Abstract
Gene duplication is an important mechanism for evolution of new genes. In plants, a special group of transposable elements, called Pack-MULEs or transduplicates, is able to duplicate and amplify genes or gene fragments on a large scale. Despite the abundance of Pack-MULEs, the functionality of these duplicates is not clear. Here, we present a comprehensive analysis of expression and purifying selection on 2809 Pack-MULEs in rice (Oryza sativa), which are derived from 1501 parental genes. At least 22% of the Pack-MULEs are transcribed, and 28 Pack-MULEs have direct evidence of translation. Chimeric Pack-MULEs, which contain gene fragments from multiple genes, are much more frequently expressed than those derived only from a single gene. In addition, Pack-MULEs are frequently associated with small RNAs. The presence of these small RNAs is associated with a reduction in expression of both the Pack-MULEs and their parental genes. Furthermore, an assessment of the selection pressure on the Pack-MULEs using the ratio of nonsynonymous (Ka) and synonymous (Ks) substitution rates indicates that a considerable number of Pack-MULEs likely have been under selective constraint. The Ka/Ks values of Pack-MULE and parental gene pairs are lower among Pack-MULEs that are expressed in sense orientations. Taken together, our analysis suggests that a significant number of Pack-MULEs are expressed and subjected to purifying selection, and some are associated with small RNAs. Therefore, at least a subset of Pack-MULEs are likely functional and have great potential in regulating gene expression as well as providing novel coding capacities.
INTRODUCTION
The duplication and divergence of existing genes plays a major role in the creation of new genes in eukaryotes (Ohno, 1970; Lynch and Conery, 2000; Zhang, 2003). This process is especially prominent in plants, where there is an abundance of gene families with members often dispersed throughout the genome (Moore and Purugganan, 2005). One mechanism to account for the origin of gene families is polyploidization, which is very common in plants (Masterson, 1994; Ramsey and Schemske, 1998; Wendel, 2000; Blanc and Wolfe, 2004; Adams and Wendel, 2005; Hancock, 2005). Alternatively, unequal crossing over can expand the number of genes in a tandem array (Zhang and Gaut, 2003). In addition, mobilization of genes by transposable elements (TEs) may provide an explanation for their dispersal to new locations (Kazazian, 2004; Bennetzen, 2005). Based on their transposition mechanisms, TEs are divided into two classes. Class I elements, or retrotransposons, use the element-encoded mRNA as the transposition intermediate; thus, the transposition is duplicative. Class II elements, or DNA transposons, transpose through a DNA intermediate and are capable of excising from their insertion site.
Retrotransposition of the human long interspersed nuclear element L1 has been shown to transduce nearby cellular genes or other genomic sequences, contributing to the duplication of 1% of the human genome (Moran et al., 1999; Pickeral et al., 2000). In plants, several families of TEs, including both transposons and retrotransposons, have been reported to acquire and mobilize host genes or gene fragments (Jiang et al., 2004; Juretic et al., 2005; Morgante et al., 2005; Wang et al., 2006). For example, the maize (Zea mays) Bs1 long terminal repeat retrotransposon carries part of a plasma membrane proton-translocating ATPase gene without its intron sequence (Bureau et al., 1994; Jin and Bennetzen, 1994). In soybean (Glycine max), an insertion by Tgm, a CACTA TE, led to the modification of flower color and seed weight, and five unrelated gene fragments were found inside the element (Zabala and Vodkin, 2005; Zabala and Vodkin, 2007). Similarly, another CACTA element, Tpn1, was found to harbor a fragment containing an HMG domain in morning glory (Ipomoea tricolor) (Takahashi et al., 1999; Kawasaki and Nitasaka, 2004). In maize, the duplication of gene fragments by Helitron elements has contributed significantly to the thousands of gene fragments that are not shared among different maize cultivars (Fu and Dooner, 2002; Morgante et al., 2005).
Mutator-like transposable elements (MULEs), a superfamily of DNA elements, were first identified in maize (Robertson, 1978; Bennetzen et al., 1984). Subsequently, Mutator elements were shown to be a ubiquitous family of transposons among plants, fungi, protozoans, and bacteria (Eisen et al., 1994; Lisch, 2002; Walbot and Rudenko, 2002; Chalvet et al., 2003; Pritham et al., 2005). Autonomous MULEs encode a transposase that can instigate the transposition of both themselves and related nonautonomous MULEs that generally do not contain functional transposase domains. Members of the Mutator superfamily share some characteristics that distinguish them from other DNA transposons. First, most of them have exceptionally long terminal inverted repeats (TIRs; 50 to 600 bp) and create 9-bp target site duplications upon insertion. Transpositionally competent MULEs, such as the Mutator and Jittery elements from maize, At-Mu1 and At-Mu6 from Arabidopsis thaliana, and Hop from fungi, have highly similar or identical TIRs (Bennetzen, 1996; Singer et al., 2001; Chalvet et al., 2003; Xu et al., 2004). As elements lose their activity, the sequence between TIRs becomes more divergent. As a result, the age of elements can be roughly estimated by the sequence identity between their TIRs. Second, promoters can be present in both TIRs of the element and may initiate transcription in convergent orientations (Lisch, 2002). Third, for a single family, all members share TIR sequences but harbor a diverse array of sequences between their TIRs. For example, many nonautonomous MULEs are not simple deletion derivatives of their cognate autonomous elements, but instead, host (nontransposon) sequences may be found within nonautonomous elements (Bennetzen and Springer, 1994). This phenomenon was initially documented with the maize autonomous MuDR element and its eight nonautonomous members (named Mu1-Mu8; reviewed in Lisch, 2002). In particular, the internal sequence of the Mu1/Mu2 subfamily was likely derived from a nontransposon maize gene called MRS-A (Talbert and Chandler, 1988). Due to the recent availability of genomic sequences, many more examples of gene capture by MULEs have been identified, and this type of nonautonomous MULEs are designated Pack-MULEs or transduplicates (Yu et al., 2000; Turcotte et al., 2001; Jiang et al., 2004; Juretic et al., 2005; Ohtsu et al., 2005; Holligan et al., 2006; Wang and Dooner, 2006). Strikingly, there are thousands of these elements present in rice (Oryza sativa; a monocot) and Lotus japonicus (a dicot), suggesting that the mechanism of gene duplication by MULEs predates the monocot-dicot split and is likely widespread among flowering plants (Holligan et al., 2006).
Despite the abundance of Pack-MULEs, it is not clear whether these elements have any functional roles. Studies based on cDNA sequences have shown that gene fragments inside Pack-MULEs are transcribed in both sense and antisense orientations (Jiang et al., 2004; Juretic et al., 2005), and it was speculated that the antisense transcripts may affect the expression of the parental genes (Juretic et al., 2005; Lisch, 2005). However, there is no evidence demonstrating an effect of acquired fragments on the expression of host genes. In addition, there is a debate as to whether or not Pack-MULEs encode functional proteins. Our previous studies indicated that a small subset of Pack-MULEs in rice may have been subjected to functional constraints (Jiang et al., 2004), whereas another study argued that rice duplicates mediated by MULEs represent pseudogenes that are incapable of encoding functional proteins (Juretic et al., 2005; Hoen et al., 2006). To test whether Pack-MULEs have a functional role in the genome, we performed a detailed analysis of 2809 Pack-MULEs and their parental genes in rice. Our analysis indicates that Pack-MULEs are frequently expressed and many are subject to significant functional constraint. The novel features of Pack-MULEs revealed by our analyses facilitates our understanding of the function and impact of these unusual elements and provides new insights into the evolutionary process of gene fragments duplicated by Pack-MULEs.
RESULTS
An Inventory of Pack-MULEs in the Rice Genome
Following the procedures described in Methods, 2809 Pack-MULEs were detected in the rice genome (based on pseudomolecules with a size of 370 Mb). The positions and other information about the 2809 Pack-MULEs on each pseudomolecule are described in Supplemental Data Set 1 online. The total size of these Pack-MULEs sums to 6.3 Mb, accounting for 1.6% of the rice genome. Among the 12 chromosomes, chromosome 1 harbors the greatest number of Pack-MULEs and chromosome 9, the least (378 versus 149; Table 1). The frequency of Pack-MULE insertion is generally lower on the chromosomes with large blocks of condensed chromatin or heterochromatin (chromosomes 4, 7, 9, 11, and 12; Table 1) (Fukui and Iijima, 1991). According to the most recent estimate, the genome size of japonica rice (cv Nipponbare) is 389 Mb (International Rice Sequencing Project, 2005). If the insertion density of Pack-MULEs in sequencing gaps is comparable to that in the sequenced regions, there are ∼2953 Pack-MULEs genome-wide. The sequences of Pack-MULE TIRs belong to 133 families (classified based on sequence similarities using TIR sequences; see Supplemental Data Set 1 online). There are nine families with 100 or more Pack-MULEs, contributing a total of 1665 (59%) elements. The most abundant TIR family is Os0037, a group of 606 related elements that accounts for 22% of the total number of elements. These findings indicate that a small number of Pack-MULE TIR families are involved in duplicating rice genes more frequently than others.
Table 1.
Number of Pack-MULEs and Parental Genes on Each Chromosome
| Chromosome | No. of Pack-MULEs | Insertion Density (PMs/Mb) | No. of Parental Genes | Percentage of Total Genes |
|---|---|---|---|---|
| 1 | 378 | 8.7 | 213 | 4.0 |
| 2 | 301 | 8.4 | 182 | 4.2 |
| 3 | 277 | 7.6 | 182 | 4.0 |
| 4 | 207 | 5.9 | 128 | 3.5 |
| 5 | 257 | 8.6 | 145 | 4.4 |
| 6 | 269 | 8.6 | 117 | 3.4 |
| 7 | 195 | 6.6 | 111 | 3.4 |
| 8 | 209 | 7.4 | 91 | 3.1 |
| 9 | 149 | 6.5 | 82 | 3.4 |
| 10 | 197 | 8.6 | 85 | 3.5 |
| 11 | 178 | 6.2 | 78 | 2.7 |
| 12 | 192 | 7.0 | 87 | 3.3 |
| Total | 2809 | 7.6 | 1501 | 3.7 |
PM, Pack-MULE.
To determine which genes were acquired by Pack-MULEs, the internal sequences of the Pack-MULEs were used to search the genomic sequence of rice; those sequences with the highest score but not flanked by MULE TIRs were considered the parental copies of the Pack-MULE internal sequences. Among the parental copies, 1501 were annotated as unique, non-TE genes, accounting for 3.7% of all the non-TE genes in the rice genome. As shown in Supplemental Figure 1 online, more than half (58%) of the parental genes were acquired or duplicated (through transposition) only once; the remainder of the genes were duplicated multiple times. As a result, there are 2768 acquisition or duplication events.
As mentioned above, Helitrons are another class of DNA transposons that acquire gene fragments on a large scale. In maize, it appears that gene fragments (from different genes) in Helitrons are in the same orientation with respect to the direction of transcription (Brunner et al., 2005). The same is true of the Tpn1 elements (Kawasaki and Nitasaka, 2004). To test whether gene fragments in Pack-MULEs orient in the same manner, we examined the relative orientation of genes inside Pack-MULEs. There are 656 (23% of the total) Pack-MULEs that have captured portions of two or more genes. Among the 551 Pack-MULEs that are derived from just two genes, 284 (52%) were found to carry genes in the same orientation and 267 (48%) in a different orientation, which is not significantly different from the random expectation (P > 0.5, χ2 test). Thus, it appears either that Pack-MULEs arrange or acquire genes using a different mechanism from that employed by Helitrons or that selection operates on transduced sequences differently in these two transposon superfamilies.
Expression of Pack-MULEs
The Fraction of Transcribed Pack-MULEs
The transcriptional profile of rice has been studied by several genome-wide approaches. These include a collection of full-length cDNAs (fl-cDNAs) (Kikuchi et al., 2003), the sequencing of mRNAs and small RNAs (sRNAs) through massively parallel signature sequencing (MPSS) (Nobuta et al., 2007), and the definition of transcriptionally active regions (TARs) based on tiling microarrays (Li et al., 2007). These efforts provide a unique opportunity for a comprehensive analysis of the expression profile of Pack-MULEs and their possible impact on parental genes.
Based on currently available data sets, 274 (10%) Pack-MULEs have perfect and unique matches to one or more fl-cDNA sequences, and 459 (16%) Pack-MULEs contain unique MPSS signatures. In addition, 1519 (54%) Pack-MULEs overlap with one or more TARs from the published rice tiling array experiment. Based on the cDNA collection and MPSS signatures, there are 613 (22%) expressed Pack-MULEs. When all data are considered, there are 1764 (63%) expressed Pack-MULEs. However, a closer examination reveals that almost all TARs (95%) inside Pack-MULEs have multiple copies (E < 10−5) in the genome, raising the question as to whether or not the substantially higher percentage representation of Pack-MULEs by TARs is due to cross-hybridization. To be cautious, we consider the Pack-MULEs associated with TARs but not other types of expression evidence as possibly expressed Pack-MULEs, and those are excluded from either expressed or nonexpressed Pack-MULEs. As a comparison, we also examined the proportion of expressed MULEs containing transposase domains (referred to as auto-MULEs). We identified only 53 out of 670 (8%) auto-MULEs that have cDNA or MPSS matches, which is significantly less than the proportion of Pack-MULEs (22%) with matches in these two data sets (P < 2.2e-16, Fisher's exact test).
The Transcription Level and Tissue Specificity of Pack-MULEs and Their Parental Genes
Using the MPSS data set derived from 18 libraries, we compared the tissue specificity and transcription level of Pack-MULEs and their parental genes. Overall, the expression level of Pack-MULEs is significantly lower than that of the parental genes (P < 0.001, Wilcoxon rank sum [WRS] test), and they are also less ubiquitous (P < 0.001, WRS test). On average, each signature from a Pack-MULE was detected in 2.5 libraries, and the relative expression level was 15 transcripts per million per transcription case (a transcription case is defined as one signature detected in one library). By contrast, each signature from a parental gene is expressed in 3.6 libraries with a relative expression level of 64 transcripts per million per transcription case, four times higher than that of Pack-MULEs. Nonetheless, in 29% of the cases, Pack-MULEs are transcribed at a higher level than their cognate parental genes.
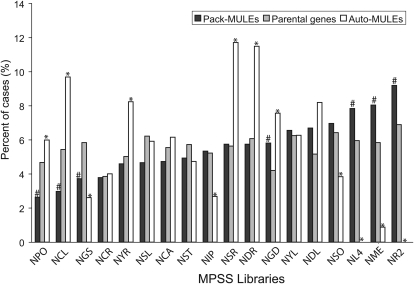
If both Pack-MULE and the parental gene are transcribed, are they transcribed in the same tissue or in different tissues? To answer this question, we compared each Pack-MULE to its parental copy in a pairwise fashion. In a total of 4228 transcription cases, 2749 (65%) are specific to parental genes (only transcripts from parental genes are detected in the relevant tissue), 814 (19%) are specific to Pack-MULEs, and only 665 (16%) overlap between parental copies and Pack-MULEs. This suggests that the tissue (or library) specificity is often divergent between Pack-MULEs and their parental copies. The overall expression pattern is significantly different between Pack-MULEs and their parental genes (P < 0.001, χ2 test). Interestingly, for Pack-MULEs, the fewest expression events (39 out of 1479 or 2.6%; Figure 1) were detected in mature pollen (NPO), a tissue in which transposons normally express at high levels (Nobuta et al., 2007). The tissue specificity of Pack-MULEs is also dramatically different from that of auto-MULEs (Figure 1). Strikingly, auto-MULEs have no or few expression events in the tissues or conditions where Pack-MULEs have the most expression events (Figure 1). Auto-MULEs have most expression events in tissues under stress, including young roots stressed in NaCl and drought (NSR and NDR) and young leaves in cold (NCL), much more frequently than Pack-MULEs. This suggests that the expression pattern of Pack-MULEs is distinct from that of other TEs, including the auto-MULEs.
Figure 1.
Tissue Specificity of Transcription for Pack-MULEs, Their Parental Genes, and MULEs That Encode Transposases (Auto-MULEs).
The percentage of cases was calculated using the number of MPSS tags detected for a particular sequence class (Pack-MULE, parental genes, or auto-MULEs) in each library divided by the number of total MPSS tags for the sequence class in question among all libraries. The abbreviations for the libraries are as follows: NPO, mature pollen; NCL, 14 d, young leaves stressed in 4°C cold for 24 h; NGS, 3 d, germinating seed; NCR, 14 d, young roots stressed in 4°C cold for 24 h; NYR, young roots; NSL, 14 d, young leaves stressed in 250 mM NaCl for 24 h; NCA, 35 d, callus; NST, stem; NIP, 90 d, immature panicle; NSR, 14 d, young roots stressed in 250 mM NaCl for 24 h; NDR, 14 d, young roots stressed in drought for 5 d; NGD, 10 d, germinating seedlings grown in dark; NYL, young leaves; NDL, 14 d, young leaves stressed in drought for 5 d; NSO, ovary and mature stigma; NL4, leaf; NME, 60 d, crown vegetative meristematic tissue; NR2, root. The pound sign indicates libraries in which difference in percentage of cases is significant (q < 0.05) between Pack-MULEs and parental genes. The asterisk indicates libraries in which difference in percentage of cases is significant (q < 0.05) between Pack-MULEs and auto-MULEs.
Translation of Pack-MULEs
To determine how many Pack-MULEs are translated, we collected peptide sequences that represent 3230 rice proteins from recent proteomic analyses in rice (Koller et al., 2002; Chitteti and Peng, 2007; Lee et al., 2007; Tan et al., 2007; Li et al., 2008). The 3230 proteins represent 6% of all of the annotated genes (56,278 genes) based on information from release 5 of rice pseudomolecules from the J. Craig Venter Institute (The Institute for Genome Research [TIGR]). A total of 28 (1%) Pack-MULEs were found to have perfect and unique matches with the peptide sequences in the database (see Supplemental Table 1 online). In addition to these 28 matches, many matches were excluded because when the translated region overlaps with the acquired region, the peptide sequences are often identical between Pack-MULEs and their parental genes. As a result, the number of translated Pack-MULEs is an underestimate. If we use the sequence of parental genes to search the same database with the same criteria, 6% (93 out of 1501) of the parental genes have unique matches, a proportion that coincides with the ratio of the number of available protein sequences to the number of annotated genes.
The Orientation of Expression and the Transcription of Chimeric Pack-MULEs
Since MULEs have potential promoters in both TIR sequences, it is possible for them to induce transcription in both sense and antisense orientations, with respect to the transcriptional direction of the parental genes. If an acquired fragment was transcribed or translated in one orientation (sense or antisense, referred to as unidirectional), it was considered as one event in the orientation in question (see Methods for details). As shown in Table 2, there are significantly more sense events than antisense events for both Pack-MULEs and parental genes. This is true for each type of expression evidence. For a few Pack-MULEs, the same acquired region is transcribed in both directions (bidirectional; Table 2). In all cases but one, the sense and antisense transcripts are detected in different libraries or conditions. Moreover, there are 13 cases where paralogous regions in different Pack-MULEs are transcribed in opposite orientations. Interestingly, among the elements with peptide matches, some peptides are generated through the translation of the sense strand of the acquired regions, while others are generated on the antisense strand. Assuming that these translated products are functional, this suggests that Pack-MULEs can recycle genomic sequence in many different ways, including generating novel proteins using coding capacity on the antisense strand. In contrast with Pack-MULEs, there is no antisense peptide detected for parental genes. Nonetheless, there are antisense or bidirectional transcripts for parental genes, especially for transcripts detected by MPSS, yet the ratio of antisense transcripts is much lower than that for Pack-MULEs (Table 2).
Table 2.
Sense and Anti-sense Expression Events among Pack-MULEs and Parental Genes
| Evidence for Expression | Peptide | cDNA | MPSS | Total | |
|---|---|---|---|---|---|
| Pack-MULEs | Sense | 11 | 147 | 106 | 229 |
| Antisense | 4 | 85 | 68 | 140 | |
| Bidirectional | 0 | 12 | 7 | 25 | |
| Sense/antisensea | 2.75 | 1.64 | 1.51 | 1.54 | |
| P < (χ2 test)b | 0.10 | 10−4 | 0.001 | 10−5 | |
| Parental genes | Sense | 93 | 946 | 690 | 784 |
| Antisense | 0 | 23 | 34 | 25 | |
| Bidirectional | 0 | 42 | 456 | 506 | |
| Sense/antisensea | 15.20 | 2.34 | 2.43 | ||
| P < (χ2 test)b | 10−6 | 10−6 | 10−6 | 10−6 |
Bidirectional events were considered as one sense and one antisense event.
P value to test whether number of sense events is equal to that of antisense events.
As mentioned above, there are 656 Pack-MULEs containing acquired fragments from two or more genes. When the percentage of expressed Pack-MULEs is examined, it is clear that the elements with acquired sequences from multiple genes are much more likely to be expressed than those only with sequence from one gene (P < 10−6, χ2 test; Table 3). Furthermore, if the genes are in the same orientation, the percentage of expressed elements is slightly higher than those with genes in different orientation (Table 3). Although the difference is not statistically significant (possibly due to small number of samples), it may suggest that there is a preference for the expression of acquired fragments that are in the same orientation. For elements with fragments in different orientations, the number of antisense transcripts is reduced by a limitation of the transcribed region to the fragment from one gene (see Supplemental Results online for details). As a result, although the arrangement of gene fragments inside Pack-MULEs is largely random (see above), the transcription of those fragments is likely subject to selection.
Table 3.
Variation of Expression of Pack-MULEs with Number of Acquired Genes
| From Two Genes
|
From Three or More Genes
|
||||
|---|---|---|---|---|---|
| Type of Pack-MULEs | Fragment from One Gene | Same Orientation | Different Orientation | Same Orientation | Different Orientation |
| No. of elements | 1328 | 284 | 267 | 18 | 87 |
| Expressed | 279 | 91 | 73 | 9 | 36 |
| Percentage expressed | 21 | 32 | 27 | 50 | 41 |
The Relationship between Expression and TIR Sequence and Identity
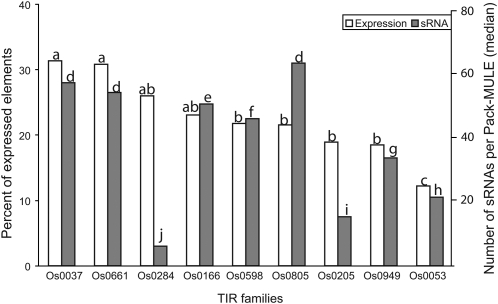
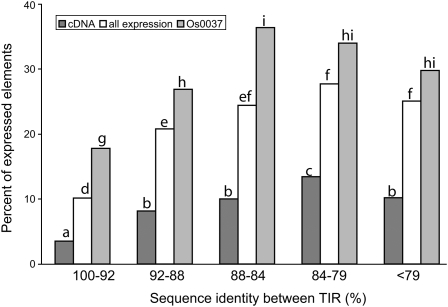
Since the promoters of Mutator elements are located inside TIRs and most transcripts from Pack-MULEs are initialized from or close to TIRs (Lisch, 2002; Walbot and Rudenko, 2002; Jiang et al., 2004), it is expected that the expression of Pack-MULEs is influenced by the sequence of TIRs. Figure 2 diagrams the percentage of expressed elements for the nine most abundant TIR families; there is a threefold variation in the percentage of expressed elements with different TIR sequences. Moreover, the percentage of expressed elements appears to vary with the sequence identity between the TIRs for a single element. Among all the Pack-MULEs, 2504 (89%) have TIRs longer than 100 bp. These 2504 elements were divided into different groups based on the degree of sequence identity between the two TIRs. As shown in Figure 3, when the sequence identity is higher than 79%, the fraction of expressed elements increases as the identity decreases. Regardless of which expression evidence is considered, the percentage of expressed elements is significantly lower in the group of elements with sequence identity of 92% or higher. These elements are possibly active or were active recently. To test whether the variation relative to TIR sequence identity is an artifact resulting from combining different TIR families in each group, the same analysis was performed with a single TIR family, Os0037. A similar trend was observed (Figure 3), indicating there is a correlation between expression and the sequence identity of the TIRs.
Figure 2.
The Variation in Percentage of Expressed Pack-MULEs and Association with sRNA Signatures (All sRNAs) among Pack-MULEs with Different TIR Sequences.
For each type of data (expression or small RNAs), if two columns have different letters (a, b, c, etc.), the difference between the relevant families is significant (χ2 test, P < 0.05); otherwise, the difference is not significant.
Figure 3.
The Relationship between Percentage of Expressed Pack-MULEs and Sequence Identity between TIRs.
For cDNA, only elements associated with fl-cDNA sequences are considered expressed. For all expression, elements with any type of expression evidence (cDNA, MPSS, peptide) are considered. For Os0037, all expression data are used. The letters on the columns indicate the significance of difference (see legend for Figure 2).
Pack-MULE Small RNAs and Their Role in Gene Expression
Small RNAs and Gene Expression
To determine whether or not the presence of sRNAs correlates with the lower levels of expression of Pack-MULEs, we examined sRNAs associated with Pack-MULEs and their cognate parental genes using MPSS sRNA sequences (Nobuta et al., 2007). For convenience, the small RNA signatures involved in this study are classified into three types: “unique (or self) sRNA signatures” are those with only a single match in the rice genome and their origins are clear. “Shared sRNA signatures” are the nonunique sRNA signatures that match both Pack-MULEs and their parental genes. The remaining sRNA signatures are called “other nonunique sRNA signatures.” More than half of the Pack-MULEs (1722 out of 2809; 61%) match unique sRNA signatures, which indicates that at least 61% of Pack-MULEs are directly involved in the generation of sRNAs. Almost all Pack-MULEs (2755; 98%) are associated with nonunique sRNAs, and 2168 (77%) share sRNAs with the parental genes. A total of 228 Pack-MULEs are associated only with other nonunique sRNAs. The expression level of these 228 Pack-MULEs is significantly higher than all other types of Pack-MULEs except those without any sRNAs (P < 10−4, WRS test; Table 4). This suggests that the presence of unique sRNAs or shared sRNAs has negatively affected the abundance of transcripts from relevant Pack-MULEs.
Table 4.
The effect of sRNAs on Transcription of Pack-MULEs and Parental Genes
| PMs | Without sRNA | With Only Self sRNAs | With Only Shared sRNAs | With Both sRNAs | PMs with Other sRNAs | |
|---|---|---|---|---|---|---|
| Total elements | 26 | 387 | 833 | 1335 | 228 | |
| Expressed | 5 (19.2) | 58 (15.0) | 137 (16.4) | 231 (17.3) | 28 (12.3) | |
| Median | 11 | 6 | 6 | 6 | 14 | |
| Mean | 16 | 13 | 13 | 13 | 33 | |
| Pa | 0.226 | 10−4 | 10−4 | 10−5 | ||
| sRNA (median) | 2 | 2 | 2 | 2 | ||
| sRNA (mean) | 5 | 6 | 6 | 6 | ||
| PGs | Without sRNA | With Only Self sRNAs | With Only Shared sRNAs | With Both sRNAs | PGs with Other sRNAs | |
| Total genes | 227 | 45 | 869 | 216 | 144 | |
| Expressed | 162 (71.4) | 33 (73.3) | 691 (79.5) | 181 (83.8) | 113 (78.5) | |
| Median | 14 | 16 | 13 | 13 | 12 | |
| Mean | 112 | 91 | 53 | 64 | 49 | |
| Pb | 0.250 | 0.033 | 0.1018 | 0.008 | ||
| sRNA (median) | 2 | 3 | 3 | 2 | ||
| sRNA (mean) | 5 | 31 | 7 | 4 | ||
| Pc | 0.300 | 10−4 | 10−6 |
PM, Pack-MULE; PG, parental gene.
P value to test whether the entire distribution of expression levels of all cases are different (WRS test) from that of Pack-MULEs with other sRNAs.
P value to test whether the entire distribution of expression levels of all cases are different (WRS test) from that of parental genes without sRNAs.
P value to test whether the entire distribution of sRNA levels are different (WRS test) from that of other parental genes.
Unlike Pack-MULEs, only a small portion (261 out of 1501; 17%) of the parental genes is associated with unique sRNAs; however, 1085 (72%) of the parental genes share sRNAs with Pack-MULEs. The shared sRNAs account for more than half (51%) of all sRNAs associated with parental genes (see Supplemental Table 2 online). In addition, the number of sRNA signatures associated with each Pack-MULE is 6- (mean) to 11-fold (median) higher than that for a parental gene (see Supplemental Table 2 online), which may explain why the expression level of Pack-MULEs is significantly lower than that of the parental genes (see above). Compared with the parental genes not associated with any sRNAs, the expression level of the parental genes with shared sRNAs is significantly lower (P < 0.05, WRS test; Table 4). By contrast, if the parental genes are associated with unique sRNAs, the difference between those parental genes and the genes without sRNAs is not as significant (P = 0.250, WRS test; Table 4). This is possibly because the abundance of unique sRNAs from parental genes is significantly lower than that of shared sRNAs (Table 4).
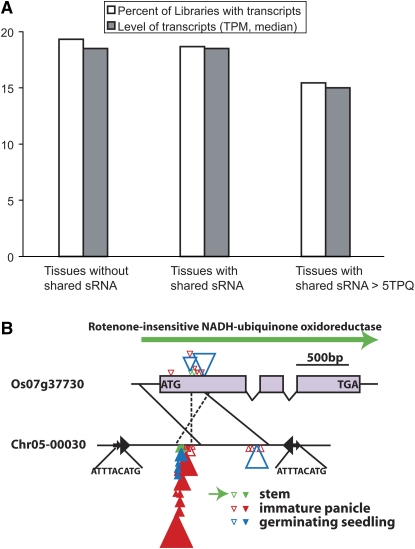
If the shared sRNAs have an impact on the expression of the parental genes, one would expect the tissues with shared sRNAs are associated with reduced transcription from parental genes. To test this hypothesis, we examined the three libraries (stem, immature panicle, and germinating seedling) for which there is both sRNA and mRNA data available. Among the tissues with transcripts, the transcription levels of parental genes are slightly lower in the tissues with shared sRNAs; however, the difference is not significant (WRS test, P = 0.076 when sRNA > 5 transcripts per quarter million [TPQ]; Figure 4A). If the percentage of libraries with transcripts is compared, the value is lower among the tissues with shared sRNAs, and the difference is more dramatic when the sRNA level increases (χ2 test, P = 0.006 when sRNA > 5 TPQ; Figure 4A). Figure 4B diagrams a parental gene that is transcribed in stem tissues. All sRNAs associated with the gene are located in the region that was duplicated by the relevant Pack-MULE. The absence of transcripts from panicles and germinating seedlings was accompanied by the accumulation of a large amount of sRNAs in the acquired region, including shared sRNAs with Pack-MULEs and sRNAs generated by Pack-MULEs (unique sRNAs) (Figure 4B). The reduced occurrence of transcripts of parental genes in the libraries with shared sRNAs suggests that it is very likely that Pack-MULEs suppress the expression of their parental genes through the formation of sRNAs.
Figure 4.
The Impact of Pack-MULE–Related Small RNAs on the Tissue-Specific Transcription of Their Parental Genes.
(A) Transcription level and percentage of libraries with transcripts from parental genes vary with the presence and level of shared sRNAs. TPM, transcripts per million; TPQ, transcripts per quarter million.
(B) A parental gene (Os07g37730) that is possibly influenced by sRNAs caused by the corresponding Pack-MULE (chr05-00030). Pack-MULE TIRs are shown as black arrowheads, and black horizontal arrows indicate target site duplications with their sequences shown underneath. Exons are depicted as purple boxes and introns as the lines connecting exons; other sequences are shown as horizontal lines. The gene structure was based on annotation provided by TIGR. Homologous regions are associated with solid or dashed lines. The long arrow above the gene indicates transcripts from the gene. Unique sRNAs are shown as solid triangles and nonunique sRNAs as empty triangles. The sizes of triangles are proportional to the level of sRNAs. Green triangles indicate sRNAs from stem, with red ones from immature panicle and blue from germinating seedlings. For the gene, all sRNAs in the relevant tissues are shown. For the Pack-MULE, only those in the acquired region are shown.
Inverted Repeats and Small RNAs
Inverted repeats are known to produce hairpin transcripts that trigger the formation of sRNAs. One feature distinguishing MULEs from other TEs is the presence of long TIRs, in addition to short inverted repeats in the subterminal regions. In some cases, the acquired region may form an inverted repeat as well. Among the 4656 unique sRNAs derived from Pack-MULEs, 2671 (57.4%) signatures are associated with inverted repeats. The total size of inverted repeats accounts for 21.6% of the entire length of the Pack-MULEs. Consequently, sRNAs are enriched five times [57.4% × (1 − 21.6%)/(1 − 57.4%)/21.6% = 4.9] in the inverted regions compared with noninverted regions, consistent with an earlier report (Nobuta et al., 2007). Although sRNAs are frequently associated with inverted repeats, few of the inverted repeats overlapped with the regions acquired by Pack-MULEs. Among the 2809 Pack-MULEs, 142 (5.1%) have a 20 bp or longer overlap between the inverted repeat and the acquired region, and such overlaps are associated with only 49 (∼1% of 4308) unique sRNAs. Accordingly, although acquired regions can be duplicated and inverted within Pack-MULEs, it is not a frequent event or is selected against, and these inversions are not a major source of sRNAs from Pack-MULEs.
Since the percentage of expressed Pack-MULEs varies depending on TIR sequences as well as the sequence identity between TIRs, it seems reasonable that these features could affect the quantity of sRNAs produced. If that was the case, a more frequent occurrence of expression would be expected to be associated with fewer sRNAs. As shown in Figure 2, the number of sRNAs associated with Pack-MULEs does vary among TIR families. However, there is no obvious relationship. For example, Os0037, the family with the highest percentage of expressed elements, ranks second based on number of total sRNAs per element. Therefore, the frequency of association with sRNAs does not seem to be the main factor responsible for the variation of expression among Pack-MULEs with different TIRs.
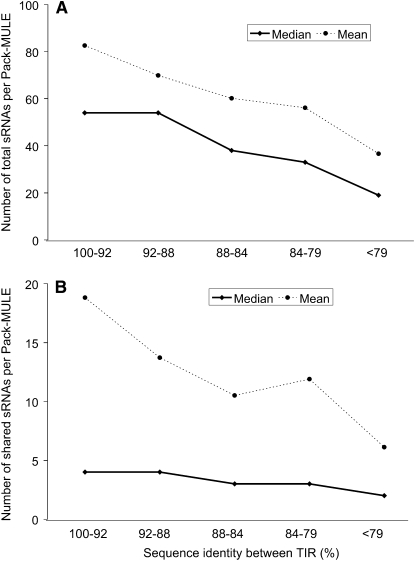
Unlike the situation for different TIR families, the enhanced expression demonstrated by elements with less similar TIRs does seem to be negatively correlated with the abundance of sRNAs. If all sRNAs (unique, shared, and other nonunique sRNAs) are considered, Pack-MULEs with higher TIR similarity match many more sRNA signatures (Figure 5A). Similarly, Pack-MULEs with higher TIR similarity share more sRNAs with their parental genes (Figure 5B). There was evidence of a strong negative correlation between expression and abundance of sRNAs (Rs = −0.7 between percentage of expressed elements and number of total sRNAs per element; Spearman's Rank test). As a result, it is clear that the abundance of sRNAs is responsible, at least partially, for the less frequent expression of Pack-MULEs with high TIR similarity (Figure 3).
Figure 5.
The Relationship between TIR Similarity and Abundance of Small RNAs.
(A) Total sRNAs.
(B) Shared sRNAs (with parental genes).
Purifying Selection on Pack-MULEs
Functional elements, including genes, are typically subject to strong selective constraints. A functional coding sequence is expected to undergo stronger selective constraints on nonsynonymous sites (mutations that alter amino acid sequence) than for synonymous ones (those that do not alter amino acid sequence) (Li, 1997; Makalowski and Boguski, 1998). To assess the degree of functional constraint on the protein coding sequences within Pack-MULEs, Jiang et al. (2004) calculated the ratio of nonsynonymous (Ka) and synonymous (Ks) substitution rates between Pack-MULEs and their parental genes. For a small subset of Pack-MULEs (>10%), the ratio of Ka/Ks is significantly < 1. Therefore, the authors concluded that some of the Pack-MULEs have been functionally constrained. A later study, however, concluded that Pack-MULEs (or transduplicates, as called in that study) represent pseudogenes that lack coding capacities, and no putatively functional protein-coding Pack-MULE was identified (Juretic et al., 2005). Their conclusion was reached based on the following observations or reasoning: (1) there are premature stop codons or frame shifts in the acquired region of Pack-MULEs, or the acquired region is not in the largest open reading frame; (2) the functional constraint reflected by Ka/Ks value could be solely due to the constraint on the parental genes; (3) the distribution of Ka/Ks between Pack-MULEs and their parental copies is similar to that of human pseudogenes created by retrotransposition. To address these issues, we conducted a detailed analysis on the nature of the selective constraints experienced by the coding sequences inside Pack-MULEs.
In-Frame Coding Sequence inside Pack-MULEs
We first examined the 125 Pack-MULEs with cDNA matches in sense orientation or bi-orientations. Detailed information for 55 Pack-MULEs, in which >100 bp (ranging from 104 to 862 bp) of the acquired region is in frame, is provided in Supplemental Table 3 online. Among these Pack-MULEs, there are 15 cases where the entire acquired region (coding region) falls into an open reading frame without a frame shift or a premature stop codon compared with the frame of the parental gene. In addition, there are 21 cases where the stop or start codon is near the border of the acquired region, or a frame shift is followed by a reversion of the frame shift. In those cases, at least 70% of the acquired region is in frame. The fact that a considerable amount of the acquired region is in frame suggests that some of the Pack-MULEs may have retained at least part of the coding capacity of their parental genes. In addition, the frame shifts or stop codons may have resulted in an altered but still functional protein.
Selection for Coding Capacity Revealed by Ka/Ks Values
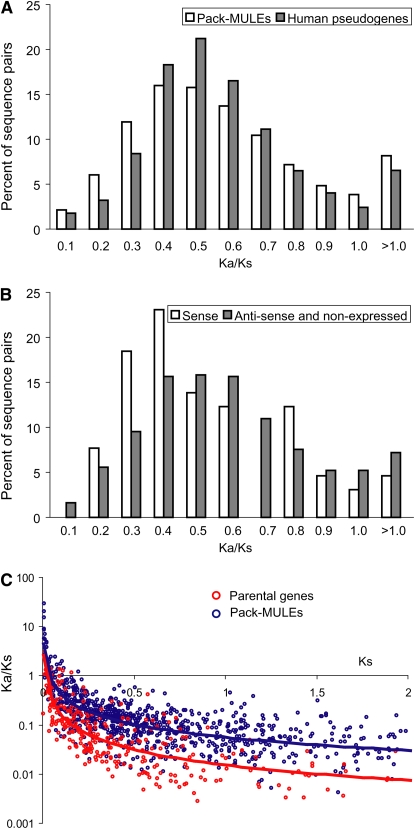
To test whether Pack-MULEs are different from pseudogenes in the strength of purifying selection experienced, we calculated the Ka/Ks value for each Pack-MULE–parental gene pair where there is a continuous alignment encoding at least 50 amino acids. These results were compared with those for human pseudogenes and their paralogous genes. The human pseudogenes were examined because similar data from plants are not available. For reliability, only 1408 sequence pairs (between Pack-MULE and their parent genes) with Ks > 0.05 were considered. If the distribution of Ka/Ks values is similar between Pack-MULEs and human pseudogenes, one would expect that the percentage of Pack-MULE sequence pairs in each Ka/Ks bin, which refers to a certain range of Ka/Ks value, would be largely equivalent to that for human pseudogene and their parental gene pairs. Among human pseudogene-parental pairs, most Ka/Ks values fall between 0.4 and 0.7, with a peak near 0.5 (Figure 6A; Zhang et al., 2003). The overall distribution of Ka/Ks values for Pack-MULEs and human pseudogenes is significantly different (P < 0.01, Kolmogorov-Smirnov test). There are relatively fewer Pack-MULE–parental pairs with Ka/Ks values from 0.4 to 0.7 (Figure 6A). Most importantly, Pack-MULE–parental gene pairs are significantly overrepresented compared with human pseudogene-parental pairs (P < 10−6, χ2 test) in the bins where Ka/Ks is 0.3 or less, suggesting that more Pack-MULEs than human pseudogenes are subject to strong purifying selection.
Figure 6.
Purifying Selection on Pack-MULEs Reflected by Ka/Ks Values.
(A) The comparison of Ka/Ks distribution between Pack-MULE–parental sequence pairs and human pseudogene-parental sequence pairs.
(B) The distributions of Ka/Ks ratios for Pack-MULE–parental sequence pairs for Pack-MULEs with sense, antisense, and no expression.
(C) Relationships between Ka/Ks and Ks values in the Pack-MULE and the parental gene lineages.
Realizing that the values for Ka/Ks from organisms as divergent as rice and human are influenced by many other factors in addition to selective constraints (for example, codon usage bias), we sought alternative ways to evaluate the level of selective constraints experienced by Pack-MULEs. If Pack-MULEs with relatively lower Ka/Ks values tend to be those that experienced functional constraints, we would expect Pack-MULEs with lower Ka/Ks values to be overrepresented among elements that are expressed in the sense orientation relative to those that are not expressed or expressed only in the antisense orientation. To determine if this is the case, we compared the Ka/Ks distribution for the two groups of Pack-MULEs, based on the presence/absence of peptide and fl-cDNA evidence. As shown in Figure 6B, in the bins with Ka/Ks < 0.5, the sequence pairs where Pack-MULEs are expressed in the sense orientation greatly outnumber the ones with no expression and those expressed in the antisense orientation (P < 0.02, χ2 test)
The Partition of Purifying Selection Pressure between Pack-MULEs and Parental Genes
To further distinguish whether the purifying selection as reflected by lower Ka/Ks among Pack-MULE–parental gene pairs is due to selection solely on the parental genes or on Pack-MULEs as well, a putative ancestral sequence for each Pack-MULE and parental gene pair was inferred by a maximum likelihood method (Yang et al., 1995). This approach allows us to calculate Ka/Ks values for the branches leading to the Pack-MULE and to its parental gene independently. For both the Pack-MULEs and the parental genes, the Ka/Ks values decline as Ks values increase, but the Ka/Ks values for parental genes are significantly lower (P < 0.001, WRS test) than those for Pack-MULEs (Figure 6C). Thus, it appears that parental sequences have been subject to stronger purifying selection than Pack-MULEs. To determine if any Pack-MULE lineage has experienced selective constraints at the protein level, we conducted statistical tests to see if the Pack-MULE lineage Ka/Ks values are significantly < 1. We found that ≥15% of the Pack-MULE lineages have Ka/Ks values significantly < 1 (Table 5), suggesting functional constraints at the amino acid level on a substantial number of Pack-MULEs. Taken together, the lineage-based analysis indicates that both Pack-MULEs and parental genes have been subject to purifying selection, although the pressure on Pack-MULEs is relatively relaxed.
Table 5.
Purifying Selection on Pack-MULEs and Their Parental Genes
| Pack-MULEs | Parental Copies | |
|---|---|---|
| Available cases | 1255 | 1255 |
| Cases without significant purifying selection | 888 (70.1%) | 500 (39.8%) |
| Cases without change | 27 (2.2%) | 411 (32.7%) |
| Total cases Ka/Ks < 1 (P < 0.05) | 340 (27.1%) | 344 (27.4%) |
| Total cases Ka/Ks < 1 (P < 0.01) | 191 (15.2%) | 281 (22.4%) |
DISCUSSION
Pack-MULEs Provide Raw Materials for the Evolution of Coding Regions
With the availability of large quantities of genomic sequences, it is now clear that TEs are involved in various aspects of genome evolution. Prior to the discovery that DNA elements such as MULEs contributed to duplicated genes on a large scale, the formation of retrogenes was considered a major source of duplicated genes. Unlike the genes duplicated by DNA elements, retrogenes lack introns and promoter regions. Despite this, some retrogenes are expressed and functional, possibly because they have been inserted in the region downstream of a promoter sequence (Long, 2001; Emerson et al., 2004). Given the fact that most Pack-MULEs carry gene fragments but not entire genes, it is not surprising that many Pack-MULEs are likely pseudogenes due to truncation in their coding regions or misregulation. The question is whether gene duplication by Pack-MULEs produces only pseudogenes or whether the formation and retention of some Pack-MULEs lead to the generation of novel functional components.
High Frequency of Expression
Based on the fl-cDNA sequences and MPSS data, 22% of the Pack-MULEs are transcribed under at least some conditions. Again, this is an underestimate because only unique MPSS signatures were considered. The number of expressed Pack-MULEs is significantly higher than that of the putative autonomous MULEs (8%) as well as that of rice pseudogenes (2.5 to 5%) (based on cDNA and MPSS evidence, C. Zhou and S.-H. Shiu, unpublished data). Aside from the fact that Pack-MULEs are much better represented by transcriptional evidence than auto-MULEs, two additional features in expression set Pack-MULEs and auto-MULEs apart. First, whereas other TEs have an unusually high percentage of expression cases in pollen (Nobuta et al., 2007), the fraction of expression cases for Pack-MULEs is lowest in pollen. Second, the putatively autonomous MULEs have the highest expression level under stressed conditions, which resembles the fact that TEs are often activated by stresses (Wessler, 1996; Grandbastien et al., 2005; Lukens and Zhan, 2007). However, this is not observed among Pack-MULEs.
In addition to transcripts associated with Pack-MULEs, 28 Pack-MULEs (1%) have perfect, unique matches in a database containing peptide sequences that represent 3230 proteins (6% of the all annotated genes including TEs and non-TEs). With the same search criteria, 6% of the parental genes are found to have hits in the same database. If we assume the peptides are representative and most of the parental genes encode functional proteins, the false negative rate is ∼94% when using the proteomic data set available to estimate the rice protein repertoire. Thus, after correcting for the high false negative rate of the available proteomic data sets, there could be hundreds of proteins contributed by Pack-MULEs.
Selection for Coding Capacity
Given the presence of noncoding genes and the production of antisense transcripts from regular genes, there is no reason to believe that all transcripts must encode functional proteins. However, the selective constraints (measured by Ka/Ks values) on the Pack-MULEs that are transcribed from the sense orientation are significantly stronger than those that are not expressed or are transcribed only from the antisense strand (Figure 6B). This finding suggests that at least some expressed Pack-MULE transcripts have a coding capacity that has been under purifying selection. In addition, there are many more Pack-MULEs that express in the sense orientation than those that express in the antisense orientation. There are at least two mutually nonexclusive explanations for this observation. One explanation is that the antisense transcripts have negative impacts on the expression of the parental genes, resulting in selection against them (Kawasaki and Nitasaka, 2004). Alternatively, it is possible that the Pack-MULEs with sense transcripts are more likely to retain at least some functions of parental genes. This is supported by the observation that Pack-MULEs with sense transcripts are associated with lower Ka/Ks values. If all Pack-MULEs were pseudogenes, it would be very difficult to explain why their Ka/Ks values would be different depending on the status and orientation of their expression.
One argument for the notion that all Pack-MULEs are pseudogenes is that the low Ka/Ks ratios between Pack-MULEs and parental genes could simply reflect the selection on parental genes, not on Pack-MULEs. However, a substantial number of Pack-MULEs have significantly lower Ka/Ks values than human pseudogenes, suggesting that some Pack-MULEs likely have experienced strong purifying selection. We also calculated the Ka/Ks values on a per lineage basis by constructing a putative ancestral sequence for each Pack-MULE–parental gene pair. Although the purifying selection pressure is indeed stronger for parental genes than that for Pack-MULEs, >15% of Pack-MULEs have experienced functional constraints. Even if the Ka/Ks value is ∼1, or if there is a premature stop codon or frame shift, this does not necessarily indicate that the Pack-MULE is nonfunctional; it could have evolved novel functions that no longer require the complete coding sequence and/or be functional at the RNA level. Considering that Pack-MULEs frequently harbor gene fragments from more than one locus and chimeric Pack-MULEs are more likely to be expressed (Table 3), the evolution of novel functions is probably more important than the retention of original functions. Such a process should be promoted by the relaxed selective pressure on Pack-MULEs that we have observed.
The Regulation of Gene Expression by Pack-MULEs
Many TEs, including MULEs, insert into genic regions and affect the expression of nearby genes. Because they share sequences with their parental genes, Pack-MULEs may have the potential to control the expression of genes in trans. In this study, we provide evidence that Pack-MULEs may influence the expression of their parental genes, likely through the action of small RNAs. First, the sRNAs shared with Pack-MULEs make up the single largest source (51%) of sRNAs for parental genes. Second, parental genes with shared sRNAs have lower expression levels compared with genes without associated with sRNAs (Table 4). Finally, in the tissues (or libraries) where shared sRNAs are >5 TPQ, the number of cases with detectable transcripts is reduced (Figure 4). Although the origin of shared sRNAs is not clear, they are more likely derived from Pack-MULEs than from the parental genes, given the fact that Pack-MULEs are more frequently associated with the generation of sRNAs. As such, Pack-MULEs should play a role in the regulation of expression of their parental genes.
The frequent association of Pack-MULEs with sRNAs could be partially attributed to the presence of inverted repeats, where the sRNAs are five times more enriched than other regions in Pack-MULEs. For other regions inside Pack-MULEs, the formation of sRNAs might be enhanced by promoters in the TIRs that could lead to the formation of double-stranded RNAs if both promoters are active simultaneously. Alternatively, double-stranded RNAs could be formed through transcripts in different orientations from Pack-MULEs that share internal sequences. Both cases were recorded in this study, but the complementary transcripts were rarely detected from same library (see Results). This is likely due to the fact that if they are from the same library, the formation of double-stranded RNAs may lead to rapid degradation of both mRNAs so that they become undetectable.
One interesting observation made in this study is that all of the features related to expression seem to be correlated with the similarity between the TIRs of individual Pack-MULEs. Elements having the highest TIR similarities are least likely to be expressed (Figure 3). Our analysis indicated that this is largely because more sRNAs, including those shared with parental genes, are associated with those Pack-MULEs (Figure 5). In other words, Pack-MULEs with higher TIR similarity are targeted by more sRNAs so that they are more intensively silenced. If we consider that higher TIR similarity is linked to recent activity, this phenomenon is readily understandable. This is because recently active TEs are more similar to each other and more likely to share sRNAs. In the case of Pack-MULEs, there are more chances for recent elements to share sRNAs with other Pack-MULEs as well as their parental genes. In addition, recently active TEs are also more likely to be inserted into regions where they are inappropriately expressed, and there has not been sufficient time for them to be eliminated by selection.
In summary, our data suggest that the expression of Pack-MULEs and the relationship between Pack-MULEs and their parental genes vary with the age of the elements. Pack-MULEs may negatively regulate the expression of their parental genes, and such an effect would be expected to be strongest when the elements are young, when they share more sRNAs with their parental genes and are more likely to produce sRNAs. Meanwhile, Pack-MULEs themselves are subject to suppression by the same mechanism so that young Pack-MULEs are less frequently expressed. As the elements age, the suppressive effect is gradually relieved, and the surviving Pack-MULEs are more likely to be expressed. Thus, through a long-term evolution and selection process, some Pack-MULEs may fulfill the transition from selfish genetic element to functional host gene.
Pack-MULEs: Past, Present, and Future
The phenomenon of gene capturing by Mutator-like elements was reported 20 years ago, yet little is known about how those elements evolve and what their impact is on genome evolution. The generation of Pack-MULEs is a mutagenic process that produces new combinations of coding and regulatory sequences. Like all other mutagenic processes, it is likely to be random with respect to selective advantage upon the generation of the mutation. Therefore, it is expected that the vast majority of Pack-MULEs would be nonfunctional. However, unlike other forms of mutagenesis, such as point mutations or random rearrangements, the formation of Pack-MULEs produces new, potentially functional stretches of DNA. In this study, we identified 2809 Pack-MULEs in the rice genome. Due to the stringent search criteria and the fact that the life cycle of TEs is only a few million years, it is conceivable that many more Pack-MULEs have been generated during the evolution of the rice genome and many of them are no longer recognizable. Given their prevalence, if only a small fraction of them are functional, Pack-MULEs may have had a significant impact on the repertoire of duplicated genes in the genome.
The analysis presented here does not provide unambiguous evidence for the function of any individual Pack-MULE, which requires experimental approaches, such as gene knockouts or overexpression. However, several lines of evidence suggest that a subset of Pack-MULEs have been subject to selection at different levels. Based on this evidence, we suggest that it is very likely that some Pack-MULEs represent functional genes. If we consider only protein coding genes, it is apparent that the 28 (or more) Pack-MULEs that are associated with the generation of proteins should be considered legitimate genes, albeit ones whose function is not clear. From this point of view, the amplification of Pack-MULEs provides a large reservoir for coding as well as regulatory sequences, with great potential to modify the genome, both genetically and epigenetically.
Given the widespread distribution of MULEs and Pack-MULEs in plants, our study demonstrates the need for experimental analysis of Pack-MULE functions. On the one hand, plants with active MULEs, such as maize, may be used to identify new acquisitions and test their direct impact on the expression of their parental genes, since newly formed Pack-MULEs seem to be associated with most sRNAs. On the other hand, knockout and overexpression experiments can be applied to test the function of proteins generated by Pack-MULEs. Through these experiments, it will be intriguing to test how often Pack-MULEs interfere with the preexisting genetic networks and how often they encode entirely new functions.
METHODS
Annotation of All Pack-MULEs in Rice and Identification of Parental Genes
The procedure for the annotation of Pack-MULEs in the rice (Oryza sativa) genome was similar to that described previously with modifications (Jiang et al., 2004). The sequences for 12 rice (Nipponbare) pseudomolecules were downloaded from TIGR (currently named the J. Craig Venter Institute; http://www.tigr.org/tdb/e2k1/osa1/, release 3, with a total size of 370 Mb). The genomic sequences was masked with RepeatMasker (version 07/07/2001, default parameters) using a library containing sequence of all MULE-TIRs identified so far, which was based on a library of repetitive sequences built using RECON (Bao and Eddy, 2002). Based on RepeatMasker output, all possible pairs of MULE TIRs located within 40 kb were examined and considered as Pack-MULEs if they satisfy the criteria for Pack-MULEs (see Supplemental Methods online). Among the 2809 Pack-MULEs, 2800 of them have TIRs within 20 kb of each other. After all Pack-MULEs were identified, they were remapped to psuedomolecules of release 5 from TIGR, and all subsequent analyses were based on the information from release 5 except that with TARs (Li et al., 2007), which was based on release 3. The sequence identity between TIRs was estimated by alignment of the terminal sequence in reversed orientation by DIALIGN2-2 (Morgenstern, 1999), followed by calculation of identical nucleotides in the alignable region. Additional inverted repeat sequences were identified by a self-comparison with element sequence (BLASTN e-value < 10−5; Altschul et al., 1990). The sequences of putative autonomous elements were recovered using the sequence of autonomous MULEs in the RECON library to mask the rice genome. If a MULE matches the sequence in the library and is longer than 3 kb, it is considered an autonomous element. As a result, this group of elements includes both true autonomous elements and their large derivatives.
To identify the origin of the sequences captured by Pack-MULEs, the internal regions of Pack-MULEs were masked using non-MULE TEs and then used to query the rice genome for similar sequence (BLASTN e-value < 1.0E-10). For an individual Pack-MULE, the sequence with highest score but not associated with MULE TIR was considered as the parental copy of the Pack-MULE. If the position of the parental copy overlapped with the coding region of a non-TE gene in psuedomolecules of release 5 from TIGR (http://www.tigr.org/tdb/e2k1/osa1/), the relevant gene was considered as the parental gene for this Pack-MULE.
Expression Analysis
The full-length cDNA data set was downloaded from http://cdna01.dna.affrc.go.jp/cDNA/ on August 24, 2008. A Pack-MULE is considered to have a cDNA match if (1) sequence similarity (BLASTN; Altschul et al., 1990) between the Pack-MULE and the cDNA is higher than 99.5% over the entire length of the cDNA, and (2) the chromosomal position of the Pack-MULE is consistent with the genomic position of the particular cDNA provided by the rice full-length cDNA consortium (Kikuchi et al., 2003). The MPSS data set is based on published results (Nobuta et al., 2007), and a signature for mRNA or sRNA was assigned to a Pack-MULE or parental gene if it is inside the Pack-MULE or the parental gene based on its position. Similarly, positions of TARs were downloaded based on information from Li et al. (2007) and assigned to Pack-MULEs in the same way. The comparison of expression level using the MPSS data set was performed using R Package (http://www.r-project.org; see Supplemental Methods online for details). For each individual signature and each library, only a single expression level was used for analysis (no duplicate available). The relative orientation of transcription by Pack-MULEs was determined by comparison of the orientation of cDNA, MPSS signature, and the open reading frame for parental genes. If there are two fl-cDNA sequences that fall into the same acquired region and both are in the sense orientation, it is counted as one sense event. If a transcript covers portions from two genes that are in opposite orientation, it is counted as two events: one sense and one antisense event.
Peptide sequences or accession numbers representing 3230 proteins were downloaded from the relevant references (Koller et al., 2002; Chitteti and Peng, 2007; Lee et al., 2007; Tan et al., 2007; Li et al., 2008) used as queries to search (TBLASTN; Altschul et al., 1990) against all Pack-MULE sequences. Peptide sequences that generated perfect hits with Pack-MULE sequences were then used to search the rice genomic database. A particular Pack-MULE sequence was considered to have a peptide match if it was the only perfect hit in the genome.
Evolutionary Rate Analysis
To identify potential coding regions of Pack-MULEs, each Pack-MULE sequence was searched against the rice protein sequences in GenBank with BLASTX (Altschul et al., 1990; e < 10−5). Based on the amino acid alignment generated by the search, we constructed pairwise nucleotide sequence alignment between each Pack-MULE and its top scoring rice protein coding sequence (regarded as the putative parental gene) to match the coding frames. Nucleotide positions involved in frame shifts and stops were excluded from the nucleotide sequence alignment to facilitate evolutionary rate calculation. To reduce the sampling errors in substitution rate estimation, we analyzed only those Pack-MULEs with alignment lengths > 50 codons. To calculate the substitution rates on a per-lineage basis, we first searched for a putative outgroup coding sequence (SO) for a Pack-MULE coding sequence (SM) and its parental copy (SP) using RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/). To find the most recently diverged outgroup sequence, for each potential SO, Ks values were calculated between SO and SP (KsO-P), between SO and SM (KsO-M), and between SP and SM (KsP-M) by the modified Nei and Gojobori method (Zhang et al., 1998). For each potential SO, when both KsO-P and KsO-M is > KsP-M, and KsO-P is the smallest among all possible RefSeq matches, this SO was defined as the outgroup for the SP-SM pair in question. Using the coding sequences of each SP-SM-SO trio, we first inferred the ancestral sequence between SM and SP using the Maximum Likelihood method implemented in PAML (Yang et al., 1995). Lineage-based Ka and Ks values between reconstructed ancestral sequences and SM or SP were then calculated by the modified Nei and Gojobori method (Zhang et al., 1998). χ2 tests were conducted to determine if the Ka/Ks value of any lineage is significantly smaller than one (Nei and Kumar, 2000).
The evolutionary rate distribution for human pseudogenes was based on published results (Zhang et al., 2003). The human pseudogenes analyzed included those with frame disruptions and those without frame disruptions. To generate a fraction value for a certain bin of Ka/Ks value for human pseudogenes, the fraction value for both types (with and without frame disruption) of pseudogenes was combined based on their genomic fraction provided by Zhang et al. (2003). The Kolmogorov-Smirnov test between human pseudogenes and rice Pack-MULEs was based on the distribution of individual Ka/Ks values of duplicated human pseudogenes with their parental genes and that for Pack-MULEs and their parental genes. In all Ka/Ks analysis, sequence pairs with Ks < 0.05 were excluded due to the relatively large errors in rate estimation.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Frequency of Duplication of Parental Genes.
Supplemental Table 1. Peptides Associated with Pack-MULEs.
Supplemental Table 2. Small RNAs Associated with Pack-MULEs and Their Parental Genes.
Supplemental Table 3. In-Frame Sequence within Pack-MULEs.
Supplemental Table 4. Transcription Orientation of Pack-MULEs Containing Fragments from Two Genes.
Supplemental Methods. Criteria for Pack-MULEs; Comparison of Transcription Level and Ubiquity of Transcription; and Comparison of Percentage of Transcription Cases in Each Library as well as Overall Tissue Specificity.
Supplemental Results. The Transcription of Chimeric Pack-MULEs.
Supplemental Data Set 1. Pack-MULEs in the Genome of Rice.
Supplementary Material
Acknowledgments
We thank Cheng Zou (Michigan State University) for sharing unpublished data and Kevin Childs and Robin Buell (Michigan State University) for technical assistance. This study is supported by grants from the USDA Cooperative State Research, Education, and Extension Service–National Research Initiative Plant Genome Program and Genetic Mechanism Program to N.J. and National Science Foundation Awards DBI-0638591 and MCB-0749634 to S.-H.S, DBI-0321437 to B.C.M, and DBI-031726 to D.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ning Jiang (jiangn@msu.edu).
Online version contains Web-only data.
References
- Adams, K.L., and Wendel, J.F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8 135–141. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Bao, Z., and Eddy, S.R. (2002). Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. (1996). The Mutator transposable element system of maize. Curr. Top. Microbiol. Immunol. 204 195–229. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2005). Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15 621–627. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Springer, P.S. (1994). The generation of mutator transposable element subfamilies in maize. Theor. Appl. Genet. 87 657–667. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., Swanson, J., Taylor, W.C., and Freeling, M. (1984). DNA insertion in the first intron of maize Adh1 affects message levels: Cloning of progenitor and mutant Adh1 alleles. Proc. Natl. Acad. Sci. USA 81 4125–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, S., Pea, G., and Rafalski, A. (2005). Origins, genetic organization and transcription of a family of non-autonomous helitron elements in maize. Plant J. 43 799–810. [DOI] [PubMed] [Google Scholar]
- Bureau, T.E., White, S.E., and Wessler, S.R. (1994). Transduction of a cellular gene by a plant retroelement. Cell 77 479–480. [DOI] [PubMed] [Google Scholar]
- Chalvet, F., Grimaldi, C., Kaper, F., Langin, T., and Daboussi, M.J. (2003). Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol. Biol. Evol. 20 1362–1375. [DOI] [PubMed] [Google Scholar]
- Chitteti, B.R., and Peng, Z. (2007). Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza sativa) roots. J. Proteome Res. 6 1718–1727. [DOI] [PubMed] [Google Scholar]
- Eisen, J.A., Benito, M.I., and Walbot, V. (1994). Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22 2634–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, J.J., Kaessmann, H., Betran, E., and Long, M. (2004). Extensive gene traffic on the mammalian X chromosome. Science 303 537–540. [DOI] [PubMed] [Google Scholar]
- Fu, H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui, K., and Iijima, K. (1991). Somatic chromosome map of rice by imaging methods. Theoretical and Applied Genetics 81 589–596. [DOI] [PubMed] [Google Scholar]
- Grandbastien, M.A., et al. (2005). Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet. Genome Res. 110 229–241. [DOI] [PubMed] [Google Scholar]
- Hancock, J.F. (2005). Contributions of domesticated plant studies to our understanding of plant evolution. Ann. Bot. (Lond.) 96 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen, D.R., Park, K.C., Elrouby, N., Yu, Z., Mohabir, N., Cowan, R.K., and Bureau, T.E. (2006). Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol. Biol. Evol. 23 1254–1268. [DOI] [PubMed] [Google Scholar]
- Holligan, D., Zhang, X., Jiang, N., Pritham, E.J., and Wessler, S.R. (2006). The transposable element landscape of the model legume Lotus japonicus. Genetics 174 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Eddy, S.R., and Wessler, S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573. [DOI] [PubMed] [Google Scholar]
- Jin, Y.K., and Bennetzen, J.L. (1994). Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic, N., Hoen, D.R., Huynh, M.L., Harrison, P.M., and Bureau, T.E. (2005). The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 15 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, S., and Nitasaka, E. (2004). Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol. 45 933–944. [DOI] [PubMed] [Google Scholar]
- Kazazian, H.H., Jr. (2004). Mobile elements: Drivers of genome evolution. Science 303 1626–1632. [DOI] [PubMed] [Google Scholar]
- Kikuchi, S., et al. (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379. [DOI] [PubMed] [Google Scholar]
- Koller, A., Washburn, M.P., Lange, B.M., Andon, N.L., Deciu, C., Haynes, P.A., Hays, L., Schieltz, D., Ulaszek, R., Wei, J., Wolters, D., and Yates III, J.R. (2002). Proteomic survey of metabolic pathways in rice. Proc. Natl. Acad. Sci. USA 99 11969–11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.G., Ahsan, N., Lee, S.H., Kang, K.Y., Lee, J.J., and Lee, B.H. (2007). An approach to identify cold-induced low-abundant proteins in rice leaf. C. R. Biol. 330 215–225. [DOI] [PubMed] [Google Scholar]
- Li, G., Nallamilli, B.R., Tan, F., and Peng, Z. (2008). Removal of high-abundance proteins for nuclear subproteome studies in rice (Oryza sativa) endosperm. Electrophoresis 29 604–617. [DOI] [PubMed] [Google Scholar]
- Li, L., et al. (2007). Global identification and characterization of transcriptionally active regions in the rice genome. PLoS One 2 e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.-H. (1997). Molecular Evolution. (Sunderland, MA: Sinauer Associates).
- Lisch, D. (2002). Mutator transposons. Trends Plant Sci. 7 498–504. [DOI] [PubMed] [Google Scholar]
- Lisch, D. (2005). Pack-MULEs: Theft on a massive scale. Bioessays 27 353–355. [DOI] [PubMed] [Google Scholar]
- Long, M. (2001). Evolution of novel genes. Curr. Opin. Genet. Dev. 11 673–680. [DOI] [PubMed] [Google Scholar]
- Lukens, L.N., and Zhan, S. (2007). The plant genome's methylation status and response to stress: Implications for plant improvement. Curr. Opin. Plant Biol. 10 317–322. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Makalowski, W., and Boguski, M.S. (1998). Synonymous and nonsynonymous substitution distances are correlated in mouse and rat genes. J. Mol. Evol. 47 119–121. [DOI] [PubMed] [Google Scholar]
- Masterson, J. (1994). Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science 264 421–424. [DOI] [PubMed] [Google Scholar]
- Moore, R.C., and Purugganan, M.D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8 122–128. [DOI] [PubMed] [Google Scholar]
- Moran, J.V., DeBerardinis, R.J., and Kazazian, H.H., Jr. (1999). Exon shuffling by L1 retrotransposition. Science 283 1530–1534. [DOI] [PubMed] [Google Scholar]
- Morgante, M., Brunner, S., Pea, G., Fengler, K., Zuccolo, A., and Rafalski, A. (2005). Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37 997–1002. [DOI] [PubMed] [Google Scholar]
- Morgenstern, B. (1999). DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15 211–218. [DOI] [PubMed] [Google Scholar]
- Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. (New York: Oxford University Press).
- Nobuta, K., Venu, R.C., Lu, C., Belo, A., Vemaraju, K., Kulkarni, K., Wang, W., Pillay, M., Green, P.J., Wang, G.L., and Meyers, B.C. (2007). An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 25 473–477. [DOI] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (New York: Springer-Verlag).
- Ohtsu, K., Hirano, H.Y., Tsutsumi, N., Hirai, A., and Nakazono, M. (2005). Anaconda, a new class of transposon belonging to the Mu superfamily, has diversified by acquiring host genes during rice evolution. Mol. Genet. Genomics 274 606–615. [DOI] [PubMed] [Google Scholar]
- Pickeral, O.K., Makalowski, W., Boguski, M.S., and Boeke, J.D. (2000). Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham, E.J., Feschotte, C., and Wessler, S.R. (2005). Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol. Biol. Evol. 22 1751–1763. [DOI] [PubMed] [Google Scholar]
- Ramsey, J., and Schemske, D.W. (1998). Pathways, mechanisms and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29 477–501. [Google Scholar]
- Robertson, D.S. (1978). Characterization of a Mutator system in maize. Mutat. Res. 51 21–28. [Google Scholar]
- Singer, T., Yordan, C., and Martienssen, R.A. (2001). Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S., Inagaki, Y., Satoh, H., Hoshino, A., and Iida, S. (1999). Capture of a genomic HMG domain sequence by the En/Spm-related transposable element Tpn1 in the Japanese morning glory. Mol. Gen. Genet. 261 447–451. [DOI] [PubMed] [Google Scholar]
- Talbert, L.E., and Chandler, V.L. (1988). Characterization of a highly conserved sequence related to mutator transposable elements in maize. Mol. Biol. Evol. 5 519–529. [DOI] [PubMed] [Google Scholar]
- Tan, F., Li, G., Chitteti, B.R., and Peng, Z. (2007). Proteome and phosphoproteome analysis of chromatin associated proteins in rice (Oryza sativa). Proteomics 7 4511–4527. [DOI] [PubMed] [Google Scholar]
- Turcotte, K., Srinivasan, S., and Bureau, T. (2001). Survey of transposable elements from rice genomic sequences. Plant J. 25 169–179. [DOI] [PubMed] [Google Scholar]
- Walbot, V., and Rudenko, G.N. (2002). MuDR/Mu transposable elements of maize. In Mobile DNA II, N. Craig, R. Craigie, M. Gellert, and A. Lambowitz, eds (Washington, DC: America Society of Microbiology Press), pp. 533–564.
- Wang, Q., and Dooner, H.K. (2006). Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl. Acad. Sci. USA 103 17644–17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., et al. (2006). High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42 225–249. [PubMed] [Google Scholar]
- Wessler, S.R. (1996). Turned on by stress. Plant retrotransposons. Curr. Biol. 6 959–961. [DOI] [PubMed] [Google Scholar]
- Xu, Z., Yan, X., Maurais, S., Fu, H., O'Brien, D.G., Mottinger, J., and Dooner, H.K. (2004). Jittery, a Mutator distant relative with a paradoxical mobile behavior: Excision without reinsertion. Plant Cell 16 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Kumar, S., and Nei, M. (1995). A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z., Wright, S.I., and Bureau, T.E. (2000). Mutator-like elements in Arabidopsis thaliana. Structure, diversity and evolution. Genetics 156 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala, G., and Vodkin, L. (2007). Novel exon combinations generated by alternative splicing of gene fragments mobilized by a CACTA transposon in Glycine max. BMC Plant Biol. 7 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala, G., and Vodkin, L.O. (2005). The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily. Plant Cell 17 2619–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. (2003). Evolution by gene duplication: An update. Trends Ecol. Evol. 18 292–298. [Google Scholar]
- Zhang, J., Rosenberg, H.F., and Nei, M. (1998). Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., and Gaut, B.S. (2003). Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 13 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Harrison, P.M., Liu, Y., and Gerstein, M. (2003). Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13 2541–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.