Abstract
One of the central questions in eukaryotic transcription is how activators can transmit their signal to stimulate gene expression in the context of chromatin. The multisubunit SAGA coactivator complex has both histone acetyltransferase and deubiquitination activities and remodels chromatin to allow transcription. Whether and how SAGA is able to regulate transcription at specific loci is poorly understood. Using mass spectrometry, immunoprecipitation, and Western blot analysis, we have identified human SPT20 (hSPT20) as the human homologue of the yeast Spt20 and show that hSPT20 is a bona fide subunit of the human SAGA (hSAGA; previously called TFTC/STAGA/PCAF) complex and that hSPT20 is required for the integrity of the hSAGA complex. We demonstrate that hSPT20 and other hSAGA subunits, together with RNA polymerase II, are specifically recruited to genes induced by endoplasmic reticulum (ER) stress. In good agreement with the recruitment of hSAGA to the ER stress-regulated genes, knockdown of hSTP20 hampers ER stress response. Surprisingly, hSPT20 recruitment was not observed for genes induced by another type of stress. These results provide evidence for a direct and specific role of the hSPT20-containing SAGA complex in transcriptional induction of ER stress-responsive genes. Thus, hSAGA regulates the transcription of stress-responsive genes in a stress type-dependent manner.
The ordered assembly of a functional preinitiation complex is a prerequisite for the transcription of RNA polymerase II (Pol II). Besides the general transcription factors, the presence of transcriptional activators and cofactors is necessary to make the transcription initiation a tightly regulated process. In addition, activators and repressors have to communicate with the promoter-bound factors in the context of chromatin. To achieve this, chromatin has to be opened, a process which is carried out by ATP-dependent chromatin remodelers and promoted by enzymes catalyzing posttranslational modifications of histone tails (23, 40). One of the most widely studied reactions leading to the loosening up of the chromatin structure is histone acetylation. The activity of histone acetyltransferases (HATs) is thought to increase the accessibility of DNA for transcription factors (23, 29). The first enzyme proven to possess HAT activity was Gcn5 (10). Since then, many other HATs have been identified, most of them functioning in large multiprotein complexes (26). One of these HAT-containing complexes is the yeast SAGA (ySAGA; Spt-Ada-Gcn5 acetyltransferase), comprising the Gcn5 HAT and the Ubp8 deubiqitinase enzymes together with TATA binding protein (TBP)-associated factors (TAFs), Tra1, Ada, and Spt family proteins (2). The ySAGA complex shares a high degree of similarity with the human TFTC/STAGA/PCAF complexes in subunit content, structure, and function (references 34 and 52 and references therein). The exact composition of the human complexes is still not known; however, recently, several new mammalian homologues of ySAGA subunits have been described (25, 57, 59). These reports further demonstrate that the ySAGA and human TFTC/STAGA/PCAF complexes are functionally and structurally equivalent entities and that a SAGA-type complex can be purified from species all along the evolutionary scale (34).
ySpt20 (also called Ada5) was first described as a suppressor of Ty element transposition that alters correct transcription initiation (39). ySpt20 was later shown to be a bona fide subunit of ySAGA and proved to play a role in the structural integrity of the complex, as no intact SAGA could be purified in spt20Δ strains (17, 44, 53). Genomewide analysis suggested that only ∼10% of genes are affected when SAGA-specific subunits were deleted or inactivated by thermosensitive mutations (19, 27). Interestingly, ySAGA-dependent genes were suggested to be mostly stress inducible, while yTFIID-regulated genes seem to fulfill housekeeping functions (19). SAGA, and especially ySpt20, was found to be indispensable under endoplasmic reticulum (ER) stress conditions that trigger the unfolded protein response (UPR). It was demonstrated that spt20Δ yeast cells undergoing ER stress fail to carry out a proper UPR. In stressed spt20Δ cells, no expression of genes, which normally get induced by ER stress, was detected (47, 49). All these results hinted at the involvement of yeast Spt20 (ySpt20) in the regulation of stress-induced gene transcription. However, whether or not ySAGA was directly or indirectly involved in the regulation of ER stress-induced genes has not been studied.
Until now, no human homologue of ySpt20 has been identified, and its putative role in transcriptional coactivation has not been established. In this study, we describe human SPT20 (hSPT20) as an integral component of the human SAGA (hSAGA) complex by using mass spectrometry (MS) and a variety of biochemical approaches. Moreover, we investigated the cellular role of hSPT20 in the regulation of stress-activated gene transcription. Indeed, we show that hSPT20 is recruited to the promoter of ER stress-induced genes, but not to arsenite stress-regulated genes, suggesting that hSAGA regulates the transcription of stress-responsive genes in a stress type-dependent manner.
MATERIALS AND METHODS
Antibodies. (i) SPT20 antibody 2487.
The (C)RPPKRKYLSSGRKSVFQ peptide corresponding to hSPT20 amino acids 20 to 37 was synthesized, coupled to ovalbumin, and used for generation of rabbit polyclonal antibody. The serum was purified by Sulfolink coupling gel (Pierce) as suggested by the manufacturer. The characterization of the antibody is shown in Fig. S5 in the supplemental material.
(ii) SGF29 antibody 2461.
Polyclonal rabbit serum was developed against the peptide HATNKYEVDDIDEEGKERH(C) corresponding to human SGF29 amino acids 185 to 203.
Antibodies against the following proteins have been described earlier: GNC5, 2GC2C11 (6); TAF10, 23TA1H8 (50); TRRAP, 2TRR2D5 (18); TAF5, 1TA1C2 (16); TAF6, 25TA (4); TBP, 3G3 (9); ATXN7, 3SCA1C1 (18); TAF4, 20TA1B12 (32); TAF1, Santa Cruz sc-735; USP22, 2391 (59); ATXN7L3, 2325 and 1ATX2D7 (59); SPT20 (p38IP), 4112 (60); and SPT3 (31), H2BUb (a kind gift from M. Oren).
Immunoprecipitation was carried out as described earlier (14, 28).
Cell growth condition and stress treatments.
HeLa cells were grown in Dulbecco's modified Medium supplemented with 1 g/liter glucose, 5% fetal calf serum, and gentamicin. Stress treatments were carried out using 250 μM Na-arsenite (Sigma), 100 nM thapsigargin (Sigma), or dimethyl sulfoxide (DMSO).
Chromatin immunoprecipitation (ChIP).
HeLa cells were grown up to 90% confluence, washed with phosphate-buffered saline, and cross-linked with 1% formaldehyde for 10 min at room temperature. The reaction was stopped with 0.125 M glycine, and cells were washed with ice-cold phosphate-buffered saline supplemented with 0.5 mM phenylmethylsulfonyl fluoride (PMSF), scraped, and resuspended in swelling buffer (25 mM HEPES, pH 7.8, 1.5 mM MgCl2, 10 mM KCl, 0.1% NP-40, 1 mM dithiothreitol [DTT], 0.5 mM PMSF, protease inhibitor cocktail [PIC], Amersham). Cells were broken with a Dounce homogenizer, and the nuclear fraction was resuspended in sonication buffer (50 mM HEPES, pH 7.8, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.5% sodium dodecyl sulfate [SDS], 0.5 mM PMSF, PIC). The chromatin was sonicated with a Bioruptor (Diagenode) sonicator into 200- to 500-bp fragments and centrifuged to avoid any remaining cell debris. The supernatant was used for further immunoprecipitation after dilution to a final concentration of 0.1% SDS, supplementing it with sonicated salmon sperm DNA (20 μg/ml) and bovine serum albumin (1 mg/ml). Protein A (for polyclonal rabbit antibodies)- or protein G (for monoclonal mouse antibodies)-Sepharose beads were washed and saturated with bovine serum albumin and single-stranded DNA. The precleared chromatin samples were shaken overnight at 4°C with the antibody, and then beads were added for 4 h to the samples to pull down specific protein-DNA complexes. The following washes were carried out at 4°C: twice with sonication buffer (0.1% SDS), twice with buffer A (50 mM HEPES, pH 7.8, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 0.5 mM PMSF, PIC), twice with buffer B (20 mM Tris, pH 8, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 0.5 mM PMSF, PIC), and finally twice with Tris-EDTA buffer (10 mM Tris, pH 7.5, 1 mM EDTA). Bound fraction of the chromatin was eluted with 200 μl of elution buffer (50 mM Tris, pH 8, 1 mM EDTA, 1% SDS) at 65°C, RNase A treated (5 μg/ml), reverse cross-linked (200 mM NaCl) at 65°C overnight, and finally incubated with proteinase K. DNA was phenol-chloroform extracted and precipitated by ethanol and then used for quantitative PCR (qPCR) analysis using a Roche LightCycler 480 with Sybr green (Qiagen) master mix.
As a negative control, we immunoprecipitated the cross-linked material with an irrelevant monoclonal antibody (anti-glutathione S-transferase) or preimmune serum. The background level detected in the immunoprecipitation reached 0.06% input as a maximum (data not shown), and subtracting the corresponding levels from the specific immunoprecipitation would not change the recruitment profile significantly. The ChIP experiments were repeated at least twice, and all the qPCR reactions were done in triplicates. The results from one representative experiment are shown with the standard deviation (SD).
RNA purification, reverse transcription, and qPCR.
Total RNA was purified using Trizol reagent (Invitrogen), reverse transcribed by Moloney murine leukemia virus reverse transcriptase using random hexamers, and analyzed by the qPCR machine Roche LightCycler 480 with Sybr green (Qiagen) master mix. All the detected values represented in the manuscript have been normalized to cyclophilin B.
Small interfering RNA (siRNA) negative control (reference number D-001810-10), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) siRNA (reference number D-001830-10), or anti-hSPT20 siRNA (reference number L-013820-00) from Dharmacon was transfected using Lipofectamine 2000 and OptiMEM serum-free medium, following the recommendations of the supplier.
Nuclear extract (NE) preparation and size exclusion chromatography were described in reference 14. MS was performed using an LTQ-FT mass spectrometer (Thermo Electron, Bremen, Germany) essentially as described previously (59).
Deubiquitination (deUb) assay.
Histones were prepared from HeLa cells by lysing the cells in 10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1.5 mM PMSF, 10 mM N-ethylmaleimide supplemented with 0.2 M HCl. Samples were incubated on ice for 30 min and centrifuged, and the supernatant was dialyzed first against 0.1 M acetic acid and then against water. Substrate (∼2 μg) was incubated with immunopurified complexes in 100 mM Tris-HCl, pH 8, 5% glycerol, 1 mM EDTA, 3 mM DTT for 2 h at 37°C. Samples were then divided into two; one part was analyzed using Coomassie brilliant blue-stained SDS-polyacrylamide gel electrophoresis, and the other was analyzed using Western blotting.
Peptide acetylation assay.
Peptide (1.2 μg; corresponding to the N-terminal tail of histone H3 at positions 1 to 20) was added to the immunopurified protein sample together with 1 μl of H3 acetyl coenzyme A (50 μCi/ml; Amersham) in the reaction buffer (50 mM Tris-HCl, pH 8, 20 mM KCl, 5 mM DTT, 4 mM EDTA) and incubated at 30°C for 1 h. Samples were dropped on Whatman P81 nitrocellulose filters, washed three times for 10 min in ice-cold 50 mM NaHCO3 (pH 9) buffer, and finally dried. Filters were then dropped into 5 ml of ReadySafe liquid scintillation cocktail (Beckman Coulter), and radioactivity was quantified by an LS6000SC Beckman counter.
RESULTS
Identification of hSPT20 as a new subunit of hSAGA.
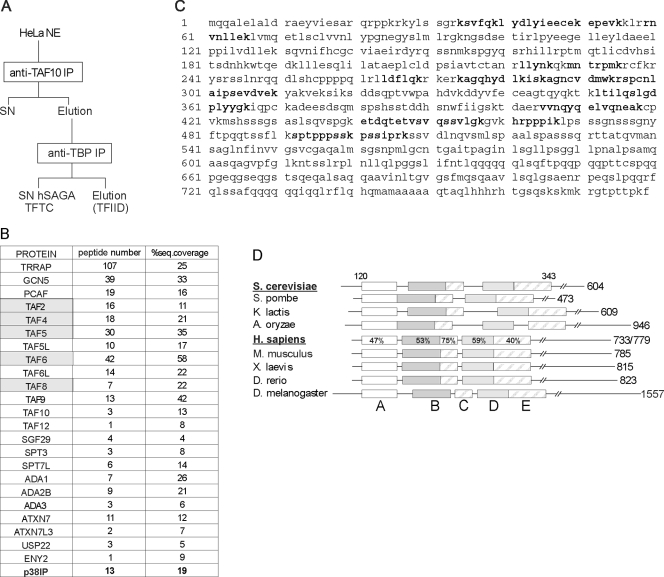
To find potential novel subunits in hSAGA (previously called TFTC), we analyzed by MS the hSAGA complex obtained by immunopurification (IP) of TAF10 containing complexes further depleted of TFIID by anti-TBP IP (7, 50) (Fig. 1A; see Fig. S1 in the supplemental material). Apart from the subunits already described (reference 34 and references therein), a new protein was identified in the sample, with 13 peptides resulting in 19% coverage (Fig. 1B and C). This protein (called p38IP) has been reported to be an interactor of the p38 mitogen-activated protein kinase (MAPK) and was shown to be required for proper embryogenesis (60). Psi-BLAST searches and multiple alignments revealed that the protein is the closest human homologue of ySpt20, a subunit of ySAGA. Thus, this human protein is called hSPT20. Detailed multiple alignments indicate that Spt20 homologues can be identified in many species and that they all share five conserved regions on their N-terminal part (Fig. 1D; see Fig. S2 in the supplemental material).
FIG. 1.
The newly identified hSAGA component shares sequence similarity with ySpt20. (A) Schematic representation of purifying TFTC (hSAGA) with a TAF10 IP followed by a TBP IP. hSAGA is in the final supernatant fraction (SN hSAGA). (B) Results of an MS analysis of the hSAGA/TFTC sample, purified as shown in panel A (for silver nitrate staining, see Fig. S1 in the supplemental material), are presented. The newly identified subunit, p38IP (called SPT20 further in this study) is bold; TAFs copurifying with the complex (see text) are marked with gray shading. (C) The complete amino acid sequence of the hSPT20 (p38IP) protein identified by MS in human hSAGA preparation is shown. Bold peptides were found in the MS sample. (D) Schematic representation of the multiple alignments highlighting the sequence homology between ySpt20 and the newly identified protein. The alignment covers amino acid positions 120 to 343 in the ySpt20 sequence, as marked on the top of the figure. Five evolutionarily conserved boxes were identified (represented with different shadings); the level of similarity between the yeast and human proteins is marked as a percentage. The length of each protein is marked on the right of the figure. For the human protein, we marked the two putative splice variants (see Fig. S3 in the supplemental material). S. cerevisiae, Saccharomyces cerevisiae; S. pombe, Schizosaccharomyces pombe; K. lactis, Kluyveromyces lactis; A. oryzae, Aspergillus oryzae; H. sapiens, Homo sapiens; M. musculus, Mus musculus; X. laevis, Xenopus laevis; D. rerio, Danio rerio; D. melanogaster, Drosophila melanogaster. A to E labeling of the boxes refers to Fig. S2 in the supplemental material, where the detailed sequence alignments of the boxes are shown. Note that the spacing between the conserved boxes is not conserved.
Our Psi-BLAST searches also identified a second gene in the human genome homologous to SPT20 that we called hSPT20L (Table 1). hSPT20L is potentially encoding a protein highly similar to hSPT20 (see Fig. S3 in the supplemental material), but no hSPT20L peptide was identified in our MS analysis of hSAGA. In good agreement, hSPT20L transcript(s) could not be detected in HeLa cells or in 13 other cell lines, while hSPT20 was highly expressed in the same cells (see Table S1 in the supplemental material).
TABLE 1.
Description of the two genes encoding hSPT20 and hSPT20La
| Gene | mRNA accession no. | Chromosome | Exon | Transcript length (nt) | Protein accession no. | Polypeptide length (aa) | Predicted mass (kDa)b |
|---|---|---|---|---|---|---|---|
| SPT20 | NM_001014286 | 13 | 26 | 2,944 | NP_001014308 | 779 | 85.78 |
| NM_017569 | 13 | 25 | 2,843 | NP_060039 | 733 | 80.1 | |
| SPT20L | XM_093087 | X | 6 | 2,919 | XP_093087 | 973 | 106.97 |
| XM_293352 | X | 6 | 2,661 | XP_293352 | 887 | 95.97 |
nt, nucleotides; aa, amino acids.
Predictions were made using the ExPASy site (http://www.expasy.ch/tools/pi_tool.html).
Polyclonal antibodies raised against hSPT20 (developed in this study or previously published [60]) detected a polypeptide at the predicted molecular mass (∼100 kDa) both in HeLa cell NE and in the TFTC/supernatant hSAGA (SN hSAGA) preparation (Fig. 2B, lanes 1 and 2; see Fig. S4 in the supplemental material). We previously showed that this preparation contains hSAGA and other molecular entities that are free of TBP but contain TAF10 (14). Hence, we purified the complex in a different way, by carrying out two consecutive IPs and then directly eluting the immunopurified hSAGA complex (Fig. 2A). We detected hSTP20 in this complex together with other hSAGA subunits (e.g., TRRAP, GCN5, and TAF10), while no TAF5 and TAF6 were copurified with the sample (Fig. 2B, lane 3; see Discussion).
FIG. 2.
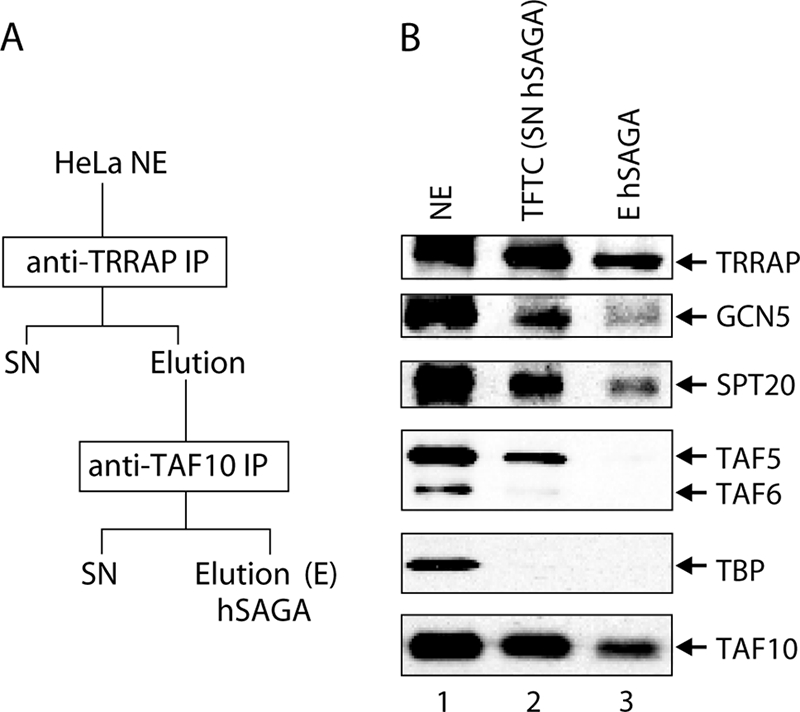
SPT20 is stably associated with hSAGA independently from the preparation method used. (A) Schematic representation of purifying endogenous hSAGA from HeLa NE by means of two consecutive IPs. (B) Western blot analysis of the two hSAGA preparations (see panel A and Fig. 1A) shows that hSPT20 is present in both preparations (eluted [E] or supernatant [SN]). However, TAF5 and TAF6 are not integral components of the complex as they copurify only with the SN hSAGA (previously called TFTC) (Fig. 1A).
Similarly, when we carried out IPs using polyclonal antibodies developed against the recently identified hSAGA subunits USP22 and ATXN7L3 (59), hSPT20 was found to be associated with these two subunits together with other components, such as TRRAP, GCN5, SGF29, and TAF10 (Fig. 3A). When proteins associated with hSPT20 were immunopurified from HeLa NE, we found that known hSAGA subunits coprecipitated with hSPT20 (e.g., TRRAP, GCN5, ATXN7, and TAF10) (Fig. 3B). In the same anti-hSPT20 IP experiment, no TFIID-specific subunits, such as TBP, TAF1, TAF4, TAF5, and TAF6, could be detected (Fig. 3C; see Discussion).
FIG. 3.
The newly identified protein is a bona fide hSAGA subunit. (A) hSPT20, together with other hSAGA subunits, is present in the anti-ATXN7L3 and anti-USP22 IP samples. Proteins from HeLa NEs were immunoprecipitated with the indicated rabbit polyclonal antibodies and eluted with the corresponding peptides. Proteins were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting using the indicated antibodies. (B) Immunoprecipitation of hSPT20 results in the coprecipitation of hSAGA subunits. (C) No TFIID-specific proteins copurify with hSPT20. IPs were carried out with purified rabbit polyclonal serum developed against hSPT20 or with the preimmune serum as a control. In this negative control IP, we could not detect any of the proteins tested (lane 2 in panels B and C). (D) The anti-SPT20 IP sample is active in the acetylation assay. Acetylation of a synthetic histone H3 tail peptide was measured by liquid scintillography. Both the SN hSAGA (TFTC) purification and the anti-SPT20 IP possessed similar HAT activities in the test. (E) The complex purified by an anti-SPT20 IP deubiquitinated histones. Core histones purified from HeLa cells were subjected to a deUb assay by different complexes (as indicated). Both the anti-SPT20 immunoprecipitated sample and the SN hSAGA (TFTC) preparation efficiently deubiquitinated H2B, as shown on the Western blot (WB). The Coomassie brilliant blue (CBB)-stained loading control of histones is also presented.
To show that our anti-hSPT20 IP protocol indeed purifies the enzymatically active hSAGA complex, we tested our purified SPT20-containing complex in HAT and deUb assays. In the HAT assay, the complex purified by an anti-SPT20 IP efficiently acetylated the synthetic H3 peptide, which is a known substrate of hGCN5 (Fig. 3D). The efficiency was comparable to that of TFTC (SN hSAGA) that we used as a positive control in the assay. In addition, the SPT20-containing complex possessed activity similar to that of the TFTC (SN hSAGA) purification (Fig. 1A) in our in vitro deUb assay when measuring the decrease in the monoubiquitinated H2B level (Fig. 3E). In summary, our results clearly show that the hSPT20 protein described above is an integral component of the fully active hSAGA complex.
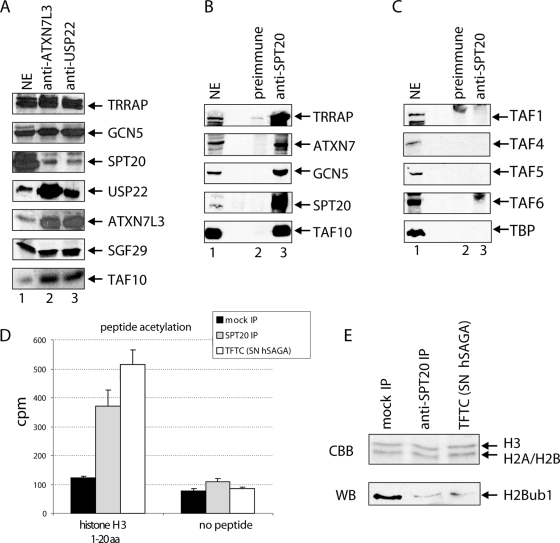
hSPT20 is present in high-molecular-mass protein assemblies.
To further analyze the presence of hSPT20 in hSAGA, an anti-hSPT20 IP has been carried out and the eluted sample was then fractionated by a Superose 6 gelfiltration chromatography column separating molecular entities in the 5 MDa to 5 kDa range. We could detect hSAGA subunits (TRRAP, GCN5, and TAF10) in fractions 15 to 17 corresponding to the expected size of the hSAGA complex (∼2 MDa) (34) (Fig. 4A). This observation provides further evidence that hSPT20 is a component of the 2 MDa hSAGA coactivator complex.
FIG. 4.
hSPT20 is present in high-molecular-mass protein complexes in the cell. (A) hSPT20 copurifies with other hSAGA components that fractionate at the 2-MDa size range. An anti-SPT20 sample was separated on a Superose 6 size exclusion column. Fractions 11 to 21 are shown, where hSAGA subunits were detected by Western blot analysis. Fraction numbers are marked on the top of the figure together with the 2-MDa-molecular-mass marker elution pattern; INP, input. (B) hSPT20 elutes at the 1- to 2-MDa size range in size exclusion chromatography. HeLa cell NE was fractionated on a Superose 6 size exclusion column, and odd number fractions were analyzed by Western blotting with the indicated antibodies. Fraction numbers are indicated together with the elution profiles of known molecular mass proteins.
To analyze whether hSPT20 is present also in other molecular assemblies in the nucleus, we fractionated HeLa NE on the same size exclusion chromatography column as described above. hSPT20 cofractionated with other hSAGA subunits at the molecular size range corresponding to the hSAGA together with GCN5, USP22, SGF29, and TAF10, four other subunits of the complex (Fig. 4B). We detected no significant hSPT20 in low-molecular-mass fractions. However, as a weak signal was also detected in 1- to 2-MDa-sized fractions (fractions 17 to 20), we cannot exclude that hSPT20 is also present in other, smaller-sized multiprotein complexes or simply in partially assembled hSAGA complexes.
hSPT20 is recruited to the promoter of ER stress-induced genes.
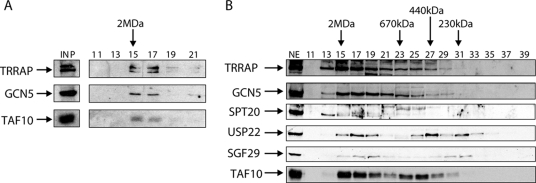
In yeast it was shown that spt20Δ cells subjected to ER stress fail to trigger the UPR pathway. However, a direct involvement of ySpt20 in ER stress response was not demonstrated. To investigate the possible role of hSPT20 and hSAGA in the induction of ER stress-responsive genes, human HeLa cells were treated with either thapsigargin or DMSO as a control. Thapsigargin is known to deplete calcium from the ER by blocking the ATPase pump and thus leads to the accumulation of improperly folded proteins in the ER lumen (42), which is the signal for the UPR. To study the role of hSAGA in the ER stress response, first we analyzed the mRNA level of GRP78, HERPUD, CHOP, and ERP70, four ER stress-induced genes (22, 35, 38, 42). Cells responded to the thapsigargin treatment within 2 to 3 h with a prominent transcriptional upregulation of CHOP and ERP70 genes and a lower but significant upregulation of GRP78 and HERPUD mRNA levels (Fig. 5A). Meanwhile, the mRNA level of hSPT20 did not change upon thapsigargin treatment (Fig. 5A).
FIG. 5.
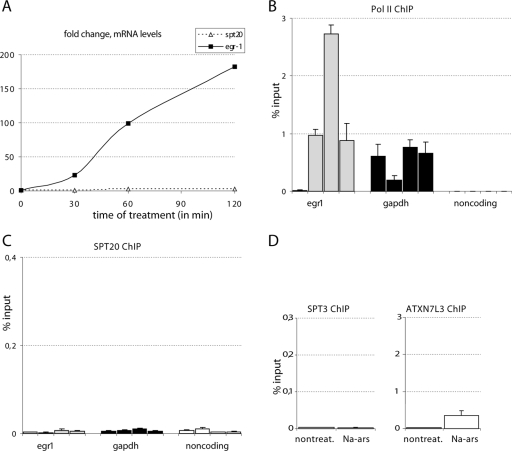
hSPT20/hSAGA is recruited to the promoter of ER stress-responsive genes after thapsigargin induction. (A) qPCR analysis of reverse transcribed RNA shows a significant activation of the four ER stress-responsive genes. mRNA levels were normalized to cyclophilin B and are presented as change compared to the control DMSO-treated sample. Samples were analyzed 1, 2, 3, and 4 h after treatment. (B to E) Time course ChIP results obtained with RNA Pol II (B), SPT20 (C), SPT3 (D), and ATXN7L3 (E) antibodies are shown as a percentage of input genomic DNA. Different shadings represent the promoter sequences of the corresponding ER stress-inducible genes analyzed by qPCR (indicated below the axis). The results obtained in five different conditions (nontreated sample; 1 h, 2 h, 3 h, and 4 h of thapsigargin treatment) are represented from left to right for each promoter. The promoter of GAPDH and a noncoding sequence serve as controls. Similar results were obtained in two biological replicates. Results obtained in a representative experiment are shown with the SD calculated from qPCR triplicates for each time point.
Next we analyzed the recruitment of Pol II and several hSAGA subunits, including hSPT20, to the promoters of GRP78, HERPUD, CHOP, and ERP70 by using ChIP experiments. As a control, we analyzed the presence of these proteins at the promoter of GAPDH and also at a noncoding region. For the ChIP, primers were designed to amplify a region close to the transcriptional start site of the investigated genes (see Table S2 in the supplemental material). The amplified sequences also contained an ER stress-response element that was previously shown to be bound by several transcription factors (3, 15). A clear increase in the Pol II level on the promoter of the four ER stress-induced genes was detected after 1 h of treatment, a signal which persisted for about 3 h and started to decrease at the 4-h time point. No significant changes were detected on the control GAPDH promoter or at the noncoding region. A similar recruitment pattern to that observed for Pol II (Fig. 5B) was measured for three hSAGA subunits, hSPT20, hSPT3, and hATXN7L3 (Fig. 5C, D, and E). Interestingly, the recruitment kinetics of ATXN7L3 (a subunit of the deUb module of the complex) was somewhat different from the other two hSAGA components tested. While hSPT20 and hSPT3 reached their maximum occupancy already at 1 h, ATXN7L3 seemed to peak slightly later at the promoter of the tested ER stress-regulated genes, suggesting that a “free” form of the deUb module may also exist in the cells in addition to the full hSAGA complex (compare Fig. 5C and D with E; see Discussion). Alternatively, the complex may change conformation, which could lead to a more efficient cross-linking of ATXN7L3 to the DNA at later time points. The control DMSO treatment had no significant effect on the recruitment of any of these transcription factors (not shown). Increased occupancy of hSAGA subunits and Pol II at these genes was not due to an increase in the corresponding protein levels, as they remained unchanged upon thapsigargin treatment (see Fig. S5 in the supplemental material). These results together indicate that the transcriptional coactivator hSAGA complex is recruited to ER stress-induced genes and probably plays a direct role in their induction.
hSPT20 is not recruited to the promoter of other stress-induced genes.
Evidence has suggested that hSPT20/SAGA may play a role not only in the UPR, but also in other stress-induced responses in the cell (see the introduction). Moreover, the hSPT20 protein was shown to interact with the p38 MAPK (60). Thus, we decided to test whether hSPT20 is implicated in the induction of known p38 MAPK-regulated genes at the transcriptional level. To this end, HeLa cells were treated with Na-arsenite, a known p38 MAPK cascade inducer, and were harvested at 30 min, 1 h, and 2 h after induction (Fig. 6A). Time course experiments showed a strong induction in the expression of the EGR-1, an immediate early gene induced by the p38 MAPK pathway (33) (Fig. 6A), while the mRNA level of SPT20 itself did not change (Fig. 6A). The protein levels of neither Pol II nor the tested hSAGA subunits changed due to the treatment (see Fig. S5 in the supplemental material). The ChIP experiments showed a highly increased occupancy of the EGR-1 promoter by Pol II already at 30 min (Fig. 6B). On the other hand, we could not detect any increase in the recruitment of SPT20 at the tested time points at this promoter (Fig. 6C). These results show that the increased mRNA level of a p38 MAPK-induced gene and the higher Pol II occupancy at the promoter of this gene do not correlate with an increased recruitment of hSAGA when monitored with the presence of the SPT20 subunit. When we investigated the presence of other hSAGA subunits at the promoter of the Na-arsenite-induced EGR-1 gene, we failed to show a significant recruitment of SPT3, and the increase in the occupancy level for ATXN7L3 was also very small compared to that of the ER stress-induced genes (compare Fig. 5E and 6D). Altogether, the data strongly suggest that hSAGA does not play a direct role in the transcriptional stimulation of the Na-arsenite-induced genes.
FIG. 6.
hSPT20 occupancy of a p38MAPK-induced gene does not increase after Na-arsenite induction. (A) mRNA quantification by RT qPCR shows that Na-arsenite efficiently induces the expression of EGR-1. mRNA levels normalized to cyclophilin B are shown as change compared to the nontreated sample on the figure at the indicated times (in minutes). (B to C) Time course ChIP results obtained with RNA Pol II (B) and SPT20 (C) antibodies are shown. Different shadings mark the different promoter sequences analyzed by qPCR, as indicated below the axis. The results obtained at four time points (nontreated sample; 30 min, 1 h, and 2 h of Na-arsenite treatment) are shown from left to right for each promoter. The promoter of GAPDH and a noncoding sequence serve as controls (see text). The amplified fragment of the EGR-1 promoter contains the cyclic AMP response element known to play a positive role in the upregulation of the gene (33). Similar results were obtained in two biological replicates. Results of a representative experiment are shown with error bars calculated from qPCR triplicates for each time point. (D) ChIP results obtained with antibodies raised against the indicated hSAGA subunits after 1 h of Na-arsenite treatment on the EGR-1 promoter.
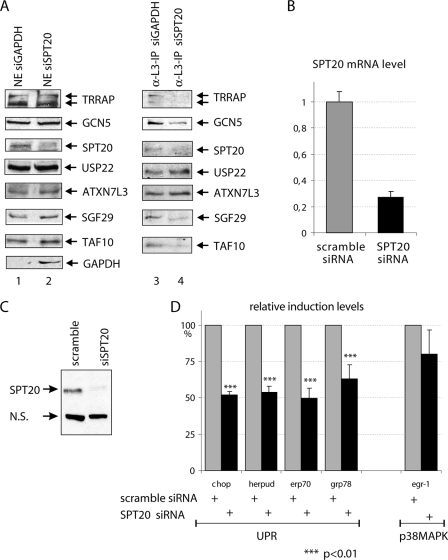
SPT20 knockdown leads to the decrease of correctly assembled hSAGA complex and to consequent defects in the ER stress-induced gene expression.
To further test the involvement of hSPT20 and the hSAGA complex in different stress responses, we have knocked down the expression of hSPT20 and analyzed the effect of the decrease of this SAGA subunit on the integrity of the complex. To this end, HeLa cells were transfected with siRNAs against either GAPDH, as a control, or hSPT20. Forty-eight hours after transfection, cells were harvested and NEs were prepared. In these extracts, we observed an ∼70% decrease in the hSPT20 and a 95% decrease in the GAPDH control protein levels by Western blot analysis (Fig. 7A), indicating that the knockdowns were efficient. Interestingly, in these NEs we have not observed any change in the expression level of the other hSAGA subunits tested (Fig. 7A). Next, we attempted to isolate hSAGA complexes from these NEs by anti-ATXN7L3 IP. While we were able to immunopurify comparable amounts of ATXN7L3 from both extracts, the presence of the tested core SAGA subunits (such as TRRAP, GCN5, SGF29, and TAF10) showed a serious decrease in the complexes isolated from the SPT20 knockdown sample compared to SAGA immunopurified from the control sample (Fig. 7A, compare lanes 3 and 4). This result shows that the integrity of the hSAGA complex requires hSPT20. Interestingly, however, the anti-ATXN7L3 IP copurified equal amounts of USP22 from both extracts (another subunit of the deUb module of SAGA), indicating that the integrity of the deUb module is independent of the cellular levels of SPT20. This observation may further suggest that the deUb module may exist also in a “free” form (see Discussion).
FIG. 7.
In vivo knockdown of hSPT20 results in impaired SAGA assembly and defective ER stress response but has no significant effect on p38MAPK signaling. (A) HeLa cells were transfected with siRNAs against either GAPDH (as a control) or SPT20. Forty-eight hours after transfection, cells were collected and NEs were prepared. The presence of the different proteins in these extracts was detected by Western blot analysis as indicated (lanes 1 and 2). From these extracts, an anti-ATXN7L3 IP was carried out and the coimmunopurified proteins tested by Western blot as indicated (lanes 3 and 4). (B and C) The anti-SPT20 siRNA transfection leads to an ∼70% decrease in the level of hSPT20 mRNA (B) and an ∼65% drop in protein level (C) as judged by reverse transcription-qPCR or Western blot analysis quantified by densitometry, respectively. (D) In anti-SPT20 siRNA-treated cells, the induction of the four tested ER stress-induced genes is significantly affected, while no significant effect in the Na-arsenite-induced stress response could be detected. Results are represented as percent relative induction levels compared to the control (scramble) siRNA-treated sample (100% induction) in each case. Average results of five independent experiments are shown with the SD. P values are marked above the bars with asterisks.
Next we wanted to analyze how the disintegration of hSAGA in the absence of SPT20 influences the regulation of the previously tested stress-regulated genes. Thus, we performed RNA interference knockdown of SPT20 and treated the cells with different stress agents. We observed, as above, an ∼70% decrease in the level of the hSPT20 mRNA (Fig. 7B) and hSPT20 protein (Fig. 7C) compared to that of the control siRNA-transfected cells. Following thapsigargin treatment in SPT20 siRNA knockdown cells, the transcriptional activation of the genes GRP78, HERPUD, CHOP, and ERP70 was significantly reduced (to about 50%) (Fig. 7D). In contrast, the induction of the EGR-1 expression was only weakly affected in SPT20 knockdown cells after Na-arsenite treatment (Fig. 7D). Altogether, these results show that the hSPT20-containing hSAGA is recruited to the promoters and that the complex is essential for the induction of the specific ER stress-regulated genes. In contrast, hSAGA dos not seem to be present and required for the regulation of genes induced by Na-arsenite.
DISCUSSION
In this study, we identify hSPT20 as an integral component of the hSAGA-type complexes and show that hSPT20 is required for the integrity of the hSAGA complex. In addition, we show that hSPT20 is recruited to the promoter of ER stress-induced genes and is also necessary for their activation but not for that of a Na-arsenite stress-regulated gene. These results indicate that hSAGA plays a role in ER stress response and provide the first experimental evidence for the direct involvement of hSAGA in the transcriptional regulation of a group of stress-inducible genes in mammalian cells. Indeed, our siRNA experiments showed that hSPT20 knockdown impaired the induction of ER stress-responsive genes but not those induced by the Na-arsenite stress.
Evolutionary conservation of SAGA-type complexes.
Although results from recent years suggested that the SAGA, TFTC, STAGA, or PCAF coactivator complexes isolated from several different species are highly conserved both in structure and function (2, 34), differences still remained between them, especially in their respective polypeptide compositions. Some subunits of ySAGA were not identified in the corresponding human complexes. However, since during the last year five new subunits have been identified in TFTC/STAGA, SPT20 (this work), USP22, ATXNL3, ENY2 (57, 59), and SGF29 (25), the differences between the metazoan and yeast complexes seem to disappear. In addition to the structural and functional similarities between the yeast and human complexes (reference 34 and references therein), these identifications led us to the conclusion that the 2-MDa multifaceted complexes are almost identical. Our observation that hSPT20 is required for the integrity of the hSAGA complex, as is ySpt20 for the ySAGA complex, further underlines this conserved similarity between the yeast and mammalian complexes. The only remaining difference between the yeast and human complexes is that no homologue of ySpt8 has been found in the human complexes. This may suggest that the human complex is more similar to yeast SLIK (SAGA like) or SALSA (SAGA altered Spt8 absent) (37, 43) also lacking Spt8. This latter complex was reported to contain a variant of ySpt7 lacking its bromo domain (43). In this respect, it is interesting to note that human SPT7L does not contain a bromo domain either (14).
Another difference between the human TFTC and the other human complexes (STAGA, GCN5, and PCAF complexes) was the presence of TAF2, TAF4, TAF5, and TAF6 in TFTC preparations (31, 50) (Fig. 1B). Here we clearly demonstrate by coimmunoprecipitation, using antibodies against newly identified hSAGA-specific subunits that TAF4, TAF5, and TAF6 are not associated with hSAGA (Fig. 2B and 3C). Several other IP experiments further indicate that a TAF core, probably containing the seven-TAF complex (14, 51), copurifies with the hSAGA (data not shown). These TAFs are not integral components of the 2-MDa complex. This suggests that all the human and probably metazoan 2-MDa SAGA-type complexes containing either the GCN5 or the highly homologous PCAF HAT subunits are similar, and thus we can call all these complexes SAGA for simplicity (36).
hSAGA and ER stress.
Cells subjected to ER stress change their metabolic pathways, reach a translational attenuation, and upregulate the expression of molecular chaperones (5, 58). In spt20Δ yeast cells, the UPR is defective due to the lack of this induced gene expression (47-49). The transcriptional stimulation of the ER stress-responsive genes requires the recruitment of the general transcription factors and Pol II to their promoters. Increase of positive histone marks and consequent changes in the occupancy of promoters by Pol II can be detected within a few hours after thapsigargin treatment in human cells (15). The acetylation level of histone H3 on K9 and K14 increases tremendously (15), which strongly suggests the requirement of hGCN5 (or PCAF) for these promoters. In good agreement, our present results show that the hSAGA complex is recruited to the ER stress-regulated promoters, thus bringing the hGCN5 (or PCAF) HAT in vicinity of the transcription initiation sites for chromatin remodeling. The signaling pathway leading to the regulated occupancy of hSAGA at specific sites on the genome is not yet well understood. One of the exciting questions is how the hSAGA complex gets recruited only to a set of promoters but not to all active genes. Sequence-specific activators are likely playing an active role in the process. In the case of ER stress-responsive genes, one of the sequence-specific transcription factors binding to and having a positive effect on the expression of many ER stress-inducible genes is NFY. NFY was shown to bind directly and constitutively to the ER stress-response element sequences in promoters of the induced genes (46, 54) and was also reported to interact with GCN5 (13). A dominant negative version of NF-YA was reported to lead to defects in the transcription of genes normally associated with H3K4me3 and H3K9-14ac active marks in HeLa cells (11). Thus, all these data together with our results show that both NFY and SAGA are indispensable for normal stimulation of ER stress-responsive genes. The reported contact between NFY and GCN5 suggests that in vivo hSAGA might be recruited to the ER-responsive sequences via an interaction with NFY.
Interestingly, after a rapid recruitment of the two hSAGA core subunits (hSPT20 and SPT3) (Fig. 5C and D), ATXN7L3, a subunit of the deUb module (21, 59), continues to accumulate for up to 3 h of treatment (Fig. 5E). Moreover, SPT20 knockdown does not seem to influence the formation of the deUb module (Fig. 7A). Thus, our time course analysis and IP experiments from SPT20 knockdown cells suggest that the “full” hSAGA complex and the deUb module, composed of the USP22 enzyme, ATXN7L3, and ENY2, may exist in the cells independently. It is conceivable that the putative “free” deUb subcomplex is additionally recruited to the promoters. Further experiments will be needed to investigate the existence, role, and substrate specificity of the SAGA-independent deUb module.
hSAGA and the p38 MAPK pathway.
The proper adaptation to extracellular stimuli requires regulation of gene expression by the MAPKs (20, 56). The classical model system for analyzing MAPK signaling in yeast is the observation of osmostress-induced changes. ySAGA mutants show growth defects in high osmolarity conditions (8). Directed phosphorylation taking place on transcription activators mediated by Hog1 (the yeast homologue of p38 MAPK) and its strong binding to chromatin together lead to osmostress response (1, 8). ySAGA and the Mediator complex are essential for the proper upregulation of osmostress-induced genes. Hog1 was recently shown to interact with the elongating RNA Pol II via SAGA (55). Although the ySAGA subunit interacting with Hog1p was not identified, one could imagine that it would be ySpt20, as their human homologues (hSPT20/p38IP and the p38 MAPK) were shown to have direct contact (60).
While the mammalian SPT20 (p38IP) was shown to interact with the p38 MAPK, when hSPT20 was knocked down by siRNA in human cells, no impairment in the activation of the p38 MAPK pathway was detected (24, 41), which is in good agreement with our observations (Fig. 6C and 7D). Moreover, if hSPT20 (p38IP) acted upstream of p38 MAPK, one would expect hSPT20 to have a cytoplasmic localization. In contrast, by using immunofluorescent imaging techniques, we have detected hSPT20 (p38IP) exclusively in the nucleus, similarly to all of the tested hSAGA-specific subunits (see Fig. S6 in the supplemental material). In addition, the C-terminal end of SPT20, which has been reported to interact with p38 MAPK, is not conserved through evolution. From the evolutionary conservation observed among all SPT20 homologues, one would expect that the functionally important regions of the SPT20 proteins reside in their N-terminal conserved half (Fig. 1C). The fact that we failed to detect p38 MAPK in our hSAGA or anti-SPT20 IP samples even after p38 MAPK induction (data not shown) suggests that if hSPT20 and p38 MAPK interact in vivo, this interaction is dynamic and is not strong enough to resist our stringent copurification assays. However, we cannot exclude the possibility that the interaction takes place under special conditions in certain cell types.
Stimuli activating the MAP kinase cascades elicit a rapid phosphorylation of H3 on serine 10. This modification, restricted to rapidly inducible genes poised for transcription, correlates with gene induction, suggesting a role in their activation of transcription (30). Data show that kinase and HAT enzymes function independently on the histone H3 tail, leading to phosphoacetylation (45). The signal leading to enhanced acetylation and phosphorylation of the same H3 histone tails is presently unclear (12). Our results show no significant recruitment of hSPT20, hSPT3, and ATXN7L3 to the EGR-1 promoter after induction of the EGR-1 gene by Na-arsenite; thus, hSAGA does not seem to play any direct role in the activation of this immediate early gene. Our new findings support the theory that GCN5, as a subunit of hSAGA, plays no role in the induced histone acetylation at the promoter of Na-arsenite-responsive genes.
In conclusion, our results clearly show that in human cells, hSAGA functions in stress-induced transcriptional regulation in a stress type-dependent manner. SPT20 has primordial function in the assembly of the hSAGA complex and in ER stress response but seems dispensable for the p38 MAPK pathway stimulated by Na-arsenite. Its direct role in ER stress-induced cellular response opens novel directions of research in many diseases, as ER stress has been implicated in immune response, diabetes, obesity, and neurodegeneration (58).
Supplementary Material
Acknowledgments
We are grateful to M. Brand for purifying the original TFTC fraction, to R. Mantovani for advice, to R. Roeder for the SPT3 antibody, to G. Duval for generating antibodies, and to D. Devys, F. Coin, A. Krebs, and M. E. Torres Padilla for critically reading the manuscript. We also thank the IGBMC cell culture facility for providing cells.
Z.N. and M.O. were supported by a fellowship from the European Community grant (HPRN-CT-00504228), Z.N. was supported by a fellowship from the Fondation pour la Recherche Médicale (FRM), and A.R. was supported by a fellowship of the Alsace Region. This work was supported by funds from CNRS, INSERM, Universite Louis Pasteur, the FRM, the Fonds National de La Science ACI, ANR (05-BLAN-0396-01; Regulome), and European Community (HPRN-CT 00504228, STREP LSHG-CT-2004-502950, and EUTRACC LSHG-CT-2007-037445) grants.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alepuz, P. M., E. de Nadal, M. Zapater, G. Ammerer, and F. Posas. 2003. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 222433-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. P., and P. A. Grant. 2007. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 265329-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumeister, P., S. Luo, W. C. Skarnes, G. Sui, E. Seto, Y. Shi, and A. S. Lee. 2005. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol. Cell. Biol. 254529-4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, B., E. Scheer, and L. Tora. 2001. Identification of hTAF(II)80 delta links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8591-600. [DOI] [PubMed] [Google Scholar]
- 5.Bernales, S., F. R. Papa, and P. Walter. 2006. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22487-508. [DOI] [PubMed] [Google Scholar]
- 6.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 203187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 27418285-18289. [DOI] [PubMed] [Google Scholar]
- 8.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 2591760-1763. [DOI] [PubMed] [Google Scholar]
- 9.Brou, C., S. Chaudhary, I. Davidson, Y. Lutz, J. Wu, J. M. Egly, L. Tora, and P. Chambon. 1993. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84843-851. [DOI] [PubMed] [Google Scholar]
- 11.Ceribelli, M., D. Dolfini, D. Merico, R. Gatta, A. M. Vigano, G. Pavesi, and R. Mantovani. 2008. The histone-like NF-Y is a bifunctional transcription factor. Mol. Cell. Biol. 282047-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clayton, A. L., C. A. Hazzalin, and L. C. Mahadevan. 2006. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell 23289-296. [DOI] [PubMed] [Google Scholar]
- 13.Currie, R. A. 1998. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J. Biol. Chem. 2731430-1434. [DOI] [PubMed] [Google Scholar]
- 14.Demény, M. A., E. Soutoglou, Z. Nagy, E. Scheer, A. Janoshazi, M. Richardot, M. Argentini, P. Kessler, and L. Tora. 2007. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS ONE 2e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donati, G., C. Imbriano, and R. Mantovani. 2006. Dynamic recruitment of transcription factors and epigenetic changes on the ER stress response gene promoters. Nucleic Acids Res. 343116-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrovskaya, V., A. C. Lavigne, I. Davidson, J. Acker, A. Staub, and L. Tora. 1996. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 153702-3712. [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 111640-1650. [DOI] [PubMed] [Google Scholar]
- 18.Helmlinger, D., S. Hardy, S. Sasorith, F. Klein, F. Robert, C. Weber, L. Miguet, N. Potier, A. Van-Dorsselaer, J. M. Wurtz, J. L. Mandel, L. Tora, and D. Devys. 2004. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 131257-1265. [DOI] [PubMed] [Google Scholar]
- 19.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13573-585. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, G. L., and R. Lapadat. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2981911-1912. [DOI] [PubMed] [Google Scholar]
- 21.Köhler, A., M. Schneider, G. G. Cabal, U. Nehrbass, and E. Hurt. 2008. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 10707-715. [DOI] [PubMed] [Google Scholar]
- 22.Kokame, K., K. L. Agarwala, H. Kato, and T. Miyata. 2000. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J. Biol. Chem. 27532846-32853. [DOI] [PubMed] [Google Scholar]
- 23.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 24.Kunze, D., S. Fuessel, A. Meye, M. P. Wirth, and U. Schmidt. 2006. Functional analyses of C13orf19/P38IP in prostate cell lines. Oncol. Rep. 151599-1604. [PubMed] [Google Scholar]
- 25.Kurabe, N., K. Katagiri, Y. Komiya, R. Ito, A. Sugiyama, Y. Kawasaki, and F. Tashiro. 2007. Deregulated expression of a novel component of TFTC/STAGA histone acetyltransferase complexes, rat SGF29, in hepatocellular carcinoma: possible implication for the oncogenic potential of c-Myc. Oncogene 265626-5634. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. K., and J. L. Workman. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8284-295. [DOI] [PubMed] [Google Scholar]
- 27.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405701-704. [DOI] [PubMed] [Google Scholar]
- 28.Leurent, C., S. L. Sanders, M. A. Demeny, K. A. Garbett, C. Ruhlmann, P. A. Weil, L. Tora, and P. Schultz. 2004. Mapping key functional sites within yeast TFIID. EMBO J. 23719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 30.Mahadevan, L. C., A. C. Willis, and M. J. Barratt. 1991. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65775-783. [DOI] [PubMed] [Google Scholar]
- 31.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 27323781-23785. [DOI] [PubMed] [Google Scholar]
- 32.Mengus, G., M. May, L. Carre, P. Chambon, and I. Davidson. 1997. Human TAF(II)135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 111381-1395. [DOI] [PubMed] [Google Scholar]
- 33.Mignacca, R. C., H. J. Lee, E. M. Kwon, and K. M. Sakamoto. 1996. Mechanism of transcriptional activation of the immediate early gene Egr-1 in response to PIXY321. Blood 88848-854. [PubMed] [Google Scholar]
- 34.Nagy, Z., and L. Tora. 2007. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 265341-5357. [DOI] [PubMed] [Google Scholar]
- 35.Oyadomari, S., and M. Mori. 2004. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11381-389. [DOI] [PubMed] [Google Scholar]
- 36.Pijnappel, W. W., and H. T. Timmers. 2008. Dubbing SAGA unveils new epigenetic crosstalk. Mol. Cell 29152-154. [DOI] [PubMed] [Google Scholar]
- 37.Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy, R. G. Cook, J. L. Workman, J. R. Yates III, and P. A. Grant. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 228774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy, R. K., C. Mao, P. Baumeister, R. C. Austin, R. J. Kaufman, and A. S. Lee. 2003. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J. Biol. Chem. 27820915-20924. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, S. M., and F. Winston. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 163206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7437-447. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, U., S. Fuessel, M. Haase, K. Kraemer, A. Meye, and M. P. Wirth. 2005. Quantification of C13orf19/P38IP mRNA expression by quantitative real-time PCR in patients with urological malignancies. Cancer Lett. 225253-260. [DOI] [PubMed] [Google Scholar]
- 42.Schröder, M., and R. J. Kaufman. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74739-789. [DOI] [PubMed] [Google Scholar]
- 43.Sterner, D. E., R. Belotserkovskaya, and S. L. Berger. 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 9911622-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1986-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomson, S., A. L. Clayton, and L. C. Mahadevan. 2001. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell 81231-1241. [DOI] [PubMed] [Google Scholar]
- 46.Ubeda, M., and J. F. Habener. 2000. CHOP gene expression in response to endoplasmic-reticular stress requires NFY interaction with different domains of a conserved DNA-binding element. Nucleic Acids Res. 284987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welihinda, A. A., W. Tirasophon, S. R. Green, and R. J. Kaufman. 1997. Gene induction in response to unfolded protein in the endoplasmic reticulum is mediated through Ire1p kinase interaction with a transcriptional coactivator complex containing Ada5p. Proc. Natl. Acad. Sci. USA 944289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welihinda, A. A., W. Tirasophon, and R. J. Kaufman. 1999. The cellular response to protein misfolding in the endoplasmic reticulum. Gene Expr. 7293-300. [PMC free article] [PubMed] [Google Scholar]
- 49.Welihinda, A. A., W. Tirasophon, and R. J. Kaufman. 2000. The transcriptional co-activator ADA5 is required for HAC1 mRNA processing in vivo. J. Biol. Chem. 2753377-3381. [DOI] [PubMed] [Google Scholar]
- 50.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393187-191. [DOI] [PubMed] [Google Scholar]
- 51.Wright, K. J., M. T. Marr II, and R. Tjian. 2006. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. USA 10312347-12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15199-208. [DOI] [PubMed] [Google Scholar]
- 53.Wu, P. Y., and F. Winston. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 225367-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2001. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol. Cell. Biol. 211239-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zapater, M., M. Sohrmann, M. Peter, F. Posas, and E. de Nadal. 2007. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol. Cell. Biol. 273900-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zarubin, T., and J. Han. 2005. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 1511-18. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, X. Y., M. Varthi, S. M. Sykes, C. Phillips, C. Warzecha, W. Zhu, A. Wyce, A. W. Thorne, S. L. Berger, and S. B. McMahon. 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 29102-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, L., and S. L. Ackerman. 2006. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 18444-452. [DOI] [PubMed] [Google Scholar]
- 59.Zhao, Y., G. Lang, S. Ito, J. Bonnet, E. Metzger, S. Sawatsubashi, E. Suzuki, X. Le Guezennec, H. G. Stunnenberg, A. Krasnov, S. G. Georgieva, R. Schule, K. I. Takeyama, S. Kato, L. Tora, and D. Devys. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 2992-101. [DOI] [PubMed] [Google Scholar]
- 60.Zohn, I. E., Y. Li, E. Y. Skolnik, K. V. Anderson, J. Han, and L. Niswander. 2006. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 125957-969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.