycin as described. (A) RT-PCR for Meis1 and Gapdh (housekeeping gene). (B) Bar graph depicting cell growth after Meis1 shRNA transduction compared with vector control, the latter being 100%. (C) By light microscopy, Meis1 knockdown resulted in monocytic differentiation. Staining for the monocyte-specific marker F4/80 showed increased uptake with Meis1 knockdown. Slides were viewed with a Nikon Eclipse 80i microscope (Nikon, Melville, NY) at a 10×/0.3 NA, WD 16.0 mm objective with fixed cells imaging medium. Images were acquired using a Nikon Digital Sight DS-5M-L1 camera and were processed using Nikon DS-L1 image acquisition software. Image adjustment was performed using Adobe Photoshop CS2 (Adobe Systems, San Jose, CA).
The growth potential of myeloid progenitors was assessed by their ability to form colonies in semisolid media.13 In 7-day cultures, M26-transduced cells formed significantly fewer colonies compared with vector-transduced cells (Figure S2A). Colonies formed by the MLL-fusion gene carrying hematopoietic cells display a dense morphology, indicative of a differentiation block.9,13 Meis1 knockdown resulted in reversal of this differentiation block in 4166 cells, as evidenced by a decrease in dense colonies compared with controls (Figure S2B). By light microscopy, Wright-Giemsa–stained 4166 cells reveal a blast-like morphology with a high nuclear-to-cytoplasmic ratio. With Meis1 knockdown, we observed appearance of monocyte-like morphology with an increase in cell size and a distinct increase in the amount of granular cytoplasm (Figure 1C left panel). These large cells stained positive for the monocyte marker F4/80, whereas control cells were negative (Figure 1C right panel). Overall, these results demonstrate that Meis1 is critical to the maintenance of an undifferentiated state in leukemic cells.
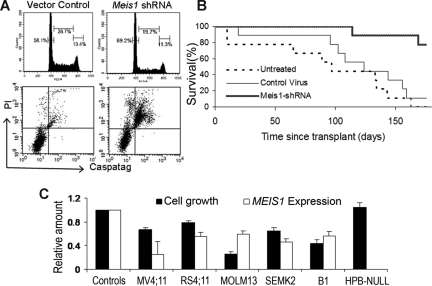
To discern the mechanisms by which Meis1 inhibition leads to reduced cell growth, we performed cell-cycle and apoptosis analyses. Flow cytometric analysis of PI-stained nuclei showed that Meis1 knockdown led to a cell-cycle arrest in the G0/G1 phase. As shown in Figure 2A, M26-transduced cells showed an increase in G0/G1 and a decrease in the proportions of S phase and G2/M nuclei. Meis1 knockdown also resulted in increased apoptosis, as evidenced by increased uptake of PI and a stain for activated caspases (CaspaTag) by M26-transduced cells compared with control cells (Figure 2A bottom panel). These results indicate that Meis1 is required for proliferation and survival of 4166 leukemia cells.
Figure 2.
Meis1 knockdown induces cell-cycle arrest and apoptosis and delays establishment of leukemia in vivo. The 4166 cells were transduced with M26 or control virus as described and cultured for 48 hours without puromycin. (A) Top panel: Nuclei were extracted and stained with PI. Analysis of DNA content by flow cytometry showed an increase in the proportion of G0/G1 nuclei (left peak) in M26-transduced cells compared with control virus. Bottom panel: Increased apoptosis was evident by an increase in the uptake of PI and the pan-activated caspase maker CaspaTag. (B) Lethally irradiated mice were given 4166 cells that were either untreated or transduced with M26 or control virus as described. Mice were killed when showing signs of distress. The graph shows a survival analysis comparing the 3 groups (9 mice per group). Mice receiving M26-transduced cells had significantly prolonged survival (P < .001). (C) MEIS1 knockdown inhibits growth of human MLL-fusion gene leukemia cell lines. The cell lines indicated were transduced with MEIS1 shRNA lentivirus or control lentivirus for 3 to 5 days as described at MOI of 10 to 50. Bars represent relative cell growth and MEIS1 expression compared with controls (mean ± SE; n = 3; P < .05 except for HPB-NULL).
We next investigated the role of Meis1 in the initiation and maintenance of leukemia in vivo. Lethally irradiated mice were transplanted with 4166 cells untreated or 1 day after transduction with Meis1-shRNA or vector control lentiviruses. Mice were killed when displaying signs of leukemic progression. At necropsy, mice showed leukocytosis and splenomegaly, indicating an infiltrative acute leukemia. As shown in Figure 2B, most of the mice receiving untreated (9 of 9) or control vector-transduced (8 of 9) cells died of leukemia, whereas 80% of those receiving Meis1-shRNA–transduced cells (7 of 9) were alive and well at day 170 after transplantation. A survival analysis showed that Meis1-shRNA significantly delayed the development of leukemia in recipient mice (Figure 2B; P < .001, log rank test).
To characterize the molecular effects of Meis1 knockdown, we performed gene expression profiling of leukemia cells with and without Meis1 expression (see the “Gene expression analysis” section in Document S1). The results showed that Meis1 knockdown resulted in decreased expression of 808 genes (1053 probe sets, Table S1). Of note were genes regulating DNA replication and cell-cycle entry, including Cdk2, Cdk6, Cdkn3, Ccna2, Cdc7, Cdc42, Rbl1, and Wee1.14–17
To assess the significance of MEIS1 in human MLL-rearranged leukemia, we performed lentivirus-shRNA–mediated knockdown experiments with human leukemia cell lines with MLL rearrangements. As shown in Figure 2C, 5 different MLL-rearranged cell lines (MV4;11, RS4;11, MOLM13, SEMK2, and B1) showed decreased cell growth with MEIS1 knockdown, whereas the MEIS1-negative cell line HPB-NULL did not show growth inhibition.
Overall, our results demonstrate that Meis1 is central to the maintenance of murine Mll-AF9 and human MLL-rearranged leukemias. We also show that inhibition of Meis1 induces cell-cycle arrest as well as cell death in these leukemia cells. Inhibition of MEIS1-regulated pathways is a promising approach to the development of future therapy.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01-CA087053, J.H.K.; K08-CA122191, A.R.K.), the Leukemia Research Fund (A.R.K.), and the Children's Cancer Research Fund (J.H.K., A.R.K.). The Masonic Cancer Center shared resources in Biostatistics/Informatics and Flow Cytometry, and the Minnesota Supercomputing Institute was critical to this project.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.R.K. designed experiments, performed data analysis, and wrote the manuscript; Q.L. performed lentivirus and cell growth experiments; W.A.H. performed mouse transplantations; W.C. performed colony assays; T.S. performed microarray and RT-PCR experiments; Q.Y. performed cell-cycle and apoptosis assays; E.A.L. and B.J.K. performed real-time PCR experiments; B.W. performed statistical analysis of microarray data; and J.H.K. supervised the research and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John Kersey, 554E Masonic Cancer Research Building, 425 East River Road, Minneapolis, MN 55455; e-mail: kerse001@umn.edu.
References
- 1.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 2.Rozovskaia T, Ravid-Amir O, Tillib S, et al. Expression profiles of acute lymphoblastic and myeloblastic leukemias with ALL-1 rearrangements. Proc Natl Acad Sci U S A. 2003;100:7853–7858. doi: 10.1073/pnas.1132115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 4.Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- 5.Zeisig BB, Milne T, Garcia-Cuellar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W, Kumar AR, Hudson WA, et al. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13:432–440. doi: 10.1016/j.ccr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobson CL, Warren AJ, Pannell R, et al. The Mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 1999;18:3564–3574. doi: 10.1093/emboj/18.13.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson JJ, Chen W, Hudson W, et al. Prenatal and postnatal myeloid cells demonstrate stepwise progression in the pathogenesis of MLL fusion gene leukemia. Blood. 2003;101:3229–3235. doi: 10.1182/blood-2002-05-1515. [DOI] [PubMed] [Google Scholar]
- 10.Stong R, Kersey J. In vitro culture of leukemic cells in t(4;11) acute leukemia. Blood. 1985;66:439–443. [PubMed] [Google Scholar]
- 11.Yao Q, Nishiuchi R, Li Q, Kumar AR, Hudson WA, Kersey JH. FLT3 expressing leukemias are selectively sensitive to inhibitors of the molecular chaperone heat shock protein 90 through destabilization of signal transduction-associated kinases. Clin Cancer Res. 2003;9:4483–4493. [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005;24:2909–2915. doi: 10.1038/sj.onc.1208618. [DOI] [PubMed] [Google Scholar]
- 15.Woo RA, Poon RY. Cyclin-dependent kinases and S phase control in mammalian cells. Cell Cycle. 2003;2:316–324. [PubMed] [Google Scholar]
- 16.Blanchard JM. Cyclin A2 transcriptional regulation: modulation of cell cycle control at the G1/S transition by peripheral cues. Biochem Pharmacol. 2000;60:1179–1184. doi: 10.1016/s0006-2952(00)00384-1. [DOI] [PubMed] [Google Scholar]
- 17.Narumiya S, Oceguera-Yanez F, Yasuda S. A new look at Rho GTPases in cell cycle: role in kinetochore-microtubule attachment. Cell Cycle. 2004;3:855–857. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.