Abstract
In plants, cell wall placement during cytokinesis is determined by the position of the preprophase band (PPB) and the subsequent expansion of the phragmoplast, which deposits the new cell wall, to the cortical division site delineated by the PPB. New cell walls are often incorrectly oriented during asymmetric cell divisions in the leaf epidermis of maize (Zea mays) discordia1 (dcd1) mutants, and this defect is associated with aberrant PPB formation in asymmetrically dividing cells. dcd1 was cloned and encodes a putative B'' regulatory subunit of the PP2A phosphatase complex highly similar to Arabidopsis thaliana FASS/TONNEAU2, which is required for PPB formation. We also identified alternative discordia1 (add1), a second gene in maize nearly identical to dcd1. While loss of add1 function does not produce a noticeable phenotype, knock down of both genes in add1(RNAi) dcd1(RNAi) plants prevents PPB formation and causes misorientation of symmetric and asymmetric cell divisions. Immunolocalization studies with an antibody that recognizes both DCD1 and ADD1 showed that these proteins colocalize with PPBs and remain at the cortical division site through metaphase. Our results indicate that DCD1 and ADD1 function in PPB formation, that this function is more critical in asymmetrically dividing cells than in symmetrically dividing cells, and that DCD1/ADD1 may have other roles in addition to promoting PPB formation at the cortical division site.
INTRODUCTION
Plant cells are surrounded by rigid walls that constrain cell movement. Consequently, the shapes of plant cells and organs are determined solely by cell division and cell expansion. Both of these processes depend on distinct microtubule arrays associated with different stages of the cell cycle. The interphase cortical microtubule array regulates the direction of cell expansion, while the plane of division in most plant cells is determined by the sequential formation and placement of three other cytoskeletal structures: the preprophase band (PPB), the mitotic spindle, and the phragmoplast. In a process unique to plants, the plane of cell division is specified prior to mitosis and is revealed by the position of the PPB (Pickett-Heaps and Northcote, 1966a, 1966b; Gunning, 1982). PPBs are cortical rings of parallel microtubules and actin microfilaments that circumscribe the future plane of division in most somatic plant cells. The PPB is thought to modify the mother cell cortex to create a cortical division site that is later recognized by the cytokinetic apparatus, the phragmoplast (Mineyuki, 1999; Van Damme and Geelen, 2008; Wright and Smith, 2008). At the end of prophase, the PPB is disassembled as the mitotic spindle forms. The spindle forms so that its axis is perpendicular to that of the former PPB and the eventual division plane of the cell. After separation of the chromosomes, the phragmoplast, which is composed of two antiparallel arrays of microtubules and microfilaments, arises between the daughter nuclei and expands centrifugally toward the mother cell cortex directing the deposition of the new cell plate (Jürgens, 2005). The new cell plate fuses with the mother cell cortex at the former location of the PPB. Although the basic role of these three cytoskeletal structures is clear, many questions remain regarding the molecular cues that regulate their formation, spatial positioning, and function.
In addition to microtubules and microfilaments, a small number of proteins involved in the spatial regulation of cytokinesis have been identified in Arabidopsis thaliana and maize (Zea mays). Four proteins, FASS/TONNEAU2 (FASS), TONNEAU1A and TONNEAU1B (TON1), and MICROTUBULE ORGANIZATION1 (MOR1) are known to promote PPB formation. Arabidopsis FASS encodes a putative regulatory B'' subunit of the Thr/Ser phosphatase PP2A, and fass mutants completely lack PPBs (Traas et al., 1995; McClinton and Sung, 1997; Camilleri et al., 2002). Similarly, Arabidopsis mutants lacking TON1A/B, two similar proteins related to a human centrosomal protein, also fail to make PPBs (Azimzadeh et al., 2008). MOR1 encodes a microtubule binding protein that promotes microtubule lengthening, and a subset of cells in Arabidopsis mor1 mutants lacks PPBs (Whittington et al., 2001; Kawamura et al., 2006). The absence of PPBs in fass and ton1 mutants is associated with randomly oriented cell divisions, supporting the hypothesis that the PPB is critical for division plane establishment (Traas et al., 1995; Camilleri et al., 2002; Azimzadeh et al., 2008). However, the nature of the cortical division site and the role of the PPB in establishment of this site are poorly understood. Currently, only two negative markers and two positive markers of the cortical division site are known. Both actin and an Arabidopsis kinesin called KCA1 are localized to the cortex in dividing cells but are depleted at the cortical division site during mitosis and cytokinesis (Cleary et al., 1992; Liu and Palevitz, 1992; Sano et al., 2005; Vanstraelen et al., 2006). By contrast, TANGLED (TAN) and RanGAP1 positively mark the cortical division site throughout mitosis and cytokinesis in Arabidopsis (Walker et al., 2007; Xu et al., 2008). Misorientation of cell divisions in Arabidopsis and maize tan mutants and in Arabidopsis plants lacking RanGAP1 function supports a role for these proteins in maintaining the identity of the cortical division site after breakdown of the PPB (Smith et al., 1996; Cleary and Smith, 1998; Walker et al., 2007). Two functionally redundant Arabidopsis kinesins, PHRAGMOPLAST-ORIENTING KINESIN1 (POK1) and POK2, are also required for the spatial regulation of cytokinesis. POK1 interacts with Arabidopsis TAN1 and RanGAP1 and is required for their localization at the cortical division site (Müller et al., 2006; Xu et al., 2008). Together, these observations suggest that proteins initially associated with the PPB modify the cortex at the cortical division site so that the region can be identified by the expanding phragmoplast during cytokinesis.
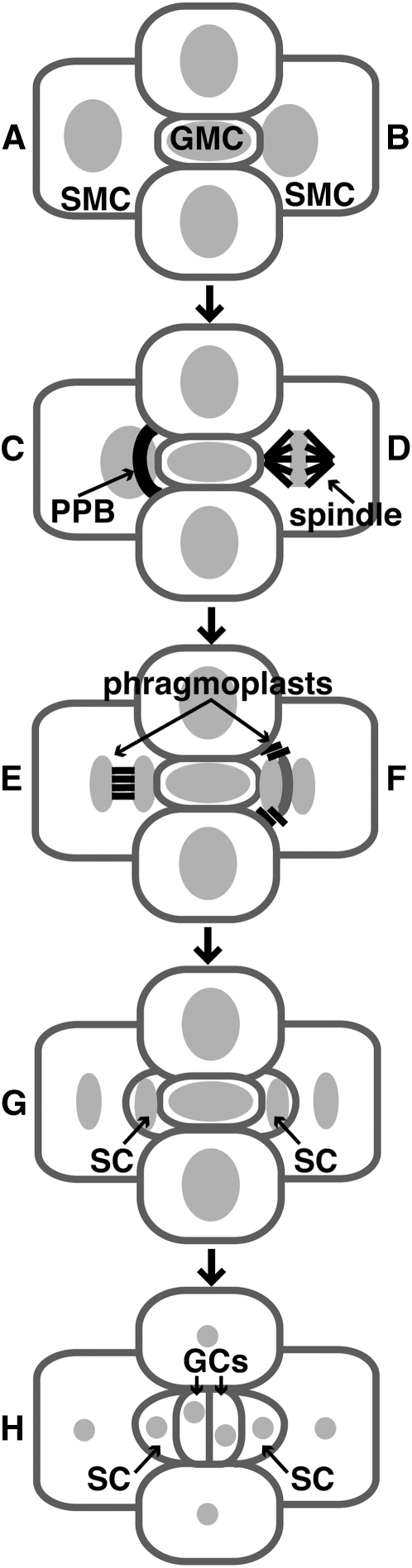
To further elucidate mechanisms governing the spatial regulation of cytokinesis, we isolated mutations disrupting normal planes of cell division in the maize leaf epidermis and characterized their phenotypes. Two such mutations, discordia1 (dcd1) and dcd2, specifically misorient asymmetric divisions, including those that produce stomatal complexes (Gallagher and Smith, 1999). Stomatal complex formation in maize is initiated with the formation of a guard mother cell (GMC; Figure 1A). The GMC apparently induces the polarization of each of its lateral neighbors, the subsidiary mother cells (SMCs), causing the migration of the SMC nucleus to a position adjacent to the GMC (Figure 1B) and subsequent formation of an asymmetric PPB that predicts the future SMC division plane (Figure 1C). As this PPB disassembles, the spindle forms and is orientated so that one pole remains associated with the cortex adjacent to the GMC (Figure 1D). After chromosome separation, the phragmoplast arises between the daughter nuclei (Figure 1E) and expands around the daughter nucleus adjacent to the GMC to reach the cortical division site previously occupied by the PPB separating the new subsidiary cell from its sister pavement cell (Figures 1F and 1G). Following the asymmetric SMC divisions, the GMC divides symmetrically producing the guard cells and completing the formation of the four cells that make up each stomatal complex (Figure 1H; Stebbins and Shah, 1960; Cho and Wick, 1989; Gallagher and Smith, 1999; Galatis and Apostolakos, 2004; Panteris et al., 2007). In dcd1 and dcd2 mutants, 30 to 50% of the SMCs divisions are misoriented, producing abnormally shaped stomatal subsidiary cells (Gallagher and Smith, 1999). Analysis of the mitotic microtubule arrays in dcd1 and dcd2 mutants showed that PPBs were present and oriented normally, whereas phragmoplasts were often misguided, suggesting a function for these genes in guidance of phragmoplasts to the former PPB site in asymmetrically dividing cells (Gallagher and Smith, 1999).
Figure 1.
Stomatal Complex Formation in Maize.
(A) The incipient stomatal complex consists of a GMC flanked by two SMCs.
(B) The SMC nucleus migrates to the lateral cell wall adjacent to the GMC.
(C) A PPB forms predicting an asymmetric cell division.
(D) A mitotic spindle separates chromosomes.
(E) and (F) An early phragmoplast forms (E) and is guided to the former site of the PPB during phragmoplast expansion (F).
(G) New cell walls define subsidiary cells (SCs).
(H) The GMC divides symmetrically to produce two equally sized guard cells (GCs). For illustrative purposes only, the stages are shown alternating between two SMCs, but in actuality, all SMCs progress through all stages.
Here, we report that dcd1 and a highly similar gene, alternative discordia1 (add1), are the maize homologs of Arabidopsis FASS. Consistent with the phenotype of Arabidopsis fass mutants, maize plants deficient in both DCD1 and ADD1 have a severe phenotype characterized by defects in PPB formation. DCD1 and ADD1 colocalize with PPBs and remain at the cortical division site through metaphase, suggesting that they may have roles in addition to promoting PPB formation.
RESULTS
dcd1 and add1 Encode Nearly Identical Proteins Related to Arabidopsis FASS
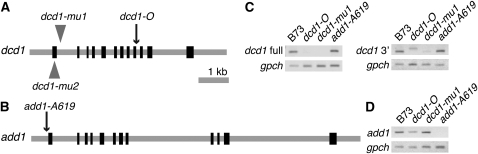
The originally characterized dcd1 mutant (dcd1-O) was identified in a screen of ethyl methanesulfonate–mutagenized maize plants (Gallagher and Smith, 1999). To facilitate cloning of the gene, we isolated two Mutator1 (Mu1) insertion alleles of dcd1 (dcd1-mu1 and dcd1-mu2) via a noncomplementation screen. The dcd1 gene was cloned by transposon tagging using the dcd1-mu1 allele. To confirm that dcd1 had been correctly identified, the gene was fully sequenced in all three mutant backgrounds. Mu1 insertions were found in the first intron in dcd1-mu1 and the first exon in dcd1-mu2, while dcd1-O changes the splice donor site immediately following exon 9 (Figure 2A). Failure to splice out intron 9 introduces 17 incorrect amino acids and a premature stop codon resulting in a truncated protein (324 versus 486 amino acids). To determine if any of these alleles represent a complete loss of function, RT-PCR was used to assess the presence or absence of dcd1 mRNA in plants homozygous for the alleles used in this study. Primers that amplify the full-length message in wild-type plants did not detect any message in the dcd1-mu1 mutants, and only very low levels of a larger-than-normal message in the dcd1-O mutants consistent with a splicing defect (Figure 2C). Primers located downstream of the Mu1 insertion detected reduced message levels in dcd1-mu1 plants compared with the wild type and revealed more clearly the larger-than-normal dcd1-O mRNA (Figure 2C). These results suggest that the dcd1 mutations severely compromise dcd1 function by encoding truncated and/or defective mRNAs that accumulate at much reduced levels.
Figure 2.
Gene Models and RT-PCR Analysis of dcd1 and add1.
(A) and (B) Gray lines indicate genome sequence, and black boxes represent exons 1 to 12. Triangles and arrows indicate the locations of the genetic lesions in dcd1 and add1.
(A) Gene model of maize dcd1.
(B) Gene model of maize add1.
(C) RT-PCR of dcd1 and gpch in the indicated genetic backgrounds. dcd1 full primers amplify the full-length message, while dcd1 3′ primers amplify only exons 8 to 12.
(D) RT-PCR of add1 and gpch in the indicated genetic backgrounds. add1 primers amplify exons 11 and 12. gpch is a 419-bp fragment of glyceraldehyde-3-phosphate dehdrogenase subunit C and serves as an amplification control.
BLAST analysis of the predicted DCD1 protein sequence revealed that it encodes a predicted B'' regulatory subunit of a PP2A phosphatase that is 86% identical to Arabidopsis FASS (Altschul et al., 1990; Camilleri et al., 2002). fass mutants are dwarfed and sterile with defects in cell division orientation associated with a complete lack of PPBs in all cell types examined (Traas et al., 1995; Camilleri et al., 2002). This phenotype differs dramatically from that observed in dcd1 mutants, which make PPBs and are morphologically normal aside from misoriented asymmetric cell divisions. However, in the course of assembling the genomic sequence of dcd1, we identified a closely related gene we named add1 (Figure 2B). At the amino acid level, ADD1 is 96% identical to DCD1, suggesting these proteins might be functionally redundant, providing a possible explanation for the mild phenotype of the dcd1 maize mutants compared with the fass mutants of Arabidopsis.
dcd1 and add1 Function Redundantly to Promote PPB Formation in Symmetrically and Asymmetrically Dividing Cells
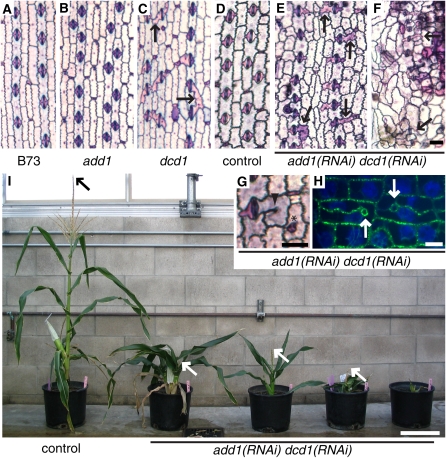
We sought a loss-of-function allele of add1 to permit functional analysis of this gene. We first examined our collection of mutants with phenotypes similar to dcd1, hypothesizing that an add1 mutant would have a dcd1-like phenotype. We sequenced the add1 coding region and used RT-PCR to evaluate mRNA levels in two such mutants, dcd2 (Gallagher and Smith, 1999) and dcd3 (unpublished data, L.G. Smith). Although we found that add1 does not correspond to either dcd2 or dcd3, this analysis led to the fortuitous discovery that the A619 wild-type inbred background contains an allele of add1 that produces no detectable mRNA (Figure 2D). The add1-A619 allele contains a G deletion 33 bp upstream of the add1 start site relative to the B73 allele, though it is not clear whether this single base pair insertion by itself affects add1 expression. Analysis of the cell pattern of the add1 leaf epidermis did not reveal any cell division defects (cf. Figures 3A and 3B, Table 1), indicating that DCD1, but not ADD1, is sufficient for correctly oriented asymmetric cell divisions, while DCD1 and ADD1 are both sufficient for correctly orientated symmetric cell divisions.
Figure 3.
Phenotype of dcd1 Mutants, add1 Mutants, and add1(RNAi) dcd1(RNAi) Plants.
(A) to (G) Epidermal peels stained with Toluidine Blue O to highlight cell outlines. Select abnormally shaped or divided subsidiary cells are indicated with black arrows. (A) to (F) are on the same scale.
(A) B73 (wild type) with normally shaped subsidiary cells.
(B) add1-A619 with normally shaped subsidiary cells.
(C) dcd1-mu1 with a mixture of normally and abnormally shaped subsidiary cells.
(D) Herbicide marker-only transformed control with normally shaped subsidiary cells.
(E) add1(RNAi) dcd1(RNAi) showing a relatively weak phenotype with normally and abnormally shaped subsidiary cells.
(F) add1(RNAi) dcd1(RNAi) showing a strong phenotype with abnormally shaped pavement and subsidiary cells.
(G) add1(RNAi) dcd1(RNAi) cells with a cell wall stub (arrowhead) and a cell within a cell (asterisk).
(H) Tubulin immunofluorescence (green) highlights the outlines of add1(RNAi) dcd1(RNAi) cells with incomplete walls that terminate in loops (white arrows); DNA stained with propidium iodide (false-colored blue).
(I) Mature control and add1(RNAi) dcd1(RNAi) plants. Herbicide marker-only transformed control plant is on the left; the remainder are add1(RNAi) dcd1(RNAi) plants that show increasingly severe growth phenotypes from left to right. Arrows indicate the top of the tassel (when visible).
Bars = 50 μm in (F) and (G), 10 μm in (H), and 20 cm in (I).
Table 1.
dcd1 Mutants Have Defects in PPB Formation in Dividing SMCs but Not in Symmetrically Dividing Cells
| Abnormal SMC PPBs
|
||||
|---|---|---|---|---|
| Genotype | Abnormal Sym. PPBsa | Completeb | Incompletec | Abnormal Mature SCsd |
| B73 | 4.4% n = 45 | 1.9% n = 52 | 1.9% n = 52 | 0.3% n = 1489 |
| add1-A619 | 6.1% n = 49 | 4.4% n = 46 | 0% n = 46 | 0.13% n = 1485 |
| dcd1-0 | 2% n = 50 | 16.6%* n = 48 | 35.4%** n = 48 | 25.5%** n = 1991 |
| dcd1-mu1 | 0% n = 50 | 15.5%* n = 71 | 39.4%** n = 71 | 32.3%** n = 1655 |
All cells analyzed were in prophase with condensed chromosomes. Tabulated data were obtained from two (symmetric), three (SMC), or five (SC) independent plants. P values were determined by comparing B73 values to mutant values using Fisher's exact test. *Associated P values < 0.05, indicating significant difference from the wild type. **Associated P values < 0.001, indicating significant difference from wild type.
PPBs were weak or sparse.
PPBs were frayed or bundled, but encircled the SMC completely (e.g., Figure 6C, 2).
PPBs were frayed or bundled and failed to encircle the SMC completely (e.g., Figure 6C, 3).
Determined by counting abnormally shaped SMCs in Toluidine Blue O–stained epidermal leaf tissue.
To determine the phenotype of plants lacking both dcd1 and add1, we generated dcd1 add1 double mutants by crossing dcd1-O and A619 plants. Of the 32 dcd1 homozygotes identified in the F2 generation, none were homozygous for the add1-A619 allele, suggesting that the double homozygotes are not viable. Thus, we were unable to use these double mutants to analyze the roles of DCD1 and ADD1 during cell divisions in adult leaves. As an alternative approach to generating plants depleted for dcd1 and add1, an RNA interference (RNAi) construct targeting add1 and a separate construct containing an herbicide marker were cobombarded into maize callus. Due to the high similarity at the nucleotide level between dcd1 and add1 in the targeted region (96% identity), we anticipated that the RNAi construct would reduce both add1 and dcd1 message levels. Since the severity of RNAi phenotypes typically varies from plant to plant, we expected that this experiment would result in a phenotypic series that would allow some plants to survive past embryogenesis. Sixteen independently transformed lines, each represented by multiple plants derived from the same callus, were obtained that showed the phenotypes described below. Those with the most severe phenotypes could not be propagated, so our phenotypic analyses used T0 plants to sample the full range of RNAi phenotypes.
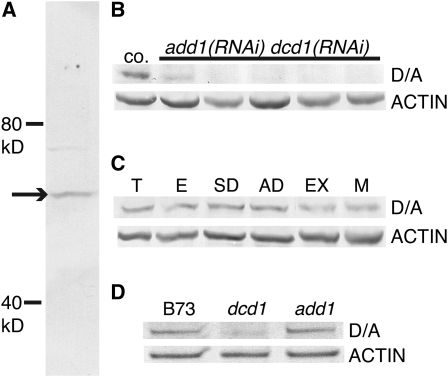
To determine whether the add1 RNAi construct was reducing ADD1 and DCD1 protein levels, we raised an antibody against a peptide that corresponds to identical sequences at the C termini of both DCD1 and ADD1. On immunoblots, this antibody predominantly recognizes a single protein band of ∼55 kD, the predicted molecular mass of DCD1 and ADD1 (Figure 4A). Analysis of five independent add1(RNAi) dcd1(RNAi) transformants with strong phenotypes showed that this protein band was depleted in one and undetectable in the other four (Figure 4B). In addition to confirming the action of the RNAi construct, the immunoblot experiment also demonstrates the specificity of the peptide antibody for DCD1/ADD1.
Figure 4.
Immunoblot Analysis of DCD1 and ADD1 in Extracts from Maize Leaf Tissue Enriched in Dividing Cells.
(A) The affinity-purified anti-DCD1/ADD1 antibody recognizes a single 55-kD band (arrow) in B73.
(B) to (D) For each immunoblot, all lanes were loaded with the same amount of protein. The blots were probed first with the DCD1/ADD1 antibody and then with an actin antibody as a loading/transfer control.
(B) DCD1/ADD1 protein (D/A) is detectable in an herbicide marker-only transformed plant (co.) but is depleted or undetectable in add1(RNAi) dcd1(RNAi) plants with strong phenotypes (phenotypic severity increases from left to right).
(C) DCD1/ADD1 proteins accumulate in immature tassel (T), immature ear (E), immature leaf tissue containing symmetrically dividing cells (SD), immature leaf tissue containing asymmetrically dividing cells (AD), leaf tissue containing expanding cells (EX), and mature leaf tissue (M).
(D) dcd1 mutants have less DCD1/ADD1 protein than B73 (wild type) and add1 mutants.
add1(RNAi) dcd1(RNAi) plants show defects in plant growth, cell growth, and epidermal cell pattern, with these defects varying in severity from plant to plant (Figures 3E to 3I). Plants in all 16 add1(RNAi) dcd1(RNAi) lines analyzed were shorter than controls (transformed only with the herbicide marker) due to a reduction in internode length. The average height at maturity of 19 add1(RNAi) dcd1(RNAi) plants representing nine independent transformants was 17 cm (range 4.3 to 27.4 cm; Figure 3I), while the average height of seven mature control plants was 100 cm (range 75.8 to 114 cm; Figure 3I). Additionally, many add1(RNAi) dcd1(RNAi) plants had leaves that were dramatically reduced in length and somewhat reduced in width. The tassels of the add1(RNAi) dcd1(RNAi) plants were shorter and less branched than those in control plants. Toluidine Blue O (TBO) staining of juvenile and mature leaf tissue revealed that leaf epidermal cells in add1(RNAi) dcd1(RNAi) plants were smaller than those in control plants (cf. Figures 3F and 3D). Together, these observations suggest that DCD1/ADD1 promote cell elongation.
add1(RNAi) dcd1(RNAi) plants also have defects in the epidermal cell pattern indicative of aberrantly oriented cell divisions. Some transformants had epidermal phenotypes similar to the dcd1 phenotype with abnormally shaped subsidiary cells but otherwise a relatively normal cell pattern (cf. Figures 3C and 3E). In add1(RNAi) dcd1(RNAi) plants with more pronounced growth defects, the epidermal cell pattern was also more severely affected. In addition to abnormal subsidiary cells, the unspecialized pavement cell patterns were also disrupted (Figure 3F). This suggests that in addition to causing defects in asymmetric SMC divisions, loss of DCD1 and ADD1 also disrupts the proliferative, symmetric cell divisions in the maize leaf epidermis.
Tubulin immunostaining was used to visualize microtubules arrays in dividing cells at the base of immature leaves in nine strongly affected add1(RNAi) dcd1(RNAi) plants (the DCD1/ADD1 protein levels of five of these plants are reported in Figure 4B). Since PPB formation normally occurs well before chromosome condensation (Gunning and Sammut, 1990), wild-type cells in prophase with condensed chromosomes invariably have well-defined PPBs in both symmetrically dividing cells and asymmetrically dividing SMCs (Figures 5A and 5G). However, in add1(RNAi) dcd1(RNAi) plants, many symmetrically and asymmetrically dividing cells with condensed chromosomes lacked PPBs (Table 2, Figures 5D and 5J). Typically, these cells had a surplus of microtubules on the nuclear surface (cf. Figures 5A to 5D and 5G to 5J). Other defects in PPB structure in add1(RNAi) dcd1(RNAi) cells included weak/sparse PPBs in symmetrically dividing cells and abnormal PPBs in asymmetrically dividing SMCs (Table 2, Figure 6D). These results indicate that dcd1 and add1 function redundantly to promote PPB formation during both symmetric and asymmetric cell division in maize and that their function is analogous to that of FASS in Arabidopsis.
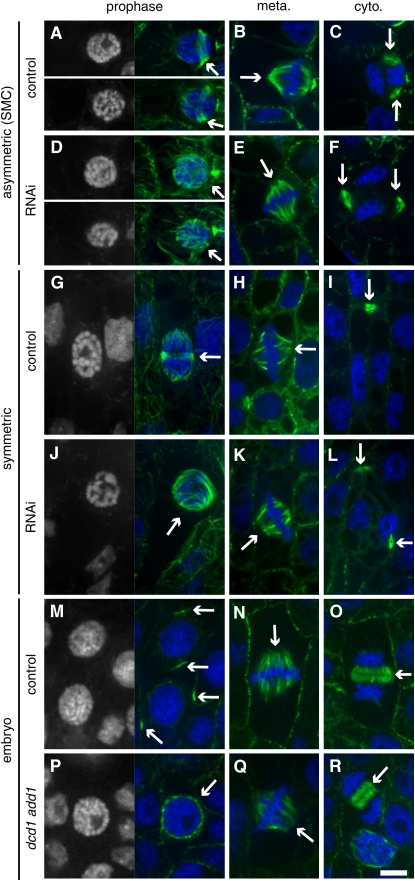
Figure 5.
Microtubule Structures in add1(RNAi) dcd1(RNAi) Plants, dcd1 add1 Double Mutant Embryos, and Corresponding Controls.
Microtubules were detected by tubulin indirect immunofluorescence (green), and DNA was stained with propidium iodide (false-colored blue). For prophase cells ([A], [D], [G], [J], [M], and [P]), the DNA image alone is shown on the left for easy visualization of condensed chromosomes.
(A) to (C) Control SMCs from plants transformed with the herbicide marker; adjacent guard mother cell is to the right of each SMC (partially visible).
(A) Prophase cells with PPBs (arrows).
(B) Metaphase cell with a spindle (arrow).
(C) Expanded control phragmoplast with two visible edges (arrows) during cytokinesis.
(D) to (F) add1(RNAi) dcd1(RNAi) SMCs; adjacent guard mother cell is to the right of each SMC (partially visible).
(D) Prophase cells lacking PPBs (arrows).
(E) Metaphase cell with a spindle (arrow).
(F) Expanded, misoriented phragmoplast with two visible edges (arrows) during cytokinesis.
(G) to (I) Control symmetrically dividing cells from plants transformed with the herbicide marker.
(G) Prophase cell with a PPB (arrow).
(H) Metaphase cell with a spindle (arrow).
(I) Expanded phragmoplast with only one visible edge (arrow) during cytokinesis.
(J) to (L) add1(RNAi) dcd1(RNAi) symmetrically dividing cells.
(J) Prophase cell with no PPB (arrow).
(K) Metaphase cell with a spindle (arrow).
(L) Off-track, expanded phragmoplast with two visible edges at a right angle to each other (arrows) during cytokinesis.
(M) to (O) Cells from dcd1-O (add1-A619)/+ control embryos.
(M) Prophase cells with PPBs (arrows).
(N) Metaphase cell with a spindle (arrow).
(O) Expanding phragmoplast during cytokinesis (arrow).
(P) to (R) Cells from dcd1-O add1-A619 double mutant embryos.
(P) Prophase cell with no PPB (arrow).
(Q) Metaphase cell with a spindle (arrow).
(R) Expanding phragmoplast during cytokinesis (arrow). Bar = 10 μm.
Table 2.
Quantitative Analysis of PPB and Phragmoplast Defects in Symmetrically Dividing Cells and Asymmetrically Dividing SMCs of add1(RNAi) dcd1(RNAi) Plants
| PPBsa
|
Phragmoplastsb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Cell Type | n | Normal | Abnormalc | Absent | n | Normal | Off-Trackd |
| Control | Symmetric | 30 | 100% | 0% | 0% | 19 | 95% | 5% |
| SMC | 11 | 91% | 9% | 0% | 15 | 87% | 13% | |
| add1(RNAi) | Symmetric | 87 | 21% | 17% | 62% | 74 | 32% | 68% |
| dcd1(RNAi) | SMC | 22 | 9% | 18% | 73% | 20 | 35% | 65% |
Data were compiled from nine plants representing seven independent transformation events.
Cells analyzed were in prophase with condensed chromosomes.
Phragmoplasts had extended beyond the width of the daughter nuclei.
In symmetrically dividing cells, abnormal PPBs were weak or sparse; in SMCs, abnormal PPBs were frayed, aberrantly bundled, or incomplete.
In symmetrically dividing cells, off-track phragmoplasts were curved or tilted; in SMCs, off-track phragmoplasts were not lined up with the correct division plane.
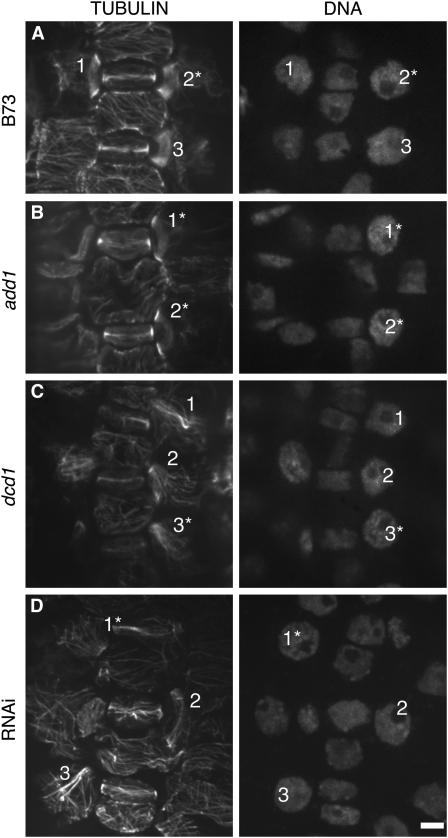
Figure 6.
PPBs in B73, add1, dcd1, and add1(RNAi) dcd1(RNAi) SMCs.
Left panels show tubulin indirect immunofluorescence; right panels show nuclei stained with propidium iodide. Identical numbers within each panel label PPBs of interest and their corresponding nuclei in the left and right panels. Cells in prophase (as indicated by chromosome condensation) are marked with an asterisk.
(A) Normal PPBs in B73 (wild type) SMCs (1 to 3).
(B) Normal PPBs (1 and 2) in add1 SMCs.
(C) Incomplete PPBs (1 and 3) and a complete but frayed PPB (2) in dcd1 SMCs.
(D) Incomplete PPBs (1 and 3) and a relatively normal PPB (2) in add1(RNAi) dcd1(RNAi) SMCs. Bar = 10 μm.
GMC cells form cortical, transverse microtubule rings during interphase that resemble PPBs. At preprophase, these rings disappear when longitudinal PPBs form (Galatis, 1982). Interestingly, formation of the interphase GMC rings is not disrupted in the add1(RNAi) dcd1(RNAi) cells, suggesting that their formation does not require DCD1/ADD1 (see Supplemental Figure 1 online).
To confirm that the PPB defect observed in the RNAi plants was solely due to the loss of dcd1 and add1 function, we examined microtubule structures in dcd1 add1 double mutant embryos. dcd1-O/dcd1-O add1-A619 /+ plants were selfed, and 13- to 19-d-old kernels were dissected to separate the embryo, which was fixed, from the endosperm, which was used for genotyping. The kernels identified as dcd1-O add-A619 double homozygotes had noticeably smaller embryos that were processed for whole-mount tubulin immunolabeling. PPBs were readily observed in surface layers of wild-type but not double mutant embryos. Instead, as in add1(RNAi) dcd1(RNAi) plants, cells were observed in double mutant embryos that had condensed chromosomes characteristic of prophase and a high density of microtubules on the nuclear surface but lacked PPBs (cf. Figures 5M to 5P). However, structurally normal spindles and phragmoplasts were present in both the double mutant and control embryos (cf. Figures 5N and 5O with 5Q and 5R). These observations confirm that DCD1 and ADD1, in combination, are required for PPB formation.
dcd1 Alone Is Needed for Proper PPB Formation in Asymmetrically Dividing Cells
Earlier analysis of the dcd1-O single mutant phenotype indicated that dcd1 was not needed for PPB formation and suggested that aberrantly oriented SMC divisions in this mutant are due to a defect in phragmoplast guidance to the former PPB site (Gallagher and Smith, 1999). To reconcile the results of this earlier study with our present findings indicating a crucial role for DCD1 and ADD1 in PPB formation, PPBs were reexamined in dcd1-O single mutants and examined for the first time in dcd1-mu1 and add1-A619 mutants. To ensure we were comparing cells at similar stages of division and that minor deficiencies in PPB formation were not being overlooked, the results presented in Table 1 represent only SMCs with condensed chromosomes, though some PPBs shown in Figure 6 are associated with preprophase cells with noncondensed chromosomes. This analysis revealed that although SMCs in both dcd1 mutants do consistently form PPBs, a substantial percentage of these PPBs are abnormal (Table 1). Some abnormal PPBs were complete but frayed in appearance, with a portion of the PPB microtubules splaying across the cell surface instead of being confined to a tight ring (Figure 6C, 2). In other SMCs, abnormal, incomplete PPBs failed to encircle the cell and instead extended out across the middle of the SMC (Figure 6C, 1 and 3). Similar PPB abnormalities were observed in SMCs of add1(RNAi) dcd1(RNAi) plants (Figure 6D, 1 and 3). In wild-type cells, fraying PPBs were occasionally observed in preprophase SMCs (26% of SMCs with noncondensed chromosomes [n = 40]) but rarely in prophase SMCs (4% of SMCs with condensed chromosomes [n = 50]; Cho and Wick, 1989). Consistent with the lack of SMC division defects in add1-A619 mutants, no significant differences from the wild type were seen in add1-A619 SMC PPBs (cf. Figures 6A and 6B, Table 1). Moreover, no PPB imperfections were seen in symmetrically dividing cells of any genotype consistent with the lack of symmetric division defects in dcd1 and add1 single mutants (Table 1).
Quantitative analysis of abnormal PPBs in SMCs of dcd1 mutants suggests that they can account for the aberrant subsidiary cell divisions in these mutants (Table 1). The percentage of aberrant SMCs is lower than that of all abnormal PPBs (complete + incomplete) but is not significantly different from the percentage of incomplete PPBs (for dcd1-O, P = 0.08, and for dcd1-mu1, P = 0.13; Fisher's exact test). This correlation suggests that aberrant divisions occur in SMCs that had incomplete PPBs. In this regard, it is interesting that new cell walls in aberrantly divided subsidiary cells are usually connected at the correct location at one edge but at a random location at the other edge (Gallagher and Smith, 1999). This parallels our observations of incomplete PPBs in dcd1 mutants, which are well organized at one edge but not the other (Figure 6C). Thus, the cortical area where the PPB was well organized may function correctly as a cortical division site, attracting the expanding phragmoplast, while the area where the PPB was not present may not, leading to a random path of phragmoplast expansion on that side.
Other Cell Division Abnormalities in add1(RNAi) dcd1(RNAi) Plants Can Potentially Be Explained by the Absence of PPBs
Formation of an asymmetric PPB is one manifestation of polarity in premitotic SMCs, but there are several others, and these were also examined in add1(RNAi) dcd1(RNAi) plants. An early event in SMC polarization is migration of the nucleus to the site of GMC contact, where it remains throughout prophase (Stebbins and Shah, 1960; Kennard and Cleary, 1997). The spindle remains polarized throughout mitosis, and after nuclear division, the phragmoplast and associated daughter nuclei remain in this polarized position throughout cytokinesis. We evaluated the positions of propidium iodide–labeled nuclei in control and add1(RNAi) dcd1(RNAi) SMCs throughout the cell cycle (including prophase, mitotic, and cytokinetic cells) and found that 100% (n = 40) were correctly polarized in controls cells compared with 79% in add1(RNAi) dcd1(RNAi) cells (n = 48). The lack of a strong polarization defect fits with earlier findings that actin, but not microtubules, are needed for nuclear migration in premitotic SMCs (Kennard and Cleary, 1997; Panteris et al., 2007). However, these studies also suggested that microtubules are needed to maintain nuclear polarization. The few unpolarized nuclei observed may have polarized initially but later moved away from the GMC contact site due to the lack of a PPB.
Previous studies have suggested that PPBs inhibit microtubule nucleation at perinuclear regions adjacent to PPBs, thereby promoting bipolar spindle formation (Wick and Duniec, 1984; Mineyuki et al., 1991; Nogami et al., 1996; Chan et al., 2005; Yoneda et al., 2005). The overabundance of microtubules on the nuclear surface in RNAi and double mutant preprophase and prophase cells lacking PPBs also supports this idea (Figures 5D, 5J, and 5P). To further explore this possibility, we compared the distribution of microtubules on the nuclear surface of symmetrically dividing control and add1(RNAi) dcd1(RNAi) cells with condensed chromosomes. Twenty-eight of thirty control cells with PPBs had a bipolar microtubule distribution on the nuclear surface (e.g., Figure 5G), while in the add1(RNAi) dcd1(RNAi) cells, 34 of 67 prophase cells lacking a PPB also lacked a bipolar distribution of microtubules on the nuclear surface (Figure 5J). These observations support the idea that the presence of a PPB promotes a timely formation of a bipolar spindle, although a more direct role for DCD1/ADD1 in promotion of spindle bipolarity is also possible. In mitotic cells of add1(RNAi) dcd1(RNAi) plants, spindles appeared structurally normal and no evidence of mitotic defects was observed, arguing against a direct role for DCD1/ADD1 in spindle organization or function (cf. Figures 5B to 5E and 5H to 5K).
Analysis of cells undergoing cytokinesis in add1(RNAi) dcd1(RNAi) plants revealed that phragmoplasts appear structurally normal but that the path of phragmoplast expansion was often disrupted (cf. Figures 5C to 5F and 5I to 5L). The position of mid- and late-stage phragmoplasts (those that have expanded beyond the width of the nuclei or are associated with nuclei that contain decondensed chromosomes) was evaluated in SMCs, where the cortical target of each phragmoplast can be accurately predicted. In control cells, 13 of 15 phragmoplasts were on track to meet the mother cell cortex at the correct position, while in add1(RNAi) dcd1(RNAi) cells, only 7 of 20 phragmoplasts were on track. Of the late add1(RNAi) dcd1(RNAi) phragmoplasts only 2 of 13 were on track. These results suggest that in the add1(RNAi) dcd1(RNAi) cells, most of which fail to form PPBs (Table 2), the phragmoplasts do not recognize a cortical division site and expand in random routes to the cell cortex. Occasionally, SMC division walls failed to connect with the mother cell wall on one side, producing cell wall stubs with terminating loops that are presumably formed when the phragmoplast attaches to the newly forming cell wall (Figures 3G and 3H). In some cases, aberrant SMC divisions formed a cell within a cell, which most likely results when the phragmoplast edges fuse together instead of connecting with the mother cell cortex (Figure 3G). Similar defects were seen in dcd1 single mutants, although at a lower frequency (Gallagher and Smith, 1999, 2000). Assuming that the PPB is essential for establishment of a cortical division site that guides phragmoplast expansion during cytokinesis, the phragmoplast orientation defects observed in the add1(RNAi) dcd1(RNAi) cells can be explained by defects in PPB formation.
DCD1 and ADD1 Colocalize with PPBs and Remain at the Cortical Division Site during Mitosis
Immunoblot analysis using the validated DCD1/ADD1 peptide antibody indicated that DCD1/ADD1 are expressed in all zones of the maize leaf, including those where epidermal cells are mostly undergoing symmetric cell divisions, asymmetric cell divisions, cell expansion, and cell maturation. In addition, DCD1 and ADD1 were also detected in immature ears and tassels that contain many cells undergoing active division (Figure 4C).
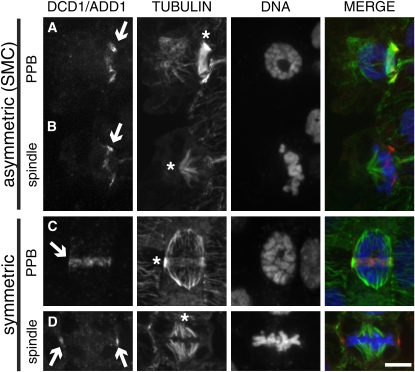
DCD1/ADD1 and tubulin antibodies were used in immunostaining experiments to determine the intracellular localization of the DCD1 and ADD1 proteins in dividing cells. In both asymmetrically and symmetrically dividing cells, the antibody labels a ring coinciding with the PPB (Figures 7A and 7C; see Supplemental Figure 2 online). This ring was not observed in any premitotic cells lacking a microtubule PPB, suggesting that the formation of the DCD1/ADD1 ring does not precede PPB formation. Interestingly, following PPB breakdown, DCD1/ADD1 rings persist into mitosis in both asymmetrically and symmetrically dividing cells (Figures 7B and 7D; see Supplemental Figure 2 online). While visible in cells in metaphase, cortical DCD1/ADD1 rings were not detected in anaphase cells or during cytokinesis when phragmoplasts are expanding, suggesting that DCD1/ADD1 are transient components of the cortical division site established by the PPB. Alternatively, DCD1/ADD1 may remain at the cortical division site at levels too low to be detected via immunostaining. In a previous study, FASS–green fluorescent protein (GFP) expressed from the constitutive 35S promoter in cultured tobacco (Nicotiana tabacum) BY-2 cells localized to the spindle and phragmoplast (its localization in preprophase/prophase cells was not described; Van Damme et al., 2004). However, we observed no specific labeling of spindles or phragmoplasts with anti-DCD1/ADD1. Possibly, overexpression of FASS-GFP caused its mislocalization in BY-2 cells. Alternatively, there may be a low level of DCD1/ADD1 in the spindle and/or phragmoplast that we could not detect by immunolocalization.
Figure 7.
Localization of DCD1/ADD1 in Dividing Maize Leaf Epidermal Cells.
DCD1/ADD1 (left panel, shown in red in the merged image) and tubulin (middle-left panel, green in the merged image) were detected by indirect immunofluorescence, while DNA (middle-right panel, blue in the merged image) was stained with SYTOX blue. The merge is shown in the right panel.
(A) Surface view of a SMC showing colocalization of DCD1/ADD1 (arrow) and a PPB (asterisk).
(B) Surface view of a SMC after PPB breakdown with an early metaphase spindle (asterisk) and a cortical ring of DCD1/ADD1 (arrow).
(C) Surface view of a symmetrically dividing epidermal cell showing colocalization of DCD1/ADD1 (arrow) and a PPB (asterisk).
(D) Mid-plane view of a symmetrically dividing epidermal cell with a metaphase spindle (asterisk). DCD1/ADD1 is visible on two points of the cortex coincident with the presumed cortical division site (arrows). Bar = 10 μm.
The growth defects and small cells observed in add1(RNAi) dcd1(RNAi) plants clearly suggest a role for DCD1/ADD1 in expanding cells. Consistent with this, immunoblot analysis showed that DCD1/ADD1 are present not only in all tissues tested where cells are dividing but also in regions of the developing maize leaf where cells are expanding postmitotically (Figure 4C). However, no specific labeling of DCD1/ADD1 was observed by immunolocalization in other interphase cells. Most likely, our immunolocalization method is not sufficiently sensitive to detect DCD1/ADD1 protein in most nondividing cells. However, although the RNAi phenotype suggests that dcd1 and add1 are not required for the formation of interphase cortical rings in GMCs, DCD1/ADD do localize to these rings (see Supplemental Figure 1 online).
Although DCD1 and ADD1 are 96% identical, dcd1 mutants have observable phenotypes, while add1 mutants do not. One possible explanation for this is that DCD1 and ADD1 might be expressed and/or localized differently. To investigate this possibility, anti-DCD1/ADD1 labeling was evaluated in dcd1-mu1 mutants (which only have ADD1) and add1-A619 mutants (which only have DCD1). The staining pattern in the leaf epidermis of both mutants was identical to the wild type, indicating that DCD1 and ADD1 are both expressed and localized to the cortical division site in all dividing cells (data not shown). Antibody staining was consistently fainter in the dcd1-mu1 mutants than in add1-A619 mutants, however, suggesting that ADD1 is less abundant than DCD1. To investigate this further, DCD1/ADD1 protein levels in leaf extracts from B73, dcd1-mu1, and add1-A619 plants were compared by immunoblotting. B73 (expressing both DCD1 and ADD1) and add1-A619 (expressing only DCD1) have equivalent amounts of protein detected by the DCD1/ADD1 antibody, while the intensity of this band was reduced in dcd1-mu1 (expressing only ADD1; Figure 4D), potentially explaining why a mild phenotype is observed in dcd1 but not add1 single mutants.
DISCUSSION
The PPB plays a central role in determining the plane of division in plant cells, but the mechanisms governing its formation, positioning, and function are poorly understood. Our analysis of DCD1 and ADD1 proteins in maize has provided new insights regarding the roles played by these proteins in PPB formation and function.
Maize dcd1 mutants have defects in the orientation of asymmetric cell divisions. We cloned dcd1 and its close relative, add1, and found them to be homologous to Arabidopsis FASS. All three genes encode proteins with homology to regulatory subunits of PP2A phosphatase complexes. These complexes are composed of three subunits: a catalytic subunit, C, a scaffolding subunit, A, and a regulatory subunit, B. Regulatory B subunits appear to function by targeting PP2A complexes to appropriate substrates or subcellular locations. There are currently three identified classes of B regulatory subunits, B, B', and B'', which are unrelated to each other at the amino acid level (Janssens and Goris, 2001; Sontag, 2001). The C-terminal halves of DCD1, ADD1, and FASS are homologous to the C terminus of the founding member of the B'' class, the human protein PR72 (Hendrix et al., 1993; Camilleri et al., 2002). The N-terminal domains of DCD1, ADD1, and FASS are related to each other and other vertebrate proteins but are unrelated to the corresponding region of PR72. Nevertheless, these plant proteins likely function as phophatase subunits since FASS binds PP2A scaffolding subunits in yeast two-hybrid experiments (Camilleri et al., 2002). Moreover, the Caenorhabditis elegans FASS/DCD1/ADD1 homolog RSA-1 coprecipitates with PP2A catalytic and scaffolding subunits (Schlaitz et al., 2007), supporting the conclusion that this divergent class of B'' subunits functions in phosphatase regulation.
PPBs fail to form when DCD1 and ADD1 are simultaneously depleted in both add1(RNAi) dcd1(RNAi) plants and in dcd1 add1 double mutants. Lack of PPB formation was also observed in Arabidopsis fass mutants, suggesting that the function of these proteins is conserved between monocots and dicots. One of the longstanding questions about PPB formation is how the assembly of microtubules becomes limited to a narrow band encircling the mother cell, when previously during interphase, the entire cortex was capable of supporting microtubule nucleation. A partial explanation for this is offered by the specific localization of DCD1 and ADD1 to the site of PPB assembly. A likely scenario is that localization of DCD1 and ADD1 to the cortical division site targets its associated PP2A complex to that region of the cortex where it dephosphorylates as yet unidentified target proteins to promote localized microtubule assembly and/or stabilization necessary for PPB formation. A potential target of DCD1/ADD1 is the maize homolog of TON1. Like Arabidopsis fass mutants, Arabidopsis ton1 mutants also fail to form PPBs, and the proteins are known to be phosphorylated in Arabidopsis (Benschop et al., 2007; Azimzadeh et al., 2008; Sugiyama et al., 2008). RSA-1, the C. elegans homolog of DCD1/ADD1, localizes to the centrosome, a site of microtubule assembly in animal cells, where it interacts with proteins that bind microtubules, suggesting that the association of this class of B'' regulatory subunits with microtubule nucleation and stability has been maintained in both plant and animal cells (Schlaitz et al., 2007).
Phenotypic analysis of single mutants showed that both DCD1 and ADD1 are sufficient for proper PPB formation and division orientation in symmetrically dividing cells. However, the frayed and incomplete PPBs observed in SMCs of dcd1 single mutants show that only DCD1 is sufficient and necessary for proper PPB formation and orientation of division in asymmetrically dividing SMCs. DCD1 and ADD1 proteins are both associated with PPBs in SMCs, but immunoblot analysis showed that the DCD1/ADD1 pool is depleted by more than half in dcd1 single mutants, suggesting that DCD1 is more abundant in wild-type SMCs than is ADD1. PPBs that form in SMCs are distinct from those in symmetrically dividing cells in that they do not encircle the entire diameter of a cell. Instead, they are confined to one side of the SMC, tightly encircling a small protrusion of the SMC that joins it to the GMC (Panteris et al., 2006). Most likely, these PPBs are more susceptible to depletion of the DCD1/ADD1 protein pool simply because their unusual characteristics make them more dependent on stabilization by DCD1/ADD1-dependent dephosphorylation. Alternatively, although the 96% sequence identity between DCD1 and ADD1 argues against the possibility that these proteins are functionally divergent, DCD1 protein may be better suited than ADD1 for supporting PPB formation in SMCs.
DCD1 and ADD1 persist at the cortical division site after PPB breakdown and are the only other proteins besides Arabidopsis TAN and RanGAP1 that show this localization to date (Walker et al., 2007; Xu et al., 2008). TAN is required for the spatial regulation of cytokinesis but does not appear to play a role in PPB formation. It colocalizes with PPBs and then persists at the cortical division site throughout mitosis and cytokinesis. Thus, TAN has been proposed to function as a marker of the division plane that helps guide phragmoplast expansion (Walker et al., 2007). RanGAP1 is a GTPase activating protein of Ran that aids in creating populations of RanGTP and RanGDP at particular subcellular localizations. When RanGAP1 and a related protein, RanGAP2, were depleted in Arabidopsis roots, misoriented cell divisions and cell wall stubs are observed. However, since technical limitations have prevented examining the effects of RanGAP1 and RanGAP2 depletion on cytoskeletal structures during cell division, it is currently unknown if loss of these proteins affects PPB formation or phragmoplast guidance (Xu et al., 2008).
In contrast with TAN and RanGAP1, DCD1 and ADD1 are observed at the cortical division site only during mitosis, arguing against a direct role for these proteins in phragmoplast guidance during cytokinesis. Moreover, PPB abnormalities in both add1(RNAi) dcd1(RNAi) plants and dcd1 single mutants are apparently sufficient to explain their division plane defects without any need to propose a direct role for DCD1/ADD1 in phragmoplast guidance. Thus, rather than serving as a marker of the division plane, DCD1/ADD1 may function during prophase and metaphase to target phosphatase activity to other division site components left behind after PPB disassembly that are important for phragmoplast guidance. Notably, FASS is required for the localization of Arabidopsis TAN and RanGAP1 at the cortical division site (Walker et al., 2007; Xu et al., 2008). Since microtubules are also required for recruitment of TAN to the division site, and FASS is required for PPB formation, TAN and/or RanGAP1 localization may depend on FASS simply because they depend on PPBs. Alternatively, TAN and/or RanGAP1 may depend more directly on FASS for their localization. Indeed, TAN and/or RanGAP1 may need to be dephosphorylated by a FASS-associated phosphatase to be recruited to and/or maintained at the cortical division site during the early stages of mitosis. Supporting the idea that there are separate mechanisms for initial recruitment of proteins to the PPB and subsequent maintenance at the cortical division site, RanGAP1 initially localizes to the PPB but then fails to be maintained at the cortical division site in pok1/pok2 mutants (Xu et al., 2008). Additionally, the TAN ring sharpens at the onset of cytokinesis, slightly after DCD1 and ADD1 apparently disappear from the cortex (Walker et al., 2007). Thus, TAN ring sharpening might reflect a transition from a FASS-dependent mechanism to a FASS-independent mechanism for maintenance of TAN at the cortical division site.
Genetic disruption of SMC divisions in the maize leaf epidermis has previously provided insights into the establishment of cell polarity as well as its impact on the division plane and subsequent cellular differentiation (e.g., Gallagher and Smith, 2000; Frank et al., 2003). Our work has shed new light on global mechanisms governing PPB formation and function and suggests that the unique characteristics of the polarized SMC division cause it to be more susceptible than other divisions to mild disruptions in these mechanisms. The unique features of SMC divisions, in combination with the growing collection of mutants affecting this cell type, make maize SMCs a useful focus for studies addressing basic questions about cell polarity, cell division, and cellular differentiation in plants.
METHODS
Maize Material
dcd1-O arose in an ethyl methanesulfonate–mutagenized background (Gallagher and Smith, 1999) and was backcrossed to B73 at least four times. dcd1-mu1 and dcd1-mu2 were isolated in a noncomplementation screen using a B73 Mu line and were backcrossed to B73 twice or once respectively. add1-A619 is the inbred A619 line.
Toluidine Blue O staining of epidermal peels was performed as described by Gallagher and Smith (1999). Plant tissue used in the following protocols was obtained from plants grown in parallel with 8 to 12 visible leaves. Outer leaves were removed until the leaf with an expanded sheath <0.5 cm was uncovered, and then the remainder of the plant was processed as described in the below protocols.
Cloning of dcd1 and add1
A 3-kb, Mu1-containing BamHI restriction fragment that cosegregated with the mutant phenotype in the dcd1-mu1 background was identified using standard methods of DNA isolation and DNA gel blot analysis (Chen and Dellaporta, 1994; Warren and Hershberger, 1994). dcd1-mu1 genomic DNA was cut with BamHI, size fractionated, and self-ligated using the protocol of Earp et al. (1990). Inverse PCR using a Mu1-specific primer (MuEnd 5′-AGAGAAGCCAACGCCAACGCCTCCATTTCGTC-3′) amplified an 1142-bp fragment of the dcd1 gene. The complete sequences of dcd1 and add1 were assembled using DNA sequence available in public databases at the time (primarily MaizeGDB) along with new sequence generated by PCR to fill in the gaps. The dcd1 intron and exon boundaries were determined by sequencing cDNAs (0021C16 and 0083G13) from the maize (Zea mays) ZM_BFa library obtained from the Arizona Genomics Institute. The add1 intron and exon boundaries were determined using EST sequences available in public databases.
RT-PCR
RNA was extracted from the basal 4 cm of each plant prepped as above using Trizol (Invitrogen). First-strand cDNA was generated using the Retroscript kit (Ambion). Primers used in the amplification of dcd1 and add1 were dcd1for (5′-CATGCCGTGGGTCCGCAAT-3′), dcd13′for (5′-TGAGGAAGAGGTAACCGATAC-3′), dcd1rev (5′-GTACCTGGTGCCTCAAAATTG-3′), add1for (5′-CAGTCGCGAGATGGACTTC-3′), and add1rev (5′-CTGATTGAATGCTCACAGCC-3′). For a complete description of the RT protocol and gpch primer sequences, see http://www.chromatinconsortium.org/docs/rt-pcr_protocol.pdf (McGinnis et al., 2005). Total amplification amount was maintained below 100 ng per reaction, and DNA was visualized with ethidium bromide.
Generation of add1(RNAi) dcd1(RNAi) Plants
The add1 gene was amplified from B73 cDNA using dcd1ATG (5′-ATGAGCACCGCCTCTGGC-3′) and 23676f (5′-CTGATTGAATGCTCACAGCC-3′) and cloned into pGEM-T Easy vector (Promega). A 700-bp fragment of add1 was amplified from this construct using altRNAfor (5′-NNACTAGTGGCGCGCCTGAAGTTTGAGAAGGATGAC-3′) and altRNArev (5′-NNGCGATCGCCCTAGGGTGGATGTCTGCAGTTGTAAG-3′), which introduce AcsI/AvrII and SpeI/SgfI sites into the ends of the amplified products. Matching restriction sites in the pMCG161 vector allowed two copies of the add1 fragment to be sequentially ligated into the vector in opposite orientations creating an add1 hairpin. The pMCG161 vector and complete cloning procedure are described by McGinnis et al. (2005).
The Iowa State Plant Transformation Facility cotransformed the add1 RNAi construct and a vector carrying the herbicide resistance bar gene into Hi Type II hybrid maize callus. Two to twelve regenerated plants from 34 independent lines were screened for cell division defects.
Antibody Generation
A peptide consisting of amino acids 461 to 480 (DVRGFWAHDNRENLLQEEEE) from DCD1 was made, conjugated to KLH, and used to generate two rabbit polyclonal antibodies (Genemed Synthesis). The antibodies were affinity purified using Sulfolink resin and Gentle Ag/Ab binding and elution buffers according to the manufacturer's protocols (Pierce/ThermoFisher Scientific).
Immunoblot Analysis
The basal 4 cm from plants prepped as above was homogenized in TBS, 10% sucrose, 1% protease inhibitor cocktail for plants (Sigma-Aldrich), 5 mM EDTA, 5 mM EGTA, 0.3% β-mercaptoethanol, and 0.5% Triton using an Omni TH homogenizer. To remove cell debris, the extracts were centrifuged at 750g and then 2100g. The protein concentration of each extract was determined using Bio-Rad protein assay. After electrophoresis on 10% polyacrylamide gels, separated proteins were transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was blocked in TBS with 5% BSA, probed for 2 h with anti-DCD1/ADD1 antibody (1 μg/mL) in TBS with 0.01% BSA, washed in TBS with 0.05% Tween, and then incubated in anti-rabbit IgG alkaline phosphatase conjugate (Promega; 250 μg/mL) for 90 min. The membrane was treated with Nitro-Blue Tetrazolium Chloride and 5-Bromo-4-Chloro-3′-Indolyphosphate p-Toluidine Salt (Promega) to visualize the antibody-bound proteins. The same blots were subsequently probed with an anti-actin antibody (0.1 μg/mL MAB1501R; Chemicon) and anti-mouse IgG alkaline phosphatase conjugate (Promega; 250 μg/mL) to show equal loading and transfer of the protein samples.
For the developmental immunoblot, developing ears (2 cm long), developing tassels (4.5 cm long), and leaf samples were taken from B73 plants and processed as above. The leaf samples spanned 0.5 to 2 cm (symmetric cell divisions), 2 to 3.5 cm (SMC cell divisions), 5 to 6.5 cm (expanding cells), and 8 to 9.5 cm (maturing cells) from the base of plants prepped as above.
Immunolocalization of Tubulin and DCD1/ADD1 and DNA Staining
Tissue strips (2 mm wide) were cut from the basal 0 to 1.5 cm (symmetric cell divisions) and 1.5 to 3 cm (asymmetric cell divisions) of plants prepped as above. Strips or embryos 13 to 19 d after pollination were fixed in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 4 mM MgCl2, pH 6.9) with 4% formaldehyde and 0.1% Triton X-100 for 2 h and then washed with PHEM with 0.05% Triton X-100 (PHEMt). Cell walls were permeabilized by digestion in 1% Driselase (Sigma-Aldrich) and 0.5% Pectolyase Y-23 (MP Biomedicals) in water for 15 min and then washed with PHEMt. The strips were extracted in PHEM with 1% Triton X-100 and 1% DMSO for 1 h, rinsed in PBS, and then blocked for 30 min in PBS with 5% normal goat serum. To visualize microtubules, the strips were incubated overnight in an anti-α-tubulin antibody (clone B-5-1-2; Sigma-Aldrich) diluted to 0.5 μg/mL in blocking solution and then washed with PBS with 0.05% Triton X-100 (PBSt). The localized tubulin antibodies were labeled with 100 μg/mL Alexa Fluor-488–conjugated anti-mouse IgG (Invitrogen) in blocking solution for 3 to 4 h at room temperature and then washed with PBSt.
For double immunolabeling of DCD1/ADD1 and tubulin, leaf strips were fixed, blocked, and incubated in 10 μg/mL anti-DCD1/ADD1 antibody (G4620) diluted in PBS. G4620 was detected using a 488 tyramide signal amplification kit following the manufacturer's protocol (Invitrogen). The leaf strips were reblocked, washed with PBSt, and incubated in tubulin antibody and Alexa Fluor-568–conjugated anti-mouse IgG as above except the antibody incubations were done in PBS. To label nuclei, leaf strips or embryos were incubated in 10 μg/mL propidium iodide (Sigma-Aldrich) in PBS or 500 nM SYTOX-blue (Invitrogen) in water for 10 min and washed in PBSt. Leaf strips and embryos were mounted in Vectashield (Vector Laboratories) for observation.
Immunoblot and Microtubule Analysis of add1(RNAi) dcd1(RNAi) Plants
Severely affected RNAi plants (6 to 25 cm tall) and a similarly aged control plant (34 cm tall) were stripped of leaves as described above. The basal 3 cm of each plant was cut in half lengthwise with one half used for antibody staining and the other for protein extraction.
Confocal Microscopy, Image Processing, and Analysis
Alexafluor 488, Alexafluor 568, propidium iodide, and SYTOX-blue were excited at the appropriate wavelengths with an argon (488-nm line), argon/krypton laser (568-nm line), or violet blue laser (440-nm line). Fluorescence was visualized using a Nikon TE-200U microscope equipped with a ×60 1.2 numerical aperture water immersion objective, a Yokogawa Nipkow spinning disk confocal head, Chroma HQ525/50 (for Alexafluor 488), HQ620/60 (for Alexafluor 568 and propidium iodide), and Chroma HQ480/40 (for SYTOX-blue) band-pass emission filters, and a Roper CascadeII 512b EM CCD camera using on-chip gain and reading off at 5 MHz. The confocal system was controlled using MetaMorph software version 7.0r1 (Universal Imaging). Z-projections of selected slices from stacks were assembled using Metamorph. Image processing was performed using Adobe Photoshop 10.0 applying only linear adjustments to pixel values.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ469779 (ADD1) and FJ469780 (DCD1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DCD1/ADD1 Localizes to the Interphase Cortical Rings in GMCs but Is Not Needed for Their Formation.
Supplemental Figure 2. Localization of DCD1/ADD1 in Wild-Type (B73) Dividing Maize Leaf Epidermal Cells.
Supplementary Material
Acknowledgments
We thank members of the Smith Lab for their contributions, particularly Michelle Nolasco for cosegregation analysis, EiThander Win for assistance with RT-PCR, and Carolyn Rasmussen and John Humphries for critical reading of the manuscript. This work was supported by USDA Grant 2006-35304-17342 and National Institutes of Health Grant R01 GM53137 to L.G.S. and a University of California, San Diego/San Diego State University Institutional Research and Academic Career Development Award (NIH GM 68524) to A.J.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Amanda J. Wright (ajwright@post.harvard.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Azimzadeh, J., Narcy, P., Christodoulidou, A., Drevensek, S., Camilleri, C., Amiour, N., Parcy, F., Pastuglia, M., and Bouchez, D. (2008). Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20 2146–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop, J., Mohammed, S., O'Flaherty, M., Herk, A., Slijper, M., and Menke, F. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6 1198–1214. [DOI] [PubMed] [Google Scholar]
- Camilleri, C., Azimzadeh, J., Pastuglia, M., Bellini, C., Grandjean, O., and Bouchez, D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Calder, G., Fox, S., and Lloyd, C. (2005). Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in Arabidopsis suspension cells. Plant Cell 17 1737–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., and Dellaporta, S. (1994). Urea-based plant DNA miniprep. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 526–527.
- Cho, S.-O., and Wick, S.M. (1989). Microtubule orientation during stomatal differentiation in grasses. J. Cell Sci. 92 581–594. [Google Scholar]
- Cleary, A.L., Gunning, B.E.S., Wasteneys, G.O., and Hepler, P.K. (1992). Microtubule and F-actin dynamics at the division site in living Tradescantia stamen hair cells. J. Cell Sci. 103 977–988. [Google Scholar]
- Cleary, A.L., and Smith, L.G. (1998). The Tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell 10 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earp, D., Lowe, B., and Baker, B. (1990). Amplification of genomic sequences flanking transposable elements in host and heterologous plants: a tool for transposon tagging and genome characterization. Nucleic Acids Res. 18 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, M.J., Cartwright, H.N., and Smith, L.G. (2003). Three Brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130 753–762. [DOI] [PubMed] [Google Scholar]
- Galatis, B. (1982). The organization of microtubules in guard cell mother cells of Zea mays. Can. J. Bot. 60 1148–1166. [Google Scholar]
- Galatis, B., and Apostolakos, P. (2004). The role of the cytoskeleton in the morphogenesis and function of stomatal complexes. New Phytol. 161 613–639. [DOI] [PubMed] [Google Scholar]
- Gallagher, K., and Smith, L.G. (1999). discordia mutations specifically misorient asymmetric cell divisions during development of the maize leaf epidermis. Development 126 4623–4633. [DOI] [PubMed] [Google Scholar]
- Gallagher, K., and Smith, L.G. (2000). Roles for polarity and nuclear determinants in specifying daughter cell fates after an asymmetric cell division in the maize leaf. Curr. Biol. 10 1229–1232. [DOI] [PubMed] [Google Scholar]
- Gunning, B.E.S. (1982). The cytokinetic apparatus: Its development and apatial regulation. In The Cytoskeleton in Plant Growth and Development, C.W. Lloyd, ed (London/New York: Academic Press), pp. 229–292.
- Gunning, B.E.S., and Sammut, M. (1990). Rearrangements of microtubules involved in establishing cell division planes start immediately after DNA synthesis and are completed just before mitosis. Plant Cell 2 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix, P., Mayer-Jaekel, R.E., Cron, P., Goris, J., Hofsteenge, J., Merlevede, W., and Hemmings, B.A. (1993). Structure and expression of a 72-kDa regulatory subunit of protein phosphatase 2A. Evidence for different size forms produced by alternative splicing. J. Biol. Chem. 268 15267–15276. [PubMed] [Google Scholar]
- Janssens, V., and Goris, J. (2001). Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353 417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens, G. (2005). Plant cytokinesis: Fission by fusion. Trends Cell Biol. 15 277–283. [DOI] [PubMed] [Google Scholar]
- Kawamura, E., Himmelspach, R., Rashbrooke, M.C., Whittington, A.T., Gale, K.R., Collings, D.A., and Wasteneys, G.O. (2006). MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard, J., and Cleary, A. (1997). Pre-mitotic nuclear migration in subsidiary mother cells of Tradescantia occurs in G1 of the cell cycle and requires F-actin. Cell Motil. Cytoskeleton 36 55–67. [DOI] [PubMed] [Google Scholar]
- Liu, B., and Palevitz, B.A. (1992). Organization of cortical microfilaments in dividing root cells. Cell Motil. Cytoskeleton 23 252–264. [Google Scholar]
- McClinton, R.S., and Sung, Z.R. (1997). Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201 252–260. [DOI] [PubMed] [Google Scholar]
- McGinnis, K., Chandler, V., Cone, K., Kaeppler, H., Kaeppler, S., Kerschen, A., Pikaard, C., Richards, E., Sidorenko, L., Smith, T., Springer, N., and Wulan, T. (2005). Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 392 1–24. [DOI] [PubMed] [Google Scholar]
- Mineyuki, Y. (1999). The preprophase band of microtubules: Its function as a cytokinetic apparatus in higher plants. Int. Rev. Cytol. 187 1–49. [Google Scholar]
- Mineyuki, Y., Marc, J., and Palevitz, B.A. (1991). Relationship between the preprophase band, nucleus, and spindle in dividing Allium cotyledon cells. J. Plant Physiol. 138 640–649. [Google Scholar]
- Müller, S., Han, S., and Smith, L.G. (2006). Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr. Biol. 16 888–894. [DOI] [PubMed] [Google Scholar]
- Nogami, A., Suzaki, T., Shigenaka, Y., Nagahama, Y., and Mineyuki, Y. (1996). Effects of cycloheximide on preprophase bands and prophase spindles in onion (Allium cepa L.) root tip cells. Protoplasma 192 109–121. [Google Scholar]
- Panteris, E., Apostolakos, P., and Galatis, B. (2006). Cytoskeletal asymmetry in Zea mays subsidiary cell mother cells: A monopolar prophase microtubule half-spindle anchors the nucleus to its polar position. Cell Motil. Cytoskeleton 63 696–709. [DOI] [PubMed] [Google Scholar]
- Panteris, E., Galatis, B., Quader, H., and Apostolakos, P. (2007). Cortical actin filament organization in developing and functioning stomatal complexes of Zea mays and Triticum turgidum. Cell Motil. Cytoskeleton 64 531–548. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps, J.D., and Northcote, D.H. (1966. a). Organization of microtubules and endoplasmic reticulum during mitosis and cytokinesis in wheat meristems. J. Cell Sci. 1 109–120. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps, J.D., and Northcote, D.H. (1966. b). Cell division in the formation of the stomatal complex of the young leaves of wheat. J. Cell Sci. 1 121–128. [DOI] [PubMed] [Google Scholar]
- Sano, T., Higaki, T., Oda, Y., Hayashi, T., and Hasezawa, S. (2005). Appearance of actin microfilament 'twin peaks' in mitosis and their function in cell plate formation, as visualized in tobacco BY-2 cells expressing GFP-fimbrin. Plant J. 44 595–605. [DOI] [PubMed] [Google Scholar]
- Schlaitz, A.-H., et al. (2007). The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell 128 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.G., Hake, S., and Sylvester, A.W. (1996). The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122 481–489. [DOI] [PubMed] [Google Scholar]
- Sontag, E. (2001). Protein phosphatase 2A: The Trojan Horse of cellular signaling. Cell. Signal. 13 7–16. [DOI] [PubMed] [Google Scholar]
- Stebbins, G., and Shah, S. (1960). Developmental studies of cell differentiation in the epidermis of monocotyledones. II. Cytological features of stomatal development in the Gramineae. Dev. Biol. 2 477–500. [Google Scholar]
- Sugiyama, N., Nakagami, H., Mochida, K., Daudi, A., Tomita, M., Shirasu, K., and Ishihama, Y. (2008). Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylatin in Arabidopsis. Mol. Syst. Biol. 4 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375 676–677. [Google Scholar]
- Van Damme, D., Bouget, F.-Y., Van Poucke, K., Inzé, D., and Geelen, D. (2004). Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J. 40 386–398. [DOI] [PubMed] [Google Scholar]
- Van Damme, D., and Geelen, D. (2008). Demarcation of the cortical division zone in dividing plant cells. Cell Biol. Int. 32 178–187. [DOI] [PubMed] [Google Scholar]
- Vanstraelen, M., Van Damme, D., De Rycke, R., Mylle, E., Inzé, D., and Geelen, D. (2006). Cell cycle-dependent targeting of a kinesin at the plasma membrane demarcates the division site in plant cells. Curr. Biol. 16 308–314. [DOI] [PubMed] [Google Scholar]
- Walker, K., Müller, S., Moss, D., Ehrhardt, D.W., and Smith, L.G. (2007). Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17 1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, C., and Hershberger, J. (1994). Southern blots of maize genomic DNA. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 566–568.
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411 610–613. [DOI] [PubMed] [Google Scholar]
- Wick, S.M., and Duniec, J. (1984). Immunofluorescence microscopy of tubulin and microtubule arrays in plant cells. II. Transition between the pre-prophase band and the mitotic spindle. Protoplasma 122 45–55. [Google Scholar]
- Wright, A., and Smith, L. (2008). Division plane orientation in plant cells. In Cell Division Control in Plants, D.P.S. Verma and Z. Hong, eds (Berlin/Heidelberg, Germany: Springer), pp. 33–57.
- Xu, X.M., Zhao, Q., Rodrigo-Peiris, T., Brkljacic, J., He, C.S., Müller, S., and Meier, I. (2008). RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc. Natl. Acad. Sci. USA 105 18637–18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda, A., Akatsuka, M., Hoshino, H., Kumagai, F., and Hasezawa, S. (2005). Decision of spindle poles and division plane by double preprophase bands in a BY-2 cell line expressing GFP-tubulin. Plant Cell Physiol. 46 531–538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.