Abstract
The plant-specific DYW subclass of pentatricopeptide repeat proteins has been postulated to be involved in RNA editing of organelle transcripts. We discovered that the DYW proteins CHLORORESPIRATORY REDUCTION22 (CRR22) and CRR28 are required for editing of multiple plastid transcripts but that their DYW motifs are dispensable for editing activity in vivo. Replacement of the DYW motifs of CRR22 and CRR28 by that of CRR2, which has been shown to be capable of endonucleolytic cleavage, blocks the editing activity of both proteins. In return, the DYW motifs of neither CRR22 nor CRR28 can functionally replace that of CRR2. We propose that different DYW family members have acquired distinct functions in the divergent processes of RNA maturation, including RNA cleavage and RNA editing.
INTRODUCTION
Pentatricopeptide repeat (PPR) proteins form a large protein family that is particularly prevalent in land plants and includes 450 members in Arabidopsis thaliana (Lurin et al., 2004; O'Toole et al., 2008). The family members are defined by a tandem array of PPR motifs, each of which is a highly degenerate unit consisting of 35 amino acids and expected to fold into a pair of antiparallel helices (Small and Peeters, 2000). Most PPR proteins are predicted to localize to plastids or mitochondria (Lurin et al., 2004). PPR proteins are involved in almost all stages of gene expression, including transcription (Pfalz et al., 2006), splicing (Schmitz-Linneweber et al., 2006; de Longevialle et al., 2007, 2008), RNA cleavage (Hashimoto et al., 2003; Meierhoff et al., 2003; Hattori et al., 2007), RNA editing (Kotera et al., 2005; Okuda et al., 2007, 2008; Chateigner-Boutin et al., 2008; Zhou et al., 2008), translation (Fisk et al., 1999; Williams and Barkan, 2003), and RNA stabilization (Yamazaki et al., 2004; Beick et al., 2008). The most probable explanation for these divergent roles is that they are sequence-specific RNA binding adaptors recruiting effector enzymes to the target RNA (Delannoy et al., 2007). The PPR protein family is divided into P and PLS subfamilies (Lurin et al., 2004); the latter accounts for roughly half of the members in Arabidopsis and is specific to land plants (O'Toole et al., 2008). Based on differences in C-terminal motifs, the PLS subfamily is further classified into PLS, E, and DYW subclasses (Schmitz-Linneweber and Small, 2008).
In RNA editing, specific cytidine nucleotides are altered to uridine in RNA in mitochondria and plastids of higher plants (Shikanai, 2006). Thirty-four sites are edited in 18 transcripts of Arabidopsis plastids (Chateigner-Boutin and Small, 2007), and >450 sites are edited in almost all protein-encoding transcripts of Arabidopsis mitochondria (Giege and Brennicke, 1999; Bentolila et al., 2008; Zehrmann et al., 2008). In vivo approaches using plastid transformation and in vitro RNA editing assays clarified that a cis-element consisting of <30 nucleotides surrounding the editing site is essential for site recognition (Chaudhuri and Maliga, 1996; Hirose and Sugiura, 2001; Sasaki et al., 2006). The case is similar in mitochondria (Takenaka et al., 2004). Nucleus-encoded factors responsible for specific RNA editing have been identified by genetic studies. Expression of the Arabidopsis genes CHLORORESPIRATORY REDUCTION4 (CRR4) and CRR21 are required for RNA editing of sites 1 (ndhD-1) and 2 (ndhD-2) in ndhD transcripts, respectively (Kotera et al., 2005; Okuda et al., 2007). ndhD encodes a subunit of the chloroplast NAD(P)H dehydrogenase (NDH) complex involved in photosystem I cyclic electron flow (Shikanai et al., 1998). CHLOROPLAST BIOGENESIS19 (CLB19) is required for RNA editing of rpoA and clpP transcripts (Chateigner-Boutin et al., 2008). All three of these PPR proteins belong to the E subclass (Kotera et al., 2005; Okuda et al., 2007; Chateigner-Boutin et al., 2008). We previously demonstrated that CRR4 binds to the sequence surrounding its target site (Okuda et al., 2006). It is generally accepted that a PPR protein is a trans-factor essential for recognizing the RNA editing site. However, none of these PPR proteins appear to possess a domain likely to catalyze the editing reaction, suggesting that the unknown editing enzyme is a different protein.
Salone et al. (2007) proposed that PPR proteins of the DYW subclass, named for the highly conserved C-terminal DYW tripeptides (Asp, Tyr, and Trp), might carry the catalytic function required for RNA editing in plant organelles. The DYW subclass is defined by the characteristic DYW motif (Lurin et al., 2004), which contains invariant Cys and His residues matching the active site of cytidine deaminases, including the human RNA editing enzyme APOBEC1 (Salone et al., 2007). In addition, the phylogenetic distribution of the DYW motif is strictly correlated with the presence of RNA editing (Salone et al., 2007), an idea that was supported by more extensive analysis of transcripts encoding DYW proteins in early-diverging bryophytes (Rüdinger et al., 2008). More recently, genetic evidence about a link between the DYW subclass and RNA editing was provided. The Arabidopsis yellow seedling1 (ys1) mutant, defective in a gene encoding a DYW subclass PPR protein, is specifically impaired in RNA editing of rpoB-1 (Zhou et al., 2008). In contrast with ys1, the Arabidopsis crr2 mutant, which is defective in a different DYW gene, is specifically impaired in intercistronic RNA cleavage of rps7/ndhB transcripts but not in RNA editing (Hashimoto et al., 2003). Furthermore, the DYW motif of At2g02980 was shown to have endoribonuclease activity in vitro (Nakamura and Sugita, 2008). The At2g02980 product targets to mitochondria, and its defect causes a severe dwarf phenotype, but its molecular function is unknown (Nakamura and Sugita, 2008). Similarly, endoribonuclease activity was also shown in the DYW motif of Os05g30710 in the same study (Nakamura and Sugita, 2008). These results suggest that the DYW motif serves as a sequence-specific endoribonuclease with the aid of PPR motifs. Thus, the function of the DYW subclass is still controversial. Of the 90 DYW genes in Arabidopsis, only the function of two genes, CRR2 and YS1, is known. To clarify the role of the DYW subclass, it is essential to analyze its members more extensively.
Here, we report that the DYW proteins CRR22 and CRR28 are involved in multiple RNA editing events. We also demonstrate that the DYW motifs of CRR22 and CRR28 are functionally distinct from that of CRR2 as an endoribonuclease by in vivo and in vitro analyses. Based on these results, we discuss the evolution of the DYW subclass and RNA editing in plant organelles.
RESULTS
Arabidopsis crr22 and crr28 Mutants Are Defective in DYW Genes Required for NDH Activity
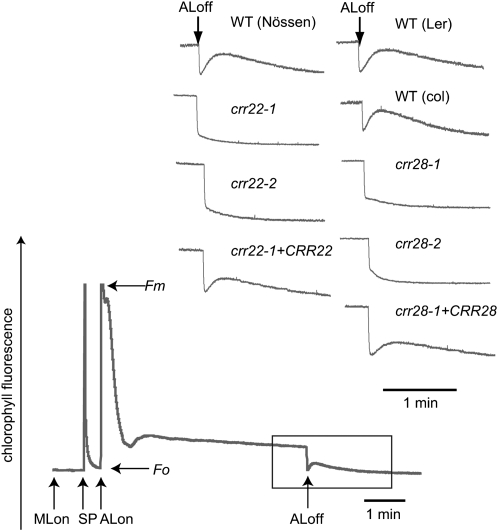
Chloroplast NDH catalyzes electron donation to plastoquinone from the stromal electron pool, and its activity can be monitored as a transient increase in chlorophyll fluorescence reflecting plastoquinone reduction after turning off actinic light (Shikanai et al., 1998). We focused on this fluorescence change to identify Arabidopsis crr mutants with impaired NDH activity (Hashimoto et al., 2003). Both crr22 and crr28 mutants were isolated by screening of Ds transposon tagged-lines by pulse amplitude modulation fluorometry (Okuda et al., 2007). Figure 1 shows a typical trace of the chlorophyll fluorescence level in the wild type. In crr22 and crr28, the postillumination increase was suppressed (Figure 1), indicating impaired NDH activity.
Figure 1.
Monitoring NDH Activity Using Chlorophyll Fluorescence Analysis.
The curve shows a typical trace of chlorophyll fluorescence in the wild type. Leaves were exposed to actinic light (50 μmol photons m−2 s−1) for 5 min. Actinic light was turned off, and the subsequent change in chlorophyll fluorescence level was monitored. The transient increase in chlorophyll fluorescence is due to the plastoquinone reduction based on NDH activity. Insets are magnified traces from the boxed area. The fluorescence levels were normalized by the maximum fluorescence at closed photosystem II centers in the dark (Fm) levels. ML, measuring light; SP, saturating pulse of white light; crr22-1+CRR22, crr22-1 complemented by introduction of the wild-type genomic CRR22; crr28-1+CRR28, crr28-1 complemented by introduction of the wild-type genomic CRR28.
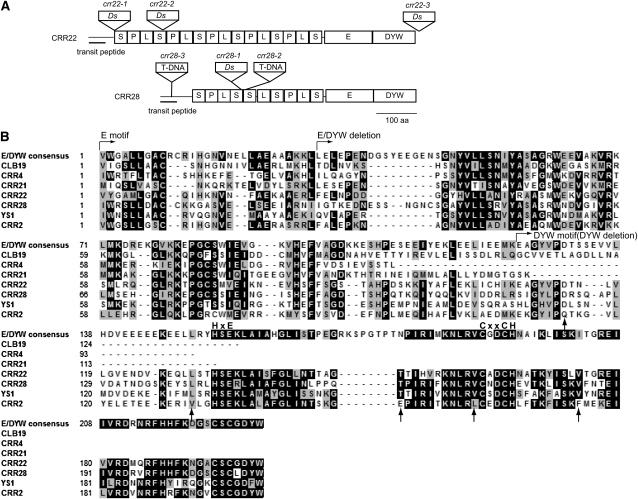
In crr22-1, crr22-2, and crr22-3, At1g11290 was disrupted by independent insertions of Ds, while At1g59720 was disrupted in crr28-1 (Figure 2A). In crr28-2 (SALK_115133) and crr28-3 (SALK_012455), At1g59720 was disrupted by T-DNA insertions (Figure 2A). Both crr22-1 and crr28-1 are recessive mutations, and their crr phenotype cosegregated with the Ds insertions. The wild-type genomic DNAs covering each gene were introduced into crr22-1 and crr28-1, respectively, resulting in full restoration of the postillumination increase in fluorescence (Figure 1). We conclude that the crr22 and crr28 phenotypes are due to the disruption of At1g11290 and At1g59720, respectively. The CRR22 and CRR28 genes do not contain introns and encode putative PPR proteins consisting of 809 and 638 amino acids, respectively. The program ChloroP 1.1 (Emanuelsson et al., 1999) predicted that the first 46 amino acids of CRR22 and the first 40 amino acids of CRR28 were plastid targeting signals (Figure 2A). CRR22 and CRR28 contain 16 and 10 PPR or PPR-like (P, L, L2, and S) motifs, respectively (Figure 2A). Both CRR22 and CRR28 contain E and DYW motifs and are thus classified in the DYW subclass.
Figure 2.
Predicted Motif Structure of CRR22 and CRR28.
(A) PPR (or PPR-like), E, and DYW motifs are depicted as boxes with letters. The designation of the P, L, and S corresponds to the PPR motif, PPR-like S (for short) motif, and PPR-like L (for long) motif, respectively, proposed by Lurin et al. (2004). The putative plastid transit peptides are underlined. Sites of Ds or T-DNA insertions in mutant alleles are indicated.
(B) Comparison of the E and DYW motifs among CLB19, CRR4, CRR21, YS1, and CRR2. Alignment was performed using the ClustalW program. The consensus sequence of the E and DYW motifs according to Lurin et al. (2004) is shown at the top. Amino acids that are fully or semiconserved are shaded black and gray, respectively. The invariant Cys and His residues in the DYW motif (Salone et al., 2007) are indicated above the sequences. Amino acids that are conserved among CRR22, CRR28, and YS1 but not in CRR2 are indicated by arrows. The point at which the sequences were truncated in the CRR2ΔDYW, CRR22ΔDYW, CRR28ΔDYW, CRR22ΔEDYW, and CRR28ΔEDYW constructs is specified.
crr22 and crr28 Are Impaired in RNA Editing of Several Specific Sites
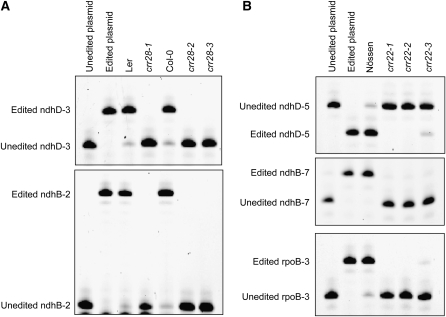
PPR proteins are generally involved in gene expression in organelles, including RNA editing in higher plants (Andres et al., 2007), implying that the gene expression of chloroplast ndh gene(s) would be impaired in crr22 and crr28. We systematically examined the editing status of chloroplast transcripts using a new high-resolution melting screen (Chateigner-Boutin and Small, 2007) (see Supplemental Figure 1 online). Among 34 RNA editing sites present in Arabidopsis plastids (Tillich et al., 2005; Chateigner-Boutin and Small, 2007), we identified defects in RNA editing of ndhB-7, ndhD-5, and rpoB-3 in crr22, and ndhB-2 and ndhD-3 in crr28. These defects were confirmed by more sensitive poisoned primer extension assays (Figure 3). crr22-3 is considered to be a weak allele since the Ds transposon inserts into 3′-untranslated region sequences (Figure 2A). This can explain the weak editing activity remaining in crr22-3 (Figure 3B). Other sites were edited correctly as in the wild type.
Figure 3.
Editing Defects in crr28 and crr22 Mutants.
Poisoned primer extension assays were conducted on the editing sites ndhD-3 (116290) and ndhB-2 (96698) for crr28 (A) and ndhD-5 (116281), ndhB-7 (96419), and rpoB-3 (25779) for crr22 (B). The editing sites are specified relative to the nucleotide sequence of the complete Arabidopsis chloroplast genome (Genbank Accession number AP000423). RT-PCR products were obtained with primers surrounding the editing sites and serve as templates for the extension reaction from a 5′-labeled 6-carboxyfluorescein primer that anneals next to the target editing site (a forward poisoned primer extension primer was used for all sites except for ndhD-5, for which we used a reverse primer). The extension is stopped by the incorporation of ddCTP at the location of the editing site for unedited molecules producing a short unedited product. The extension is stopped at the next C/G for the edited molecules producing a longer edited product. For ndhD-5, the extension is stopped by the incorporation of ddATP giving a longer product for the unedited ndhD-5.
The defects in RNA editing may be secondarily caused by aberrant RNA processing. To test this possibility, the levels and patterns of transcripts were analyzed by RNA gel blots. Supplemental Figure 2 online shows that there are no obvious alterations in ndhB, ndhD, and rpoB transcripts either in crr22 or crr28. Exceptionally, ndhB transcripts accumulate more in crr22 than in the wild type. We cannot eliminate the possibility that this change influences the RNA editing efficiency by titration of trans-factors (Chaudhuri et al., 1995), but the other sites present in the same transcript were completely edited, contrasting with the complete loss of editing in ndhB-7. We conclude that crr22 and crr28 are primarily defective in multiple RNA editing events in the ndhB, ndhD, and rpoB transcripts.
NDH Activity Was Specifically Impaired in crr22 and crr28
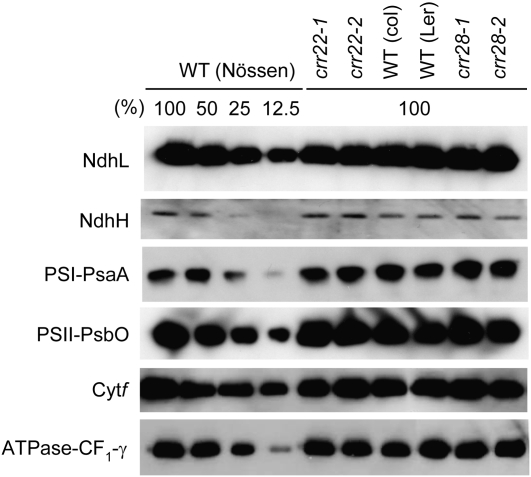
RNA editing at ndhB-2 and ndhB-7 converts Pro-156 to Leu and Ser-249 to Phe, respectively, while editing at ndhD-3 and ndhD-5 converts Ser-293 to Leu and Phe-296 to Leu. Thus, defects in RNA editing in the crr22 and crr28 mutants will result in amino acid changes that may destabilize NdhB and NdhD. To assess whether the subunits stably accumulate in vivo, protein blots were analyzed using antibodies against NdhH and NdhL. The NDH complex is unstable without NdhB or NdhD (Peng et al., 2008), and the antibodies can be used to monitor accumulation of the complex. In crr22 and crr28, neither NdhH nor NdhL levels were substantially affected (Figure 4). Because the editing at the NdhB and NdhD sites in these mutants was below detection levels (Figure 3), probably all of the NdhB and NdhD accumulating in the mutants was translated from unedited RNA. These results suggest that Ser-249 and Phe-289 in NdhB and Leu-293 and Leu-296 in NdhD are not essential for stabilizing NDH, but some or all of them are essential for NDH activity. Editing of these transcripts results in the restoration of codons for amino acids conserved in other land plants (see Supplemental Figure 3 online).
Figure 4.
Protein Blot Analysis of the NDH Complex and the Major Photosynthetic Complexes.
Immunodetection of NDH subunits, NdhH and NdhL; a subunit of the Cytb6f complex, Cytf; a subunit of photosystem I, PsaB; a subunit of photosystem II, PsbO; and the γ-subunit of the chloroplast F0F1-ATPase, CF1-γ. The proteins were extracted from thylakoid membrane fractions. Lanes were loaded with protein samples corresponding to 0.5 μg chlorophyll for Cytf, PsaB, PsbO, and CF1-γ, 1 μg chlorophyll for NdhL, and 5 μg chlorophyll for NdhH (100%) and the series of dilutions indicated.
The protein blots were also analyzed to monitor the accumulation of major photosynthetic complexes by probing their subunits. Levels of subunits in both photosystems, cytochrome (Cyt) b6f complex, and chloroplast F0F1-ATPase were not substantially affected in crr22 or crr28 (Figure 4). Consistent with these results, crr22 and crr28 did not show any phenotypic changes in photosynthetic electron transport except for NDH activity (see Supplemental Figure 4 online).
In addition to ndhB-7 and ndhD-5, crr22 is impaired in editing of rpoB-3, which converts Ser-184 to Leu in RpoB, one of the core subunits of the plastid-encoded RNA polymerase (PEP). Leu-184 is generally produced by RNA editing in both dicots and monocots (Tsudzuki et al., 2001). ΔrpoB tobacco (Nicotiana tabacum) plants completely lacking PEP are defective in pigment accumulation and photosynthesis (Allison et al., 1996), which results from defective transcription in chloroplasts, notably from a drastic decrease in transcription of photosynthesis-related genes (Hajdukiewicz et al., 1997). If Leu-184 was essential for PEP, crr22 would exhibit highly perturbed transcript patterns, such as those observed in ΔrpoB tobacco and also in Arabidopsis mutants defective in PEP, such as ptac2, clb19, and ys1 (Pfalz et al., 2006; Chateigner-Boutin et al., 2008; Zhou et al., 2008). Hierarchical clustering of crr22 plastid transcript profiles shows that their RNA profile is similar to that of wild-type plants and other crr mutants but not to that of PEP mutants (see Supplemental Figure 5 and Supplemental Table 1 online). Furthermore, we showed that RpoA protein levels are not affected in crr22 (see Supplemental Figure 6 online). Consistent with the lack of an observed PEP phenotype in crr22, the residue corresponding to Leu-184 in Escherichia coli RpoB is located in Dispensable Region I, which can be deleted without any significant effect on the fidelity of the RNA polymerase (Borukhov et al., 1991). We conclude that under the growth conditions employed, both crr22 and crr28 are specifically defective in NDH.
The DYW Motif Is Essential for CRR2 but Not for CRR22 and CRR28
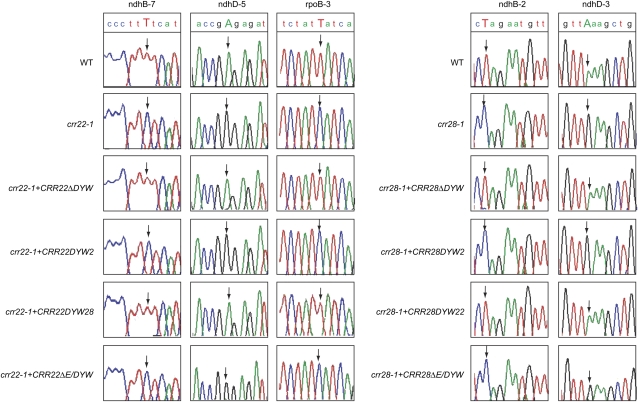
CRR22 and CRR28 are involved in multiple RNA editing events (Figure 3), which is consistent with the hypothesis that the DYW motif might be characteristic of an RNA editing enzyme (Salone et al., 2007). However, the DYW-containing CRR2 protein is required for RNA cleavage, crr2 mutants have no discernable editing defects (Hashimoto et al., 2003), and DYW motifs of At2g02980 and Os05g30710 have endoribonuclease activity in vitro (Nakamura and Sugita, 2008). To study the reason for this discrepancy, we focused on the function of the DYW motifs of CRR2, CRR22, and CRR28. First, we examined whether the DYW motifs of CRR22 and CRR28 are essential for RNA editing in vivo. CRR22 and CRR28, in which their DYW motifs were deleted and the truncated proteins were fused with the HA-tag, were expressed in crr22-1 and crr28-1, respectively, and both transgenes were driven by their own promoters. Both truncated genes could completely restore RNA editing at all five sites (Figure 5). We conclude that the DYW motifs of CRR22 and CRR28 are dispensable for RNA editing in vivo.
Figure 5.
The Effects on RNA Editing of Deleting or Swapping the DYW Motifs in CRR22 and CRR28.
Nucleotide sequences including the RNA editing sites of ndhB-2, ndhB-7, ndhD-3, ndhD-5, and rpoB-3 are shown as sequencing chromatograms. Editing sites are indicated by arrows pointing to the corresponding peaks. crr22-1+CRR22ΔDYW, crr22-1 transformed with CRR22 lacking the DYW motif; crr28-1+CRR28ΔDYW, crr28-1 transformed with CRR28 lacking the DYW motif; crr22-1+CRR22DYW2, crr22-1 transformed with CRR22, in which the DYW motif was replaced by that of CRR2; crr28-1+CRR28DYW2, crr28-1 transformed with CRR28, in which the DYW motif was replaced by that of CRR2; crr22-1+CRR22DYW28, crr22-1 transformed with CRR22, in which the DYW motif was replaced by that of CRR28; crr28-1+CRR28DYW22, crr28-1 transformed with CRR28, in which the DYW motif was replaced by that of CRR22; crr22-1+CRR22ΔE/DYW, crr22-1 transformed with CRR22 lacking the E and DYW motifs; crr28-1+CRR28ΔE/DYW, crr28-1 transformed with CRR28 lacking the E and DYW motifs.
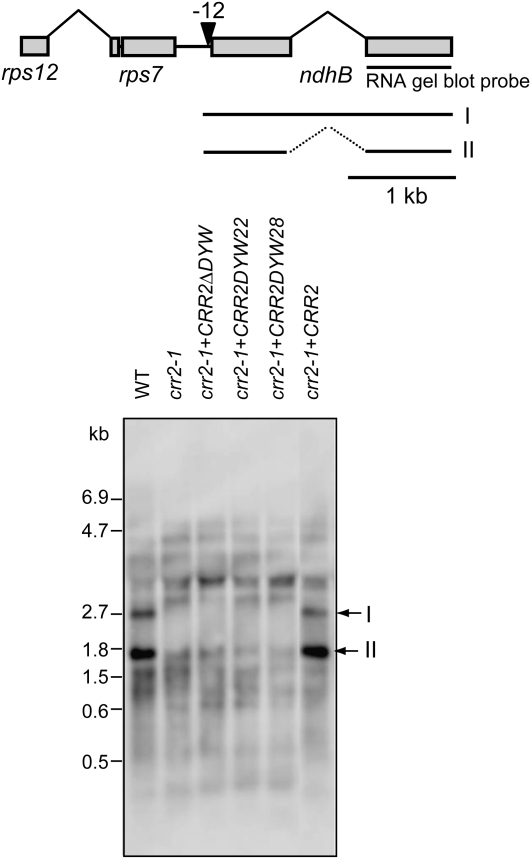
Next, we introduced CRR2 that lacks its DYW motif and is fused with the HA-tag into crr2-1. Again, the transgene was expressed under the control of its native promoter. The signal corresponding to the mature form of ndhB transcript was detected in the wild type and in crr2-1 transformed with full-length CRR2 fused with the HA-tag, indicating that the HA-tag does not interfere with CRR2 function (Figure 6). By contrast, the mature ndhB transcript was not detectable in crr2-1 expressing CRR2 lacking its DYW motif (Figure 6). Thus, in contrast with the results for CRR22 and CRR28 (Figure 5), the DYW motif is essential for CRR2 function in vivo. Although the proteins were fused with an HA-tag, we could not detect them in protein blots even for full-length CRR2-HA, probably due to the low accumulation level of PPR proteins involved in the expression of ndh genes. We cannot eliminate the possibility that CRR2 lacking its DYW motif is unstable in vivo, but similar truncations did not affect the function of CRR22 and CRR28 (Figure 5).
Figure 6.
The Effects of Deleting or Swapping the DYW Motif in CRR2.
The rps7-ndhB region is shown schematically. The arrowhead indicates the site that is not cleaved in crr2; this site is located at position −12 with respect to the ndhB translation initiation codon. Total RNA (5 μg) isolated from leaves of 4-week-old wild-type and transgenic plants was analyzed by RNA gel blot and hybridization. The probe used for the experiments is indicated by a bar beneath the 3′ exon of ndhB. The signal identities (I and II) were based on previous analysis (Hashimoto et al., 2003). The migration of RNA size markers is indicated at the left. crr2-1+CRR2ΔDYW, crr2-1 transformed with CRR2 lacking the DYW motif; crr2-1+CRR2DYW22, crr2-1 transformed with CRR2, in which the DYW motif was replaced by that of CRR22; crr2-1+CRR2DYW28, crr2-1 transformed with CRR2, in which the DYW motif was replaced by that of CRR28; crr2-1+CRR2, crr2-1 complemented by introduction of the wild-type genomic CRR2.
The DYW Motifs of CRR22 and CRR28 Are Functionally Distinct from That of CRR2
CRR22 and CRR28 do not exhibit high sequence similarity to CRR2 with respect to PPR motifs, consistent with the idea that the tandem array of PPR motifs determines RNA binding specificity (Okuda et al., 2006; Delannoy et al., 2007). By contrast, their DYW motifs are well conserved (Figure 2B). The DYW motif of CRR2 shows 48 and 52% sequence identity (60 and 61% similarity) to that of CRR22 and CRR28, respectively. The DYW motif contains invariant His and Cys residues conserved in human APOBEC1 (Salone et al., 2007), and these Cys residues are critical for the endoribonuclease activity of the DYW motif from At2g02980 (Nakamura and Sugita, 2008). These residues are completely conserved in the DYW motifs of CRR22 and CRR28 (Figure 2B). We clarified that the DYW motif is essential in vivo for CRR2 but not for CRR22 and CRR28 (Figures 5 and 6), suggesting that the function of the DYW motif is not equivalent among PPR proteins. To examine this possibility, the DYW motif of CRR2 was replaced by that of CRR22 and CRR28, and the chimeric genes were introduced into crr2-1. The transgenes were expressed by their own promoters. Despite the high similarity, the DYW motifs of CRR22 and CRR28 could not complement the function of the DYW motif in CRR2 (Figure 6).
Reciprocally, the DYW motifs of CRR22 and CRR28 were exchanged with that of CRR2, and the chimeric genes were introduced into crr22-1 and crr28-1, respectively. The transgenes were expressed by their own promoters. The chimeric genes did not restore CRR22 or CRR28 function (Figure 5). This is inconsistent with the fact that the DYW motif is dispensable in CRR22 and CRR28 (Figure 5). This result could be explained by the effect of the putative endoribonuclease activity of the CRR2 DYW motif on the target transcripts. However, the hybridization patterns of ndhB, ndhD, and rpoB transcripts were identical between the transgenic lines with chimeric genes and their original mutants (see Supplemental Figure 7 online).
Although the DYW motif was not interchangeable between CRR2 and CRR22, nor between CRR2 and CRR28, we tested whether the DYW motif could be functionally interchanged between CRR22 and CRR28. The relevant chimeric genes were introduced into crr22-1 and crr28-1, respectively. The transgenes were expressed by their own promoters. Both chimeric genes could complement the function of the original gene (Figure 5). Taken together, the DYW motifs of CRR22 and CRR28 have identical functions that, despite the high sequence similarity, are distinct from the function of the DYW motif of CRR2.
The E Motifs of CRR22 and CRR28 Are Essential for RNA Editing
The DYW motif is dispensable in vivo for CRR22 and CRR28 (Figure 5). This result is consistent with the fact that CRR4, CRR21, and CLB19 lacking the DYW motif are also RNA editing factors (Kotera et al., 2005; Okuda et al., 2007; Chateigner-Boutin et al., 2008). We previously showed that the E motif is essential for RNA editing and its function is common between CRR4 and CRR21 (Okuda et al., 2007). This motif may be involved in recruiting an unknown editing enzyme and shows high similarity among CRR4, CRR21, CLB19, CRR22, and CRR28 (Figure 2B), suggesting that the function of E motifs may also be conserved in the DYW proteins CRR22 and CRR28. To test this possibility, CRR22 and CRR28 lacking both their E and DYW motifs were expressed in crr22-1 and crr28-1, respectively. The transgenes were expressed by their own promoters. The editing function of neither CRR22 nor CRR28 was complemented by the introduction of the truncated genes (Figure 5). Since the deletion of the E motif is unlikely to affect RNA binding (Okuda et al., 2007), we believe that the E motifs of CRR22 and CRR28 are essential for the RNA editing reaction.
The DYW Motif of CRR2 Has Higher Endoribonuclease Activity Than That of CRR22
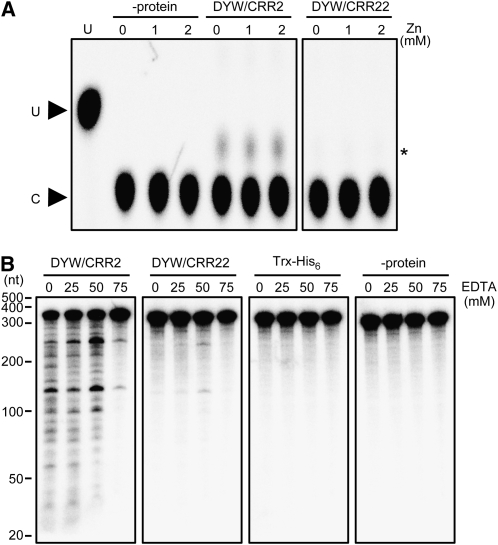
To test the possibility that the DYW motifs of CRR2 and CRR22 have different activities, the respective recombinant proteins were purified from E. coli (see Supplemental Figure 8 online) and used in RNA editing and cleavage assays in vitro. So far, we have been unable to express the DYW motif of CRR28. As a substrate, a 318-nucleotide RNA containing the 5′ exon of ndhB was generated by in vitro transcription, and cytidine residues were labeled by 32P. If any cytidine, including that targeted by CRR22 in vivo, was edited, it would be detected as a spot of uridine in thin layer chromatography. Our assay was sensitive enough to detect 1% of the C-to-U conversion in any C residues. However, we did not detect any radioactivity in the position corresponding to uridine using either DYW motif in the presence or absence of zinc ions, which are required for the activity of cytidine deaminases (Navaratnam et al., 1995) (Figure 7A). At least under these experimental conditions, the DYW motifs do not show any cytidine deaminase activity.
Figure 7.
Activity Assay of the DYW Motif in CRR22.
(A) The in vitro RNA editing assay. The [α-32P]CTP–labeled NB2 RNA (0.2 nM) was incubated with the indicated proteins (100 nM) or without proteins (−protein) in the presence or absence of zinc ions (Zn; 0 to 2 mM). The RNA was digested into mononucleotides and separated by thin layer chromatography. The position of the U spot was confirmed using [α-32P]UTP–labeled RNA. The spots indicated by an asterisk are probably pCp residues resulting from RNA cleavage by DYW/CRR2.
(B) The in vitro RNA cleavage assay. The indicated protein (100 nM) was incubated with 0.2 nM 32P-labeled NB2 RNA in buffer containing 10 mM MgCl2. The reaction was performed in the absence or presence of metal ion chelator, EDTA (0 to 75 mM). The reaction was also performed with recombinant thioredoxin-Hisx6 tag protein (Trx-His6) or without any protein (−protein). The 32P-labeled RNAs were extracted and then separated by denaturing PAGE.
The substrate RNA was incubated with the recombinant DYW motifs and subjected to polyacrylamide gel electrophoresis to assay endoribonuclease activity. The DYW motif of CRR2 cleaved the substrate RNA (Figure 7B) like that of At2g02980 and Os05g30710 (Nakamura and Sugita, 2008). This result is consistent with the fact that CRR2 is involved in intercistronic RNA cleavage (Hashimoto et al., 2003). Compared with the endoribonuclease activity detected in the DYW motif of CRR2, the recombinant DYW motif of CRR22 showed significantly lower endoribonuclease activity (Figure 7B). We believe that this trace level of activity depends on the DYW motif of CRR22 since the activity was significantly higher than that of the negative controls (Trx-His6 and -protein) and also since it has a similar dependency on EDTA and a similar cleavage pattern to that seen with the DYW motif of CRR2 (Figure 7B).
DISCUSSION
Like CLB19 (Chateigner-Boutin et al., 2008), CRR22 and CRR28 recognize multiple RNA editing sites (Figure 3). In vitro RNA editing assays have suggested that the cis-elements recognized by the same trans-factor show high sequence identity (Kobayashi et al., 2007). Consistent with this idea, the nucleotides surrounding the ndhB-2 and ndhD-3 sites are highly conserved (see Supplemental Figure 9 online), as has been observed in several other cases where a single PPR protein recognizes two RNAs (Schmitz-Linneweber et al., 2005). By contrast, the putative cis-regions of ndhB-7, ndhD-5, and rpoB-3 (−40 to +10 surrounding the editing sites) show no obvious similarity (see Supplemental Figure 9 online). Although the putative cis-elements of RpoB-3 and Rps2-1 sites are also partially conserved in tobacco and are hypothesized to be detected by the same trans-factor (Chateigner-Boutin and Hanson, 2002), the Rps2-1 site is encoded by T in the Arabidopsis genome. Moreover, little sequence similarity was found between the putative cis-elements of the rpoA and clpP sites recognized by CLB19 (Chateigner-Boutin et al., 2008). It is conceivable that different sets of PPR motifs within a single protein may independently recognize unrelated cis-sequences.
We previously demonstrated that the E motif common to almost all PLS family PPR proteins is essential for RNA editing in vivo by CRR4 and CRR21, presumably via an association with an unknown RNA editing enzyme (Okuda et al., 2007). The E motifs of CRR22 and CRR28 are equally essential for RNA editing (Figure 5). Salone et al. (2007) postulated that the unknown enzyme is the DYW motif, a hypothesis at first sight reinforced by the fact that CRR22, CRR28, and YS1 (Zhou et al., 2008) all contain DYW motifs. However, the in vitro assay using the recombinant DYW motif of CRR22 did not detect any cytidine deaminase activity under our assay conditions (Figure 7A), although we attempted to optimize the concentration of ATP and zinc ions based on information from in vitro chloroplast RNA editing systems (Hirose and Sugiura, 2001) and the assay for human APOBEC1 (Navaratnam et al., 1995). A negative result may not be very informative given the many trivial reasons why an in vitro assay might fail (i.e., the chloroplast in vitro editing reactions only works with a few substrates) (Sasaki et al., 2006) and may require a number of factors that are not supplied in this reaction, although the DYW motifs of CRR22 and CRR28 were dispensable in vivo (Figure 5). Hence, CRR22 and CRR28 behave exactly like the editing specificity factors CRR4, CRR21, and CLB19 despite the presence of the additional DYW motif. Since the DYW motif is dispensable for editing, it may have been lost in members of the E subclass, including CRR4, CRR21, and CLB19.
The loss of the DYW motif appears to have occurred frequently during the evolution of the PPR family in angiosperms. In Physcomitrella patens, all PPR proteins with an E motif also possess a C-terminal DYW motif (O'Toole et al., 2008). In rice (Oryza sativa) and Arabidopsis, the majority of PPR proteins with an E motif lack a DYW motif (O'Toole et al., 2008), and there are several pairs of putative orthologs between the two plants where the rice protein has a DYW motif and the Arabidopsis protein lacks it (for example, OsPPR_10g39460/AtPPR_3g25060, OsPPR_01g08120/AtPPR_3g15930, and OsPPR_01g74600/AtPPR_3g16610). Presumably, at least in Arabidopsis, DYW motifs are not always essential for the function of the protein. These results do not support the idea that the DYW motif has RNA editing activity (Salone et al., 2007). However, we cannot eliminate the possibility that a trans-acting DYW motif complements the function of the DYW motifs present in CRR22 and CRR28 in vivo. E. coli cytidine deaminase forms homodimers with the active sites at the interface of the subunits (Betts et al., 1994). Navaratnam et al. (1998) proposed that like E. coli cytidine deaminase, the catalytically active form of APOBEC-1 is an asymmetric homodimer, one site of which is bound to a U downstream of the edited C, and that this interaction is essential for the C deamination catalyzed by another active site. Both CRR22 and CRR28 may form heterodimers by interaction with other proteins, perhaps via their E motif sequences. We cannot eliminate the possibility that CRR22 and CRR28 interact with other PPR proteins with the DYW motif and the loss of a single DYW motif is complemented by another DYW domain of the partner protein.
In contrast with the unproven function of the DYW motif in CRR22 and CRR28, it is clear that the DYW motif of CRR2 has endoribonuclease activity that is essential for CRR2 function in vivo. The idea was strongly supported by both in vivo and in vitro results (Figures 6 and 7B) and is consistent with the crr2 phenotype (Hashimoto et al., 2003). We conclude that CRR2 is a sequence-specific endoribonuclease, in which the N-terminal PPR motifs are likely to determine its sequence specificity by analogy with CRR4 (Okuda et al., 2007), while the C-terminal DYW motif is a catalytic domain. The DYW motif is essential for CRR2 in vivo and cannot be replaced by the apparently equivalent motifs from CRR22 and CRR28 (Figure 6). This implies there is something specific about the DYW motif of CRR2 that cannot be complemented in trans by other DYW proteins in vivo or even in cis by other DYW motifs. There are no obviously significant differences between the respective DYW motifs of CRR2, CRR22, CRR28, and YS1. All of the Cys and His residues believed to be catalytically essential (Salone et al., 2007) are conserved among all four (Figure 2B). Several amino acids are conserved among CRR22, CRR28, and YS1 but not in CRR2 (Figure 2B), which may affect endoribonuclease activity. The endoribonuclease activity of the DYW motif in CRR22 is weaker than that in CRR2 in vitro (Figure 7B).
What might be the relationship between RNA editing and RNA cleavage? RNA editing is a process of cytidine deamination or transamination in plant mitochondria and does not involve base excision or cleavage of the phosphate backbone (reviewed in Takenaka et al., 2008), so a direct link between endonuclease activity and editing is unlikely. However, alterations in cofactor binding can convert RNA binding proteins into ribonucleases; a particularly interesting example is provided by a zinc finger peptide that efficiently cleaves RNA but only in the absence of zinc (Lima and Crooke, 1999). This is reminiscent of the EDTA-dependent cleavage shown by the DYW motifs in vitro (Figure 7B) and may provide a clue as to how the divergent function of the DYW motif in CRR2 has arisen.
METHODS
Plant Material
Arabidopsis thaliana ecotypes Columbia (Col-0), Landsberg erecta (Ler), and Nössen were used in this study. crr22-1, crr22-2, crr22-3 (Nössen), and crr28-1 (Ler), were mutagenized by Ds transposon insertion (Kuromori et al., 2004; Ito et al., 2005). crr28-2 (SALK_115133, Col-0) and crr28-3 (SALK_012455, Col-0) were obtained from the ABRC Stock Center.
Chlorophyll Fluorescence Analysis
Chlorophyll fluorescence was measured using a MINI-PAM portable chlorophyll fluorometer (Walz). The transient increase in chlorophyll fluorescence after turning off actinic light was monitored as previously described (Shikanai et al., 1998).
Analysis of RNA Editing
High-resolution melting analysis of amplicons was performed as previously described (Chateigner-Boutin and Small, 2007) except that the primers used for the PCR were designed to give shorter amplicons than in the previous study. Poisoned primer extension of RT-PCR products was performed as previously described (Chateigner-Boutin and Small, 2007). For analysis of RNA editing in a series of transgenic plants, total RNA was isolated from rosette leaves using an RNeasy plant mini kit (Qiagen) and treated with DNase I (Invitrogen). DNA-free RNA (2.5 μg) was reverse transcribed with random hexamers. Sequences including the editing sites were amplified by PCR. The RT-PCR products were sequenced directly. The primers are listed in Supplemental Table 2 online.
Immunoblot Analysis
Chloroplasts were isolated from the leaves of 4-week-old plants as previously described (Okuda et al., 2007). The amount of samples was standardized by measuring chlorophyll concentration. The protein samples were separated by 12.5% SDS-PAGE. After electrophoresis, the proteins were transferred onto a Hybond-P membrane (GE Healthcare) and incubated with specific antibodies. The signals were detected using an ECL Advance Western Blotting Detection Kit (for NdhH) (GE Healthcare) or an ECL Plus Western Blotting Detection Kit (for the others) (GE Healthcare) and visualized by a LAS1000 chemiluminescence analyzer (Fuji Film).
RNA Preparation and RNA Gel Blot Analysis
The DNA fragments corresponding to the ndhB 3′ exon region were amplified by PCR (primers given in Supplemental Table 2 online) and labeled with a PCR DIG Probe Synthesis Kit (Roche). Total RNA was isolated from the leaves of 4-week-old plants using an RNeasy plant mini kit (Qiagen) and treated with DNase I (Invitrogen). Total RNA (5 μg) was electrophoresed and then transferred onto a nylon membrane, and hybridized bands were detected using a Gene Image CDP-Star detection kit (GE Healthcare) as previously described (Okuda et al., 2007).
Constructs and Plant Transformation
Plasmids for transgenic plants were constructed by standard techniques. Full details are provided in the Supplemental Methods online. These constructs were cloned into pBIN19 or pGWEB-NB1 binary vectors and introduced into crr22 or crr28 mutant plants via Agrobacterium tumefaciens MP90 or ASE.
Overexpression and Purification of the Recombinant Proteins
The sequences corresponding to the DYW motifs of CRR2 and CRR22 were amplified by PCR using the primers CRR2_H4-F and CRR2_F_R, or DYW22-OX-FW and DYW22-OX-RV, respectively (see Supplemental Table 2 online). The PCR products were ligated in frame into the pBAD/Thio-TOPO vector (Invitrogen). The recombinant proteins were overexpressed in Escherichia coli LMG194 strain as a fusion with thioredoxin and six histidine residues at the N and C termini, respectively. The expressed proteins were purified successively by Probond N resin (Invitrogen) and a His-HP column in an AKTA system (GE Healthcare).
Preparation of RNA Probes
To prepare the 318-nucleotide NB2 RNA probes, the ndhB sequence was amplified using the primers NB2-F and NB2-R (see Supplemental Table 2 online). The PCR product contains the promoter sequence of T7 RNA polymerase and sequences forming stem-loop structures at each end to prevent attack by exonucleases. The PCR product was then in vitro transcribed by T7 RNA polymerase to produce a [α-32P]CTP– or [α-32P]UTP–labeled NB2 RNA.
Assay of Cytidine Deaminase Activity
[α-32P]CTP–labeled NB2 RNA (0.2 nM, 5000 cpm/fmol) was incubated for 30 min at 25°C with the recombinant protein (100 nM) in 10 mM Tris-HCl, pH 7.9, 30 mM KCl, 6 mM MgCl2, 2 mM ATP, 2 mM DTT, 8% glycerol, and 0 to 2 mM ZnCl2 (Hirose and Sugiura, 2001). The RNA was extracted and digested at 37°C for 3 h into 5′ mononucleotides by 1 μg of nuclease P1 and 120 units of S1 nuclease (Takara) in the presence of 50 mM ammonium acetate, pH 4.8. The resultant mononucleotides were separated on a cellulose thin layer chromatography plate using isopropanol/hydrochloride/water (70:15:15). The separated 32P-mononucleotides were visualized by a FLA-5000 phosphor imager (Fuji Film).
RNA Cleavage Assay
Internal [α-32P]CTP–labeled NB2 RNA (0.2 nM, 5000 cpm/fmol) and the recombinant protein (100 nM) were incubated for 15 min at 25°C in 10 mM Tris-HCl, pH 7.9, 30 mM KCl, 6 mM MgCl2, 2 mM DTT, and 8% glycerol in the presence or absence of EDTA (0 to 75 mM). After incubation, the 32P-RNA was extracted by phenol/chloroform followed by ethanol precipitation and then analyzed on a 6% polyacrylamide gel containing 6 M urea.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CLB19 (AT1G05750, Q9MA50), CRR2 (AT3G46790, Q9STF3), CRR4 (AT2G45350, O22137), CRR21 (AT5G55740, Q9FM64), CRR22 (AT1G11290, Q3E6Q1), CRR28 (AT1G59720, Q0WQW5), and YS1 (AT3G22690, Q9LUJ2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. High-Resolution Melting Screen of crr22 and crr28 Mutants.
Supplemental Figure 2. Transcripts Profiles of Genes with Editing Defects in crr22 and crr28.
Supplemental Figure 3. Partial Sequence Alignments of NdhB, NdhD, and RpoB around the Amino Acids Affected by RNA Editing.
Supplemental Figure 4. In Vivo Analysis of Electron Transport Activity.
Supplemental Figure 5. Hierarchical Clustering of crr22 and crr28 Mutants Based on Their Plastid RNA Profiles Assessed by Quantitative RT-PCR.
Supplemental Figure 6. Protein Blot Analysis of the Plastid-Encoded RNA Polymerase.
Supplemental Figure 7. Patterns of rps7-ndhB, psaC-ndhD, and rpoB Transcripts.
Supplemental Figure 8. Purified Recombinant Proteins Used in This Study.
Supplemental Figure 9. Comparison of the Nucleotide Sequences in the Regions Surrounding the Editing Sites Affected in crr22 and crr28.
Supplemental Table 1. Comparison of Chloroplastic Transcript Abundance.
Supplemental Table 2. Oligonucleotide Primers Used.
Supplemental Methods.
Supplemental References.
Supplemental Dataset 1. Text File of Alignment in Figure 1B.
Supplemental Dataset 2. Text File of the Alignment in Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank Asako Tahara (Kyoto University, Kyoto, Japan) and Misato Hirose (Kyushu University, Fukuoka, Japan) for skilled technical support. We also thank Tsuyoshi Endo (Kyoto University, Kyoto, Japan), Amane Makino (Tohoku University, Sendai, Japan), Toru Hisabori (Tokyo Institute of Technolgy, Yokohama, Japan), Yuzuru Tozawa (Ehime University, Matsuyama, Japan), and Kensuke Kusumi (Kyushu University, Fukuoka, Japan) for giving us antibodies. We are also grateful to Tsuyoshi Nakagawa (Shimane University, Matsue, Japan) for giving us the binary vector pGWB-NB1. This work was supported by Grant-in-Aid 16085206 for Scientific Research on Priority Areas, Grant 17GS0316 for Creative Science Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, GPN0008), a grant from the Australian Research Council (CE0561495), and support from the Western Australian State Government and the University of Western Australia.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Toshiharu Shikanai (shikanai@pmg.bot.kyoto-u.ac.jp).
Online version contains Web-only data.
References
- Allison, L.A., Simon, L.D., and Maliga, P. (1996). Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Andres, C., Lurin, C., and Small, I. (2007). The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant. 129 14–22. [Google Scholar]
- Beick, S., Shumitz-Linneweber, C., Williams-Carrier, R., Jensen, B., and Barkan, A. (2008). The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28 5337–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila, S., Elliott, L.E., and Hanson, M.R. (2008). Genetic architecture od mitochondrial editing in Arabidopsis thaliana. Genetics 178 1693–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts, L., Xiang, S., Short, S.A., Wolfenden, R., and Carter, C.W.J. (1994). Cytidine deaminase. The 2.3 Å crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 235 635–656. [DOI] [PubMed] [Google Scholar]
- Borukhov, S., Severinov, K., Kashlev, M., Lebedev, A., Bass, I., Rowland, G.C., Lim, P.P., Glass, R.E., Nikforov, V., and Goldfarb, A. (1991). Mapping of trypsin cleavage and antibody-binding sites and delineation of a dispensable domain in the beta subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 15 23921–23926. [PubMed] [Google Scholar]
- Chateigner-Boutin, A.L., and Hanson, M.R. (2002). Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol. Cell. Biol. 22 8448–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin, A.L., Ramos-Vega, M., Guevara-Garcia, A., Andrés, C., de la Luz Gutiérrez-Nava, M., Cantero, A., Delannoy, E., Jiménez, L.F., Lurin, C., Small, I., and León, P. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56 590–602. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin, A.L., and Small, I. (2007). A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 35 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, S., Carrer, H., and Maliga, P. (1995). Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J. 15 2951–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, S., and Maliga, P. (1996). Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 15 5958–5964. [PMC free article] [PubMed] [Google Scholar]
- Delannoy, E., Stanley, W.A., Bond, C.S., and Small, I.D. (2007). Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 35 1643–1647. [DOI] [PubMed] [Google Scholar]
- de Longevialle, A.F., Hendricson, L., Taylor, N.L., Delannoy, E., Lurin, C., Badger, M., Millar, A.H., and Small, I. (2008). The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 56 157–168. [DOI] [PubMed] [Google Scholar]
- de Longevialle, A.F., Meyer, E.H., Andrés, C., Taylor, N.L., Lurin, C., Millar, A.H., and Small, I.D. (2007). The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege, P., and Brennicke, A. (1999). RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 96 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P.T., Allison, L.A., and Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M., Endo, T., Peltier, G., Tasaka, M., and Shikanai, T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36 541–549. [DOI] [PubMed] [Google Scholar]
- Hattori, M., Miyake, H., and Sugita, M. (2007). A pentatricopeptide repeat protein is required for RNA processing of clpP pre-mRNA in moss chloroplasts. J. Biol. Chem. 282 10773–10782. [DOI] [PubMed] [Google Scholar]
- Hirose, T., and Sugiura, M. (2001). Involvement of site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J. 20 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Motohashi, R., Kuromori, T., Noutoshi, Y., Seki, M., Kamiya, A., Mizukado, S., Sakurai, T., and Shinozaki, K. (2005). A resource of 5,814 dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol. 46 1149–1153. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Matsuo, M., Sakamoto, A., Wakasugi, T., Yamada, K., and Obokata, J. (2007). Two RNA editing sites with cis-elements of moderate sequence identity are recognized by an identical site-recognition protein in tobacco chloroplasts. Nucleic Acids Res. 36 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera, E., Tasaka, M., and Shikanai, T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433 326–330. [DOI] [PubMed] [Google Scholar]
- Kuromori, T., Hirayama, T., Kiyosue, Y., Takabe, H., Mizukado, S., Sakurai, T., Akiyama, K., Kamiya, A., Ito, T., and Shinozaki, K. (2004). A collection of 11,800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 37 897–905. [DOI] [PubMed] [Google Scholar]
- Lima, W.F., and Crooke, S.T. (1999). Highly efficient endonucleolytic cleavage of RNA by a Cys2His2 zinc-finger peptide. Proc. Natl. Acad. Sci. USA 96 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., and Schuster, G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15 1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T., and Sugita, M. (2008). A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett. 582 4163–4168. [DOI] [PubMed] [Google Scholar]
- Navaratnam, N., Bhattacharya, S., Fujino, T., Patel, D., Jamuz, A.L., and Scott, J. (1995). Evolutionary origins of apoB mRNA editing: Catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell 81 187–195. [DOI] [PubMed] [Google Scholar]
- Navaratnam, N., Fujino, T., Bayliss, J., Jarmuz, A., How, A., Richardson, N., Somasekaram, A., Bhattacharya, S., Carter, C., and Scott, J. (1998). Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J. Mol. Biol. 275 695–714. [DOI] [PubMed] [Google Scholar]
- Okuda, K., Habata, Y., Kobayashi, Y., and Shikanai, T. (2008). Amino acid sequence variations in Nicotiana CRR4 orthologs determine the species-specific efficiency of RNA editing in plastids. Nucleic Acids Res. 36 6155–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, K., Myouga, F., Motohashi, R., Shinozaki, K., and Shikanai, T. (2007). Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. USA 104 8178–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, K., Nakamura, T., Sugita, M., Shimizu, T., and Shikanai, T. (2006). A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 281 37661–37667. [DOI] [PubMed] [Google Scholar]
- O'Toole, N., Hattori, M., Andres, C., Iida, K., Lurin, C., Schumitz-Linneweber, C., Sugita, M., and Small, I. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25 1120–1128. [DOI] [PubMed] [Google Scholar]
- Peng, L., Shimizu, H., and Shikanai, T. (2008). The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 83 34873–34879. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K.J., and Oelmuller, R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18 176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdinger, M., Polsakiewicz, M., and Knoop, V. (2008). Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol. Biol. Evol. 25 1405–1414. [DOI] [PubMed] [Google Scholar]
- Salone, V., Rüdinger, M., Polsakiewicz, M., Hoffmann, B., Groth-Malonek, M., Szurek, B., Small, I., Knoop, V., and Lurin, C. (2007). A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 581 4132–4138. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Yukawa, Y., Wakasugi, T., Yamada, K., and Sugiura, M. (2006). A simple in vitro RNA editing assay for chloroplast transcripts using fluorescent dideoxynucleotides: Distinct types of sequence elements required for editing of ndh transcripts. Plant J. 47 802–810. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., and Small, I. (2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13 663–670. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17 2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Williams-Carreier, R.E., Williams-Voelker, P.M., Kroeger, T.S., Vichas, A., and Barkan, A. (2006). A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T. (2006). RNA editing in plant organelles: Machinery, physiological function and evolution. Cell. Mol. Life Sci. 63 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T., Endo, T., Hashimoto, T., Yamada, Y., Asada, K., and Yokota, A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95 9705–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif - A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25 46–47. [DOI] [PubMed] [Google Scholar]
- Takenaka, M., Neuwirt, J., and Brennicke, A. (2004). Complex cis-elements determine an RNA editing site in pea mitochondria. Nucleic Acids Res. 32 4137–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka, M., Verbitskiy, D., van der Merwe, J.A., Zehrmann, A., and Brennicke, A. (2008). The process of RNA editing in plant mitochondria. Mitochondrion 8 35–46. [DOI] [PubMed] [Google Scholar]
- Tillich, M., Funk, H.T., Schmitz-Linneweber, C., Poltnigg, P., Sabater, B., Martin, M., and Maier, R.M. (2005). Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 43 708–715. [DOI] [PubMed] [Google Scholar]
- Tsudzuki, T., Wakasugi, T., and Sugiura, M. (2001). Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 53 327–332. [DOI] [PubMed] [Google Scholar]
- Williams, P.M., and Barkan, A. (2003). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36 675–686. [DOI] [PubMed] [Google Scholar]
- Yamazaki, H., Tasaka, M., and Shikanai, T. (2004). PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38 152–163. [DOI] [PubMed] [Google Scholar]
- Zehrmann, A., van der Merwe, J.A., Verbitskiy, D., Brennicke, A., and Takenaka, M. (2008). Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion 8 319–327. [DOI] [PubMed] [Google Scholar]
- Zhou, W., Cheng, Y., Yap, A., Chateigner-Boutin, A.L., Delannoy, E., Hammani, K., Small, I., and Huang, J. (December 2, 2008). The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during growth. Plant J. http://dx.doi.org/10.1111/j.1365–313X.2008.03766.x. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.