Abstract
TFIIB recognizes DNA sequence-specific motifs that can flank the TATA elements of the promoters of protein-encoding genes. The TFIIB recognition elements (BREu and BREd) can have positive or negative effects on transcription in a promoter context-dependent manner. Here we show that the BREs direct the selective recruitment of TFIIA and NC2 to the promoter. We find that TFIIA preferentially associates with BRE-containing promoters while NC2 is recruited to promoters that lack consensus BREs. The functional relevance of the BRE-dependent recruitment of TFIIA and NC2 was determined by small interfering RNA-mediated knockdown of TFIIA and NC2, both of which elicited BRE-dependent effects on transcription. Our results confirm the established functional reciprocity of TFIIA and NC2. However, our findings show that TFIIA assembly at BRE-containing promoters results in reduced transcriptional activity, while NC2 acts as a positive factor at promoters that lack functional BREs. Taken together, our results provide a basis for the selective recruitment of TFIIA and NC2 to the promoter and give new insights into the functional relationship between core promoter elements and general transcription factor activity.
Transcription of a gene by RNA polymerase II requires the assembly of the general transcription factors at the promoter to form a preinitiation complex (PIC). The eukaryotic core promoter contains DNA sequence elements that direct the assembly of the general transcription factors and provide a platform for the recruitment of the PIC (4, 29, 53, 59). These elements include the TATA box, initiator, downstream promoter element (DPE), downstream core element, and motif 10 element (41). In addition, the general transcription factor TFIIB can recognize two distinct elements within the core promoter that flank the TATA box: the upstream and the downstream TFIIB recognition elements (BREu and BREd) (reviewed in reference 17).
Interaction with BREu is mediated by a helix-turn-helix within the second direct repeat of TFIIB, and interaction with BREd is mediated through a recognition loop within the first direct repeat (16, 39, 60). These interactions serve to increase the affinity of TFIIB in forming a complex with TATA-binding protein (TBP) at the promoter. Each of the BREs is potentially present in up to 30% of human TATA-containing promoters, but the extent of their functional influence remains to be determined (16, 25, 39). BREs can also function at TATA-less promoters, but their identification in this context is less reliable and thus their potential genome-wide prevalence is difficult to estimate. The function of BREs in transcription appears to be complex. The BREs can act as both positive and negative elements (8, 16, 19, 39). Moreover, activator proteins can modulate the interaction between TFIIB and BREu (19).
At the adenovirus major late (AdML) promoter the BREs act as negative elements (8, 16, 19). The reason for this is not known, but candidates for involvement include the general transcription repressors Mot1 and NC2. Mot1 acts to prevent the interaction between TBP and the TATA box, but NC2 can assemble with TBP at the promoter and precludes the assembly of TFIIB or TFIIA, thus preventing formation of the PIC (reviewed in references 50 and 59). NC2 is composed of two subunits, α and β, also known as Dr1 and DRAP, respectively (27, 33, 34, 36, 48). TFIIA can compete with NC2 for association with TBP at the promoter, and this mutually exclusive assembly is underpinned by genetic studies of Saccharomyces cerevisiae that show a direct relationship between TFIIA and NC2 (64). In addition, assembly of NC2 with TBP at the TATA box can mobilize the complex to translocate along the DNA (52). NC2 also inhibits transcription by RNA polymerase III (pol III), but not by pol I (35, 62). A few studies over the last few years have unhinged the apparently established reciprocity between NC2 and TFIIA. Several reports have suggested a positive function for NC2 (5, 7, 11, 22, 63). In addition, the transcription-regulatory properties of NC2 exhibit core promoter-specific activities (1, 45, 63). How the positive activities of NC2 reconcile with the reciprocal nature of NC2 and TFIIA in directing transcription has not been determined.
TFIIA is composed of three subunits, α, β, and γ (reviewed in reference 30), with the α and β subunits derived from a single gene (15, 43, 49, 57, 66). Although TFIIA was originally characterized as a general transcription factor, many in vitro transcription systems do not require TFIIA, and TFIIA is perhaps better described as a general cofactor that enhances transcription activation. Indeed, transcriptional activator proteins can stimulate the assembly of a TFIIA-TFIID complex at the promoter (9, 37, 40, 42, 44, 55).
In this study we have analyzed the role of the BREs in the negative regulation of the AdML promoter in living cells. We find that the BREs direct the mutually exclusive assembly of TFIIA and NC2. Moreover, TFIIA and NC2 correlate with low and high promoter activities, respectively. Use of small interfering RNA (siRNA) to specifically deplete TFIIA and NC2 in living cells confirms a positive role for NC2 in transcription and, surprisingly, a transcriptional dampening role for TFIIA. We provide evidence that this involves overlapping recognition of the core promoter by TFIIA with the BREs.
MATERIALS AND METHODS
Plasmids, site-directed mutagenesis, and cloning.
G5ML containing nucleotides −50 to +22 from the AdML promoter downstream of five GAL4 DNA-binding sites and the plasmid driving expression of GAL4 region II activation domain linked to the GAL4 DNA-binding domain have been described before (19). All AdML promoter derivatives were produced with the QuikChange kit (Stratagene). G5ML and derivatives were subcloned into the reporter vector pGL3 basic (Promega).
Cell lines.
Human embryonic kidney 293T cells were cultured in complete medium (Sigma) as described previously (6, 47). The HEK293 stable cell lines were generated using the Flp-In system (Invitrogen). Briefly, the system uses an engineered 293 cell line derivative that contains a single copy of the Flp recombinase targeting site (FRT) under the control of a zeocin selection marker. The AdML promoter (wild type or AdMLmBREud derivative) linked to luciferase was cloned into pcDNA5/FRT, replacing the cytomegalovirus promoter within the plasmid. The resulting plasmids (pcDNA5/FRT-AdML-luc and pcDNA5/FRT-AdMLmBRE-luc) were transfected into the zeocin-resistant HEK 293 cells along with the plasmid pOG44 (Invitrogen), which expresses the recombinase. Stable cell lines were obtained from single colonies through selection with hygromycin b.
Transfection, luciferase assays, and immunoblotting.
Human embryonic kidney 293T cells were cultured in complete medium (Sigma) as described previously (6, 47). The HEK 293T cells were transfected with Lipofectamine 2000 transfection reagent (Invitrogen). Cells were harvested at 48 h posttransfection, and the luciferase activity was determined by using the luciferase and β-galactosidase dual-light detection system (Applied Biosystems) as described previously (28). Meanwhile, 20 μl of cell lysate for each transfection was used for immunoblotting, where all primary antibodies for NC2β (ab50783), TFIIAα/β (ab28176), TFIIAγ (ab37846), TFIIB (ab12094), the pol II C-terminal domain (ab24758), TBP (ab818), and Mot1 (ab5268) were purchased from Abcam. Secondary antibodies were from Jackson ImmunoResearch. Detection in Western blots was performed by enhanced chemiluminescence (GE Healthcare).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed according to the published method (54) with modifications. Briefly, cells were cross-linked with 1% formaldehyde-phosphate-buffered saline for 10 min at room temperature and quenched by the addition of glycine to a final concentration of 250 mM. The cells were washed twice with cold phosphate-buffered saline and harvested. For the ChIP assays of the AdML promoter and derivatives, the cells were Dounce homogenized. Cell lysates were then transferred to microtubes, and the nuclei were collected by brief centrifugation and then resuspended in lysis buffer (50 mM Tris-Cl, pH 8.0, 10 mM EDTA, 1% sodium dodecyl sulfate). Samples were then sonicated for 10 min at high power with a Diagenode sonicator. After sonication, the samples were centrifuged in a benchtop microcentrifuge at 4°C, and the supernatants were diluted with ChIP dilution buffer (0.01% sodium dodecyl sulfate, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-Cl, pH 8.0, 167 mM NaCl). For immunoprecipitation, the samples were precleared using preblocked protein G-Sepharose (Sigma) with 1 mg/ml bovine serum albumin and 0.1 mg/ml salmon sperm DNA. Two micrograms of each antibody was added to the samples, and samples were incubated overnight at 4°C, followed by the addition of 20 μl preblocked protein G-Sepharose to the samples and incubation for 1 h at room temperature. The remainder of the procedure was followed according to the method described previously (54) except that the washing step was prolonged to 0.5 to 1 h for each wash and that the DNA was purified with a Qiagen DNA purification kit. For endogenous promoter ChIP assays, the cells were directly lysed in nucleus lysis buffer after harvesting, followed by sonication. All other steps were the same as those used for the AdML promoter ChIP assays. The primers used in the PCRs were as follows: AdML forward, 5′-CTAGCAAAATAGGCTGTCCC-3′; reverse, 5′-TCCAGCGGATAGAATGGC-3′; Bcl2 forward, 5′-TTCCTGCATCTCATGCCAAG-3′; reverse, 5′-GGATGTACTTCATCACTATCTC-3′; insulin-like growth factor II (IGFII) forward, 5′-CTCTTGGCTCGGGTTGCGG-3′; reverse, 5′-GCAGGCGTGGGCCAGGAGG-3′.
The primers used for real time PCR were as follows: forward, 5′-GAGGATCCGTGTTCCTGAAG-3′; reverse, 5′-GAGTCAGTGAGCGAGGAA-3′. Real time PCR was performed with CYBR green master mix (Eurogentec) and a Chromo 4 thermal cycler machine (MJ Research). The program used for PCR was 94°C for 45 s, 60°C for 30 s, and 72°C for 30 s for a total of 35 to 40 cycles. The enrichment of target DNA (relative occupancy) is expressed as the ratio of the amount of promoter DNA bound to the protein of interest compared to nonpromoter DNA. The input DNA was derived from 1/10 of the volume of sample used for immunoprecipitation of the target protein. All experiments were performed at least in triplicate.
siRNA knockdown, RNA isolation, and RT-PCR.
Double-stranded siRNA oligonucleotides for TFIIAαβ, TFIIAγ, NC2α, and NC2β were purchased from Eurofins Biotech. The second TFIIAαβ siRNA (2) was obtained from Santa Cruz Biotechnology. Sequences of the siRNAs were as follows: NC2α, 5′-AGAGGAACGAAGAAGAUUACTT-3′; NC2β, 5′-UUGGAGAGCUAAGUAAGUATT-3′; TFIIAαβ, 5′-GUACUGAUGGAACUAAAAATT-3′; TFIIAγ, 5′-ACAGAUCACCCCCCAACUUTT-3′. siRNAs (10 pmol) were transfected with Lipofectamine 2000 (Invitrogen) either alone or in combination with plasmid DNA. Forty-eight hours after transfection, cells were harvested and used for either luciferase assays, Western blotting, or the preparation of RNA. Total RNA was prepared with the RNeasy minikit (Qiagen). For reverse transcription-PCR (RT-PCR), cDNAs were synthesized from 3 μg of total RNA for each sample with the Access reverse transcription kit (Promega) and subjected to PCR with primers to amplify the RNA. Primers used for RT-PCR were as follows: IGFII forward, 5′-GAGTGCTGTTTCCGCAGCTGTG-3′; reverse, 5′-CTGAACGCCTCGAGCTCCTTG-3′; P21 forward, 5′-GAACTTCGACTTTGTCACCG-3′; reverse, 5′-GGTAGAAATCTGTCATGCTG-3′; Egr1 forward, 5′-CGCAAGAGGCATACCAAGATCC-3′; reverse, 5′-CAGCTGAGGAAGGGAAGCTG-3′; BCL2 forward, 5′-GTGGTGGAGGAGCTCTTCAG-3′; reverse, 5′-GCAGAGTCTTCAGAGACAGCC-3′; 18S forward, 5′-GTAACCCGTTGAACCCCATT-3′; reverse, 5′-CCATCCAATCGGTAGTAGCG-3′.
Proteins and band shift.
Promoter DNA for wild-type AdML and AdML derivatives was labeled with [α-32P]dATP (GE Healthcare). Purified native TFIIA was obtained from Protein One Inc. NC2 protein was prepared from Escherichia coli BL21 coexpressing NC2α and NC2β from a bicistronic plasmid (described in reference 44). Ni-nitrilotriacetic acid-agarose chromatography was used to purify the NC2 via a His tag at the N terminus of NC2α. Recombinant human TBP and TFIIB were purified as described before (16, 26). Band shift assays were performed as described previously (26).
RESULTS
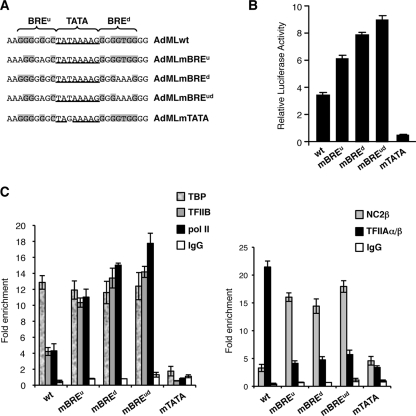
The AdML promoter contains both a BREu and BREd that flank the TATA element (Fig. 1A). Based on previous functional data we generated mutant derivatives of this promoter that lack a functional BREu (AdMLmBREu), BREd (AdMLmBREd), both BREs (AdMLmBREud), or the TATA element (AdMLmTATA) (16, 19, 39). The promoter derivatives, driving luciferase expression and controlled by five GAL4 DNA-binding sites, were transfected into human embryonic kidney 293T cells along with a plasmid driving expression of GAL4-RII (GAL4 residues 1 to 147, linked to the region II activation domain of GAL4) and a β-galactosidase internal control. Cells were harvested 48 h later, and luciferase activity was measured and presented graphically in Fig. 1B. Consistent with our previous results and those of others, mutation of either BREu or BREd caused an increase in promoter activity (8, 16, 19). Mutation of both BREs within the AdML caused a further increase in promoter activity, and mutation of the TATA element resulted in a significant reduction in promoter activity. Thus, the BREs within the AdML promoter act as negative elements either alone or in combination.
FIG. 1.
Differential recruitment of TFIIA and NC2 at the AdML promoter is dependent upon the BREs. (A) The promoter sequence of the wild-type (wt) AdML promoter is shown at top, with the TATA element underlined and BREu and BREd shaded at the bases that match their consensus sequences. (B) The promoter derivatives in panel A, linked to five GAL4 DNA-binding sites and driving luciferase transcription, were transfected into embryonic kidney 293T cells along with a vector driving expression of GAL4 activation domain II linked to the GAL4 DNA-binding domain. Forty-eight hours later luciferase activity was measured and normalized to β-galactosidase activity and is presented graphically. Each bar represents three independent experiments with standard deviation. (C) 293T cells were transfected as in panel B and subjected to ChIP with antibodies against TBP, TFIIB, and pol II (left) and TFIIAα/β or NC2β (right). The results are presented as enrichment of specific promoter DNA over nonpromoter DNA in the immunoprecipitate. Each bar represents at least three independent experiments with standard deviation. A gel-based version of these data, including the analysis of additional factors, is shown in Fig. S1 in the supplemental material. IgG, immunoglobulin G.
To determine the mechanisms that underlie the negative function of the BREs within the AdML promoter, we next assessed the pattern of assembly of key general transcription factors at the AdML promoter derivatives in living cells. The AdML promoter derivatives were transfected as described above, and then ChIP was performed with the antibodies indicated in Fig. 1C. TBP assembled at the promoters in a BRE-independent manner but showed a significant reduction in assembly at the AdMLmTATA derivative (Fig. 1C, left). TFIIB and pol II assembled at the promoter derivatives to levels that reflect their transcriptional potency, exhibiting a greater level of assembly at the AdML promoter derivatives with defective BREs (Fig. 1C, left). NC2β also showed an increase in assembly at the AdML promoters with mutant BREs (Fig. 1C, right), as did NC2α (see Fig. S1 in the supplemental material). TFIIA showed robust assembly at the wild-type AdML promoter but lower levels of assembly when either or both of the BREs were mutated (Fig. 1C, right). Taken together, these data show a strong correlation between the BREs and TFIIA/NC2 assembly. Specifically, the presence of the BREs within the AdML promoter enhances TFIIA assembly, and the converse is observed with NC2 (Fig. 1C, right). It was unexpected, however, that TFIIA assembly should correlate with the repressive function of the BREs while NC2 associates with the more active promoter derivatives.
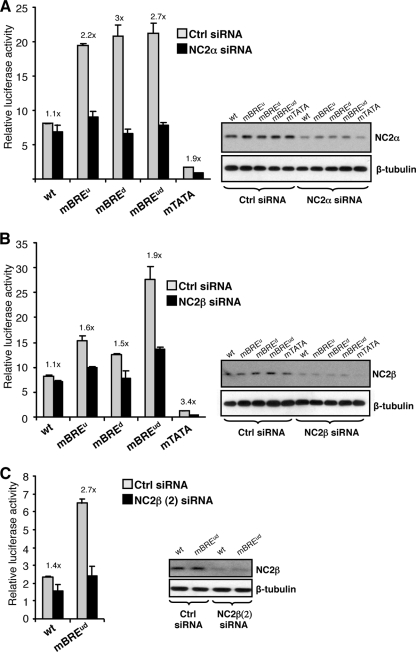
Our results so far provide a correlation between TFIIA and NC2 assembly and the presence of the BREs. Furthermore, TFIIA assembly and that of NC2 are associated with low and high promoter activities, respectively. To assess the functional relevance of the recruitment events in Fig. 1C, we next determined the contribution of TFIIA and NC2 to BRE-dependent transcription at the AdML promoter. 293T cells were transfected with siRNA targeting NC2α along with the AdML derivative promoters, a plasmid driving expression of GAL4-RII, and a β-galactosidase internal control. Forty-eight hours later cells were harvested, samples were taken for immunoblotting, and the remainder was used to measure luciferase activity (Fig. 2A). The immunoblot confirmed that NC2α was reduced in cells that were transfected with the NC2α siRNA. The graph in Fig. 2A shows luciferase activity of the AdML promoter derivatives in the presence of either control siRNA or NC2α siRNA. At the wild-type AdML promoter NC2α knockdown did not have a significant effect on promoter activity. However, knockdown of NC2α caused significant reductions in the activities of the AdML promoter derivatives in which the BREs were individually or both mutated (2.2-, 2.7-, and 3-fold). The mutant TATA AdML derivative also showed a reduction in activity in the presence of NC2α siRNA (1.9-fold). Taken together the data show that NC2α does not play a crucial role in transcription of the wild-type AdML promoter but has a positive role when either or both of the BREs are mutated. We next assessed the effect of an siRNA directed against NC2β in the same way (Fig. 2B). As we observed with NC2α, knockdown of NC2β by siRNA had a negligible effect on the wild-type AdML promoter but significantly reduced the activities of the AdML derivative promoters that contain the mutant BREs (1.5-, 1.6-, and 1.9-fold). Western blotting confirmed that the expression of NC2β was reduced by the siRNA. A second siRNA targeting NC2β, NC2β (2) siRNA, produced comparable results (Fig. 2C). Taken together, the data show that ablation of either NC2α or NC2β does not significantly affect transcription at the wild-type AdML promoter. However, when either or both of the BREs are ablated, NC2α and NC2β have a positive effect on transcription. These data are consistent with our ChIP analysis (Fig. 1C); NC2α or NC2β ablation elicits the greatest effect on the same AdML derivatives that also show greater NC2 recruitment in vivo.
FIG. 2.
NC2 acts as a positive transcription factor when the BREs in the AdML promoter are rendered nonfunctional. (A) 293T cells were transfected as for Fig. 1B except that either control siRNA or siRNA targeting NC2α was included. Forty-eight hours later cells were harvested and used to monitor luciferase activity and also for Western blotting with anti-NC2α antibodies or, as a control, anti-β-tubulin antibodies. Note that the immunoblot analysis concerns the whole cell population and therefore underestimates the extent of NC2α knockdown. The graph shows the relative luciferase activity of the AdML promoter derivatives in the presence of control (Ctrl) or NC2α siRNA. The numbers above the bars indicate the changes in activity that are caused by the NC2α siRNA. wt, wild type. (B) Same as panel A, except that siRNA targeting NC2β was used. (C) Same as panel A, except that a different siRNA targeting NC2β, NC2β (2) siRNA was used and only the wild-type AdML and AdMLmBREud reporters were tested. Each bar represents three independent experiments with standard deviation.
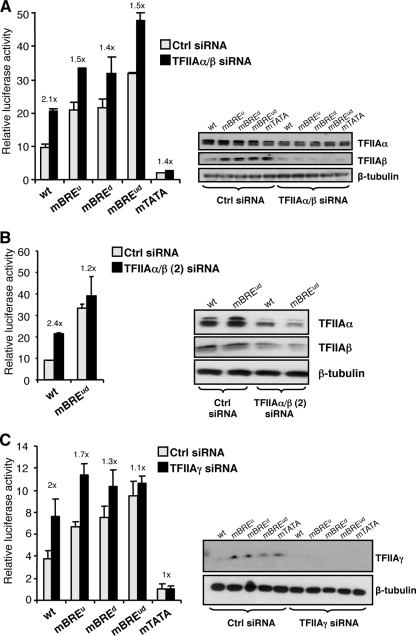
We next used siRNA to target the α and β subunits of TFIIA and monitored the activity of the AdML promoter derivatives as described above (Fig. 3A). Western blotting confirmed that TFIIAα and particularly TFIIAβ expression was knocked down by the siRNA. At the wild-type AdML promoter, knockdown of TFIIAα and TFIIAβ (TFIIAα/β) caused a 2.1-fold increase in promoter activity. At the AdML derivatives harboring individual or combined mutant BREs, however, the effect of TFIIAα/β siRNA was reduced (1.4- to 1.5-fold). The TFIIAα/β siRNA also had little effect on the AdML promoter derivative with a defective TATA element. A second siRNA targeting TFIIAα/β, TFIIAα/β (2) siRNA, produced comparable results (Fig. 3B). siRNA targeting TFIIAγ also elicited a positive effect on transcription at the wild-type AdML promoter (2-fold) but had a reduced effect on transcription at the AdML derivatives with individual (1.3- and 1.7-fold) or combined (1.1-fold) defective BREs (Fig. 3C). Also, as observed with the TFIIAα/β siRNA, the TFIIAγ siRNA did not significantly affect transcription at the AdML promoter derivative with a defective TATA element. Again, these results are consistent with the ChIP data of Fig. 1C; TFIIA recruitment is greatest at the wild-type AdML promoter, at which TFIIA ablation by siRNA also has the greatest effect. Moreover, consistent with the observation that the extent of TFIIA recruitment correlates with the least-active TATA-containing promoter, TFIIA knockdown has a positive effect on transcription at this same promoter. These results suggest that TFIIA has a dampening effect on transcription at the AdML promoter that is dependent upon the presence of the BREs.
FIG. 3.
(A) BRE-dependent function for TFIIA at the AdML promoter. 293T cells were transfected as for Fig. 1B except that either control siRNA or siRNA targeting TFIIAα/β was included. Forty-eight hours later cells were harvested and used to monitor luciferase activity and also for Western blotting with anti-TFIIAα/β antibodies or, as a control, anti-β-tubulin antibodies. Note that the immunoblot analysis concerns the whole cell population and therefore underestimates the extent of TFIIAα/β knockdown. The graph shows the relative luciferase activity of the AdML promoter derivatives in the presence of control (Ctrl) or TFIIAα/β siRNA. The numbers above the bars indicate the changes in activity that are caused by the TFIIAα/β siRNA. The difference in changes between the wild-type (wt) AdML reporter and AdMLmBREud was subjected to Student's t test and was found to be significant (P < 0.05). (B) Same as panel A, except that a different siRNA targeting TFIIAα/β, TFIIAα/β (2) siRNA, was used and only the wild-type AdML and AdMLmBREud reporters were tested. (C) Same as panel A, except that siRNA targeting TFIIAγ was used. Each bar represents three independent experiments with standard deviation.
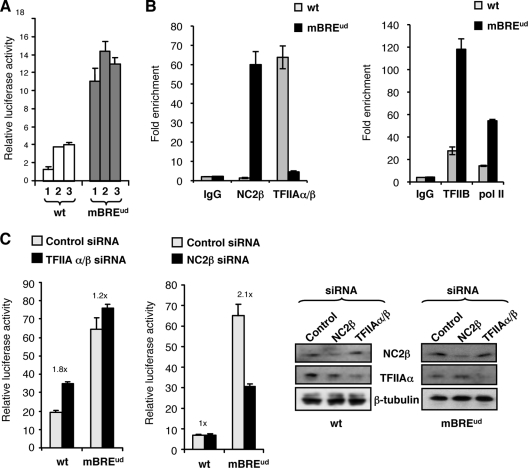
Our studies so far have used transiently transfected AdML promoter derivatives. We were interested in determining if the effects that we observed would also occur at AdML promoter derivatives that were stably integrated within the genome. For this analysis we used the Flp-In system, which employs a 293 cell line derivative containing a single FRT within the genome. Plasmids containing the wild-type AdML promoter or mutant BREud derivative linked to luciferase and flanked by FRT sequences were generated and used to create 293 cell line derivatives that contain a single copy of the AdML promoter (or BREud mutant derivative) integrated at a defined location. Three independent cell lines each for the wild-type AdML promoter or BREud mutant were derived, and the activities of the promoters were compared by luciferase assay (Fig. 4A). Consistent with our transient transfection data, all three cell line derivatives containing the AdMLmBREud promoter exhibited significantly greater luciferase activity than the cell lines generated with the wild-type AdML promoter.
FIG. 4.
Analysis of TFIIA and NC2 function at stably integrated AdML derivatives. (A) Three independent cell lines were generated for both the wild-type (wt) AdML promoter and mutant BREud derivative using the Flp-In system. Luciferase activity for each of the cell lines was assessed and is presented graphically as the mean average of three independent experiments with standard deviation. (B) The wild-type AdML and AdMLmBREud stable cell lines were used for ChIP analysis to assess the promoter occupancy of TFIIAα/β and NC2β (left) and TFIIB and pol II (right). ChIP was performed as for Fig. 1C except that detection was by real-time PCR, and the results are presented graphically. Immunoglobulin G (IgG) is used as a negative control antibody in the reaction. The results are presented as enrichment of precipitation of specific promoter DNA over nonpromoter DNA. Each bar represents at least three independent experiments with standard deviation. (C) The wild-type AdML and AdMLmBREud stable cell lines were transfected with either TFIIAα/β, NC2β, or a control siRNA. Luciferase activity was assessed and is presented graphically as the mean of three independent experiments with standard deviation. The difference in changes between the wild-type AdML and AdMLmBREud was subjected to Student's t test and found to be significant (P < 0.05). At the right is an immunoblot showing successful depletion of TFIIAα and NC2β expression by the siRNA.
We next performed ChIP by quantitative PCR with the stable AdML and AdMLmBREud cell lines to assess the occupancy of the stably integrated promoters by TFIIAα/β and NC2β (Fig. 4B). The results are in agreement with our ChIP analysis of the episomal wild-type AdML promoter and AdMLmBREud derivative (Fig. 1C). Specifically, TFIIA preferentially associates with the integrated wild-type AdML promoter and NC2 preferentially associates with the integrated AdMLmBREud derivative. Moreover, as observed with ChIP of the episomal AdML derivatives, TFIIB and pol II showed significantly higher levels of occupancy at the more active AdMLmBREud promoter than at the wild-type AdML promoter. We further analyzed the stable cell lines using siRNA to target either TFIIAα/β or NC2β expression (Fig. 4C). As before, the wild-type AdML promoter was more sensitive to TFIIA depletion, showing enhanced transcriptional function in response to the TFIIAα/β siRNA. Conversely, the AdMLmBREud derivative was more sensitive to NC2 depletion, showing reduced transcription function in response to NC2β siRNA. Thus, the suppressive effect of TFIIA and positive effect of NC2 on BRE-dependent transcription are equivalent at both episomal and stably integrated AdML promoter derivatives.
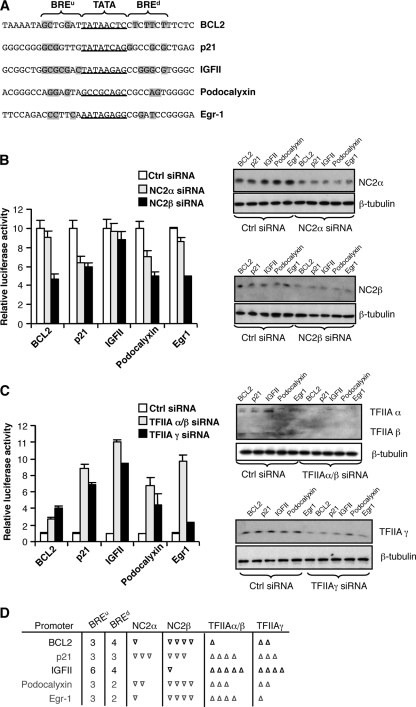
We next sought to confirm and extend our data using a series of natural promoters that contain different combinations of and levels of conformity to the consensus sequences of the BREs (Fig. 5A). Plasmids containing these promoters driving luciferase expression were transfected into 293T cells along with either NC2α or NC2β siRNA. Forty-eight hours later the cells were harvested. NC2α and NC2β expression was monitored by Western blotting, and luciferase activity was measured and is presented graphically (Fig. 5B). To aid interpretation, the effects of each of the NC2 siRNAs are presented relative to that of the control siRNA at each promoter, which has been set at an arbitrary value of 10. siRNA-mediated knockdown of NC2 caused a decrease in activity of the BCL2, p21, podocalyxin, and Egr1 promoters. At the BCL2 and Egr1 promoters, however, we note that the NC2α siRNA was consistently less effective than the NC2β siRNA. The IGFII promoter was refractory to both the NC2α and NC2β siRNAs; significantly, this promoter contains the sequence most similar to the consensus BRE sequences of all the promoters tested. Indeed, we have reported previously that the IGFII promoter BREu is contacted by TFIIB (20). In addition, band shift analysis confirmed that the BRE sequences of the IGFII promoter provided greater functionality in TFIIB recognition than the corresponding sequences of the BCL2 promoter (see Fig. S2 in the supplemental material).
FIG. 5.
Natural promoters exhibit BRE-dependent TFIIA and NC2 function. (A) The core promoter regions of five natural promoters are shown, with the TATA element underlined. The BREu and BREd regions are shown, and bases that conform to the consensus sequences are shaded. (B) The five promoters indicated, linked to luciferase, were transfected into 293T cells along with control (Ctrl) siRNA, NC2α siRNA, or NC2β siRNA. Forty-eight hours later the cells were harvested and used to monitor luciferase activity and also for Western blotting with anti-NC2α antibodies, anti NC2β antibodies, or, as a control, anti-β-tubulin antibodies. Luciferase activity is shown graphically, and each bar represents three independent experiments with standard deviation. To aid comparison of the effects of the siRNAs on the different promoters, the activity of each promoter in the presence of control siRNA was set to 10. (C) The five promoters indicated, linked to luciferase, were transfected into 293T cells as for panel B, but along with control siRNA, TFIIAα/β siRNA, or TFIIAγ siRNA. Forty-eight hours later the cells were harvested and used to monitor luciferase activity and also for Western blotting with anti-TFIIAα/β antibodies, anti TFIIAγ antibodies, or, as a control, anti-β-tubulin antibodies. Luciferase activity is shown graphically, and each bar represents three independent experiments with standard deviation. To aid comparison of the effects of the siRNAs on the different promoters, the activity of each promoter in the presence of control siRNA was set to 1. (D) Summary of the effects of NC2 and TFIIA siRNAs on the activities of the natural promoters. For each promoter the BREu and BREd match to the consensus (out of 7 bases) is shown. The transcriptional effects of the siRNAs are denoted by upward (stimulation)- and downward (repression)-pointing arrowheads. The number of arrowheads in each case is proportional to the change in luciferase activity. The BCL2 and IGFII promoters are discussed further in the text.
We next tested the effects of TFIIA ablation on the natural promoters (Fig. 5C). All of the promoters elicited a positive effect in response to TFIIAα/β and TFIIAγ siRNA. However, the greatest effect was observed with the IGFII promoter while the smallest effect was observed with the BCL2 promoter. Although the effects of NC2 and TFIIA siRNA at the natural promoters are more complex than those observed with the AdML mutant derivatives (in which targeted mutations of the BREs were performed based on previous functional data), it is clear that the IGFII promoter, which contains the sequence most similar to the consensus BRE sequences, is least dependent on NC2 for activity and is suppressed to the greatest degree by TFIIA (see summary diagram in Fig. 5D). All the other promoters show some degree of dependence on NC2 for full activity, while TFIIA is least inhibitory at the BCL2 promoter.
We next analyzed the effect of TFIIA and NC2 siRNA on the expression of the endogenous genes that correspond to the reporter plasmids used in Fig. 5. 293T cells were transfected with control, NC2α, NC2β, TFIIAα/β, or TFIIAγ siRNA, and 48 h later the cells were harvested and used to prepare whole-cell extracts or total RNA. RT-PCR was used to analyze the expression levels of IGFII, p21, Egr1, BCL2, and, as a control, pol I-transcribed 18S rRNA (Fig. 6A). Western blots of the associated whole-cell extracts with antibodies against several general transcription factors are shown. The endogenous podocalyxin gene is not expressed in 293 cells. Both TFIIAα/β and TFIIAγ siRNA caused an increase in Egr1 and IGFII mRNA. Significantly, the Egr1 and IGFII promoters were found to be the most responsive to TFIIA siRNA when the reporters were tested (Fig. 5C). The NC2β siRNA caused a reduction in the abundance of BCL2, Egr1, and p21 mRNA but had no effect on the level of IGFII mRNA. NC2α siRNA also had a negative effect on BCL2 and p21 promoter activity, but not Egr1 or IGFII promoter activity. Taken together, the results of the analysis of the endogenous genes confirm that IGFII is actively suppressed by TFIIA but is not dependent upon NC2 for activity. Conversely, the BCL2 promoter is dependent on NC2 for full activity but is not suppressed by TFIIA.
FIG. 6.
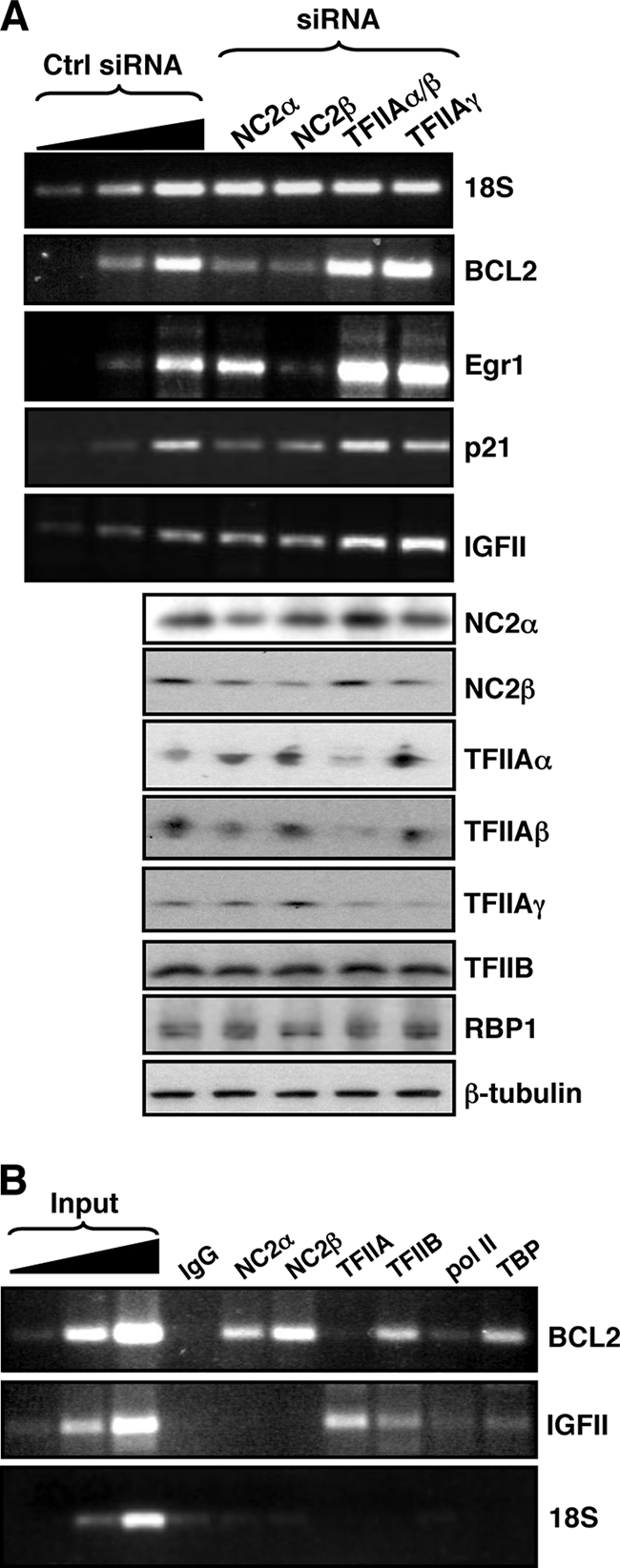
Endogenous promoters exhibit a BRE-dependent transcriptional requirement for and recruitment of TFIIA and NC2. (A) 293T cells were transfected with either a control (Ctrl) siRNA or the siRNAs indicated at the top. Forty-eight hours later the cells were harvested and both total RNA and whole-cell protein lysates were prepared. BCL2, Egr1, p21, and IGFII mRNAs were detected by RT-PCR, and assurance that the reactions were performed in the linear range was determined by a dilution profile of the RNA sample obtained from cells treated with control siRNA. Below, Western blots of the whole-cell lysates with the antibodies indicated at the right are shown. (B) 293T cells were subjected to ChIP with the antibodies indicated at the top. The core promoter regions of the BCL2 and IGFII genes were amplified by PCR, and amplification of the 18S coding region served as a negative control (bottom). Assurance that the reactions were performed in the linear range was determined by a dilution profile of the input chromatin prior to PCR. IgG, immunoglobulin G.
We next performed ChIP analysis on the endogenous BCL2 and IGFII core promoters to assess the relative assembly of the general transcription factors, including TFIIA and NC2 (Fig. 6B). The BCL2 core promoter showed robust assembly of NC2α and NC2β, but not TFIIA, consistent with its poor BRE sequences and also the effects of the NC2 and TFIIA siRNAs. In addition, the IGFII promoter showed robust TFIIA assembly but low levels of NC2α and NC2β assembly, consistent with the strong BRE consensus sequences and the effects of the NC2 and TFIIA siRNAs.
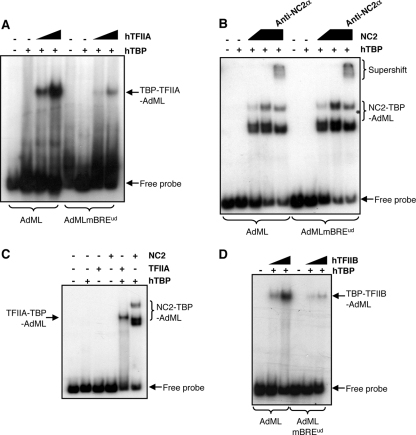
Our data so far suggest that the presence of BREs within a promoter results in the preferred assembly of TFIIA instead of NC2 at the promoter and elicits a dampening effect on transcription. In the absence of BREs (or with BREs having low similarity to the high-affinity sequences), NC2 preferentially assembles over TFIIA and elicits a positive effect on transcription. We were therefore interested to determine directly the effect of the BREs on the affinity of TFIIA and NC2 for promoter-bound TBP in the absence of other factors. The wild-type AdML promoter and mutant BRE derivative AdMLmBREud were radiolabeled and incubated with recombinant human TBP along with TFIIA, and the complexes were resolved by native electrophoresis (Fig. 7A). TFIIA-TBP-promoter complexes formed more avidly with the wild-type AdML promoter than with the AdML promoter derivative in which the BREs had been rendered nonfunctional (AdMLmBREud). We next performed a band shift reaction with purified recombinant NC2 and TBP (Fig. 7B). The NC2-TBP-AdML complexes resolved as two forms; thus, the presence of NC2 within the complexes was verified by supershifting the complex with anti-NC2α antibodies. Significantly, NC2-TBP-promoter complexes did not show any dependence on the BRE for their formation. Assembly of either TFIIA or NC2 at the AdML promoter required the presence of TBP (Fig. 7C). In addition, consistent with previous studies, TFIIB shows reduced assembly at the AdMLmBREud promoter derivative compared with that at the wild-type AdML promoter (Fig. 7D). Thus, the BREs drive formation of either TFIIA-TBP-promoter complexes or TFIIB-TBP-promoter complexes, but not NC2-TBP-promoter complexes.
FIG. 7.
TFIIA assembly at the promoter is BRE dependent. (A) The wild-type AdML promoter and mutant derivative AdMLmBREud were radiolabeled, and assembly of TBP and TFIIA was monitored by band shift analysis. Where indicated, 50 ng of human TBP (hTBP) was included in the reaction mixture. Either 50 ng or 250 ng of TFIIA was added. Free probe and TBP-TFIIA-promoter complexes are indicated at the right. (B) Same as panel A except that 10 ng and 50 ng of purified recombinant NC2 were included instead of TFIIA. Also, where indicated 500 ng of anti-NC2α antibody was included, and the supershifts are indicated. (C) TFIIA (50 ng) or NC2 (50 ng) was incubated with the radiolabeled AdML promoter either alone or along with hTBP (50 ng). Complexes were resolved as for panel A. TFIIA-TBP-AdML and NC2-TBP-AdML complexes and free probe are indicated. (D) Same as panel A except that 50 ng or 250 ng of recombinant human TFIIB was used instead of TFIIA.
DISCUSSION
The BREs can elicit either positive or negative effects on transcription. While the positive effect can be explained by the increased affinity of TFIIB for the promoter, the molecular basis for the negative effect is unclear. In this study we have shown that the presence of BREs in the AdML promoter determines the relative assembly of TFIIA and NC2. Surprisingly, however, TFIIA recruitment correlated with reduced promoter activity while NC2 recruitment correlated with high promoter activity. Treatment of cells with siRNA to reduce endogenous TFIIA and NC2 levels revealed that TFIIA suppresses transcription at the AdML promoter in a BRE-specific manner and that NC2 provides a positive transcriptional function in the absence of BREs. In vitro studies confirmed that the BREs enhance the formation of a TFIIA-TBP-promoter complex.
A positive role for NC2 in transcription has been observed before (5, 7, 11, 22, 63). How NC2 can act as a positive factor is not yet known, but it is possible that it acts by stabilizing or “priming” TFIID at the promoter, before release and replacement by factors (e.g., TFIIB) that can drive PIC assembly forward. Such a mechanism has been described before for the positive activities of Mot1 (14, 23, 24, 32). Indeed, active transcription complexes that contain Mot1 also contain TFIIB and pol II, but not TFIIA (24). We also note that RNA interference (RNAi)-mediated knockdown of NC2α frequently elicited a less pronounced effect on transcription than RNAi-mediated knockdown of NC2β, particularly with the natural promoters. This is consistent with previous reports of potentially independent activities for NC2α or NC2β (13, 33, 65), although our ChIP analysis suggests that both components generally assemble at the promoter together.
Recent genome-wide ChIP-on-chip experiments have found that NC2 associates with 25% of promoters in the genome and generally correlates with high expression, reaffirming a positive function for NC2 in transcription (1). Furthermore, this same study showed that association of NC2 with promoters shows an inverse correlation with the BRE core promoter element. Our results are entirely consistent with these findings and further suggest that this inverse correlation between the BRE and NC2 promoter occupancy arises from the binding of TFIIA to BRE-like sequences. Thus, our data raise the prospect of TFIIA recognition elements that share at least some similarity to the BREs. Indeed, previous cross-linking studies have shown that TFIIA contacts the core promoter within both the BREu and BREd elements, and it is possible that TFIIA might share overlapping DNA sequence-specific recognition with TFIIB (12, 38). The crystal structures of yeast TFIIA complexed with TBP at the AdML and CYC promoters revealed base contact upstream of the TATA element, including contact with a G 3 nucleotides upstream of the TATA element (3, 21, 58). We note that a G 3 nucleotides upstream of the TATA element forms a critical part of BREu and was replaced in our mutant AdML derivatives. In addition, several other residues in the above structure were in close proximity to the upstream DNA with potential to form direct contact. The DNA fragment used in these studies did not extend sufficiently 3′ of the TATA box to encompass the BREd region. Our band shift data, revealing BRE-dependent enhancement of TFIIA-TBP-promoter complex formation, are therefore consistent with previous studies and also with our ChIP data.
Our experiments revealed the surprising finding that reduction of cellular TFIIA enhances transcription at the wild-type AdML promoter. Moreover, BRE-directed recruitment of TFIIA at the AdML promoter correlated with reduced promoter activity. A previous study found that a highly pure TFIIA fraction repressed transcription in vitro at the AdML promoter, but not at TATA-less promoters (2). Indeed, we also observed that transcription at the AdML derivative that lacks a TATA element was not affected by the TFIIA siRNAs. However, the TATA-less podocalyxin promoter was repressed by TFIIA, suggesting that the repressive function of TFIIA is dependent on multiple factors. Indeed, Stargell et al. (56) have reported core promoter-dependent effects of TFIIA. It will therefore be important to test the other core promoter sequence elements (including the initiator, DPE, downstream core element, and the motif 10 element) with regard to TFIIA and NC2 function. Recent work by Hsu et al. (31) employed RNAi in drosophila cells to assess the role of several general transcription factors at DPE-containing versus TATA-containing core promoters. In addition to a positive function for NC2, they uncovered DPE- and TATA-dependent roles for both TBP and NC2. In addition, they found that depletion of TFIIA decreases the activity of a hybrid core promoter containing the AdML TATA region linked to the Ant P2 or E74B initiator. It is therefore likely that the core promoter context of the BREs, particularly in relation to the initiator, plays a role in determining the net contribution of TFIIA to transcriptional activity. Indeed, we have previously reported that the BREs can exert a positive or negative effect on transcription that is dependent upon the promoter context (16). Moreover, promoter-specific activities for TFIIA have also been observed in other studies (10, 36, 40, 42, 46, 56, 61). It should also be noted that, while yeast TFIIB can also contact BREd sequences (16), contact with BREu does not appear to be conserved (39), suggesting potential differences in the activities of TFIIA with respect to the BREs.
It is generally assumed that TFIIA and NC2 act in a reciprocal manner, with mutually exclusive recruitment to the promoter and effects on transcription. However, as stated previously, this has been difficult to reconcile with the numerous reports that NC2 can have a positive effect on transcription. Our findings provide an explanation that maintains the reciprocity between TFIIA and NC2, in that TFIIA and NC2 show mutually exclusive effects on transcription in a BRE-dependent manner. We suggest that, at promoters with BRE-like sequences, TFIIA assembles in preference over NC2. We did not find BRE-dependent effects in the formation of NC2-TBP-AdML promoter complexes in vitro (Fig. 7), suggesting that enhanced assembly of NC2 at non-BRE promoters in vivo is due to reduced competition from TFIIA-promoter recognition.
However, it is important to note that the BREu and BREd consensus sequences are derived from binding site selection studies and therefore represent the highest-affinity sequences. Physiologically relevant sequences will likely fall across a broad spectrum, and it is likely that many BREs might be functional for TFIIB binding but not act as affinity sites for TFIIA. In such cases TFIIA and NC2 might elicit more-competitive effects and therefore respond to ablation of both TFIIA and NC2 expression. Indeed, this is likely the case at some of the natural promoters used in this study.
It is well established that TFIIA can stabilize TFIID-promoter contacts, leading to productive PIC formation and transcription. Our data suggest that this pathway of PIC formation dominates at BRE-containing promoters. An alternative pathway employs NC2 to stabilize TFIID-promoter complexes. Although both pathways are productive, the NC2 pathway appears to be more efficient in producing transcriptionally active complexes. Thus, elimination of TFIIA by siRNA or diminished TFIIA-promoter recognition (by BRE mutation) leads to the more efficient NC2 pathway. Our previous work on mammalian cells and the work of others on yeast suggested that TFIIB most likely assembles at the promoter as a complex with pol II (18, 51). It is possible that ablation of TFIIA by RNAi might release the BREs for recognition by the TFIIB-pol II machinery, thereby enhancing transcription. TFIIA and NC2, by mutually exclusive assembly with TFIID at the promoter, could therefore provide a commitment step that can either preclude BRE function or enhance it. Thus, TFIIA might “shield” the BREs from TFIIB recognition and weaken the promoter. Conversely, if NC2 “primes” the promoter-TFIID complex but is subsequently released, this would allow recognition of the BREs by TFIIB, thereby eliciting a positive effect on transcription. Further studies will shed light on the TFIIB-TFIIA-NC2 dynamic in living cells.
Supplementary Material
Acknowledgments
We thank Bob White and members of the Oelgeschläger and Roberts labs for comments on the manuscript. We also thank Daniel Haber for the podocalyxin promoter reporter, Andrew Ward for the IGFII promoter reporter, Neil Perkins for the BCL2/p21 promoter reporters, Andy Sharrocks for the Egr1 promoter reporter, and Stephen Taylor for help with the Flp-In system.
This work was funded by the Wellcome Trust (061207/Z/00/Z/CH/TG/dr). S.G.E.R. is a Wellcome Trust Senior Research Fellow.
The authors have no competing financial interests.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albert, T. K., S. Boeing, G. Stelzer, A. Schepers, and M. Meisterernst. 2007. Global distribution of negative cofactor 2 subunit-alpha on human promoters. Proc. Natl. Acad. Sci. USA 10410000-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aso, T., H. Serizawa, R. C. Conaway, and J. W. Conaway. 1994. A TATA sequence-dependent transcriptional repressor activity associated with mammalian transcription factor IIA. EMBO J. 13435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleichenbacher, M., S. Tan, and T. J. Richmond. 2003. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J. Mol. Biol. 332783-793. [DOI] [PubMed] [Google Scholar]
- 4.Butler, J. E., and J. T. Kadonaga. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 162583-2592. [DOI] [PubMed] [Google Scholar]
- 5.Cang, Y., and G. Prelich. 2002. Direct stimulation of transcription by negative cofactor 2 (NC2) through TATA-binding protein (TBP). Proc. Natl. Acad. Sci. USA 9912727-12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter, B., K. J. Hill, M. Charalambous, K. J. Wagner, D. Lahiri, D. I. James, J. S. Anderson, V. Schumacher, B. Royer-Pokora, M. Mann, A. Ward, and S. G. E. Roberts. 2004. BASP1 is a transcriptional cosuppressor for the Wilms’ tumour suppressor protein WT1. Mol. Cell. Biol. 24537-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castano, E., P. Gross, Z. Wang, R. G. Roeder, and T. Oelgeschlager. 2000. The C-terminal domain-phosphorylated IIO form of RNA polymerase II is associated with the transcription repressor NC2 (Dr1/DRAP1) and is required for transcription activation in human nuclear extracts. Proc. Natl. Acad. Sci. USA 977184-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z., and J. L. Manley. 2003. Core promoter elements and TAFs contribute to the diversity of transcriptional activation in vertebrates. Mol. Cell. Biol. 237350-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi, T., and M. Carey. 1996. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 102540-2550. [DOI] [PubMed] [Google Scholar]
- 10.Chou, S., S. Chatterjee, M. Lee, and K. Struhl. 1999. Transcriptional activation in yeast cells lacking transcription factor IIA. Genetics 1531573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christova, R., and T. Oelgeschlager. 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 479-82. [DOI] [PubMed] [Google Scholar]
- 12.Coulombe, B., J. Li, and J. Greenblatt. 1994. Topological localization of the human transcription factors IIA, IIB, TATA box-binding protein, and RNA polymerase II-associated protein 30 on a class II promoter. J. Biol. Chem. 26919962-19967. [PubMed] [Google Scholar]
- 13.Creton, S., J. Q. Svejstrup, and M. A. Collart. 2002. The NC2 alpha and beta subunits play different roles in vivo. Genes Dev. 163265-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dasgupta, A., S. A. Juedes, R. O. Sprouse, and D. T. Auble. 2005. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 241717-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeJong, J., R. Bernstein, and R. G. Roeder. 1995. Human general transcription factor TFIIA: characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc. Natl. Acad. Sci. USA 923313-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng, W., and S. G. E. Roberts. 2005. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 192418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng, W., and S. G. E. Roberts. 2007. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 116417-429. [DOI] [PubMed] [Google Scholar]
- 18.Elsby, L. M., A. J. M. O'Donnell, L. M. Green, A. D. Sharrocks, and S. G. E. Roberts. 2006. Assembly of TFIIB at a promoter in vivo requires contact with RNA polymerase II. EMBO Rep. 7898-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans, R., J. A. Fairley, and S. G. E. Roberts. 2001. Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB. Genes Dev. 152945-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fairley, J. A., R. Evans, N. A. Hawkes, and S. G. E. Roberts. 2002. Core promoter-dependent TFIIB conformation and a role for TFIIB conformation in transcription start site selection. Mol. Cell. Biol. 226697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiger, J. H., S. Hahn, S. Lee, and P. B. Sigler. 1996. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272830-836. [DOI] [PubMed] [Google Scholar]
- 22.Geisberg, J. V., F. C. Holstege, R. A. Young, and K. Struhl. 2001. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 212736-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisberg, J. V., Z. Moqtaderi, L. Kuras, and K. Struhl. 2002. Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 228122-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisberg, J. V., and K. Struhl. 2004. Cellular stress alters the transcriptional properties of promoter-bound Mot1-TBP complexes. Mol. Cell 14479-489. [DOI] [PubMed] [Google Scholar]
- 25.Gershenzon, N. I., and I. P. Ioshikhes. 2005. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics 211295-1300. [DOI] [PubMed] [Google Scholar]
- 26.Glossop, J. A., T. R. Dafforn, and S. G. E. Roberts. 2004. A conformational change in TFIIB is required for activator-mediated assembly of the preinitiation complex. Nucleic Acids Res. 321829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goppelt, A., G. Stelzer, F. Lottspeich, and M. Meisterernst. 1996. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 153105-3116. [PMC free article] [PubMed] [Google Scholar]
- 28.Guo, B., R. E. Sallis, A. Greenall, M. M. Petit, E. Jansen, L. Young, W. J. Van de Ven, and A. D. Sharrocks. 2006. The LIM domain protein LPP is a coactivator for the ETS domain transcription factor PEA3. Mol. Cell. Biol. 264529-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn, S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoiby, T., H. Zhou, D. J. Mitsiou, and H. G. Stunnenberg. 2007. A facelift for the general transcription factor TFIIA. Biochim. Biophys. Acta 1769429-436. [DOI] [PubMed] [Google Scholar]
- 31.Hsu, J.-Y., T. Juven-Gershon, M. T. Marr II, K. J. Wright, R. Tjian, and J. T. Kadonaga. 2008. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 222353-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huisinga, K. L., and B. F. Pugh. 2007. A TATA binding protein regulatory network that governs transcription complex assembly. Genome Biol. 8R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inostroza, J. A., F. H. Mermelstein, I. Ha, W. S. Lane, and D. Reinberg. 1992. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 70477-489. [DOI] [PubMed] [Google Scholar]
- 34.Kamada, K., F. Shu, H. Chen, S. Malik, G. Stelzer, R. G. Roeder, M. Meisterernst, and S. K. Burley. 2001. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 10671-81. [DOI] [PubMed] [Google Scholar]
- 35.Kim, S., J. G. Na, M. Hampsey, and D. Reinberg. 1997. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc. Natl. Acad. Sci. USA 94820-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, T. K., Y. Zhao, H. Ge, R. Bernstein, and R. G. Roeder. 1995. TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J. Biol. Chem. 27010976-10981. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi, N., T. G. Boyer, and A. J. Berk. 1995. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol. Cell. Biol. 156465-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagrange, T., T. K. Kim, G. Orphanides, Y. W. Ebright, R. H. Ebright, and D. Reinberg. 1996. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl. Acad. Sci. USA 9310620-10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagrange, T., A. N. Kapanidis, H. Tang, D. Reinberg, and R. H. Ebright. 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence specific DNA binding by transcription factor IIB. Genes Dev. 1234-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman, P. M., J. Ozer, and D. B. Gursel. 1997. Requirement for transcription factor IIA (TFIIA)-TFIID recruitment by an activator depends on promoter structure and template competition. Mol. Cell. Biol. 176624-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim, C. Y., B. Santoso, T. Boulay, E. Dong, U. Ohler, and J. T. Kadonaga. 2004. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 181606-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, Q., S. E. Gabriel, K. L. Roinick, R. D. Ward, and K. M. Arndt. 1999. Analysis of TFIIA function in vivo: evidence for a role in TATA-binding protein recruitment and gene-specific activation. Mol. Cell. Biol. 198673-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma, D., H. Watanabe, F. Mermelstein, A. Admon, K. Oguri, X. Sun, T. Wada, T. Imai, T. Shiroya, D. Reinberg, and H. Handa. 1993. Isolation of a cDNA encoding the largest subunit of TFIIA reveals functions important for activated transcription. Genes Dev. 72246-2257. [DOI] [PubMed] [Google Scholar]
- 44.Ma, D., I. Olave, A. Merino, and D. Reinberg. 1996. Separation of the transcriptional coactivator and antirepression functions of transcription factor IIA. Proc. Natl. Acad. Sci. USA 936583-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malecova, B., P. Gross, M. Boyer-Guittaut, S. Yavuz, and T. Oelgeschlager. 2007. The initiator core promoter element antagonizes repression of TATA-directed transcription by negative cofactor NC2. J. Biol. Chem. 28224767-24776. [DOI] [PubMed] [Google Scholar]
- 46.Martinez, E., H. Ge, Y. Tao, C. X. Yuan, V. Palhan, and R. G. Roeder. 1998. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol. Cell. Biol. 186571-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKay, L. M., B. Carpenter, and S. G. E. Roberts. 1999. Regulation of the Wilms’ tumour suppressor protein transcriptional activation domain. Oncogene 186546-6554. [DOI] [PubMed] [Google Scholar]
- 48.Mermelstein, F., K. Yeung, J. Cao, J. A. Inostroza, H. Erdjument-Bromage, K. Eagelson, D. Landsman, P. Levitt, P. Tempst, and D. Reinberg. 1996. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 101033-1048. [DOI] [PubMed] [Google Scholar]
- 49.Ozer, J., P. A. Moore, A. H. Bolden, A. Lee, C. A. Rosen, and P. M. Lieberman. 1994. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 82324-2335. [DOI] [PubMed] [Google Scholar]
- 50.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 2551-14. [DOI] [PubMed] [Google Scholar]
- 51.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1349-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schluesche, P., G. Stelzer, E. Piaia, D. C. Lamb, and M. Meisterernst. 2007. NC2 mobilizes TBP on core promoter TATA boxes. Nat. Struct. Mol. Biol. 141196-1201. [DOI] [PubMed] [Google Scholar]
- 53.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72449-479. [DOI] [PubMed] [Google Scholar]
- 54.Spencer, V. A., J. M. Sun, L. Li, and J. R. Davie. 2003. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods 3167-75. [DOI] [PubMed] [Google Scholar]
- 55.Stargell, L. A., and K. Struhl. 1995. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science 26975-78. [DOI] [PubMed] [Google Scholar]
- 56.Stargell, L. A., Z. Moqtaderi, D. R. Dorris, R. C. Ogg, and K. Struhl. 2000. TFIIA has activator-dependent and core promoter functions in vivo. J. Biol. Chem. 27512374-12380. [DOI] [PubMed] [Google Scholar]
- 57.Sun, X., D. Ma, M. Sheldon, K. Yeung, and D. Reinberg. 1994. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 82336-2348. [DOI] [PubMed] [Google Scholar]
- 58.Tan, S., Y. Hunziker, D. F. Sargent, and T. J. Richmond. 1996. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381127-151. [DOI] [PubMed] [Google Scholar]
- 59.Thomas, M. C., and C. M. Chiang. 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41105-178. [DOI] [PubMed] [Google Scholar]
- 60.Tsai, F. T. F., and P. B. Sigler. 2000. Structural basis of preinitiation complex assembly on human Pol II promoters. EMBO J. 1925-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Virbasius, C. M., F. C. Holstege, R. A. Young, and M. R. Green. 2001. Promoter-specific activation defects by a novel yeast TBP mutant compromised for TFIIB interaction. Curr. Biol. 111794-1798. [DOI] [PubMed] [Google Scholar]
- 62.White, R. J., B. C. Khoo, J. A. Inostroza, D. Reinberg, and S. P. Jackson. 1994. Differential regulation of RNA polymerases I, II, and III by the TBP-binding repressor Dr1. Science 266448-450. [DOI] [PubMed] [Google Scholar]
- 63.Willy, P. J., R. Kobayashi, and J. T. Kadonaga. 2000. A basal transcription factor that activates or represses transcription. Science 290982-985. [DOI] [PubMed] [Google Scholar]
- 64.Xie, J., M. Collart, M. Lemaire, G. Stelzer, and M. Meisterernst. 2000. A single point mutation in TFIIA suppresses NC2 requirement in vivo. EMBO J. 19672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeung, K. C., J. A. Inostroza, F. H. Mermelstein, C. Kannabiran, and D. Reinberg. 1994. Structure-function analysis of the TBP-binding protein Dr1 reveals a mechanism for repression of class II gene transcription. Genes Dev. 82097-2109. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, H., S. Spicuglia, J. J. Hsieh, D. J. Mitsiou, T. Hoiby, G. J. Veenstra, S. J. Korsmeyer, and H. G. Stunnenberg. 2006. Uncleaved TFIIA is a substrate for taspase 1 and active in transcription. Mol. Cell. Biol. 262728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.