Abstract
The expression of genes involved in the pathogenesis of Staphylococcus aureus is controlled by global regulatory loci, including two-component regulatory systems and transcriptional regulators (e.g., sar family genes). Most members of the SarA family have been partially characterized and shown to regulate a large numbers of target genes. Here, we describe the characterization of sarZ, a sarA paralog from S. aureus, and its regulatory relationship with other members of its family. Expression of sarZ was growth phase dependent with maximal expression in the early exponential phase of growth. Transcription of sarZ was reduced in an mgrA mutant and returned to a normal level in a complemented mgrA mutant strain, which suggests that mgrA acts as an activator of sarZ transcription. Purified MgrA protein bound to the sarZ promoter region, as determined by gel shift assays. Among the sarA family of genes analyzed, inactivation of sarZ increased sarS transcription, while it decreased agr transcription. The expression of potential target genes involved in virulence was evaluated in single and double mutants of sarZ with mgrA, sarX, and agr. Northern and zymogram analyses indicated that the sarZ gene product played a role in regulating several virulence genes, particularly those encoding exoproteins. Gel shift assays demonstrated nonspecific binding of purified SarZ protein to the promoter regions of the sarZ-regulated target genes. These results demonstrate the important role played by SarZ in controlling regulatory and virulence gene expression in S. aureus.
Staphylococcus aureus is a human pathogen that colonizes more than 1 billion people worldwide and causes a variety of infections, ranging from cutaneous infections (impetigo, folliculitis, and carbuncles) to deep-seated infections (pneumonia, endocarditis, septicemia, and osteomyelitis), and other metastatic complications (18, 21, 27). The primary site of infection is generally the skin or soft tissues, from which the organism can spread to the bloodstream and subsequently disseminate into various tissues. Once S. aureus establishes its presence in the tissue, it produces a large number of cell surface-associated components and secreted products that include adhesins, enzymes, toxins, capsular polysaccharides, and other gene products that facilitate tissue colonization, tissue destruction, or immune evasion. The expression of many of these genes is coordinately controlled by regulatory systems, such as such as agr, arlRS, srrAB, saeRS, lytSR, vraRS, sigB, tcaRAB, sarA, and nine other sarA paralogs (i.e., sarR, sarS, sarT, sarU, sarV, rot, sarX, sarZ, and mgrA) (2-23, 28-56).
The agr system is a well-studied regulatory system that controls the synthesis of select cell surface proteins and exoproteins. The agr locus consists of two divergent transcriptional units, agr RNAII and RNAIII. RNAII encodes the sensor kinase (AgrC), response regulator (AgrA), a signal-processing protein (AgrB), and an autoinducing peptide (AIP, or AgrD). RNAIII is a 514-nucleotide RNA regulatory molecule that regulates a large number of genes, mostly at the posttranscriptional level (23, 39-41). Microarray analysis in strain RN6390 showed that 104 and 34 genes are upregulated and downregulated, respectively, due to agr inactivation (13), suggesting that agr is a global regulator in S. aureus. Several reports have shown rather contradictory roles of the agr locus in the regulation of target genes, but these conflicting results could be due to the use of different strains, growth media, and growth conditions (5, 44). Besides the autoactivation of agr by AIP and AgrA, SarA and several of the SarA paralogs, srrAB, arlSR have also been shown to modulate RNAII and RNAIII transcription (2, 3, 10, 12, 15, 20, 34, 35, 39-41, 43, 45).
The sarA locus is known to upregulate the synthesis of fibronectin and fibrinogen binding proteins, hemolysins (α, β, and δ), enterotoxins, TSST-1 toxin, and capsule biosynthesis genes and to downregulate proteases, protein A, and a collagen binding protein. SarA also binds to several regulatory and target gene promoters (e.g., agr, sarS, rot, sarV, sarT, hla, fnb, spa, cna, bap, and icaRA) to modulate gene transcription, thus implicating both agr-dependent and agr-independent pathways in SarA-mediated regulation (2, 8-12, 33, 35, 36, 45, 49, 53). Using affinity chromatography and genome sequence information, nine SarA paralogs (i.e., SarR, SarS, SarT, SarU, SarV, SarX, SarZ, Rot, and MgrA) that are involved directly or indirectly in the regulation of target genes implicated in regulation, virulence, biofilm formation, autolysis, antibiotic resistance, and metabolic processes have been identified (10). SarS (also called SarH1), a 250-residue protein, is involved in activation of spa expression (9, 42, 52). Protein A, associated with the cell surface, is an important virulence factor involved in a wide variety of interactions with various host factors during staphylococcal infection (41, 42). Inactivation of mgrA, shown to be involved in the regulation of virulence genes, as well as autolytic genes (11, 28, 29, 54), is also involved in oxidative-stress responses and indirectly affects resistance to antibiotics by controlling the expression of at least four efflux pumps (i.e., norA, norB, norC, and tet38) by repressing the expression of norG (6, 55). Previous studies have shown the SarA paralogs to be activated or repressed by other members of their own family. For example, MgrA positively and negatively regulates the expression of sarX and sarV transcripts, respectively, while SarT represses sarU transcription (32-34). The interregulatory relationship between different members of the SarA family is shown in Fig. 1. More recently, SarZ has been shown to be involved in the regulation of hemolysin production, and sarZ mutants are less virulent in silkworm and mouse infection models (20). Along with hemolysin (hla and hlb) transcripts, the sarZ mutants also showed reduced agr expression. It has also been demonstrated that the DNA binding ability of SarZ is essential for virulence in S. aureus (20). In a more recent study, sarZ has been shown to be important for biofilm formation and virulence in both Staphylococcus epidermidis and S. aureus (51, 56). Very recently, it has been shown that sarZ is involved in the activation of mgrA and the agr RNAIII molecule, and S. aureus with sarZ deleted showed increased transcription of spa, the protein A gene, while the levels of hla (the hemolysin gene) and sspA (the V8 protease gene) transcripts were decreased (51). A different study also suggested that SarZ is involved in oxidative sensing and regulation of genes involved in oxidative pathways (7).
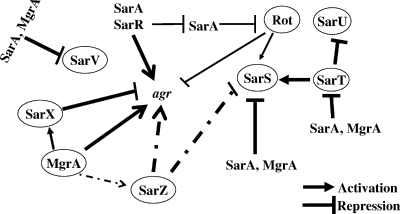
FIG. 1.
Overview of the predicted intertwined regulatory networks of 10 sar family genes and their relationship with the agr locus. This overview is mostly based on genetic and biochemical analyses in strain RN6390 under normal conditions of growth (8, 10, 28-36, 41, 49-52). Several members of the sar family of genes (e.g., sarV, sarT, and sarU) are not transcribed due to repression by other normally expressed sar family genes (32, 33, 49). The pathways identified in this study are depicted by dashed lines. The thickness of the line indicates the nature of regulation: thin, partially involved, and thick, completely involved.
In this report, we show that among the nine sarA paralog, agr, sigB, and saeRS regulatory mutant strains, expression of sarZ is activated only by mgrA. Inactivation of sarZ does not have a major effect on the transcription of most of the other members of the sarA family of genes, except for sarS. Analysis of various double mutants of sarZ with other regulatory genes suggests that sarZ has discernible effects on the expression of agr RNAII and RNAIII. As sarA family genes are involved in the regulation of virulence genes, the expression of various potential target genes (spa, hla, and sspA) in sarZ, sarX, mgrA, sarZ plus mgrA, and sarX plus sarZ mutants were evaluated. The results show that sarZ is involved in the regulation of several virulence genes, particularly those genes encoding exoproteins (proteases and hemolysins).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Phage φ11 was used as a generalized transducing phage for S. aureus strains. S. aureus strain RN4220, a restriction-deficient derivative of strain 8325-4, was used as the initial recipient for the transformation of plasmid constructs. S. aureus strains were routinely cultured in tryptic soy broth or tryptic soy agar medium supplemented with appropriate antibiotics when necessary. Luria-Bertani medium was used for growing Escherichia coli. For verification and routine maintenance, various S. aureus strains were grown with appropriate antibiotic selection as follows: 5 μg/ml of erythromycin, 50 μg/ml of kanamycin, 5 μg/ml of tetracycline, and 10 μg/ml of chloramphenicol. For E. coli, ampicillin at 50 μg/ml, chloramphenicol at 30 μg/ml, erythromycin at 200 μg/ml, and spectinomycin at 75 μg/ml were used.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| RN4220 | Restriction-deficient transformation recipient | 40 |
| RN6390 | Wild-type laboratory strain related to 8325-4, prophage-cured strain from human isolate NCTC8325 harboring an 11-bp deletion in rsbU that regulates sigB activity by activating RsbV, a factor that competitively binds to the anti-sigma factor RsbW | 40 |
| ALC488 | sarA mutant of RN6390 with sarA::ermC | 31 |
| ALC1713 | sarR mutant of RN6390 with sarR::ermC | 30 |
| ALC1927 | sarS mutant of RN6390 with sarS::ermC | 9 |
| ALC1905 | sarT mutant of RN6390 with sarT::ermC | 49 |
| ALC2272 | sarU mutant of RN6390 with sarU::ermC | 32 |
| ALC2319 | sarV mutant of RN6390 with saV::ermC | 33 |
| AM 1080 | sarX mutant of RN6390 with sarX::ermC | 34 |
| AM 1090 | sarZ mutant of RN6390 with sarZ::ermC | This study |
| ALC2530 | mgrA mutant of RN6390 with mgrA::ermC | 33 |
| AM 1102 | rot mutant of RN6390 with rot::ermC | This study |
| RN6911 | agr mutant of RN6390 with agr::tetM | 40 |
| AM1126 | sarZ agr double mutant of RN6390 with sarZ::kan and agr::tetM | This study |
| AM1175 | sarZ mgrA double mutant of RN6390 with sarZ::kan and mgrA::ermC | This study |
| AM1176 | sarZ sarX double mutant of RN6390 with sarZ::kan and sarX::ermC | This study |
| AM1194 | AM1090 complemented with 1.6-kb sarZ fragment into the geh locus on the chromosome | This study |
| AM1390 | AM1090 with pSK236 containing 1.6-kb sarZ gene at BamHI and XhoI sites | This study |
| SH1000 | Functional rsbU derivative of 8325-4 rsbU+ | 19 |
| AM1397 | sarZ mutant of SH1000 sarZ::ermC | This study |
| AM1398 | AM1397 complemented with the sarZ gene into the geh locus on the chromosome | This study |
| AM1410 | mgrA mutant of SH1000 mgrA::ermC | This study |
| AM1412 | AM1410 with pSK236 containing 1.5-kb mgrA fragment at EcoRI site | This study |
| Newman | A human clinical isolate | 37 |
| AM1419 | mgrA mutant of Newman mgrA::ermC | This study |
| AM1422 | AM1419 with pSK236 containing 1.5-kb mgrA gene at EcoRI site | This study |
| Plasmids | ||
| pUC18 | E. coli cloning vector | |
| pCR2.1 | E. coli cloning vector for direct cloning of PCR product | Invitrogen |
| pCL52.2 | Temperature-sensitive E. coli-S. aureus shuttle vector | 26 |
| pCL84 | Single-copy integration E. coli-S. aureus shuttle vector | 25 |
| pSK236 | E. coli-S. aureus shuttle vector | 16 |
| pAM927 | pUC19 containing a 2.3-kb BamHI- XhoI fragment containing sarZ region at BamHI-SalI sites | This study |
| pAM945 | pUC19 containing ∼3.3-kbDNA fragment that has a deletion of a 191-bp AflII/SwaI fragment that includes promoter and N-terminal (31-residue) regions of the sarZ gene and an insertion of the 1.2-kb ermC gene | This study |
| pAM950 | pCL52.2 containing ∼3.3-kbDNA fragment sarZ::ermC from pAM945 | This study |
| pAM1055 | Same as pAM945, but the ermC gene was replaced with 1.2-kb kanamycin gene (kan) | This study |
| pAM975 | pSK236 with a 1.6-kb DNA fragment containing the sarZ gene with 280-bp upstream sequence | This study |
| pAM980 | pCL84 with a 1.6-kb DNA fragment containing the sarZ gene with 280-bp upstream sequence | This study |
| pAM1110 | pSK236 with a 1.5-kb DNA fragment containing the mgrA gene region |
Genetic manipulations in E. coli and S. aureus.
Using a genome scan for homology with SarA, the sarZ gene product (SA2174) was initially identified in the S. aureus N315 genome database (24). To construct a sarZ mutant, the sarZ gene, together with 865-bp and 920-bp flanking sequences upstream and downstream, respectively, was amplified by PCR with the primers 5′-AACGGATCAAGTCATTTAGCA-3′ and 5′-TCTGACATCACTCAATTATATCAGA-3′, using chromosomal DNA from strain RN6390 as the template. The 2,235-bp PCR fragment was cloned into cloning vector pCR 2.1 (Invitrogen, San Diego, CA) in E. coli. The BamHI/XhoI DNA fragment-containing 2.3 kb fragment was then cloned into the BamHI and SalI sites of pUC19 (pAM927). A 191-bp fragment, comprising the promoter and N-terminal regions of the sarZ coding region, was deleted by restriction with AflII and SwaI and then replacement of the deleted fragment with an ∼1.2-kb ermC gene at these sites (pAM945). The 3.3-kb fragment containing the ermC insertion fragment within the deleted sarZ region was cloned into the temperature-sensitive shuttle vector pCL52.2 (pAM950) (26). Construction and selection of the putative sarZ mutant (tetracycline-sensitive and erythromycin-resistant colonies) was performed as described previously (30, 32-34). A phage φ11 lysate of the putative sarZ mutant was then prepared to infect fresh RN6390 and SH1000 or other clinical strains to reconstruct the sarZ mutant in an attempt to avoid any putative genomic mutations that might have occurred during the temperature shift to promote homologous recombination. To construct double mutants of sarZ with mgrA::ermC and sarX::ermC, we cloned deletion (open reading frame [ORF]) and insertion (kanamycin gene cassette) fragments (e.g., sarZ::kan) into the temperature-sensitive vector pCL52.2 (pAM1055). Construction of double mutants was performed using essentially the same protocol described for single-mutant construction, except the recipient hosts were sarX and mgrA mutants. The correct mutation was confirmed by PCR and Northern and Southern hybridization with a sarZ probe as described previously (30, 32-34).
To complement the sarZ mutation, a 1.6-kb fragment encompassing the sarZ gene and 280 bp upstream of the sarZ translation start site was amplified with PCR (using primers 5′-AACGGATCAAGTCATTTAGCA-3′ and 5′-AGAAATCGAATTACATACACGATGC-3′) and cloned into the shuttle plasmid pSK236 (pAM975) (16) or a single-copy integration vector, pCL84 (pAM980) (25). Single-copy integration into the RN6390 sarZ mutant was performed as described previously (33, 34). The recombinant pSK236 derivative plasmid (pAM975) was electroporated into RN4220 and selected for on chloramphenicol plates. The plasmid from RN4220 was then electroporated into the parental strain RN6390 and the sarZ mutant to construct trans-complemented strains.
Isolation of RNA and Northern blot hybridization.
Total RNA from S. aureus was prepared by using a Trizol isolation kit (Gibco-BRL, Gaithersburg, MD) and a reciprocating shaker, as described previously (26-31). The optical densities at 600 nm (OD600s) of various cultures were measured in a spectrophotometer (Spectronic 20). The concentration of total RNA was determined by measuring the absorbance at 260 nm. Ten micrograms of total RNA was analyzed by Northern blotting, as described previously (30, 32-36). The genes agr RNAII, agr RNAIII, sarA paralogs, spa, hla, sspB, and sspA were either amplified by PCR or excised with restriction endonucleases from the plasmids containing the desired genes. For detection of specific transcripts, gel-purified DNA probes were radiolabeled with [α-32P]dCTP by using the random-primed DNA-labeling kit (Roche Diagnostics GmbH) and hybridized under aqueous-phase conditions at 65°C. The blots were subsequently washed, exposed to a phosphorimager screen, and scanned on a phosphorimager. The intensities of the bands were quantified with Image Quant software (Amersham Life Sciences).
Purification of SarZ and MgrA proteins and production of anti-SarZ polyclonal antibodies.
The cloning and purification of a six-His tag-MgrA fusion protein was described previously (33). The 450-bp DNA fragment containing the full-length sarZ gene was amplified by PCR using chromosomal DNA from S. aureus RN6390 as the template and primers containing flanking NdeI and BamHI restriction sites (5′-ATCATATGTATGTAGAAAACAGCTAT-3′ and 5′-ATGGATCCATACTTCTGCCCATCACCTTAT-3′) to facilitate in-frame cloning into the expression vector pET14b (Novagen, Madison, WI). The recombinant plasmid containing the full-length sarZ coding region was confirmed by restriction digestion and DNA sequencing and transformed into E. coli BL21(DE3)/pLysS. His6-SarZ protein expression and purification were done in a manner similar to those for the SarA protein, as described previously (12). The purity of the His-SarZ fusion protein was checked in a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel and found to be more than 98% pure. The concentration of the purified protein was determined by using a Bio-Rad (Hercules, CA) protein estimation kit with bovine serum albumin as the standard.
Anti-SarZ sera were prepared by immunizing two (BALB/c × SJL/J)F1 mice with 100 μg per mouse of purified His-SarZ fusion protein as described previously (30). Western blotting was performed to monitor the titers of the immune sera.
Gel shift analysis.
A 270-bp DNA fragment encompassing the sarZ promoter region upstream of the translational start site (ATG) was PCR amplified by using chromosomal DNA from RN6390 with the primers 5′-AGAAATCGAATTACATACACGATGC-3′ and 5′-ACATCCAATCACTCCTTGTTAA-3′ and cloned into the pCR2.1 vector (Invitrogen, CA). The DNA fragment was excised by digestion, gel purified, and dephosphorylated. In order to determine if the recombinant MgrA protein interacted with the 270-bp sarZ promoter fragment or if SarZ interacted with the sarS (9) and sspA (33) promoter regions, gel shift assays were performed as described previously (12, 30, 32-36). Briefly, DNA fragments were end labeled with [γ32P]ATP using polynucleotide kinase. The labeled fragment (1 ng or 6 fmol) was incubated at room temperature for 20 min with various amounts of purified MgrA or SarZ protein in 25 μl of binding buffer (25 mM Tris-Cl, pH 7.5, 0.1 mM EDTA, 75 mM NaCl, 1 mM dithiothreitol, and 10% glycerol) containing 0.5 μg of calf thymus DNA (Amersham Pharmacia Biotech). The reaction mixtures were analyzed in an 8.0% nondenaturing polyacrylamide gel. The band shifts were detected by exposing the dried gels to X-ray films or a phosphorimager screen. The binding affinity of MgrA or SarZ protein to its cognate promoters was derived from the dissociation constant, which is defined as the amount of protein needed to shift 50% of the labeled probe (Kd [dissociation constant] = [protein]50% of DNA probe) (47).
Primer extension analysis.
Mapping of the 5′ end of the sarZ transcript by primer extension was performed by using the primer 5′-CATATTCCTTAAGATAGTTTGTG-3′, complementary to the sarZ coding strand and located at nucleotide positions 103 to 80 downstream from the putative proposed start codon, ATG. Primer extension was carried out by using 30 μg of total RNA isolated from the wild-type strain RN6390 in the exponential phase, as described previously (1, 30, 36). The same radiolabeled primer was used for the DNA-sequencing reactions.
Cell extract preparation and Western analysis.
Cell extracts from early exponential (OD = 0.7) and postexponential (OD = 1.7) phases of growth were prepared from RN6390 and isogenic sarZ mutant staphylococcal strains. Cells were grown in 20 ml of tryptic soy broth medium without any antibiotics. After being pelleted, the cells were resuspended in 0.5 ml of cell lysis buffer (25 mM Tris-Cl, 5 mM EDTA [pH 8.0], 100 mM NaCl) and treated with lysostaphin. The lysostaphin-treated cells were briefly sonicated and centrifuged at 40,000 rpm for 30 min at 4°C to remove cell debris. The concentration of total proteins from clear lysates was determined by using the Bradford method (Bio-Rad, Hercules, CA).
Equal amounts of total cellular proteins were separated in an SDS-12% polyacrylamide gel and transferred onto nitrocellulose membranes as described previously (30). The blot was incubated at room temperature with a 1:1,000 dilution of anti-SarZ polyclonal antibodies for 2 h, followed by another hour of incubation with a 1:10,000 dilution of goat anti-mouse-horseradish peroxidase conjugate (Pierce, IL). Immunoreactive bands were detected as described previously with an enhanced-chemiluminescence detection kit (Piece, IL). SeaBlue prestained protein standards (Invitrogen, San Diego, CA) were used for molecular weight estimations.
Zymographic analysis.
For the detection of extracellular protease activity, SDS-polyacrylamide gel electrophoresis-based zymographic analysis was performed as described by Rice et al. (46). The gels were subsequently photographed, and the white bands in the gels (zones of hydrolysis) indicated regions of protease activity.
RESULTS
Characterization of the sarZ gene.
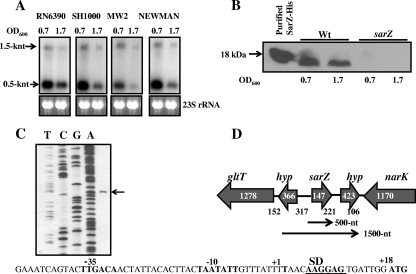
Analysis of sarZ transcription during different phases of growth was performed using Northern hybridization with total RNA isolated from various S. aureus strains, including laboratory-adapted strains (RN6390 and SH1000) and clinical isolates (Newman and MW2). A major sarZ transcript of approximately 500 nt was observed during the early exponential phase of growth (OD, ∼0.7), and there was significantly reduced transcription during the postexponential phase of growth. A minor transcript of 1,500 nt was also observed, which probably originated from a cryptic promoter (Fig. 2A). We speculated that the larger transcript may play a minor role in production of SarZ or in the regulation process; therefore, further characterization of the largest transcript was not pursued. The result indicates that sarZ transcription is growth phase dependent (Fig. 2A). The growth phase-dependent differential transcription patterns were similar in all four tested strains, including RN6390, a strain that produces less SigB due to a natural 11-bp deletion in a putative sigB activator, rsbU. The SarZ protein per se had not been shown to be expressed in S. aureus. To detect the SarZ protein in S. aureus and to investigate whether the transcription pattern correlated with the expression of the protein, Western blot analysis with the cell extracts of wild-type RN6390 and an isogenic sarZ mutant was performed (Fig. 2B). The results showed reduced expression of SarZ (17.5 kDa) in the postexponential phase of growth compared to the early exponential phase of growth. As expected, the extracts from the sarZ mutant did not react with the SarZ antiserum. These results clearly show that SarZ expression is growth phase dependent and that maximal expression occurs in the early exponential phase of growth.
FIG. 2.
Transcription, expression, and promoter analysis of the sarZ gene in S. aureus. (A) Northern analysis of the sarZ transcripts in the different wild-type strains at various phases of growth (an OD600 of 0.7 is approximately the early exponential phase of growth, and an OD600 of 1.7 is approximately the postexponential phase of growth). The blots were probed with 500-bp sarZ DNA fragments containing the entire ORF of the sarZ gene. The region of 23S rRNA of the ethidium bromide-stained gel used for blotting is also shown as a loading control. (B) Cell extracts of the RN6390 strain were immunoblotted onto nitrocellulose and probed with anti-SarZ polyclonal antibodies. A purified His tag fusion of SarZ was loaded as a positive control. Wt, wild type. (C) Primer extension of the sarZ transcript with total RNA isolated from the wild-type RN6390 at the exponential phase of growth. The nucleotide sequence with the predicted promoter region of the sarZ ribosome-binding site (SD) and the translational start codon (ATG) are indicated. (D) Location of the sarZ gene (SA2174) on the S. aureus chromosome. The sarZ gene and the other ORFs in its vicinity are depicted. The number within each ORF indicates the size of the gene (in bp), and the number below the junction of two neighboring genes is the intergenic distance (in bp) between them. hyp, hypothetical ORF with unknown function. The location of a 500-nt transcript is mapped based on primer extension results, whereas the origin of the 1,500-nt transcript is hypothetical.
In order to determine the transcriptional start site and the promoter structure of the major sarZ transcript, primer extension analysis was performed with total RNA isolated from the wild-type RN6390 (Fig. 2C). The transcription start site was mapped to a “T” located 17 nt upstream from the predicted SarZ ATG initiation codon in S. aureus (Fig. 2C). The predicted promoter regions and the ribosome binding site are also depicted in Fig. 2C. Based upon the transcriptional start site, the predicted promoter regions closely resemble the −35 and −10 consensus sequences of σA-dependent promoters. The physical location of the sarZ ORF on the staphylococcus genome is shown in Fig. 2D. The sarZ ORF is flanked by a hypothetical ORF on each side, while gltT (encoding a proton/sodium-glutamate symport protein; SA2172) and narK (encoding a nitrite transporter; SA2176) are further upstream and downstream, respectively. SarZ is highly conserved among the sequenced strains of Staphylococcus (97 to 100% identity among 11 strains of S. aureus). Orthologues of SarZ are also present in the genomes of two S. epidermidis strains (79% identity), one Staphylococcus saprophyticus strain (70% identity), and one Staphylococcus haemolyticus (65% identity) strain.
Regulatory relationship between different sarA family genes.
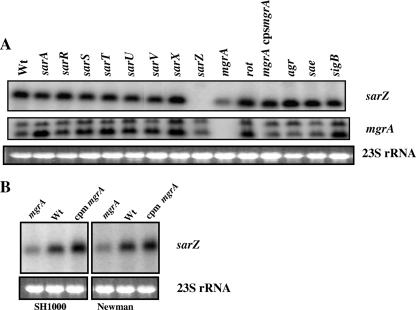
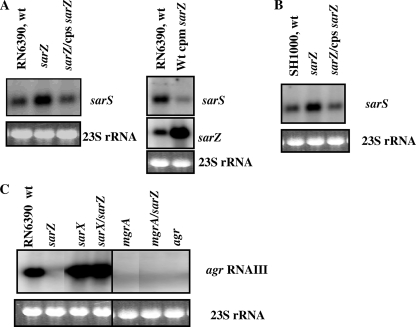
Previous characterization of sarA family genes has shown that most of these genes are regulated by other members of their own family. For example, sarV transcription, which is undetectable or very low under normal growth conditions in the wild-type strain, is significantly increased in a sarA or mgrA mutant (33). To determine the influence of sarZ on other members of the sarA family and to elucidate the phenotypic effects of sarZ inactivation, a sarZ mutant was constructed by allelic replacement as described in Materials and Methods. Northern blot assays were performed with a sarZ probe (500 bp, encompassing the coding region) with various sarA family mutants to determine if the transcription of sarZ was affected by any of the other sarA family genes. The Northern blotting results disclosed that the level of sarZ transcription was reduced threefold in the mgrA mutant, whereas transcription was not affected in the other nine sarA family mutants tested (Fig. 3A). Upon introduction of the functional mgrA gene into the mgrA mutant, the expression of the sarZ transcript returned almost to the parental level (Fig. 3A), indicating that the transcription of sarZ was activated by the mgrA gene product. To test if the above-described mode of regulation was also present in other S. aureus backgrounds, mgrA mutants were constructed in strains SH1000 and Newman. The mutants were also complemented with functional copies of the mgrA gene, and the expression of sarZ was assessed. The results showed a reduction in sarZ transcripts in both SH1000 and Newman mgrA mutants while near-normal levels of expression were observed in the complemented mutants (Fig. 3B). Therefore, the results of these analyses suggested that the expression of sarZ was activated by mgrA in all tested backgrounds in S. aureus.
FIG. 3.
(A) Northern analysis of the sarZ and mgrA transcripts in the wild type (Wt), various isogenic mutants, and a single-copy complemented strain of the mgrA mutant at exponential phase (OD600, ∼0.7) of growth. DNA fragments (500 bp and 550 bp) containing sarZ and mgrA genes, respectively, were used for hybridization. cpsmgrA indicates the complementation in single copy of the mgrA gene on the lipase locus (geh) of the mgrA mutant. (B) Northern analysis with a 500-bp sarZ DNA probe of the mgrA mutants and complemented (cpm) strains from different S. aureus strains, as indicated. The region of 23S rRNA of the ethidium bromide-stained gel used for blotting is also shown as a loading control in both panels.
Binding of MgrA to the sarZ promoter region.
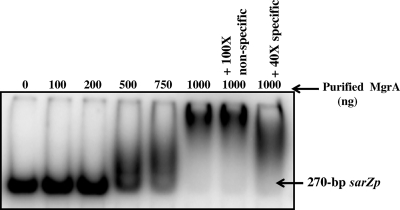
The direct interaction of MgrA with the sarZ promoter was tested in gel shift assays using purified MgrA protein and a 270-bp DNA fragment containing the sarZ promoter region (Fig. 4). Retarded protein-DNA complex could be detected with 0.5 μg of MgrA (Kd, ∼13 nM, assuming that MgrA is a dimeric protein) (47), and complete conversion occurred with 1.0 μg of MgrA. The unlabeled promoter fragment could compete with the labeled fragment, whereas a nonspecific, unrelated DNA fragment (a 185-bp fragment from the internal region of the sarX gene) did not compete with the labeled fragment (Fig. 4), indicating that DNA-protein interaction was specific. Previously, the DNA binding motif (GTTG) of the MgrA protein had been mapped on the sarV promoter region by DNase I footprinting (33). DNA sequence analysis upstream of the sarZ promoter region revealed the presence of the sequence in ACTTGACAACT, which is consistent with MgrA binding motifs (shown in bold) on promoter regions of other mgrA-regulated genes (33, 34). Interestingly, the putative MgrA binding site overlaps with the −35 promoter region of the sarZ gene (Fig. 2C). Hence, the MgrA protein can bind to the sarZ promoter region, presumably acting as an activator of sarZ transcription.
FIG. 4.
Autoradiogram of an 8.0% polyacrylamide gel showing gel shifts for purified MgrA protein with a γ-32P-labeled 270-bp sarZ promoter fragment (1 ng or 6 fmol per lane). The mobility of the band was noted in the presence of increasing amounts of MgrA protein, as indicated above the gel. The lanes containing competitor DNA, i.e., an unlabeled specific 270-bp sarZ promoter fragment (40-fold molar excess) and a nonspecific 185-bp internal DNA fragment of the sarX ORF (100-fold molar excess) are indicated.
Transcription of regulatory genes in a sarZ mutant.
To determine whether inactivation of sarZ affects other regulatory genes, particularly the members of the sarA family and agr, Northern blot analysis with probes for nine sarA paralog genes (sarA, sarR, sarS, sarT, sarU, sarV, sarX, mgrA, and rot), agr RNAII, and agr RNAIII was performed. Among the sarA family genes monitored, only transcription of sarS was considerably elevated (fourfold compared to the wild type) due to inactivation of the sarZ gene (Fig. 5A). Several members of the sarA, family such as sarA and mgrA, showed minor variation (∼20% to 30%) in transcription (data not shown), which is consistent with the slightly increased SarA expression observed in a recent study (51). Upon single-copy complementation of the wild-type sarZ gene to a sarZ mutant, the level of sarS transcription was restored to nearly the parental level; confirming that sarZ activates sarS expression. To further confirm that sarZ was indeed involved in the regulation of sarS, the sarZ gene was overexpressed in a multicopy shuttle vector in the wild-type strain RN6390 (Fig. 5A, right). The level of sarS transcription was decreased (∼6-fold) in the overexpressing sarZ strain compared to the parental strain. Hence, these results clearly indicate that the level of sarS transcription is directly regulated by the sarZ gene product. The transcription of sarS was monitored in the SH1000 sarZ mutant, which is rsbU+ and belongs to the same lineage as RN6390. As shown in Fig. 5B, sarS transcription was elevated in the SH1000 sarZ mutant, and on complementation with a functional single-copy sarZ gene, sarS transcription was restored to nearly the parental level.
FIG. 5.
Analysis of the expression of sarS and agr transcripts in various mutant strains. (A) Northern blots of sarS transcript in the wild-type (wt), sarZ mutant, and single-copy (cps) complemented strains from the mid-exponential (OD600, ∼1.1) phase of growth in an RN6390 background. (Right) Blots of sarS and sarZ transcripts in the wild type and the wild type expressing the sarZ gene in a multicopy shuttle vector, pSK236 (cpm), from the mid-exponential (OD600, ∼1.1) phase of growth. The blots were hybridized with 750-bp and 500-bp DNA fragments containing ORFs of sarS and sarZ, respectively. (B) Northern blots of sarS transcript in the wild-type, sarZ mutant, and single-copy (cps) complemented strains from the mid-exponential (OD600, ∼1.1) phase of growth in an SH1000 background. (C) Northern blots of agr RNAIII transcript in the wild type and various single and double mutants, as indicated, from the postexponential (OD600, ∼1.7) phase of growth. The blots were probed with 0.5-kb DNA fragments containing the agr RNAIII region. The region of 23S rRNA of the ethidium bromide-stained gel used for blotting is also shown as a loading control in all panels.
Previous reports showed that the agr locus was positively regulated by SarZ and MgrA (11, 20, 51), while the same locus was negatively regulated by SarX (34). To understand the interregulatory relationship between mgrA, sarX, and sarZ with respect to agr regulation, mgrA sarZ and sarZ sarX double mutants were constructed and the expression of agr transcripts was monitored. Northern analysis with a 0.5-kb agr RNAIII probe with the total RNA isolated from the wild type and various isogenic single (sarZ, sarX, and mgrA) and double (sarZ mgrA and sarZ sarX) mutants of the sarZ gene was performed (Fig. 5C). Transcription of agr RNAIII was severely diminished (∼10-fold) in the sarZ mutant but enhanced (>3-fold) in the sarX mutant compared with the wild type. Interestingly, agr RNAIII expression in the sarZ sarX double mutant was similar to that of the sarX mutant, suggesting that sarX-mediated agr repression was probably independent of sarZ. The expression of agr was virtually undetectable in the mgrA or mgrA sarZ mutant (Fig. 5C). A similar regulatory pattern was observed in the case of agr RNAII in these mutant strains (data not shown), thus demonstrating the involvement of sarZ, sarX, and mgrA gene products in agr expression.
Transcriptional regulation of the selected target genes in a sarZ mutant.
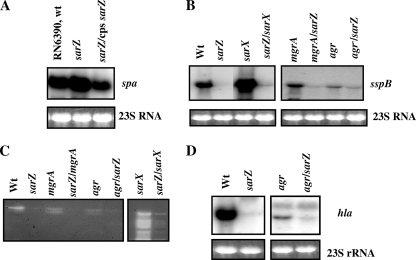
The data presented above clearly showed that inactivation of sarZ affected the expression of sarS and agr transcription in opposite ways. The expression of genes regulated by sarS and agr target genes was assessed. Transcription of spa (protein A), a gene activated by SarS, was enhanced in the sarZ mutant and on complementation; the level of spa transcription was reduced almost to parental levels (Fig. 6A). The expression of a few virulence genes, particularly those for exoproteins, known to be regulated by agr-dependent or -independent mechanisms was also monitored in the sarZ single mutant, as well as sarZ sarX, sarZ mgrA, and sarZ agr double-mutant backgrounds. Among the genes tested were those encoding proteases (a V8 protease [sspA] and a cysteine protease [sspB]) and hemolysin (e.g., hla). The protease (sspB) expression (sspA and sspB are part of the same operon) (39) was severely reduced in sarZ (∼9-fold), mgrA (3-fold), and agr (6-fold) mutants (Fig. 6B). Deletion of sarZ in an mgrA or agr mutant background led to further 9- and 3.5-fold reductions, respectively, in the expression of sspB transcript compared to the corresponding single mutant. Expression of sspB was enhanced 3-fold in the sarX mutant, while a significant reduction (∼27-fold) of the same transcript was observed in the sarZ sarX double mutant, suggesting that protease regulation is dependent on sarZ. Phenotypic characterization in a gelatin-containing gel also showed that the sarZ mutant produced lower levels of proteases than the wild type. In all the double mutants, the expression of protease was lower than that in the single mutant alone (Fig. 6C). The sarX mutant showed very high protease activity, but protease expression in the sarZ sarX double mutant was considerably lower than in the sarX single mutant, which was in good agreement with results obtained from transcriptional analysis. Similarly, a reduction of 15-fold in the transcription of α-hemolysin (hla) in the sarZ mutant with respect to the wild type was observed. The sarZ agr double mutant showed a threefold reduction in hla transcription compared to the agr mutant alone (Fig. 6D), suggesting the presence of an agr-independent mode of hla regulation by SarZ.
FIG. 6.
Analysis of target gene transcription (A, B, and D) and expression (C) in different isogenic single- and double-mutant strains of RN6390. (A) Northern blots of spa (protein A) transcript in the RN6390 wild-type (wt), sarZ mutant, and single-copy (cps) complemented strains from the mid-exponential (OD600, ∼1.1) phase of growth. (B) Northern blots of cysteine protease (sspB) transcript in the wild-type and isogenic single- and double-mutant strains, as indicated, from the postexponential phase of growth (OD600, ∼1.7). A 1.0-kb DNA fragment containing the sspB ORF was used for hybridization. In S. aureus, the V8 protease gene (sspA), the cysteine protease gene (sspB), and an unknown gene (sspC) are in a single transcriptional unit (39). (C) Gelatin zymogram of culture supernatants from various S. aureus RN6390 strains as indicated. Equal amounts of culture supernatant (OD600, ∼1.7) were used for all strains, except sarX and the sarZ sarX mutant, where one-fifth volume was applied. (D) Northern blot analysis for α-hemolysin (hla) transcript in assorted S. aureus RN6390 strains, as indicated, from the postexponential phase of growth. The Northern blot was hybridized with an hla fragment containing the coding region of the hemolysin gene. The region of 23S rRNA of the ethidium bromide-stained gel used for blotting is also shown as a loading control in panels A, B, and D.
Binding of SarZ protein to the promoter regions of various sarZ-regulated target genes.
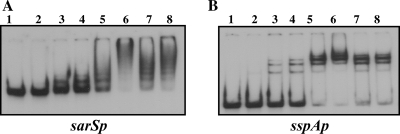
Since sarS and sspB transcription is altered in the sarZ mutant, the ability of SarZ to bind directly to the sarS or sspAB promoter regions was tested. The purified SarZ protein was used for the gel shift assays with a 287-bp sarSp- and a 250-bp sspAp-labeled DNA fragment containing the respective promoter regions (Fig. 7). Retarded DNA-protein complex was detected with as little as 200 ng or 50 ng of SarZ protein for sarSp and sspAp promoter fragments, respectively. As the concentration of SarZ increased, the retarded protein-DNA complexes became the predominant band, with complete conversion at 1.0 μg and 0.3 μg of SarZ protein for the sarS and sspA promoter fragments, respectively. Unlabeled specific competitor DNA fragments, as well as nonspecific competitor DNA, could eliminate retarded complexes equally for sarSp and sspAp promoter fragments. This indicates that SarZ has a nonspecific DNA binding activity, which is also consistent the observation made earlier by Kaito et al. (20) in gel shift assays with agr and hla promoter regions and the purified SarZ protein.
FIG. 7.
DNA binding activity of SarZ protein. Autoradiograms of 8.0% polyacrylamide gels showing the binding of SarZ protein to 287-bp and 250-bp promoter fragments (1 ng each or 5 and 6 fM, respectively) of sarS (A) and sspA (B), respectively. In panel A, lanes 1 to 5 correspond to 0 ng, 100 ng, 200 ng, 300 ng, and 500 ng, and lanes 6 to 8 correspond to 1.0 μg of purified SarZ protein. In panel B, lanes 1 to 5 correspond to 0 ng, 25 ng, 50 ng, 100 ng, and 200 ng, and lanes 6 to 8 correspond to 300 ng of purified SarZ protein. The mobilities of the bands were noted in the presence of increasing amounts of SarZ protein. A 50-fold excess (molar ratio) of specific unlabeled competitor DNA of the respective promoter fragments was used for competition in lanes 7 in both panels, whereas a 50-fold molar excess of a nonspecific competitor DNA (a 185-bp internal fragment of the sarX ORF region) is shown in lane 8 of each panel.
DISCUSSION
All living organisms process various signals through complex regulatory pathways. Thus, insight into how the behavior of a bacterial cell is controlled by its regulatory networks requires a detailed understanding of the individual regulatory components. S. aureus exhibits complex patterns of protein expression in response to various diverse environmental conditions, which are coordinately regulated by large numbers of regulatory networks (11, 18, 41). One such regulatory pathway that controls the expression of genes involved in pathogenesis, metabolic processes, antibiotic resistance, and biofilm formation involves the SarA protein family (consisting of 10 paralogous proteins) in Staphylococcus (11). In this report, we have examined the expression, regulation, and function of the sarZ gene.
The expression of the sarA paralogs is variable and in many cases depends on the phase of growth. For example, under normal laboratory growth in broth medium, sarA and mgrA transcripts are highly expressed, whereas the transcripts of sarU, sarT, and sarV are not readily detectable and sarR, rot, sarS, and sarX are expressed moderately in a growth phase-dependent manner. Although the preliminary characterization of the sarZ gene has been described previously (20, 44, 51, 56), the regulation of sarZ, especially its relationship with other members of the sar family and the characterization of sarZ transcription (e.g., transcriptional and translational start sites), were not well defined. Moreover, the actual production of SarZ protein had not been demonstrated in S. aureus. To address these issues, transcriptional analysis of sarZ transcript was performed and SarZ-specific antiserum was raised to detect the sarZ gene product in S. aureus. sarZ transcription was observed to be maximal in the early exponential phase of growth but declined in the postexponential phase. Employing the SarZ antiserum, the SarZ protein was clearly detected in whole-cell extracts of RN6390, and as with sarZ transcripts, more SarZ production was observed in the exponential phase than in the postexponential phase (Fig. 2). Indeed, expression analysis of the sarZ gene in various laboratory isolates, as well as four clinical isolates (Newman, MW2, COL, and UAMS-1), suggested a similar pattern of sarZ expression (unpublished data).
Based on the primer extension experiment, the predicted promoter was found to be σA dependent, which agrees with the observation that most of the major sar-family gene transcripts are expressed by the σA promoter (11, 30, 32-36, 52). Analysis of the sequence downstream of the sarZ gene disclosed the presence of a potential hairpin structure with an 8-bp inverted-repeat sequence, which is a rho-independent transcriptional terminator sequence. Thus, the Northern and Western studies suggest that SarZ is composed of 148 residues, as predicted in various annotations of Staphylococcus genomes. The SarZ protein is present in all of the sequenced Staphylococcus genomes and is more similar to MgrA (34% identity and 63% homology) than to SarA (28% identity and 53% homology). Among SarA family proteins, MgrA and SarZ are more similar to the MarR family of proteins among gram-negative organisms (e.g., 19% identity with E. coli MarR, 34% identity with Bacillus subtilis OhrR, and 21% identity with Pseudomonas aeruginosa MexR) and consist of six α-helices and two β-sheets.
It is well established that members of the SarA family regulate other members of their own family, as well as other regulatory systems and many target genes in S. aureus. Among 13 regulatory mutants (10 sar family mutants, agr, sae, and sigB) tested, a decrease in expression of sarZ transcript was observed only in an mgrA mutant, and transcription was restored to the parental level on complementation with the mgrA gene (Fig. 3A). The pattern of mgrA-mediated regulation was similar in another pair of laboratory (SH1000) and clinical (Newman) isolates, both of which produce more of the stress-responsive SigB factor. In gel shift studies, MgrA could bind to the sarZ promoter region. Analysis of the consensus MgrA binding motif (GTTG) suggested the presence of two similar binding motifs within the −35 region of the sarZ promoter, and it is possible that MgrA aids the binding of RNA polymerase to the promoter to enhance sarZ gene expression. Inactivation of sarZ did not have any major effect on the transcription of other sarA family genes, except for activation of sarS transcription. In fact, overexpression of sarZ in a wild-type strain led to a drastic (∼6-fold) decrease in expression of sarS transcription. These results suggest that SarZ is a repressor of sarS transcription in S. aureus, which was supported by the DNA binding ability of SarZ to the sarS promoter. However, these results should be interpreted with caution, as the ability of SarZ to bind the sarS promoter was equally inhibited by specific and nonspecific competitor DNA (Fig. 7). This suggests that in vitro SarZ has a nonspecific DNA binding activity, which is also consistent with the observation made by Kaito et al. (20). SarS is known to activate the expression of spa (protein A), and the sarZ mutant that has elevated sarS transcription showed enhanced transcription of spa. This indicates that SarZ regulates spa expression, probably via SarS, which is consistent with the inability to identify SarZ protein during an affinity pull-down assay with the spa promoter fragment and cell lysate of strain 8325-4 (42).
The results from an earlier study showed that SarX is the only negative regulator of agr (34), while MgrA (12) and SarZ are positive regulators of agr transcription. In this study, too, inactivation of sarZ drastically reduced agr expression, while sarX deletion enhanced agr expression. In the sarZ sarX double mutant, agr expression was virtually indistinguishable from that of the sarX single mutant. These results clearly suggest the involvement of sarX, mgrA, and sarZ gene products in agr regulation. It is possible that SarZ could be required to counter the repressive effects of SarX. Thus, if sarX is absent, then sarZ is not required for agr expression and enhanced agr expression is observed due to the absence of sarX-mediated repression (Fig. 5C). This raises the interesting possibility that SarZ and SarX may regulate agr by interacting with each other. Preliminary data employing in vitro protein-protein interaction assays show that SarZ can directly interact with SarX (unpublished data).
The ssp locus is important for virulence in S. aureus, and proteases have been implicated in spreading of S. aureus in host tissues (46). In addition, the proteases have been shown to contribute to in vivo growth and survival of S. aureus in different infection models. Hence, the study of ssp regulation is of particular importance in S. aureus pathogenesis. In S. aureus, agr RNAIII activates ssp expression, while several of the sar family genes repress its expression. The results presented here (Fig. 6B) clearly show that sarZ mutants have reduced agr, as well as sspB, expression. If sarZ regulates sspB expression only via agr, then sspB expression in agr single mutants or sarZ agr double mutants should remain the same. Nevertheless, there is a reduction in sspB expression (∼3.5-fold) in the double mutant compared to the single mutant. Moreover, both the sarX single mutant and the sarZ sarX double mutant produce more agr transcripts than the wild type or the sarZ mutant. However, in spite of high agr expression, the expression of sspB is reduced in the sarZ sarX double mutant. This clearly implies that sspB is regulated by sarZ independently of agr.
To summarize briefly, we have characterized the expression of the sarZ gene and have shown that MgrA is required for optimal sarZ expression. We have also shown that sarZ upregulates spa transcription by enhancing sarS expression. This suggests that the sar family genes are regulated by other members of their own family (Fig. 1). Analysis of a sarZ knockout in combination with other regulatory genes (e.g., agr and sarX) suggests that sarZ can regulate the expression of virulence genes, in particular, sspB and hla genes, in an agr-independent manner. These results are consistent with the recent finding that sarZ inactivation leads to increased transcription of spa and downregulation of hla and sspA transcripts in the RN6390 strain (51). All these results demonstrate the central regulatory role played by sarZ in controlling the regulatory and virulence genes in S. aureus. One useful way to target the bacterial cells in vivo may be to disrupt the regulatory systems, thereby minimizing the expression of cell wall-associated and extracellular proteins. Therefore, by blocking the functions of critical regulators, such as SarZ, one can expect to minimize the expression of genes involved in virulence and survival.
Acknowledgments
We thank Frank DeLeo and Ambrose Cheung for S. aureus strains MW2, Newman, and RN6390 and its various isogenic mutants, respectively. We also thank Michael Chaussee for critical reading of and comments on the manuscript.
This work was supported by the 2010 initiative startup fund and SD BRIN (2P20 RR016479) awarded to A.C.M.
Footnotes
Published ahead of print on 19 December 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Blevins, J. S., A. F. Gillaspy, T. M. Rechtin, B. K. Hurburt, and M. S. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the S. aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33317-326. [DOI] [PubMed] [Google Scholar]
- 3.Brandenberger, M., M. Tschierske, P. Giachino, A. Wada, and B. Berger-Bachi. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophy. Acta 1523135-139. [DOI] [PubMed] [Google Scholar]
- 4.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 1785810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassat, J., P. M. Dunman, E. Murphy, S. J. Projan, K. E. Beenken, K. J. Palm, S.-J. Yang, K. C. Rice, K. W. Bayles, and M. S. Smeltzer. 2006. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 1523075-3090. [DOI] [PubMed] [Google Scholar]
- 6.Chen, P. R., T. Bae, W. A. Williams, E. M. Duguid, P. A. Rice, and O. Schneewind, and C. He. 2006. An oxidative-sensing mechanism is used by the global regulator MgrA. Nat. Chem. Biol. 2591-595. [DOI] [PubMed] [Google Scholar]
- 7.Chen, P. R., S. Nishida, C. B. Poor, A. Cheng, T. Bae, L. Kuechenmeister, P. M. Dunman, D. Missiakas, and C. He. 2008. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed]
- 8.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 896462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., K. A. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 692448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y.-Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Microbiol. Lett. 16491-9. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, A. L., K. A. Nishina, M.-P., Trotonda, and S. Tamber. 2008. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien, Y.-T., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 27437169-37176. [DOI] [PubMed] [Google Scholar]
- 13.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcriptional profiling-based identification of Staphylococcus aureus genes regulated by agr and/or sarA loci. J. Bacteriol. 1837341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 1823955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41247-261. [DOI] [PubMed] [Google Scholar]
- 16.Gaskill, M. E., and S. A. Khan. 1988. Regulation of the enterotoxin B gene in Staphylococcus aureus. J. Biol. Chem. 2636276-6280. [PubMed] [Google Scholar]
- 17.Giraudo, A. L., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 16853-58. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, R. J., and F. D. Lowy. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infections. Clin. Infect. Dis. 46S350-S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinants expression and stress resistance: characterization of a functional rsbU strain of Staphylococcus aureus 8325-4. J. Bacteriol. 1845457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaito, C., D. Morishita, Y. Matsumota, K. Kurokawa, and K. Sekimizu. 2006. Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol. Microbiol. 621601-1617. [DOI] [PubMed] [Google Scholar]
- 21.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Graig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 22.Koenig, R. L., J. L. Ray, S. J. Maleki, M. S. Smeltzer, and B. K. Hurlburt. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 1867549-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 403-420. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, NY.
- 24.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K.-I. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R.-I. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 3571225-1240. [DOI] [PubMed] [Google Scholar]
- 25.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single copy integration vectors for Staphylococcal aureus. Gene 103101-105. [DOI] [PubMed] [Google Scholar]
- 26.Lee, C. Y. 1992. Cloning of genes affecting capsule expression in Staphylococcus aureus strain M. Mol. Microbiol. 61515-1522. [DOI] [PubMed] [Google Scholar]
- 27.Lowy, F. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339520-532. [DOI] [PubMed] [Google Scholar]
- 28.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. MgrA, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 1853703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luong, T. T., P. M. Dunman, E. Murphy, S. J. Projan, and C. Y. Lee. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 1881899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 1803828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manna, A. C., S. S. Ingavale, M. Maloney, W. van Wamel, and A. L. Cheung. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 1865267-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manna, A. C., and A. L. Cheung. 2006. Expression of SarX, a negative regulator of agr and exoproteins synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 1884288-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manna, A. C., and A. L. Cheung. 2006. Transcriptional regulation of the agr locus and the identification of DNA-binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol. Microbial. 601289-1301. [DOI] [PubMed] [Google Scholar]
- 36.Manna, A. C., and B. Ray. 2007. Regulation and characterization of rot transcription in Staphylococcus aureus. Microbiology 1531538-1545. [DOI] [PubMed] [Google Scholar]
- 37.McDevitt, D., P. Francois, P. Vandaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11237-248. [DOI] [PubMed] [Google Scholar]
- 38.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 1823197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 211227-1237. [DOI] [PubMed] [Google Scholar]
- 40.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 123967-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 42.Oscarsson, J., C. Harlos, and S. Arvidson. 2005. Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC8325-4. Int. J. Med. Microbiol. 295253-266. [DOI] [PubMed] [Google Scholar]
- 43.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 1862430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Queck, S. Y., M. Jameson-Lee, A. E. Villaruz, T. L. Bach, B. A. Khan, D. E. Sturdevant, S. M. Ricklefs, M. Li, and M. Otto. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rechtin, T. M., A. F. Gillaspy, M. A. Schumacher, R. G. Brennan, M. S. Smeltzer, and B. K. Hurlburt. 1999. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol. Microbiol. 33307-316. [DOI] [PubMed] [Google Scholar]
- 46.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riggs, A. D., H. Suzuki, and S. Bourgeois. 1970. Lac repressor-operator interaction. I. Equilibrium studies. J. Mol. Biol. 4867-83. [DOI] [PubMed] [Google Scholar]
- 48.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 694749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2003. sarT influences the expression of sarS in S. aureus. Infect. Immun. 715139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamber, S., and A. L. Cheung. 2008. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect. Immun. doi: 10.1128/IAI.00859-08. [DOI] [PMC free article] [PubMed]
- 52.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37398-409. [DOI] [PubMed] [Google Scholar]
- 53.Trotonda, M. P., A. C. Manna, A. L. Cheung, I. Lasa, and J. R. Penades. 2005. SarA controls Bap-dependent biofilm formation in Staphylococcus aureus. J. Bacteriol. 1875790-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Truong-Bolduc, Q. C., X. Zhang, and D. C. Hooper. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 1853127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Truong-Bolduc, Q. C., and D. C. Hooper. 2007. The transcriptional regulators NorG and MgrA modulate resistance to both quinolones and β-lactams in S. aureus. J. Bacteriol. 1892996-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, L., M. Li, D. Dong, T. H. Bach, D. E. Sturdevant, C. Vuong, M. Otto, and Q. Gao. 2008. SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J. Infect. Dis. 197254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]